Abstract
Microscopic eukaryotes are known to have important ecosystem functions, but their diversity in most environments remains vastly unexplored. Here we analyzed an 18S rRNA gene library from a subsurface iron- and sulfur-oxidizing microbial community growing in highly acidic (pH < 0.9) runoff within the Richmond Mine at Iron Mountain (northern California). Phylogenetic analysis revealed that the majority (68%) of the sequences belonged to fungi. Protists falling into the deeply branching lineage named the acidophilic protist clade (APC) and the class Heterolobosea were also present. The APC group represents kingdom-level novelty, with <76% sequence similarity to 18S rRNA gene sequences of organisms from other environments. Fluorescently labeled oligonucleotide rRNA probes were designed to target each of these groups in biofilm samples, enabling abundance and morphological characterization. Results revealed that the populations vary significantly with the habitat and no group is ubiquitous. Surprisingly, many of the eukaryotic lineages (with the exception of the APC) are closely related to neutrophiles, suggesting that they recently adapted to this extreme environment. Molecular analyses presented here confirm that the number of eukaryotic species associated with the acid mine drainage (AMD) communities is low. This finding is consistent with previous results showing a limited diversity of archaea, bacteria, and viruses in AMD environments and suggests that the environmental pressures and interplay between the members of these communities limit species diversity at all trophic levels.
The dissolution of pyritic rocks results in the formation of extremely acidic, warm, metal-rich solutions, referred to as acid mine drainage (AMD). Subaqueous and solution-air interface biofilms often develop in AMD systems. The bacterial and archaeal membership of these communities has been extensively characterized using culture-independent 16S rRNA gene surveys (6, 7, 10, 13, 15, 17), sequencing of random shotgun genomic libraries (26), and proteomics (23). This prior work has shown that a relatively small number of phylogenetically diverse archaea and bacteria inhabit these extreme environments. Much less studied are the eukaryotic members of AMD communities. Culture-independent gene surveys of AMD-impacted phototrophic communities from the Rio Tinto River in Spain (pH 2) have revealed high eukaryotic species richness (2, 3). In contrast, in preliminary work carried out within the Richmond Mine, an extremely acidic (pH < 1), subsurface AMD site at Iron Mountain in northern California, diversity was reported to be limited to Ascomycota fungi belonging to the Dothideomycetes and the Eurotiomycetes. Cultivation attempts resulted in the isolation of a member of the Dothideomycetes, “Acidomyces richmondesis,” from this site (8). Alphaproteobacterial endosymbionts have been discovered in some protists (7). Protists isolated from AMD graze on acidophilic bacteria (18, 22), suggesting that they likely play a predatory role and influence the abundances of various community members. Perhaps more importantly, eukaryotes likely play a large role in the cycling of carbon within AMD communities.
To date, three communities in the mine have been deeply sequenced using random shotgun genomic sequencing (19, 26; B. J. Baker et al., unpublished data). However, eukaryotic sequences in these data sets have not been evaluated. Therefore, the full diversity of eukaryotes is unknown, and their potential ecological roles in acidic environments remain poorly understood. In this study, a eukaryotic rRNA gene library was generated from the same sample (and DNA extraction) that was genomically characterized by Lo et al. (19). We report the design of lineage-specific fluorescent in situ hybridization (FISH) probes for each of these groups and microscopic visualization of the distribution and morphological characteristics of each population.
MATERIALS AND METHODS
Site description.
Samples were collected on 11 July 2005 from the “UBA” site (located at the back of the A drift) in the Richmond mine (19). The biofilm was growing on the surface of a pyrite pile and covered by slowly flowing AMD solutions draining from deeper regions of the mine. The pH (1.23) and temperature (38°C) were measured on site at the time of collection. Between March 2005 and 26 June 2008, the temperature at this site has been in the range of 30 to 46°C. Extensive analysis of bacterial and archaeal sequences in the UBA genomic library revealed that the sample is dominated by Leptospirillum group II (19), with lower numbers of Leptospirillum group III (D. Aliaga Goltsman et al., unpublished data) and archaea, mostly from the Thermoplasmatales lineage.
