Abstract
Many filamentous fungi produce polyketide molecules with great significance as human pharmaceuticals; these molecules include the cholesterol-lowering compound lovastatin, which was originally isolated from Aspergillus terreus. The chemical diversity and potential uses of these compounds are virtually unlimited, and it is thus of great interest to develop a well-described microbial production platform for polyketides. Using genetic engineering tools available for the model organism Aspergillus nidulans, we constructed two recombinant strains, one expressing the Penicillium griseofulvum 6-methylsalicylic acid (6-MSA) synthase gene and one expressing the 6-MSA synthase gene and overexpressing the native xylulose-5-phosphate phosphoketolase gene (xpkA) for increasing the pool of polyketide precursor levels. The physiology of the recombinant strains and that of a reference wild-type strain were characterized on glucose, xylose, glycerol, and ethanol media in controlled bioreactors. Glucose was found to be the preferred carbon source for 6-MSA production, and 6-MSA concentrations up to 455 mg/liter were obtained for the recombinant strain harboring the 6-MSA gene. Our findings indicate that overexpression of xpkA does not directly improve 6-MSA production on glucose, but it is possible, if the metabolic flux through the lower part of glycolysis is reduced, to obtain quite high yields for conversion of sugar to 6-MSA. Systems biology tools were employed for in-depth analysis of the metabolic processes. Transcriptome analysis of 6-MSA-producing strains grown on glucose and xylose in the presence and absence of xpkA overexpression, combined with flux and physiology data, enabled us to propose an xpkA-msaS interaction model describing the competition between biomass formation and 6-MSA production for the available acetyl coenzyme A.
Natural products from microbial, plant, marine, and even mammalian sources have traditionally been a major drug source and continue to play a significant role in today's drug discovery environment. Approximately 40% of new drugs introduced between the mid-1980s and 1990s originated from natural molecules, whereas 9 of the top 20 small-molecule drugs introduced in 1999 were developed from natural products (13). Polyketide natural products play an important role in the treatment of a wide range of human physiological disorders. They are widely used as antimicrobial (erythromycin, rifamycin, tetracycline), antifungal (amphotericin B), immunosuppressant (tacrolimus [TK506], rapamycin), and anticancer (doxorubicin, epothilone, geldanamycin) agents, as well as cholesterol-lowering agents (lovastatin). Current chemical technologies are in most cases unable to generate commercial quantities of most polyketide compounds. As a result, the natural sources of these compounds, usually bacteria and fungi, are employed as biocatalysts for large-scale production. Additionally, in the past two decades heterologous expression of polyketide genes has become possible by the explosive growth in the cloning and sequencing of genes responsible for polyketide production and the increase in our knowledge of the catalytic mechanisms governing the biosynthesis of polyketides. Furthermore, metabolic engineering advances associated with polyketide production can assist in making heterologous biosynthesis a reality.
An especially relevant group of enzymes in this context is the phosphoketolases (PHKs) (EC 4.1.2.9 and EC 4.1.2.22), a class of key enzymes of the PHK pathway of heterofermentative and facultatively homofermentative lactic acid bacteria, as well as the d-fructose 6-phosphate shunt of bifidobacteria (21). Xylulose-5-phosphate phosphoketolases (XPKs) (EC 4.1.2.9) are of special interest for the production of polyketides: in the presence of inorganic phosphate, d-xylulose 5-phosphate is cleaved irreversibly into acetyl phosphate and glyceraldehyde-3-phosphate. Induction of XPK in aspergilli increases the carbon flux toward the precursor acetyl coenzyme A (CoA) or acetate (20a), and consequently there is potential to improve the yields and productivities of secondary metabolites that are derived from acetyl-CoA. Recently, Panagiotou et al. (20a) identified a gene coding for XPK (xpkA) in Aspergillus nidulans and demonstrated that high flux through the pathway has substantial effects on the physiology of the cells.
The protein 6-methylsalicylic acid synthase (6-MSAS) (EC 2.3.1.165) is among the simplest and best-characterized members of the polyketide synthase family of multienzyme systems, and it uses four acetyl-CoA molecules for the biosynthesis of 6-methylsalicylic acid (6-MSA). The fungus Penicillium griseofulvum and related species produce the secondary metabolite 6-MSA (30). In previous attempts to develop a heterologous high-level expression system for fully functional polyketide synthases, Kealey et al. (16) demonstrated high-level production of the polyketide 6-MSA in both Escherichia coli and Saccharomyces cerevisiae. Up to 1.7 g of 6-MSA per liter was produced by the recombinant yeast, which was approximately twofold greater than the amount produced by the natural host, Penicillium patulum (30) and 25-fold greater than the amount produced by the heterologous host Streptomyces coelicolor harboring the 6-MSAS gene (5). Recently, using a different metabolic engineering approach targeted to increase the malonyl-CoA supply, Wattanachaisaereekul et al. (31) obtained a final concentration of 6-MSA of 554 mg/liter using S. cerevisiae as a production platform. To develop a robust host for production of 6-MSA and other related polyketides, the gene encoding 6-MSAS from P. griseofulvum (msaS) was functionally expressed in A. nidulans. Furthermore, xpkA was overexpressed since we hypothesized that this modification would lead to increased precursor (acetyl-CoA) supply and concomitantly improve the yield and productivity of 6-MSA.
To accurately test these hypotheses and evaluate the production strategy, the system was investigated at three different levels: physiology, metabolic fluxes, and transcriptional responses. In order to determine the level of 6-MSA production with and without overexpression of XPK, we grew the cells on a broad range of carbon sources (glucose, xylose, glycerol, and ethanol) (24) in well-controlled bioreactors and performed a physiological characterization analysis.
In parallel, we used 13C labeling (as described by Christensen and Nielsen [8]) to quantify the metabolic fluxes in the central carbon metabolism. This technique can add valuable information to physiological studies, especially in connection with studies of metabolite production where the aim is to direct carbon from the substrate into the metabolic product (19).
