Abstract
The complete genomes of three strains from the phylum Acidobacteria were compared. Phylogenetic analysis placed them as a unique phylum. They share genomic traits with members of the Proteobacteria, the Cyanobacteria, and the Fungi. The three strains appear to be versatile heterotrophs. Genomic and culture traits indicate the use of carbon sources that span simple sugars to more complex substrates such as hemicellulose, cellulose, and chitin. The genomes encode low-specificity major facilitator superfamily transporters and high-affinity ABC transporters for sugars, suggesting that they are best suited to low-nutrient conditions. They appear capable of nitrate and nitrite reduction but not N2 fixation or denitrification. The genomes contained numerous genes that encode siderophore receptors, but no evidence of siderophore production was found, suggesting that they may obtain iron via interaction with other microorganisms. The presence of cellulose synthesis genes and a large class of novel high-molecular-weight excreted proteins suggests potential traits for desiccation resistance, biofilm formation, and/or contribution to soil structure. Polyketide synthase and macrolide glycosylation genes suggest the production of novel antimicrobial compounds. Genes that encode a variety of novel proteins were also identified. The abundance of acidobacteria in soils worldwide and the breadth of potential carbon use by the sequenced strains suggest significant and previously unrecognized contributions to the terrestrial carbon cycle. Combining our genomic evidence with available culture traits, we postulate that cells of these isolates are long-lived, divide slowly, exhibit slow metabolic rates under low-nutrient conditions, and are well equipped to tolerate fluctuations in soil hydration.
The phylum Acidobacteria is one of the most abundantly distributed bacterial groups in the environment (32, 34, 84). Its members have been detected in numerous 16S rRNA gene surveys from environments that vary greatly in physical and biogeochemical characteristics, including soil, sediment, freshwater, marine, extreme, and polluted environments worldwide (3, 32, 34). Analyses of 3,000 acidobacterial 16S rRNA gene sequences indicated that the phylum is highly diverse, comprising 26 distinct phylogenetic subdivisions (2). Acidobacterial sequences are particularly abundant in soils and sediments, comprising 10 to 50% of the total bacterial 16S rRNA gene sequences in clone libraries (2, 5, 17, 26, 34, 39, 48, 52, 64, 76, 77), and members of different subdivisions occur in these different environments.
Members of the phylum Acidobacteria have been very difficult to isolate and culture in the laboratory, and cultured representatives of only 5 of the 26 subdivisions currently exist (2, 32, 94). Across this small collection of isolates, most are aerobic heterotrophs (16, 21, 37, 44, 61, 62, 71). One phototroph (6, 81) and two obligate anaerobes (12, 49) have also been described. Candidate names have been given to several isolates, and five genera have been formally described: Holophaga and Geothrix from subdivision 8 (12, 49) and Acidobacterium (41), Terriglobus (21), and Edaphobacter (44) from subdivision 1. The isolates from soil, all of which are members of subdivisions 1, 2, 3, and 4, do not grow on standard media. They grow very slowly, requiring several days to weeks to form visible colonies on complex, low-nutrient media (16, 21, 37, 44, 61, 81). Such culture difficulties substantially confound our ability to determine the metabolic and physiological traits of the members of this phylum.
The ubiquity and abundance of acidobacteria in soils and their ability to withstand polluted and extreme environments suggest that they serve functions that are important in the environment and that are potentially quite varied. Despite our ability to detect them in many environments by molecular methods, we know very little about their individual physiology and even less about their potential functions in, or metabolic contributions to, the environment. The goal of this project was to identify potential acidobacterial traits that suggest ecological roles for members of this phylum in soil so that these might be tested more directly in soil environments. To circumvent the culture difficulties, we took a genomics approach to identify these traits. The complete genomes of three acidobacterial strains are described here with a comparison of genome features focused on acidobacterial traits that may contribute to survival and growth in soil, including their transporters and their abilities to use carbon, cycle nitrogen, scavenge iron, and produce antimicrobial compounds.
MATERIALS AND METHODS
Acidobacterial strains.