DNA extraction and clone library construction.
Samples were stored on wet ice and transported to the laboratory. DNA was extracted ∼48 h after collection using a modified phenol-chloroform extraction method, as described by Bond et al. (10). PCR amplifications of 18S rRNA genes used the 515F (5′-GTGCCAAGCAGCCGCGGTAA-3′) and 1209R (5′-GGGCATCACAGACCTG-3′) primers. Amplification was performed for 20 cycles with an annealing temperature of 55°C using the Easy-A High-Fidelity PCR master mix from Stratagene. The PCR products were cloned into pCR-4 TOPO and sequenced by the DOE Joint Genome Institute.
Sequence analysis.
One hundred seventy-six high-quality sequences were assembled using the phred software program, resulting in 86 18S rRNA genes. These gene sequences were compared to sequences available in the nonredundant nucleotide database in the National Center for Biotechnology database using BLASTN (1). The rRNA genes from this study and closely related gene sequences from the public database were imported into the ARB software package (21). A mask consisting of 1,405 positions was generated for the SAWY clones. Trees for the Basidiomycota, Ascomycota, Heterolobosea, and acidophilic protist clade (APC) were generated using 55, 45, 37, and 50 taxa, including 910, 874, 1,072, and 1,170 character sets, respectively. The topologies of the trees were confirmed using masks with and without the hypervariable region and several different phylogenetic methods, including distance, parsimony, maximum likelihood, and bayesian. Alternative-topology full heuristic searches used 100 bootstrap replicates of random addition with a branch-swapping algorithm, tree bisection-reconnection. TREE_PUZZLE bootstrapping values were calculated in ARB with default parameters. All of the trees presented here were generated using the maximum-likelihood method (PHYLML) in ARB with a 1405 mask and HKY nucleotide substitution model. Operational taxonomic units for rarefaction analyses were defined by >95% sequence similarity.
Whole-cell rRNA FISH analysis and probe design.
Samples were fixed in 4% paraformaldehyde, washed with phosphate-buffered saline (pH 1.2), and stored at −20°C within 12 h of collection. Oligonucleotide probes were designed to target different groups identified in the clone library, as detailed elsewhere (8). Probe selection took into consideration the accessibility of the target region, as reported by Behrens et al. (9). Hybridizations were performed on fixed samples, as described previously, with incubation at 46°C and washing at 48°C for 15 min (7). Hybridizations were counterstained with a 4′,6′-diamidino-2-phenylindole dihydrochloride DNA stain. The optimal stringency for the probes was determined empirically using fixed AMD samples at 5% formamide increments from 10 to 50%. For FISH analyses, Saccharomyces cerevisiae was used as a negative control during all of the optimization hybridization experiments to confirm that we were not getting nonspecific binding. Fluorescent images (two-dimensional) were acquired using a Leica DMRX microscope at magnification ×630.
Nucleotide sequence accession numbers.
Completely sequenced clones were deposited in the National Center for Biotechnology GenBank under accession numbers DQ423685 to DQ423774.
RESULTS
Eukaryotic diversity.