For a comprehensive evaluation of genes that are modulated at the level of transcription during 6-MSA production with and without an induced PHK pathway, we analyzed gene expression in biological replicates under eight different conditions by using DNA microarrays. Despite the wide use of DNA microarrays, only a few studies have been reported transcriptome profiling of aspergilli (e.g., Aspergillus oryzae [17], Aspergillus flavus [12, 20, 26], and A. nidulans [10, 19, 28]), and the data described here are thus a significant contribution.
MATERIALS AND METHODS
Strain construction and plasmids.
A. nidulans A4 was used as the wild-type reference strain. A. nidulans strain AR1 (IBT27263) (argB2 pyrG89 veA1) was the basis for all strain construction. This strain was derived directly from G051 (9) of the Glasgow strain collection, which was derived directly from strain A4, designated G00 in the Glasgow collection, which is a natural isolate and therefore has no explicit genotype (23).
In order to overexpress XPK (xpkA) or to express 6-MSAS (msaS) in A. nidulans AR1, the genes were cloned into an integrative vector derived from pBARGPE1 (M. L. Pall and J. P. Brunelli; http://www.fgsc.net/fgn/pall.html) obtained from the FGSC, United States. For construction of plasmids, standard molecular biotechnology techniques were used as described by Sambrook and Russell (25) (see File S1 in the supplemental material), and the final constructs were verified by sequencing. The resulting strains were designated AR16msaGP74 (expressing P. griseofulvum msaS) and AR1phk6msaGP74 (overexpressing xpkA and expressing msaS).
Media.
Minimal medium used for transformation contained 10 g/liter glucose, 10 mM NaNO3, 0.52 g/liter KCl, 0.52 g/liter MgSO4, 1.52 g/liter KH2PO4, 4 × 10−4 g/liter CuSO4·5H2O, 4 × 10−5 g/liter Na2B2O7·10H2O, 8 × 10−4 g/liter FeSO4·7H2O, 8 × 10−4 g/liter MnSO4·2H2O, 8 × 10−4 g/liter Na2MoO4·2H2O, and 8 × 10−4 g/liter ZnSO4·7H2O. The concentrations of the compounds added to the minimal medium used for selection of transformants are shown in File S1 in the supplemental material.
For bioreactor cultivation, defined medium described elsewhere was used (20a). The carbon sources used were glucose, xylose, glycerol, and ethanol (20 g liter−1). Arginine (0.7 g/liter) was added by sterile filtration.
Transformation of A. nidulans.
Genetic transformation of A. nidulans protoplasts was performed as previously described (15), except that protoplasting was performed by using the enzyme Glucanex, (Novozymes A/S) at a concentration of 40 mg/ml in protoplasting buffer. The plasmid was linearized at the unique Pvul site in the ampicillin resistance gene.
The vectors are not specifically designed to integrate at a specific site in the genome, and therefore it has to be assumed that they integrate ectopically, possibly in several copies. Transformants were purified by streaking spores to obtain single colonies on selective minimal medium and incubated at 37°C for 3 to 4 days. The resulting recombinants were further purified twice by streaking spores on fresh plates containing selective medium. The selection procedure for the transformants was performed in shake flask cultures (all of the transformants were cultivated), and subsequently one transformant was selected for further analysis. For the transformants harboring the msaS gene or both the msaS and xpkA genes, 11 and 13 transformants, respectively, were evaluated, and selection was based on the final concentration of 6-MSA. The variation in 6-MSA production between the transformants was 3 to 8%.
Cultivation conditions and analysis of substrates and products.
To determine physiological characteristics and to perform gene expression analysis, cultivation was performed in well-controlled 1.5-liter bioreactors with a working volume of 1.2 liters. The bioreactors were equipped with two disk turbine impellers rotating at 350 rpm. The pH was kept constant at 5.5 by addition of 2 M NaOH or HCl, and the temperature was maintained at 30°C. Air was used for sparging the bioreactor at a constant flow rate of 1.0 volume of gas per volume of liquid per min.
When it was used, iodoacetate, an inhibitor of glyceraldehyde-3-phosphate dehydrogenase (27), was added by sterile filtration to the sterilized growth medium to a final concentration of 1 mM.
Cell dry (bio)mass was determined using nitrocellulose filters (pore size, 0.45 μm; Gelman Sciences). Fermentation samples were immediately filtered and stored at −20°C until analysis. The concentrations of glucose, xylose, glycerol, acetate, succinate, pyruvate, and 6-MSA were determined by high-performance liquid chromatography as described previously (31).
Analysis of fractional 13C enrichment and computational methods.
For the 13C labeling experiments, A. nidulans A4, AR16msaGP74, and AR1phk6msaGP74 were each cultivated with 5 g/liter of [1-13C]glucose with an aeration rate of 1.0 volume of gas per volume of liquid per min. In the late exponential phase, biomass samples were harvested from each culture, hydrolyzed, derivatized, and analyzed by gas chromatography-mass spectrometry for determination of the labeling patterns of intracellular metabolites. All fragments were derived from either glucose-6-phosphate or proteinogenic amino acids (8).
(i) Calculation of drain fluxes for biomass formation and metabolic model.
The approximate biomass composition of A. nidulans was determined by using the biomass composition of A. oryzae strain A1560 grown in glucose-limited continuous cultures (specific growth rate, 0.17 h−1) with ammonia as the nitrogen source (22). The simulation results were not sensitive to the biomass composition (results not shown). The metabolic model of Pedersen et al. (22) was used to calculate the precursor requirement for biomass formation. The nine precursors were assumed to be α-ketoglutarate, acetyl-CoA, erythrose-4-phosphate, glucose-6-phosphate, glyceraldehyde-3-phosphate, mannose, oxaloacetate, pyruvate, and ribose-5-phosphate. More details concerning the precursor calculations are shown in File S3 in the supplemental material.