Two of the sequenced acidobacterial strains represent subdivision 1: Acidobacterium capsulatum, isolated from sediments in acidic drainage from the Yanahara pyrite mine in Japan (41), and strain Ellin345, isolated from soil in ryegrass grass/clover pasture in Australia (16, 37). These subdivision 1 strains have a 16S rRNA sequence similarity of 91%. The third strain, Ellin6076, represents subdivision 3 and was also isolated from Australian ryegrass/clover pasture soil (16, 37). It has a 16S rRNA sequence similarity of 84% to A. capsulatum. The two Australian soil strains have informally been given the candidate Latin binomials “Candidatus Koribacter versatilis” Ellin345 and “Candidatus Solibacter usitatus” Ellin6076 (www.jgi.doe.gov). Here they are referred to as Ellin345 and Ellin6076, respectively.
Culture and genomic DNA.
Cells of the sequenced acidobacteria are small rods (Ellin345 and Ellin6076, approximately 0.5 by 1.0 μm; A. capsulatum, 0.5 by 2.0 μm). The two soil isolates are coated in abundant exopolysaccharide (EPS) slime. A. capsulatum DSM 11244T (A. capsulatum) was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, and its genomic DNA was extracted with the UltraClean Soil DNA kit (MoBio). Ellin345 and Ellin6076 were cultured on VL55 medium (16, 37). Their genomic DNAs were extracted with a detergent-bead mill homogenization procedure (45).
Sequencing.
The complete genome sequence of A. capsulatum was determined by the whole-genome shotgun method (83, 87). Clone libraries with insert sizes of 1.8 to 2.8 kbp (small) and 6.5 to 11 kbp (medium) were used for the random shotgun sequencing phase. Closure of physical and sequencing gaps was performed by a combination of primer walking, generation and sequencing of transposon-tagged libraries of large-insert clones, and multiplex PCR. Sequence assembly was performed with Celera Assembler (60). Repeats were identified with RepeatFinder (88), and the sequences and assembly of the repeats were confirmed by using medium-insert clones that spanned the repeat. Coverage for the A. capsulatum genome was 49,530 reads, averaging about 10-fold coverage per base.
The random shotgun method was used to sequence the genomes of Ellin345 and Ellin6076. Large (40-kb)-, medium (8-kb)-, and small (3-kb)-insert random sequencing libraries were sequenced for these genome projects. After the shotgun stage, reads were assembled with parallel Phrap (High Performance Software, LLC). Possible misassemblies were corrected with Dupfinisher (C. Han, unpublished data) or transposon bomb analysis to bridge sequences between clones. Gaps between contigs were closed by editing, custom primer walks, or PCR amplification. The completed genome sequence of Ellin345 contains 60,626 reads, while that of Ellin6076 contains 106,291 reads, with both genomes achieving an average of 10-fold sequence coverage per base with an error rate of less than 1 in 100,000.
Sequence annotation and analysis for A. capsulatum.
Identification of putative protein-encoding genes and annotation of the genome were performed as previously described (24, 82). An initial set of open reading frames (ORFs) predicted to encode proteins (coding sequence) was identified with GLIMMER. ORFs consisting of fewer than 30 codons and those containing overlaps were eliminated. Frameshifts and point mutations were corrected or designated “authentic.” Functional assignment, identification of membrane-spanning domains, determination of paralogous gene families, and identification of regions of unusual nucleotide composition were performed as previously described. Phylogenomic analysis (22, 23) was used to assist with functional predictions. Sequence alignments and phylogenetic trees were refined as described previously (74). The A. capsulatum project is registered with GenBank under project ID 28085, and a GenBank submission is in preparation.
Sequence analysis and annotation for Ellin345 and Ellin6076.
Identification of putative protein-encoding genes and initial automated annotation of the genomes were performed by the Department of Energy (DOE) Joint Genome Institute (http://compbio.ornl.gov/public/section/genome.html). Comparative analysis of the Ellin345 and Ellin6076 genomes was performed in part with the Integrated Microbial Genomes system (55) (http://img.jgi.doe.gov).
Phylogenetic reconstruction.
For each of 31 phylogenetic markers (dnaG, frr, infC, nusA, pgk, pyrG, rplA, rplB, rplC, rplD, rplE, rplF, rplK, rplL, rplM, rplN, rplP, rplS, rplT, rpmA, rpoB, rpsB, rpsC, rpsE, rpsI, rpsJ, rpsK, rpsM, rpsS, smpB, and tsf), an automated system was applied that identified the marker in query genomes, aligned the sequences, and trimmed the alignment according to a predefined mask. For each marker, seed protein sequences were aligned and manually edited and a hidden Markov model was made. A sequence mask was manually attached to each alignment to indicate the ambiguous columns to be removed. The hidden Markov models were then used with the HMMER package to accomplish steps 1 and 2 described above.