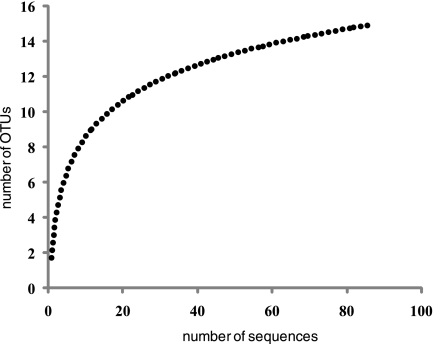
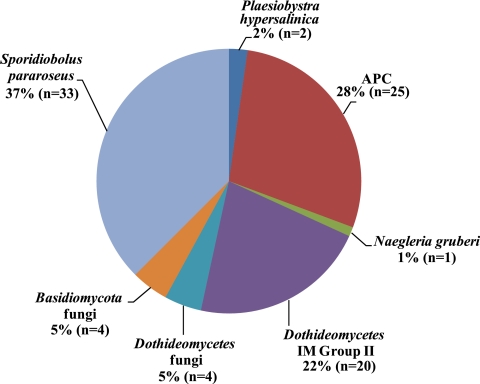
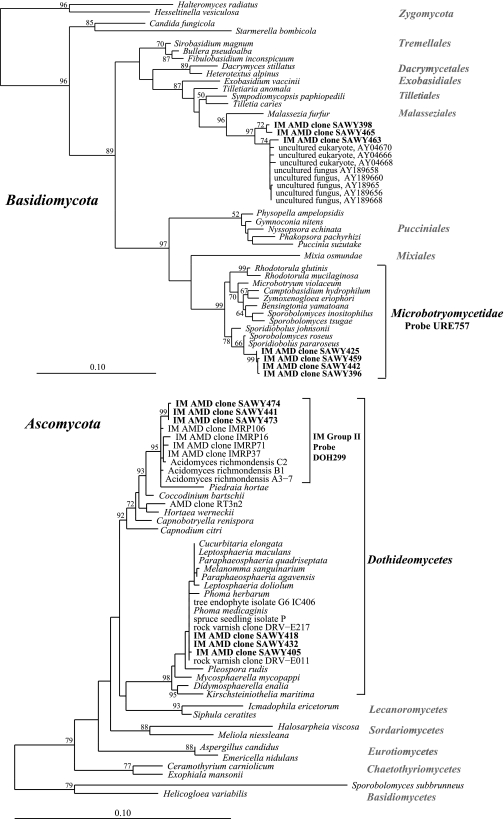
The 18S rRNA gene library was sequenced to near-saturation (Fig. 1). Therefore, excluding PCR primer and reaction biases which could result in an undersampling of the diversity, it appears that we have identified most of the eukaryotes present in that sample. The majority (68%) of the clones from the UBA 18S rRNA gene library were fungi, and 56% of these were closely related to those described by Baker et al. (8) (see Table 1. The majority of the fungal clones (33 out of 60) belonged to the Basidiomycota, class Urediniomycetes, subclass Microbotryomycetidae, order Sporidiobolales. Their closest known isolates (>99%) are Sporidiobolus pararoseus (accession no. AB021689) and Sporobolomyces roseus (accession no. X60181) (Fig. 2). Four other fungal clones (SAWY398, SAWY463, SAWY465, and SAWY503) cluster in an unclassified order in the Basidiomycota (Fig. 3, top) and are 97% similar to clones from an anoxic marine basin (25), a hospital therapy pool (5), hydrothermal sediment (20) and vent (14), and a human ear (16).
FIG. 1.
Rarefaction analyses of the 18S rRNA gene library from the UBA biofilm. The curve was smoothed using logarithmic trendline calculation from the r2 value 0.9353.
TABLE 1.
Clones from the UBA-BS 18S rRNA gene library and their taxonomic affiliations
| Clones | Closest BLAST match | % Similarity | Taxonomic affiliation |
|---|---|---|---|
| SAWY386, -388, -396, -404, -408, -425, -427, -428, -442, -387, -389, -391, -392, -414, -416, -420, -421, -430, -433, -434, -450, -453, -454, -455, -456, -457, -459, -462, -471, -475, -477, -385, -443 | Sporidiobolus pararoseus | >99 | Basidiomycota; Urediniomycetes; Sporidiobolales |
| SAWY393, -399, -413, -415, -431, -436, -441, -473, -474, -476, -423, -424, -448, -451, -458, -464, -470, -472, -438 | AMD clone IMRP57 (AY343922) | >99 | Ascomycota; Dothideomycetes, IM group II |
| SAWY394, -446, -395, -407, -400, -401, -402, -403, -406, -411, -412 -417, -422, -426, -429, -437, -449, -466, -468, -469, -444, -410, -452, -479, -440 | Nuclearia moebiusi | >84a | Novel |
| SAWY418, -432, -405, -419 | Tree endophyte isolate G6 IC406 | >99 | Ascomycota; Dothideomycetes |
| SAWY398, -463, -465, -503 | Marine isolate D149 (AY256321) | >97 | Basidiomycota |
| SAWY439, -445 | Plaesiobystra hypersalinica | >88 | Heterolobosea; Plaesiobystra |
| SAWY447 | Naegleria gruberi (M18732) | 98 | Heterolobosea; Vahlkampfiidae |
Similarity only of alignable regions; hypervariable regions excluded.