A modified version of the metabolic model of David et al. (11) was used for simulation of the metabolic fluxes. In all cases, the reaction for XpkA activity and formation of 6-MSA from 4 U of acetyl-CoA were included in the model (see File S4 in the supplemental material).
(ii) Computational method.
The fluxes were determined by in-house software based on the metabolic network analysis framework described by Wiechert (32) implemented in MatLab 7.0 (Mathwork Inc., Natick, MA). The fluxes were determined by least-squares minimization with the nonlinear Levenberg-Marquardt algorithm, where the discrepancies between measured and calculated fluxes, as well as the measured and calculated summed fractional labelings (SFLs), were minimized. Multiple initial guesses for the Levenberg-Marquardt algorithm were generated with a genetic algorithm in order to verify the existence of a global minimum and to address the significance of the simulated result. Selected metabolic fluxes are shown in Fig. 1A and 1B, while a complete list is shown in File S4 in the supplemental material.
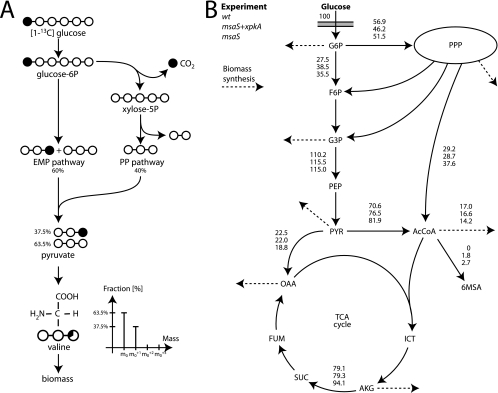
FIG. 1.
(A) Principles of metabolic network analysis with [1-13C]glucose. The 13C labeling pattern of pyruvate is dependent on the active metabolic pathways. For instance, pyruvate formed in the Embden-Meyerhof-Parnas (EMP) pathway contains 13C at position 3, while activity of the PP pathway results in loss of labeled carbon. Pyruvate is the precursor of valine, which is incorporated into biomass. Based on the labeling pattern of valine and other proteinogenic amino acids, the fluxes in central carbon metabolism can be resolved. (B) Major metabolic fluxes of the A. nidulans strains investigated, including strains A4 (wt), AR16msaGP74 (msaS), and AR1phk6msaGP74 (msaS+xpkA). All fluxes are relative to a glucose uptake rate of 100 mol (arbitrary value). The dashed arrows indicate that the metabolite is a precursor drained for biomass formation. See File S2 in the supplemental material for abbreviations used for metabolites. TCA, tricarboxylic acid; PPP, pentose phosphate pathway.
DNA microarrays. (i) Harvesting of mycelium and RNA extraction.
The cultures were harvested at the late exponential phase at a biomass concentration of 6 to 7 g (dry weight)/liter. Mycelium was harvested by filtration through sterile Miracloth (Calbiochem). The mycelium was quickly dried by squeezing and was subsequently frozen in liquid nitrogen. Samples were stored at −80°C until they were used for RNA extraction. Total RNA was isolated using a Qiagen RNeasy mini kit according to the protocol for isolation of total RNA from plants and fungi (18).
(ii) Microarray hybridization.
Fifteen micrograms of fragmented biotin-labeled cRNA was prepared from 5 μg of total RNA and hybridized to a 3AspergDTU Affymetrix GeneChip (4) as described in the Affymetrix GeneChip Expression Analysis Technical Manual (2).
(iii) Analysis of the transcriptome data.
The scanned probe array images (.DAT files) were converted into CEL data files using the GeneChip operating software from Affymetrix and were subsequently preprocessed using the statistical language and environment R (www.R-project.org), version 2.7.1. The probe intensities were normalized for background noise with the robust multiarray average method (14) using only perfect-match probes. Normalization was performed subsequently using the quantiles algorithm (7). Gene expression values were calculated using the perfect-match probes with the medianpolish summary method (14).
Statistical analysis was used to determine genes subject to differential transcription regulation using the limma package (29). To analyze 6-MSA production on glucose and xylose in the presence and absence of XpkA overexpression, a 2×2 analysis of variance (ANOVA) design was used. For all comparisons empirical Bayesian statistics were employed to moderate standard errors within each gene, and the Benjamini-Hochberg method (6) was used to adjust P values for multitesting. Unless noted otherwise, an adjusted cutoff P value of 0.05 was used to determine statistical significance.
Manual annotation.
General function (KOG/Interpro/PFAM) was assigned by comparing PFAM predictions to a manual determination of the bidirectional best hits in the Aspergillus niger ATCC 1015 genome sequence (http://genome.jgi-psf.org/Aspni1/Aspni1.home.html).
Pathway analysis.
A metabolic pathway analysis was done using an adaptation of a metabolic network map for A. niger (3). Bidirectional best BLAST hits were used to find A. nidulans homologues of metabolic genes. Different cutoff values for statistical significance were employed as described below.
Microarray accession number.
The raw microarray data and gene expression values have deposited in the GEO database under accession number GSE12859.
RESULTS
Construction of recombinant strains and physiological characterization.
The construction of an A. nidulans strain in which xpkA is overexpressed (AR1phkGP74) is described elsewhere (Panagiotou et al., submitted). In the present study, two 6-MSA-producing transformants of A. nidulans were constructed with and without concomitant overexpression of the PHK (AR1phk6msaGP74 and AR16msaGP74, respectively). Several vectors that contain the corresponding genes under the strong constitutive glycolytic promoter for glyceraldehyde-3-phosphate dehydrogenase (gpdA) of A. nidulans were also constructed (see File S1 in the supplemental material).