α-Glucuronidase sequence comparisons.
Sequences were aligned with MUSCLE (20). The aligned sequences were clustered with the SECATOR algorithm (90), which relies on BIONJ (27) to build a tree from the multiple-sequence alignment and subsequently collapses the branches from subtrees after identification of the nodes joining different subtrees (90). The neighbor-joining tree was made from the resulting distance matrix by using Blosum62 substitution parameters.
Nucleotide sequence accession numbers.
The complete genome sequences of A. capsulatum, Ellin345, and Ellin6076 have been deposited in the GenBank database under accession numbers CP001472, CP000360, and CP000473, respectively.
RESULTS AND DISCUSSION
General genome features.
The three genomes differed greatly in size (Table 1). The A. capsulatum genome was the smallest, at approximately 4.1 Mb. The Ellin345 genome was about 1.5 Mb larger than the A. capsulatum genome and contained about 50% more ORFs. The Ellin6076 genome was much larger than the other two genomes (2.4-fold larger than that of A. capsulatum), with a larger number of genes in paralogous groups. Plasmids were not identified in any of the genomes. Integrated prophages were found in the genomes of A. capsulatum (two complete regions) and Ellin345 (one complete region) but not in that of Ellin6076 (which contained only isolated integrases and transposases). The three genomes were typified by a low number of rRNA gene copies, one copy in the subdivision 1 genomes and two copies in the Ellin6076 genome. This feature is consistent with their observed slow growth and has been postulated to be a characteristic marker of slow growth and a K-selected lifestyle (42, 80). The genomes of all three isolates contained full complements of flagellar and chemotaxis genes, but flagella have only been observed in cultures of A. capsulatum and Ellin345.
TABLE 1.
General features of the acidobacterial genomes
| Strain (subdivision) | Genome size (bp) | % G+C (DNA base composition) | No. of coding sequences | % Coding | No. of rRNA operons | % of proteins similar to proteins of known function in other prokaryotes | % Hypothetical proteins | % Genes in paralogous groups | No. of paralogous groups |
|---|---|---|---|---|---|---|---|---|---|
| A. capsulatum (1) | 4,127,496 | 60.5 | 3,502 | 88.3 | 1 | 68 | 32 | 52.3 | 652 |
| Ellin345 (1) | 5,650,368 | 58.4 | 5,239 | 91 | 1 | 66 | 34 | 54.6 | 670 |
| Ellin6076 (3) | 9,965,640 | 61.9 | 8,568 | 90.8 | 2 | 65.2 | 34.8 | 67.9 | 1,128 |
Phylogeny.
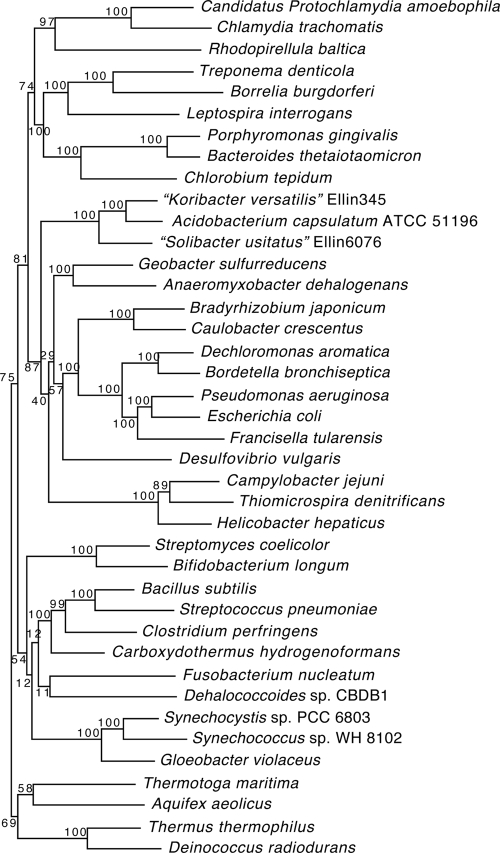
Through 16S rRNA gene sequence analysis, Acidobacteria was placed as a distinct and well-supported major phylum (32, 46, 94) but the deeper relationships to other phyla were unclear. A recent study placed two of the acidobacterial isolates within the class Deltaproteobacteria (11). However, the coherence of the class Deltaproteobacteria can be disturbed by the introduction of new taxa (53). Phylogenetic analysis with a concatenated data set of 31 proteins obtained from our genome sequence data confirmed the monophyly of the acidobacteria indicated by 16S rRNA gene analysis and placed them adjacent to the proteobacteria in 87% of the trees generated from bootstrapped data sets (Fig. 1). Single-protein phylogenies reconstructed from all of the proteins encoded by the A. capsulatum genome support the acidobacterial-proteobacterial relationship; more A. capsulatum proteins are most closely related to proteobacterial equivalents than to those from any other phylum (data not shown). Other genomic evidence suggests a shared ancestry and/or lateral transfer of genes between the acidobacteria and proteobacteria (57, 66). An acidobacterial quorum-quenching lactonase detected in a soil metagenome was found to be most closely related to rhizobial equivalents (67). In most soils, the two most abundant phyla are Acidobacteria and Proteobacteria (34) and in acid mine environments, acidobacteria and acidophilic alphaproteobacteria such as Acidosphaera spp. and Acidicaldus organovorus (35) have been found to occur together. The co-occurrence and relative abundance of these two phyla support the possibility of lateral gene transfer between their members.