FIG. 2.
Pie chart showing the percentages of clones assigned to the various groups. APC, Naegleria gruberi, and Plaesiobystra hypersalinica are all protists; the remainder are fungi. The number of clones in each group (n) is given.
FIG. 3.
Phylogenetic trees of fungal clone groups generated using the maximum-likelihood method. The tree on the top is Basidiomycota-associated and the bottom Ascomycota-associated clones. Only representative clones from those belonging to the Microbotryomycetidae and IM group II are shown (7). The taxonomic groups and FISH probe specificities are delineated on the right. Bootstrap values are shown at the respective nodes. Scale bars are equal to 0.10 changes per site.
Several clones (n = 20) from the library are very similar to a group (IM group II) of AMD fungus clones and isolates (Fig. 3) from a previous study from a different date and site in the Richmond Mine (8). Interestingly, two genomic sequences (XYG21783.b2 and XYG30961.b1) from a random shotgun library from the five-way site in the mine (26) were identified as 18S rRNA genes belonging to fungi of IM group II. One of these sequences was included in the phylogenetic tree shown in Fig. 3, “AMD CG clone XYG21783.” Three clones (SAWY405, SAWY418, and SAWY432) fall within another clade within the Dothideomycetes. This group is identical (100% at the 18S rRNA gene sequence) to Phoma medicaginis (accession no. DQ109961), a tree endophyte isolate (accession no. AY382648), a spruce tree isolate (accession no. AY275186), and environmental clones from rock varnish (accession no. AY923088).
Roughly a third of all sequences (Fig. 2) fall into four distinct protist groups. The majority (25 of 28) of these sequences form a deeply branched clade, named the APC. This group is significantly divergent from anything previously identified, with <76% similarity to any sequences in public databases. Therefore, this group represents kingdom-level novelty in the eukaryotes (12). The group is monophyletic and deeply branched within the eukaryotes (Fig. 4). There is up to 13% sequence variation between clones within the APC. This level of sequence divergence likely represents taxonomic diversity at the family level and greater. Interestingly, members of the APC are the only clones that are not closely related to organisms from neutrophilic environments.
FIG. 4.
Phylogenetic trees of protist clone groups generated using the maximum-likelihood method. The tree on the top shows the Heterolobosea, and the bottom tree shows the APC clones in relation to other major eukaryotic groups. Only representative clones are shown from those belonging to the APC. The taxonomic groups and FISH probe specificities are delineated on the right. Bootstrap values are shown at the respective nodes. Scale bars are equal to 0.10 changes per site.
Organisms belonging to the protist class Heterolobosea identified in AMD communities at Iron Mountain and Rio Tinto, Spain (2, 3, 8), fall into three different subgroups with the Vahlkampfiidae family (Fig. 4). One clone (SAWY447) is 98% similar to Naegleria gruberi. Another clone, W16 (accession no. AY394431), was previously reported (8) and has a closest match to Singhamoeba horticola (accession no. AF011456). The other two clones have 81.2% and 82.5% similarities to Plaesiobystra hypersalinica, an unclassified member of the class.
FISH analyses.