A set of experiments were carried out to examine the physiology and polyketide production of transformants AR16msaGP74 and AR1phk6msaGP74, as well as a reference wild-type strain (A4), with various carbon sources (glucose, xylose, glycerol, and ethanol). Substrate and product concentrations in the media were determined for all cultivations, as were the maximum specific growth rate and the biomass yield (Ysx) during the fully aerobic growth phase (Table 1). For each carbon source, the A4 strain and the AR16msaGP74 strain had similar maximum specific growth rates, but the Ysxs were significantly lower for the 6-MSA-producing strain. Overexpression of XPK in the 6-MSA-producing strain AR1phk6msaGP74 improved the specific growth rate compared to that of AR16msaGP74 when cells were grown on xylose and ethanol. Similarly, the Ysxs on xylose and ethanol, as well as on glucose, were higher for the AR1phk6msaGP74 strain than for the AR16msaGP74 strain. In contrast, the Ysxs for the AR1phk6msaGP74 strain were lower than those for the AR16msaGP74 strain when the strains were grown on glycerol.
TABLE 1.
Specific growth rates, Ysxs, and 6-MSA production by the A. nidulans wild-type strain and transformants cultivated with four different carbon sourcesa
| Strain | Glucose
|
Xylose
|
Glycerol
|
Ethanol
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Specific growth rate (h−1) | 6-MSA concn (mg/liter) | Ysx (g/g) | Specific growth rate (h−1) | 6-MSA concn (mg/liter) | Ysx (g/g) | Specific growth rate (h−1) | 6-MSA concn (mg/liter) | Ysx (g/g) | Specific growth rate (h−1) | 6-MSA concn (mg/liter) | Ysx (g/g) | |
| A4 | 0.23 | NDb | 0.47 | 0.16 | ND | 0.46 | 0.11 | ND | 0.42 | 0.12 | ND | 0.23 |
| AR16msaGP74 | 0.23 | 455 | 0.40 | 0.16 | 148 | 0.39 | 0.11 | 43 | 0.38 | 0.12 | 218 | 0.17 |
| AR16phkmsaGP74 | 0.23 | 310 | 0.46 | 0.19/0.14c | 142/240c | 0.46/0.47c | 0.11 | 54 | 0.32 | 0.15 | 119 | 0.20 |
The standard deviation was less than 3% for all cultivations (triplicate experiments).
ND, not detected.
Results without iodoacetate/results with iodoacetate (1 mM) added to the fermentation broth.
As described above, the hypothesis was that the PHK pathway could increase 6-MSA production by increasing the pool of the precursor acetyl-CoA. This was indeed the case when glycerol was used as the carbon source. 6-MSA production was greater in AR1phk6msaGP74 than in the AR16msaGP74 strain; however, the inverse was observed for growth on glucose and ethanol, and on xylose no significant difference was observed (Table 1). In order to further investigate the production of 6-MSA on xylose, the effect of iodoacetate (a specific inhibitor of glyceraldehyde-3-phosphate dehydrogenase) was studied; it was anticipated that this compound would redirect the carbon flux through the PHK pathway to the precursor acetyl-CoA, thus increasing 6-MSA-production. AR16msaGP74 grown in the presence of 1 mM iodoacetate did not show significant growth and 6-MSA production (data not shown), but in AR1phk6msaGP74 cultures with iodoacetate the 6-MSA production (240 mg/liter) was double that in cultures with no iodoacetate in the medium (142 mg/liter). However, growth and 6-MSA production ceased after consumption of one-half of the available carbon source (10.4 g/liter of xylose was consumed), and the yield of 6-MSA was therefore as high as 23 mg/g of xylose, the highest yield observed in this study for all carbon sources.
In vivo characterization using metabolic network analysis.
Labeling experiments were performed with strains A4, AR16msaGP74, and AR1phk6msaGP74 using d-[1-13C]glucose. The mass spectra obtained from the gas chromatography-mass spectrometry analysis of the hydrolyzed biomass were converted into SFLs (see File S2 in the supplemental material). The SFL of a molecule or a fragment of a molecule reflects the enrichment of 13C (Fig. 1). Using the SFL data, we estimated the fluxes in the central carbon metabolism by considering a simple noncompartmentalized metabolic model of A. nidulans with the Embden-Meyerhof-Parnas pathway, the pentose phosphate (PP) pathway, and the tricarboxylic acid cycle. We also evaluated the use of a more detailed metabolic model with compartmentalization of acetyl-CoA, pyruvate, and oxaloacetate, but the SFL data were only slightly better fitted. In order to avoid overfitting of the limited number of data points, we chose the simpler but more robust version of the two models (see Files S3 and S4 in the supplemental material). The measured and simulated SFLs are shown in File S2 in the supplemental material, while major fluxes are shown in Fig. 1. In general, there is a good fit between the measured and calculated SFLs, although the amino acids Asp188 and Thr175 had a greater deviation (<15%) between measured and simulated fragments in all experiments. This discrepancy may be explained by the simplicity of the model since oxaloacetate (the precursor of Asp188 and Thr175) is not compartmentalized. However, omission of these fragments had only a minor impact on the calculated flux distribution (simulation results not shown).
The flux estimation showed that the wild-type strain (A4) had a relative flux into the oxidative branch of the PP pathway of 56.9, which was reduced to 51.5 when 6-MSA was expressed (Fig. 1). The lower flux of the PP pathway for the AR16msaGP74 strain was reflected in the lower Ysxs. It is very interesting that the flux through XpkA increased from 29.2 to 37.6 when 6-MSA was the only genetic modification compared to the A4 strain, probably in order to compensate for the acetyl-CoA drained for 6-MSA production.