FIG. 1.
Maximum-likelihood tree reconstructed with the concatenated sequences of 31 proteins, confirming the phylogenetic coherence of the acidobacteria and suggesting a deep relationship to the proteobacteria.
General genome similarities.
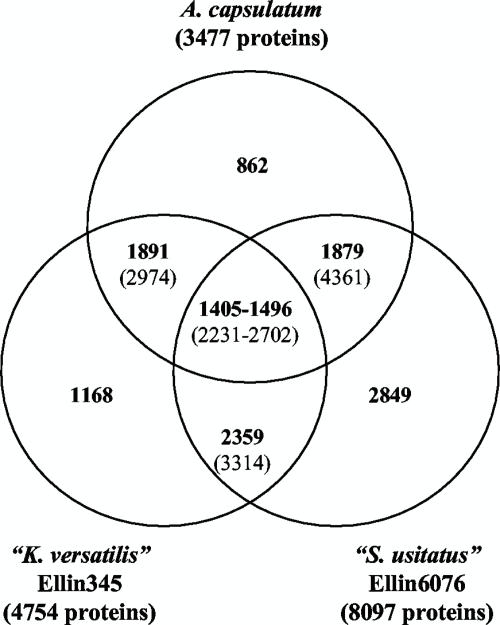
Using bidirectional best-hit analysis of all protein sequences, we compared the overall compositions of these three genomes (Fig. 2). About 25% of the protein coding sequence of A. capsulatum or Ellin345 was unique to that genome, and about 34% of the Ellin6076 genome was unique. As expected, genes common to all three strains mostly encoded housekeeping functions, while the unique genes were dominated by hypothetical proteins or proteins of unknown function. For the genes found to be common to all three genomes and in each pairwise comparison, see Table S1 in the supplemental material. Further characterization of the strain-specific genes by gene expression and protein structure analyses would provide much-needed information about the metabolic capabilities and physiology of these acidobacteria.
FIG. 2.
Common and unique genes in A. capsulatum, Ellin345, and Ellin6076 obtained by bidirectional best-hit analysis of all of the protein sequences in each genome. The values in parentheses are numbers of common genes, including paralogs. Unique genes may include some paralogs within each genome. The numbers of genes in common between any two genomes may also include those that are in common among all three genomes.
Carbohydrate use.
The ability to use a variety of carbon substrates is often used as a diagnostic characteristic of individual species and also suggests potential activities in soil. The presence of genes indicative of carbon substrate use is presented in Table 2. In some cases, this capability has also been verified by culture on that substrate (Table 2) (41, 71). All three genomes encode the ability to degrade a variety of sugars, amino acids, alcohols, and metabolic intermediates. They also encoded the ability to use complex substrates such as xylan, hemicelluloses, pectin, starch, and chitin. Polymeric carbon forms have been successful substrates for cultivating acidobacteria from soils, possibly because they allow the acidobacteria to outcompete other bacterial species unable to use these complex substrates at low concentrations (16, 37, 73, 75).
TABLE 2.