Table 2 lists the FISH probes developed to target the main eukaryotic groups identified in the Richmond Mine biofilms. Probe HLB1074 was designed to target the entire Heterolobosea group, while VAH1044 and NAE1041 target a subset of the class, as delineated in Fig. 4. The APC987 probe targets all of the APC clones that were recovered in the study.
TABLE 2.
Probes developed and used in this study
| Probe name (E. coli position) | Probe sequence (5′-3′) | Length (nta) | Tm (°C)b | Optimal formamide % | Targets |
|---|---|---|---|---|---|
| APC987 (987-998) | CCTAAYCATGTCCGGACC | 18 | 57 | 35 | AMD APC group |
| HLB1074 (1074-1092)c | AASWAGACAGATCACTCCACG | 21 | 56 | 35 | Heterolobosea |
| VAH1044 (1044-1059) | ACCATGCACCACCTTCTG | 18 | 58 | 35 | Vahlkampfiidae |
| NAE1041 (1041-1055) | TGCACTACCCACCACACA | 18 | 58 | 30 | Naegleria spp. |
| DOH299 (299-317) | CCTCTCCGGTATCGTACCC | 19 | 60 | 10 | Fungus IM group II |
nt, nucleotides.
Nearest-neighbor calculation (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/) using 50 mM NaCl and 50 μM oligonucleotide.
Requires competitor AASCAGACAGATCACTCCACG.
Microscopic analysis of samples labeled with FISH probes revealed that members of the APC range in size from 10 to 30 μm in diameter, making them the largest organisms in AMD biofilms (Fig. 5A and B). The cells highlighted with the HLB1074 and NAE1041 probes are 10 to 25 μm in diameter, and those visualized using the VAH1044 probe are the smallest protists, ranging from 5 to 10 μm in diameter (Fig. 5B).
FIG. 5.
FISH micrographs of mine communities using probes developed in this study. General eukaryote probes (EUKb-mix in fluorescein isothiocyanate [7]) are shown in green, and archaea/bacteria are blue (Cy5, ARC915, and EUB338). (A) Five-way (March 2002) community showing a dominance of Vahlkampfia-like cells hybridized to the VAH1044 probe, labeled with Cy3 (yellow). APC cells hybridized to the APC987 probe (B and C) and cells targeted by the HLB1074 probe (D) are all labeled with Cy3 (yellow) in the UBA (July 2005). Scale bars = 50 μm.
Microscopic observations of 24 archived samples using the probes revealed significant temporal and spatial variations in the abundances of eukaryotic lineages identified in the current study. For example, in the UBA sample, the most abundant protists belong to the APC, whereas the five-way sample from Tyson et al. (26) was almost completely dominated by Vahlkampfia-like cells (Fig. 5). Fungi are commonly the predominant eukaryotes in streamer biofilms that grow in flowing water and may provide the backbone for these structures.
Eukaryote sequence in shotgun genomic data sets.
Given the identification of clone SAWY447, with 98% sequence similarity to the 18S rRNA gene of Naegleria gruberi, we used the publically available genome of N. gruberi (http://genome.jgi-psf.org/Naegr1/) to search the community genomic data sets from the Richmond Mine biofilms for DNA sequences encoding eukaryotic proteins. From this we were able to identify only 98 genomic sequences that contain predicted proteins that have strong matches to eukaryote sequences available in public databases. The majority (50) of these sequences encoded hypothetical proteins, but they also included housekeeping genes (e.g., tRNA synthetases) and genes involved in transport and central carbon metabolism. This number is extremely low considering we searched a total of 473,176 genomic sequences from the three samples (UBA, 5way, and UBA BS), but this might be the result of the paucity of genome sequences for these lineages.