However, when XpkA was overexpressed in the 6-MSA production strain AR1phk6msaGP74, both the flux through the PP pathway and, surprisingly, the flux through the PHK pathway were reduced compared to the results for the wild type. Since the specific growth rates, as well as the Ysxs on glucose, are very similar for the two strains, there must be an alternative source of NADPH for the AR1phk6msaGP74 strain. The metabolic network analysis demonstrates that NADPH is provided mainly through the PP pathway, but there are alternative sources. This may occur through higher flux through the malic enzyme and/or NADP-dependent isocitrate dehydrogenase. In either case, the results demonstrate that this A. nidulans strain has a high degree of metabolic flexibility with respect to fluxes in central carbon metabolism. It is likely that overexpression of PHK enhances this flexibility as it opens an alternative route from sugars to C3 and C2 compounds.
The highest yield of 6-MSA on glucose was obtained for the strain in which heterologous expression of msaS was the sole genetic modification. The flux distribution shows that overexpression of xpkA in the 6-MSA-producing mutant results in 10% lower activity in the PP pathway and 25% lower activity in the PHK pathway compared to the results for the 6-MSA strain, indicating that both acetyl-CoA and NADPH are limiting factors for 6-MSA production in the double mutant in glucose fermentation (Table 1).
This could be further supported by the fact that when iodoacetate was added to xylose cultures, the 6-MSA yield of the double mutant was the highest yield observed in our studies (Table 1). The simultaneous high levels of both NADPH and acetyl-CoA probably led to substantial improvement in the 6-MSA production.
The higher yield of 6-MSA for AR16msaGP74 on glucose, however, was at the expense of a lower Ysx. The metabolic network analysis indicates that expression of msaS results in a high flux into the acetyl-CoA pool via both pyruvate dehydrogenase and XpkA (Fig. 1). This may be due to the higher flux toward 6-MSA and also due to the almost 20% increase in the flux through the tricarboxylic acid cycle.
Transcriptome analysis of the 6-MSAS effects.
Differential gene transcription caused by 6-MSA production in the absence and presence of xpkA overexpression was examined using the A. nidulans part of a recently described tri-Aspergillus microarray (4) containing probes for the P. griseofulvum 6-MSAS as well. The effects of the gene insertions were examined using a 2×2 ANOVA statistical analysis of strains A4, AR1phkGP74, AR16msaGP74, and AR1phk6msaGP74 grown on xylose and glucose media. Data for strains AR16msaGP74 and AR1phk6msaGP74 were generated in this study, while transcriptome data for A4 and AR1phkGP74 are described elsewhere (20a). The method used evaluates the effects of XpkA overexpression and 6-MSAS insertion and the additive effect on each of the two media. A detailed Venn diagram of the regulatory effects is shown in Fig. 2. Generally, the transcriptome results verified the expression and overexpression of msaS and xpkA, respectively. msaS was found to be the most significantly induced gene when the expression levels of the strains with the msaS gene were compared to those of the strains without the gene, as would be expected if the gene is expressed. Similarly, xpkA was found to be highly induced (>8 000-fold from a very low level).
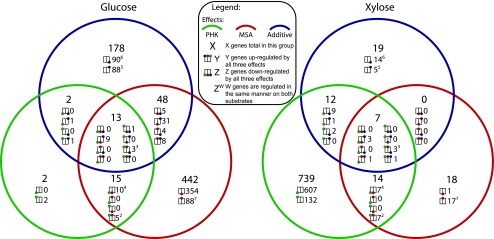
FIG. 2.
Summary of the significant effects of PHK overexpression (PHK) and 6-MSAS insertion (MSA) in A. nidulans on gene transcription levels. Each Venn diagram summarizes the transcriptome analysis performed with glucose or xylose medium. In each of the three circles are the numbers of genes affected by either PHK overexpression, 6-MSAS insertion, or the additive effect. The additive effect can be a synergetic effect (either positive or negative) of PHK overexpression and 6-MSAS. In each subdivision of the Venn diagrams the total number of significant genes is indicated (larger number), and these genes are divided into minor groups (smaller numbers); the boxes indicate the directions of significant responses to the PHK overexpression, 6-MSAS insertion, or additive effects. Superscripts indicate the numbers of genes responding in the same patterns on both carbon sources. For instance, the sixth box of the central subdivision of the glucose Venn diagram shows that there are three genes that are induced by both the PHK overexpression and 6-MSAS insertion effects, but these effects are downregulated (lessened) by the additive effect. Furthermore, these three genes are the same for both carbon sources.
An initial survey of the different effects revealed that the distributions of significantly (P < 0.05) differentially transcribed genes are very different for the two carbon sources. Only 27 genes have significant changes in expression levels with the same pattern, suggesting that the responses are highly specific to the carbon sources (Fig. 2). This is in good accordance with our study of the XpkA overexpression on four different carbon sources, where we showed that there was strong dependence of the XPK response on the carbon source (20a).
For growth on glucose, the 6-MSA and additive effects involve the largest numbers of significantly affected genes (513 and 241 genes, respectively), whereas the XPK effect in the glucose medium involves only a few genes (31 genes). As a function of the ANOVA method, this indicates that the response in AR1phk6msaGP74 diverges a lot from that in AR1phkGP74. For xylose the pattern is the opposite in that XPK overexpression has a large effect (772 genes) and few genes are influenced by the 6-MSA (39 genes) and additive (38 genes) effects. Both categories share a large percentage of their genes with the same response on glucose. It thus seems that expression of 6-MSAS on a xylose medium triggers only a minimal response and that the additive response is almost exclusively the sum of the parts, whereas far more genes are differentially expressed due to an additive effect on a glucose medium. This additive response must be due to an attempt by the cells to maintain homeostasis, since the growth-inducing effect of XPK overexpression is not seen in the double mutant and 6-MSA production is diminished, as described for the physiological characterization of AR1phk6msaGP74.