Predicted carbon and nitrogen cycling traits of acidobacteria
| Substrate or property | Pathway | Characteristic inferred from genomeb
|
||
|---|---|---|---|---|
| A. capsulatum | Ellin345 | Ellin6076 | ||
| Simple sugarsa | ||||
| Arabinose | Y | Yc | Yc | |
| l-Arabinose | Yc | Yc | Pc | |
| Cellobiose | Yc | Yc | Yc | |
| Fructose | N | Yc | Yc | |
| Galactose | Yc | Pc | Yc | |
| Galacturonate | P (1st step) | P (1st step)c | P (1st step)c | |
| β-Gentiobiose | Pc | P | P | |
| Glucose | Yc | Yc | Yc | |
| Glucuronate | P (1st step) | P (1st step)c | P (1st step)c | |
| Lactose | Yc | Yc | Yc | |
| Maltose | Pc | P | N | |
| Mannose | Pc | Pc | Pc | |
| Ribose | Y | Yc | Y | |
| Sucrose | N | Pc | Yc | |
| Amino acids | ||||
| Alanine | Y | Yc | Y | |
| Arginine | Y | Yc | Y | |
| Arginased | Y | Y | Y | |
| Arg degradation II | N | N | N | |
| Arginine decarboxylase/agmatinase | P | P | P1 | |
| Arginine decarboxylase/agmatine deiminase | P | P | P2 | |
| Arginine deiminase | P | P | P | |
| Arginase 2 | Y | Y | P | |
| Arginase 3 | P1 | P | P1 | |
| Aspartate | P | Yc | P | |
| Asparagine | Y (to OAAe) | Y (to PEPf) | Y (to OAA) | |
| Cysteine | Y | Y | N | |
| Glutamate | Y | Yc | Y | |
| Glutamate to α-ketoglutarate | Y | Y | Y | |
| Glutamate to butyrate | P | P | P | |
| Glutamate to pyruvate | P | P | P | |
| Glutamate to aspartate to fumarate | Y | Y | Y | |
| Glutamine | Y | Y | Y | |
| Glutaminase | N | N | Y | |
| Glutamate synthase | Y | Y | Y | |
| Glycine | Y | Y | Y | |
| Glycine cleavage system | Y | Y | Y | |
| Glycine degradation I glycine to CO2 through formate | P | P | P | |
| Histidine | Histidine to glutamate | P | P | P |
| Isoleucine | P1 | P1 | P1 | |
| Leucine | P1 | P1c | P1c | |
| Lysine | N | N | N | |
| Methionine | Y | Y | Y | |
| Phenylalanine | Y | Y | Y | |
| Proline | Y | Y | Y | |
| Serine | Y | Yc | Y | |
| Threonine | Y | Yc | Y | |
| Tryptophan | N | Y | Y | |
| Tyrosine | P2 | P2 | P2 | |
| Valine | P | P | P | |
| Alcohols | ||||
| Butanol | N | Yc | N | |
| Catechol | P | P | P | |
| Ethanol | P | P | P | |
| Glycerol | Y | Yc | Yc | |
| Mannitol | N | N | N | |
| Methanol | N | N | N | |
| Propanol | P1 | P1c | P1 | |
| Organic acids | ||||
| Acetate | Y | Y | Y | |
| Benzoate | P | Pc | P | |
| Butyrate | N | N | N | |
| Formate | Oxidation to CO2 | N | Y | Y |
| Glycolate | P1 | P1c | P1 | |
| Lactate | P2 | Y (l-lactate)c | P2 | |
| Tartrate | N | P1c | P1 | |
| Other | ||||
| Chitin | Y | Y | Y | |
| Pectin | Y | N | Yc | |
| Starch | Y | Y | Yc | |
| Xylan | Yc | Yc | Yc | |
| Cellulose | P | Y | Yc | |
| Urea | N | N | N | |
| Nitrogen metabolism | ||||
| N2 fixation | N | N | N | |
| Nitrate reduction | N | Y | Y | |
| Nitrite reduction | Y | Y | Y | |
| Denitrification | N | N | N | |
Unless stated otherwise, the results of testing the use of d isomers of sugars and l isomers of amino and organic acids are reported.
Y, characteristic found in the genome; N, characteristic not found in the genome; P, partial pathway found; P1, candidate genes for all steps are present, but some enzymes may have different activities even though sequences are similar; P2, gene sequences have not been identified for every step in the pathway, so there were no query sequences available for comparison.
Pathways designated according to Metacyc (http://metacyc.org).
OAA, O-acetyl acetate.