DISCUSSION
The results of the current study suggest that the dominant groups of AMD eukaryotes in the Richmond Mine are the fungi, APC, and Heterolobosea. The UBA 18S rRNA gene library achieved saturation after sequencing of a relatively small number of clones. This result indicates that the diversity of eukaryotes in the Richmond Mine biofilms is relatively low, as is the case for archaea and bacteria (6, 10, 13). Recent recovery of genomic sequences from bacteriophage and archaeal viruses from AMD biofilms also indicated a low viral diversity (4). The low viral diversity is attributed to the low bacterial and archaeal host diversity and low eukaryotic diversity to low diversity of organic substrates derived from bacteria and archaea. In turn, the dominance of AMD communities by only a few bacteria and archaea is likely related to the small number of chemoautotrophic energy sources (primarily reduced iron and sulfur compounds). This study demonstrates that the simplicity of community membership in the Richmond Mine AMD system compared to other ecosystems persists at all ecological levels. It is interesting to note that previous investigators have reported considerable species richness in the Rio Tinto system. In contrast to the self-contained subsurface Richmond Mine ecosystem, the AMD-impacted Rio Tinto system receives diverse inputs from the surrounding watershed and sunlight, which sustains a diversity of eukaryotic phototrophs.
Most bacterial and archaeal members of AMD communities form distinct lineages of strict acidophiles, suggesting that they have inhabited low-pH, metal-rich environments for hundreds of millions if not billions of years. However, the eukaryotes (especially the fungi) are closely related to neutrophiles, suggesting that they recently adapted to life in AMD communities. Heterolobosea and Dothideomycetes fungi are common to other AMD systems (2, 3, 8), and the Dothideomycetes have been cultured from sites with low pH and high metal concentrations (8, 11). Also, Naegleria spp. are commonly found in warm environments, up to 55°C (24), similar to those found in the mine. Thus, these groups may have traits that provide them with a selective advantage for survival in extremely acidic environments, such as metal resistance genes. Since the APC group appears to constitute a dominant eukaryote in the mine waters, it is likely that this group is adapted to life in the harsh AMD environment. Future studies will reveal how widespread this lineage is in the environment and what makes these organisms uniquely suited to life in AMD.
Using genomic information for an N. gruberi strain related to organisms identified in this study, we identified very little sequence from this protist population. However, the other dominant protist group, APC, is extremely divergent from anything that is in culture or has been sequenced. Thus, it is difficult to identify APC genomic sequences, despite their suspected presence in these data sets. Given that the number of N. gruberi genomic sequences is very low and that we found very few 18S rRNA genes, we do not expect many eukaryotic sequence in these libraries. This is probably due to their low cell numbers compared to archaea and bacteria.
The current study expands the number of eukaryotic lineages known to be associated with AMD to include two groups of the Basidiomycota and a novel deeply branched protist lineage and confirms that members of the Heterolobosea and fungi belonging to the Dothideomycetes are widespread in acidic environments. Low coverage of community genomic information from shotgun libraries of Vahlkampfiidae family amoebas revealed that either deeper sequencing or selected enrichments would be required to begin to characterize these organisms with a culture-independent genomic approach. The design of FISH probes for these groups enabled initial characterization of the distribution of eukaryotes in the Richmond Mine and will facilitate identification of samples for future targeted eukaryotic genomic studies. The probes also open the way for documentation of eukaryotic lineages in other AMD systems around the world.
Acknowledgments
We thank Ted Arman (president, Iron Mountain Mines Inc.) and Rick Sugarek (EPA) for access to the Richmond mine and Rudy Carver for assistance with sample collection. We thank Ian Lo for assistance with DNA extractions. Sequencing was carried out by the DOE Joint Genome Institute.