The induction of genes of metabolic pathways was examined for both sets of effects. As we were examining pathway effects, we employed two cutoff values, a strict cutoff value (P < 0.05) and a less stringent cutoff value (P < 0.25). The less stringent value allows us to identify pathways where the components are regulated uniformly in the same direction, but not all types of regulation are highly significant (see File S5 in the supplemental material for maps of the metabolic pathways described below).
Looking at genes with P values less than 0.05 for the XPK effect, we do not see any pathway-level effects on glucose medium. On xylose, we see lower expression of genes coding for enzymes involved in C5 metabolism and increased transcription of genes involved in the PP pathway, which corresponds well with increased activity of the PHK pathway, shunting carbon away from xylulose-5-phosphate toward acetate metabolism. Increasing the P cutoff value to <0.25 for the XPK effect reveals decreased transcription of genes coding for enzymes in the γ-aminobutyric acid shunt and increased expression of the genes of acetyl-CoA hydrolase and acetate metabolism in both cultures. The glyoxylate shunt and genes putatively involved in fatty acid beta-oxidization show increased expression levels on glucose medium, while the fatty acid degradation pathway genes have consistently lower expression indices due to the XPK effect in xylose cultures, indicating that regulation of the phosphoketolase pathway affects acetyl-CoA levels.
For the 6-MSA effect, we see no pathway-level trends on xylose medium for either of the cutoff levels. For glucose cultures with a P cutoff value of <0.05, steps in acetate-acetyl-CoA metabolism have lower transcription levels, which is opposite the XPK effect, suggesting that there is tighter regulation of acetate metabolism on glucose than on a xylose medium. If the cutoff value is increased to <0.25, the genes for the glyoxylate shunt and fatty acid degradation show uniformly lower expression, which, like the acetate metabolism response, is opposite the XPK effect. This implies that expression of 6-MSAS triggers a metabolic response that in some respects is opposite the transcriptional regulation caused by XPK overexpression on glucose.
When the additive effect is examined, no response at the pathway level is seen for the xylose medium and for the glucose medium at a P value of <0.05. On glucose medium, we see the following trends with a P cutoff value of <0.25: higher expression levels in the γ-aminobutyric acid shunt, oxidative phosphorylation, and fatty acid degradation and lower levels in acetate-acetyl-CoA metabolism. The last type of regulation (Fig. 3) is especially interesting, as it suggests that 6-MSA production decreases the transcriptional response to XpkA overexpression. Lower expression levels for the additive effect show that the downregulating effect is larger than the effect of 6-MSA alone. It is thus clear that the production of 6-MSA in some ways negates the cellular response to xpkA overexpression on glucose, in part due to an effect on the acetyl-CoA pool. This is consistent with the theory that a homeostatic response counteracts the acetyl-CoA-increasing effect of XpkA.
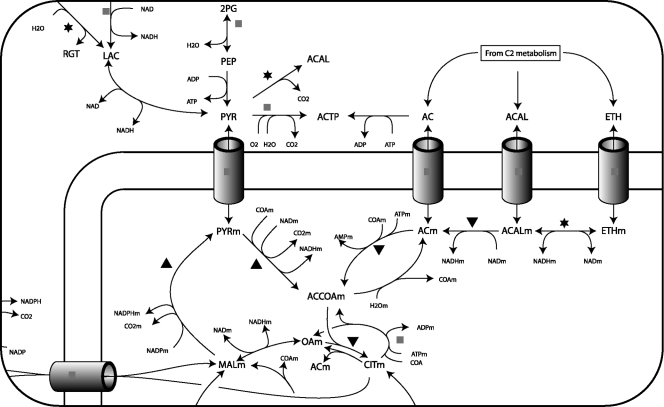
FIG. 3.
Detail from an overview of the additive effects of 6-MSAS expression and PHK overexpression. ▾, additive effect causes lower-level expression of one or more isoenzymes of the process; ▴, higher-level expression;  , the level of expression of some genes with the function is higher while that of other genes is lower;
, the level of expression of some genes with the function is higher while that of other genes is lower;  , a gene could not be assigned to the function. Adapted from a map of A. niger metabolism published by Andersen et al. (3).
, a gene could not be assigned to the function. Adapted from a map of A. niger metabolism published by Andersen et al. (3).
Following annotation of the 1,226 genes significantly regulated by these effects, single genes were examined. For the glucose medium, it is particularly noteworthy that brlA, encoding a regulatory protein inhibiting filamentous growth and involved in dampening the effects of phk overexpression on glucose (1, 20a), is significantly and strongly (194-fold) downregulated by the additive effect, indicating that there is less need for a growth-dampening response in the double mutant than in the xpkA-overexpressing strain or the msaS-expressing strain. On xylose medium, brlA expression is decreased by both the XPK and additive effects (11-fold and 27-fold, respectively). This corresponds well with the growth-arresting function, as both the specific growth rate and Ysx increase in these comparisons (Table 1). The XPK effect on the xylose medium also includes increased levels of expression of a number of putative ribosomal subunits, which is in line with the increased specific growth rates of both strains. The 6-MSAS gene does not have a significant decrease in the additive effect for either strain, indicating that precursor limitation is the more likely explanation for the lower 6-MSA production. Based on the transcriptional response, the stronger regulation seems to be affected around acetyl-CoA; however, as we saw no significant changes in the levels of expression of enzymes of the PP pathway, we can neither confirm or eliminate NADPH limitation as a possible contributor to the limited 6-MSA production based on transcriptome analysis.