PEP, phosphoenolpyruvate.
The genomes contained a large number of glycoside hydrolase-encoding genes, placing them in the top 5% of the >500 bacterial genomes surveyed in the carbohydrate-active enzyme database (www.cazy.org). The genome of Ellin6076 contained more glycoside hydrolases (n = 117) than the human genome and almost three times as many as that of Saccharomyces cerevisiae (14). Genes involved in carbohydrate transport and metabolism constituted one of the four most expanded gene families in Ellin6076 and contributed to the relatively large size of its genome.
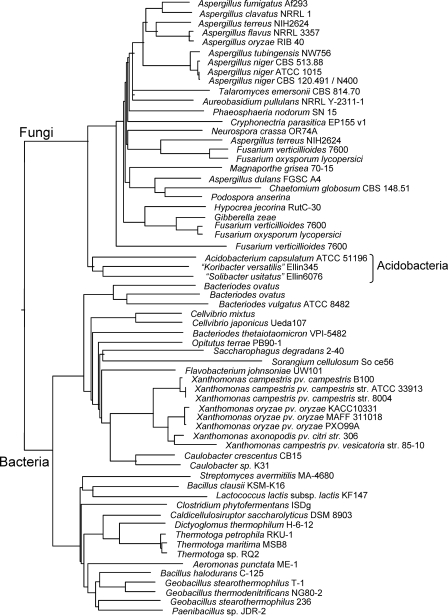
All three genomes contained candidate cellulases from glycoside hydrolase family 9. Ellin345 and Ellin6076 also contained β-glucosidases from family 1, suggesting that they are able to degrade cellulose substrates. Although none of the cellulases contained typical cellulose-binding domains, culture studies substantiate this finding (Table 2) (71). The Ellin6076 genome was also rich in enzymes capable of degrading other plant cell wall polysaccharides, with complete enzymatic pathways (polysaccharide lyases and pectin esterases) for the degradation of pectins and hemicelluloses. A. capsulatum also contained genes that encode plant cell wall-degrading enzymes, with a particularly large cluster that encodes pectin degradation. Ellin345 appeared capable of hemicellulose side chain degradation. All three acidobacterial genomes encoded a candidate α-glucuronidase from family GH67 (plant cell wall hemicellulose degradation) that appeared to share the highest sequence similarity with fungal homologs (Fig. 3). This evidence for an ancestral horizontal transfer event has not been reported for members of any other glycoside hydrolase family. Genome traits conferring carbohydrate-based carbon storage mechanisms (glycogen) and compatible solutes (trehalose) were found in all three genomes. These suggest a potential role for carbohydrates in nutritional pathways, as well as in desiccation resistance.
FIG. 3.
Neighbor-joining tree of α-glucuronidase genes from fungi and bacteria, showing the close relationship of acidobacterial and fungal genes.
The predicted polymer-degrading properties reveal acidobacteria as decomposers in the soil that potentially participate in the cycling of plant, fungal, and insect-derived organic matter. The numerical abundance of soil acidobacteria suggests that this contribution may be quantitatively significant. A recent metagenomic analysis of deep Mediterranean seawater that used 16S rRNA-targeted PCR approaches and metagenomic binning (56) found acidobacteria to be among the dominant bacterial groups. The metabolic genes were mostly related to the transport and degradation of biopolymers, suggesting that acidobacteria may also contribute to decomposition in the marine environment.
The acidobacterial genomes suggest further flexibility and novelty in their ability to metabolize carbon. Each genome contains genes predicted to encode carbon monoxide dehydrogenase, as noted previously for Ellin345 and Ellin6076 (40) but not previously reported for A. capsulatum. Given the absence of ribulose-1,5-bisphosphate carboxylase/oxygenase or other enzymes predicted to be involved in autotrophic carbon fixation, it seems likely that the acidobacteria supplement their (primarily heterotrophic) energy budget with CO rather than derive significant amounts of carbon in this way, as recently suggested for supplemental lithoheterotrophs such as Silicibacter pomeroyi (59) (referred to as “mixotrophs” in other literature [40]).