This work was supported by grants from the NSF Biocomplexity Program, the NASA Astrobiology Institute, and the DOE Genomics:GTL Program.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral Zettler, L. A., F. Gómez, E. R. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Eukaryotic diversity in Spain's river of fire. Nature 417:137. [DOI] [PubMed] [Google Scholar]
- 3.Amaral Zettler, L. A., M. A. Messerli, A. D. Laatsch, P. J. S. Smith, and M. L. Sogin. 2003. From genes to genomes: beyond biodiversity in Spain's Rio Tinto. Biol. Bull. 204:205-209. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, A. F., and J. F. Banfield. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047-1050. [DOI] [PubMed] [Google Scholar]
- 5.Angenent, L. T., S. T. Kelley, A. S. Amand, N. R. Pace, and M. T. Hernandez. 2005. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc. Natl. Acad. Sci. USA 102:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microb. Ecol. 44:1-14. [DOI] [PubMed] [Google Scholar]
- 7.Baker, B. J., P. Hugenholtz, S. C. Dawson, and J. F. Banfield. 2003. Extremely acidophilic protists host Rickettsiales-lineage endosymbionts with an intervening sequence in their 16S rRNA genes. Appl. Environ. Microbiol. 69:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker, B. J., M. A. Lutz, S. C. Dawson, P. L. Bond, and J. F. Banfield. 2004. Metabolically active eukaryotes in extremely acidic mine drainage. Appl. Environ. Microbiol. 70:6264-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behrens, S., C. Ruhland, J. Inacio, H. Huber, A. Fonseca, I. Spencer-Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bond, P. L., S. P. Smriga, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cánovas, D., C. Durán, N. Rodriguez, R. Amils, and V. de Lorenzo. 2003. Testing the limits of biological tolerance to arsenic in a fungus isolate from the River Tinto. Environ. Microbiol. 5:133-138. [DOI] [PubMed] [Google Scholar]
- 12.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Druschel, G. K., B. J. Baker, T. M. Gihring, and J. F. Banfield. 2004. Biogeochemistry of acid mine drainage at Iron Mountain, California. Geochem. Trans. 5:13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgcomb, V. P., D. T. Kysela, A. Teske, A. de Vera Gomez, and M. L. Sogin. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 99:7658-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, K. J., T. M. Gihring, and J. F. Banfield. 1999. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl. Environ. Microbiol. 65:3627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank, D. N., G. B. Spiegelman, W. Davis, E. Wagner, E. Lyons, and N. R. Pace. 2003. Culture-independent molecular analysis of microbial constituents of the healthy human outer ear. J. Clin. Microbiol. 41:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallberg, K. B., K. Coupland, S. Kimura, and D. B. Johnson. 2006. Macroscopic streamer growths in acidic, metal-rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl. Environ. Microbiol. 72:2022-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, D. B., and L. Rang. 1993. Effects of acidophilic protozoa on populations of metal-mobilizing bacteria during the leaching of pyritic coal. J. Gen. Microbiol. 139:1417-1423. [Google Scholar]
- 19.Lo, I., V. J. Denef, N. C. Verberkmoes, M. B. Shah, D. Goltsman, G. DiBartolo, G. W. Tyson, E. E. Allen, R. J. Ram, J. C. Detter, P. Richardson, M. P. Thelen, R. L. Hettich, and J. F. Banfield. 2007. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 466:537-541. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Garcia, P., H. Philippe, F. Gail, and D. Moreira. 2003. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc. Natl. Acad. Sci. USA 100:697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, Arno Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, W. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGinness, S., and D. B. Johnson. 1992. Grazing of acidophilic bacteria by a flagellated protozoan. Microb. Ecol. 23:75-86. [DOI] [PubMed] [Google Scholar]
- 23.Ram, R. J., N. VerBerkmoes, M. P. Thelen, G. W. Tyson, B. J. Baker, M. Shah, R. C. Blake II, R. Hettich, and J. F. Banfield. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915-1920. [PubMed] [Google Scholar]
- 24.Sheehan, K. B., M. J. Ferris, and J. M. Henson. 2003. Detection of Naegleria sp. in a thermal, acidic stream in Yellowstone National Park. J. Eukaryot. Microbiol. 50:263-265. [DOI] [PubMed] [Google Scholar]
- 25.Stoeck, T., G. T. Taylor, and S. S. Epstein. 2003. Novel eukaryotes from the permanently anoxic cariaco basin (Caribbean Sea). Appl. Environ. Microbiol. 69:5656-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richarson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]