DISCUSSION
We previously identified the XPK pathway in A. nidulans and demonstrated that overexpression of xpkA increased the specific growth rate significantly when cells were cultivated on xylose, glycerol, and ethanol (20a). In the present study, we evaluated A. nidulans as a host for the production of secondary metabolites, more specifically, the 6-MSA polyketide. As part of this approach, a 6-MSAS was functionally expressed in A. nidulans and xpkA was overexpressed in the 6-MSA production strain. Similar to the results of our previous study (20a), overexpression of xpkA improved the specific growth rate of the 6-MSA-producing mutant when cells were grown on xylose and ethanol, but not when cells were grown on glucose. Surprisingly, induction of the PHK pathway did not significantly increase 6-MSA production. Especially on glucose and ethanol the final concentration was decreased. Both physiological data and flux analysis suggest that a balance between NADPH and acetyl-CoA is crucial for improving 6-MSA production. If only the acetyl-CoA pool was the bottleneck for high-level production of 6-MSA, then ethanol as a carbon source would yield the highest 6-MSA levels, but this was not the case. Flux studies of the glucose cultures showed that the lower 6-MSA yield for the double mutant was due to NADPH and acetyl-CoA limitation caused by lower flux through both the PP and PHK pathways but could be remedied by regulation of the flux ratio between the lower glycolysis (by limiting or blocking the flux through the glyceraldehyde-3-phosphate dehydrogenase) and PP pathways. This knowledge can be useful for further development of a production process, where polyketide production can be induced when a sufficient biocatalytic mass has been obtained.
The transcriptome data for 6-MSAS expression verified that there is a strong additive effect of msaS expression and xpkA overexpression on glucose, while a 6-MSA and additive effect on xylose is almost nonexistent, and overexpression of xpkA enables both extra growth and 6-MSA production.
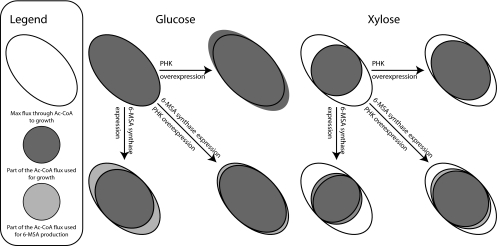
Based on the results of the physiological characterization, flux analysis, and transcription analysis, we propose a model for the response of A. nidulans to xpkA overexpression and 6-MSAS expression on glucose and xylose (Fig. 4). The model describes the competition between biomass formation and 6-MSA formation for the available acetyl-CoA in the presence and absence of XPK overexpression, which increases both acetyl-CoA availability and the growth rate. The model demonstrates that on glucose medium, the acetyl-CoA flux is already at the maximum level. The added effect of XPK overexpression destabilizes the stringent control of the acetyl-CoA levels needed at the maximum growth rate and triggers transcriptional regulation of a large number of genes (more than 400 genes, as shown in Fig. 2). The response includes upregulation of genes coding for enzymes involved in acetate metabolism to control the growth rate. 6-MSAS expression redirects some of the acetyl-CoA to 6-MSA-production, thus decreasing Ysx. The combination of the two effects decreases the regulatory response to XPK overexpression. However, in the competition between 6-MSA production and the cellular response to the overactive PHK pathway, acetyl-CoA is shunted away from 6-MSAS, meaning that XpkA overexpression on a glucose medium actually triggers a decrease in the availability of precursors, explaining the lower yield of 6-MSA. On a xylose medium, the specific growth rate is lower, and the maximum flux through acetyl-CoA does not occur. It is therefore possible to increase the specific growth rate when XpkA is overexpressed. 6-MSAS expression diverts part of the flux to 6-MSA production, thereby decreasing Ysx. This means that in the double mutant, 6-MSA production has to compete with the PHK response that increases the specific growth rate. As the cell is optimized more for growth than for 6-MSA production, the induction of the PHK pathway does not create additional precursors for 6-MSA; the precursors are simply funneled into growth. One could imagine similar models for glycerol and ethanol, but we lack the experimental evidence to validate such models.
FIG. 4.
Model of the effects of PHK overexpression and 6-MSAS insertion in A. nidulans. A tilted oval indicates a hypothetical maximum flux above which a large growth-inhibiting and flux-decreasing response is induced.
Conclusions.
A. nidulans is an important model organism for studies of fundamental eukaryotic cell biology, but because of its close relationship to A. niger and A. oryzae, studies of the metabolism in this organism are also relevant for many industrial processes. Our study shows that A. nidulans is a promising fungal platform for the production of polyketides and other secondary metabolites. Physiological studies of the 6-MSAS-expressing recombinant strains showed that there was successful production of various concentrations of 6-MSA on all carbon sources. A strategy involving the addition of iodoacetate greatly increased 6-MSA production. The transcriptional analysis showed that overexpression of xpkA seems to increase the availability of acetyl-CoA, but due to competition with biomass formation and tight regulation of acetyl-CoA levels, this does not increase the 6-MSA yield but increases the Ysx instead. Furthermore, the functional genomics analyses that we applied to A. nidulans transformants increased our understanding of how metabolism is regulated and what factors should be considered when industrial cell factories are designed.
Supplementary Material
Acknowledgments
We thank Lene Christiansen for crucial technical assistance with array preparation.
This study was financed in part by the Villum Kann Rasmussen and Lundbeck Foundations (G.P.) and by the Danish Research Agency for Technology and Production (M.R.A.).
Footnotes
Published ahead of print on 23 January 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, T., M. Boylan, and W. Timberlake. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353-362. [DOI] [PubMed] [Google Scholar]
- 2.Affymetrix. 2007. GeneChip expression analysis technical manual P/N 702232 Rev. 2. Affymetrix, Santa Clara, CA.
- 3.Andersen, M. R., M. L. Nielsen, and J. Nielsen. 2008. Metabolic model integration of the bibliome, genome, metabolome, and reactome of Aspergillus niger. Mol. Syst. Biol. 4:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen, M. R., W. Vongsangnak, G. Panagiotou, M. P. Salazar, L. Lehmann, and J. Nielsen. 2008. A tri-species Aspergillus micro array—comparative transcriptomics of three Aspergillus species. Proc. Natl. Acad. Sci. USA 105:4387-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedford, D. J., E. Schweizer, D. A. Hopwood, and C. Khosia. 1995. Expression of functional fungal polyketide synthase in the bacterium Streptomyces coelicolor A3(2). J. Bacteriol. 177:4544-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 7.Bolstad, B., R. Irizarry, M. Astrand, and T. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, B., and J. Nielsen. 2000. Metabolic network analysis of Penicillium chrysogenum using 13C labeled glucose. Biotechnol. Bioeng. 68:652-659. [PubMed] [Google Scholar]
- 9.Clutterbuck, A. J. 1974. Aspergillus nidulans, p. 447-510. In R. C. King (ed.), Handbook of genetics, vol. 1. Plenum Publishing Corp., New York, NY. [Google Scholar]
- 10.David, H., G. Hofmann, A. P. Oliveira, H. Jarmer, and J. Nielsen. 2006. Metabolic network driven analysis of genome wide transcription data from Aspergillus nidulans. Gen. Biol. 7:R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David, H., A. M. Krogh, C. Roca, M. Åkesson, and J. Nielsen. 2005. CreA influences the metabolic fluxes of Aspergillus nidulans during growth on glucose and xylose. Microbiology 151:2209-2221. [DOI] [PubMed] [Google Scholar]
- 12.Guo, B. Z., J. Yu, C. C. Holbrook, R. D. Lee, and R. E. Lynch. 2003. Application of differential display RT-PCR and EST-microarray technologies to the analysis of gene expression in response to drought stress and elimination of aflatoxin contamination in corn and peanut. J. Toxicol. 22:287-312. [Google Scholar]
- 13.Harvey, A. L. 2000. Strategies for discovering drugs from previously unexplored natural products. Drug Discov. Today 5:294-300. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry, R., B. Hobbs, F. Collin, Y. Beazer-Barclay, K. Antonellis, U. Scherf, and T. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 15.Johnstone, I. L., S. G. Hughes, and A. J. Clutterbuck. 1985. Cloning an Aspergillus nidulans developmental gene by transformation. EMBO J. 4:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kealey, J. T., L. Liu, D. V. Santi, M. C. Betlach, and P. J. Barr. 1998. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. USA 95:505-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda, H., M. Sano, Y. Maruyama, T. Tanno, T. Akao, Y. Totsuka, M. Endo, R. Sakurada, Y. Yamagata, M. Machida, O. Akida, F. Haseqawa, K. Abe, K. Guomi, T. Nakajima, and Y. Iguchi. 2004. Transcriptional analysis of genes for energy catabolism and hydrolytic enzymes in the filamentous fungus Aspergillus oryzae using cDNA microarrays and expression sequence tags. Appl. Microbiol. Biotechnol. 65:74-83. [DOI] [PubMed] [Google Scholar]
- 18.Mogensen, J., H. B. Nielsen, G. Hofmann, and J. Nielsen. 2006. Transcription analysis using high-density microarrays of Aspergillus nidulans wild type and creA mutant during growth on glucose or ethanol. Fungal Genet. Biol. 43:593-603. [DOI] [PubMed] [Google Scholar]
- 19.Nissen, T. L., U. Schulze, J. Nielsen, and J. Villadsen. 1997. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology 143:203-218. [DOI] [PubMed] [Google Scholar]
- 20.O'Brian, G. R., A. M. Fakhoury, and G. A. Payne. 2003. Identification of genes differentially expressed during aflatoxin biosynthesis in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet. Biol. 39:118-127. [DOI] [PubMed] [Google Scholar]
- 20a.Panagiotou, G., M. R. Andersen, T. Grotkjaer, T. Regueira, G. Hofmann, J. Nielsen, and L. Olsson. 2008. Systems analysis unfolds the growth effects of the phosphoketolase pathway in Aspergillus nidulans. PLoS One 3:e3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, S. J., S. Y. Lee, J. Cho, T. Y. Kim, J. W. Lee, J. H. Park, and M. J. Han. 2005. Global physiological understanding and metabolic engineering of microorganisms based on omics studies. Appl. Microbiol. Biotechnol. 68:567-579. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen, H., M. Carlsen, and J. Nielsen. 1999. Identification of enzymes and quantification of metabolic fluxes in the wild type and in recombinant Aspergillus oryzae strain. Appl. Environ. Microbiol. 65:11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 24.Ruijter, G. J. G., and J. Visser. 1997. Carbon repression in aspergilli. FEMS Microbiol. Lett. 151:103-114. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. H. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Scheidegger, K., and G. Payene. 2003. Unlocking the secrets behind secondary metabolism: a review of Aspergillus flavus from pathogenicity to functional genomics. J. Toxicol. 22:423-459. [Google Scholar]
- 27.Senac, T., and B. Hahn-Hagerdal. 1990. Intermediary metabolite concentrations in xylulose- and glucose-fermenting Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 56:120-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sims, A. H., G. D. Robson, D. C. Hoyle, S. G. Oliver, G. Turner, R. A. Prade, H. H. Russell, N. S. Dunn-Coleman, and M. E. Gent. 2004. Use of expressed sequence tag analysis and cDNA microarrays of the filamentous fungus Aspergillus nidulans. Fungal Genet. Biol. 41:199-212. [DOI] [PubMed] [Google Scholar]
- 29.Smyth, G. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article 3. [DOI] [PubMed] [Google Scholar]
- 30.Spencer, J. B., and P. M. Jordan. 1992. Purification and properties of 6-methylsalicylic acid from Penicillium patulum. Biochem. J. 288:839-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wattanachaisaereekul, S., A. E. Lantz, M. L. Nielsen, O. S. Andresson, and J. Nielsen. 2007. Optimization of heterologous production of the polyketide 6-MSA in Saccharomyces cerevisiae. Biotechnol. Bioeng. 97:893-900. [DOI] [PubMed] [Google Scholar]
- 32.Wiechert, W. 2001. 13C metabolic flux analysis. Metab. Eng. 3:195-206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.