Oxidation of CO (either experimentally determined or predicted from genome sequencing) is found in other major groups of soil organisms, such as the actinobacteria, pseudomonads, and rhizobia (40). Many of these organisms are also able to degrade complex plant polymers; it is tempting to speculate that this property and CO oxidation both arose as scavenging strategies to optimize life in a low-carbon environment. It has been estimated that CO-oxidizing bacteria remove approximately 20% of the total global CO input to the atmosphere (40). The consumption of CO by acidobacteria remains to be experimentally confirmed, but it seems plausible that acidobacteria make a quantitatively significant contribution to two major parts of the carbon cycle: degradation of complex polymers and CO oxidation.
Nitrogen metabolism.
Genomic evidence suggested that the role of acidobacteria in nitrogen cycling in soils and sediments is the reduction of nitrate, nitrite, and possibly nitric oxide. Assimilatory nitrate reductase gene sequences found in the acidobacteria are most similar to those described for the cyanobacteria (68). The A. capsulatum and Ellin6076 genomes contained candidate genes for nitrate reductase. These genes had some sequence similarity to assimilatory (narB) and respiratory (narG) nitrate reductase genes found in cyanobacteria (narB) and Thermus thermophilus (narG) (8). A homolog of these genes was not identified in Ellin345. All three genomes contain nirA, which encodes nitrite reductase, as well as ntcB and glnB, which encode nitrogen regulatory proteins that affect the transcription of narB and nirA. Ellin6076 had one candidate for another nitrogen regulator, ntcA. Only genomes from the two soil strains contained norB and potentially a norC homolog, suggesting nitric oxide reductase capability. We found no evidence of the ability to fix N2 (nifH), oxidize ammonium (amoA), or denitrify nitrous oxide (nosZ) in the three genomes. Candidate genes for nitrate transport (nrtABCD) were found in both soil strains.
Transporters.
Comparison of the major transporter families in the three genomes provided information on the potential metabolic capabilities of the acidobacterial strains and showed that the three species contained a very wide variety of individual transporters. In general, the soil strains contained a wider variety of genes that encode transporters than did A. capsulatum (Table 3). The acidobacterial genomes lacked sugar-transporting phosphotransferase system homologs, a trait shared with 22% of bacteria and one that correlates with strictly aerobic carbohydrate metabolism (1). Instead, all three genomes contained the lower-specificity major facilitator superfamily system for sugar transport, as well as high-affinity ABC transporters proposed to be advantageous in low-nutrient environments (63) (Table 3).
TABLE 3.
Transporters predicted from sequence of three acidobacterial genomes
| Transporter class | Total no. of predicted transportersa
|
||
|---|---|---|---|
| A. capsulatum | Ellin345 | Ellin6076 | |
| Ion channels | 11 (5) | 13 (7) | 12 (7) |
| ABC transporters | 39 (15) | 70 (15) | 159 (21) |
| Secondary porters (total) | 77 (37) | 101 (48) | 152 (57) |
| Major facilitators | 22 (9) | 27 (13) | 55 (17) |
| Amino acid-polyamine-organocation facilitators | 8 (3) | 17 (6) | 15 (5) |
| Resistance-nodulation-cell division transporters | 15 (3) | 12 (3) | 22 (3) |
| Drug/metabolite transporters | 5 (2) | 7 (3) | 6 (3) |
| Multidrug/oligosaccharidyl- lipid/polysaccharide transporters | 4 (3) | 4 (2) | 6 (3) |
The total number of transporter genes in a class is followed in parentheses by the total number of transporter families found in that class for each genome.
A large proportion of each genome encoded transporters (about 6% of the total coding sequence in each of the three genomes). The ion channels, ABC transporters, and different types of secondary porters (uniporters, symporters, antiporters) are summarized in Table 3. The three acidobacterial genomes contain similar numbers of ion channel proteins and P-type ATPases for cation transport. Ellin6076 contained twice as many ABC-type transporters as Ellin345 and four times as many as A. capsulatum. This difference was primarily due to a large number of macrolide exporters (n = 117) in Ellin6076.
All three genomes contained a variety of secondary porters that transport a wide variety of substrates (e.g., carbon substrates, amino acids, drugs, metals, nitrate/nitrite, fatty acids, ions, proteins) in a carrier-mediated process. Ellin6076 contained twice as many major facilitator superfamily-type carriers as Ellin345 and A. capsulatum, a difference that was largely due to an abundance of anion:cation symporters.
Iron uptake and metabolism.
There is growing evidence that acidobacteria play an important role in iron redox reactions. They have been detected in and sometimes found to dominate 16S rRNA gene sequence libraries from iron-rich mine environments (e.g., see references 4, 43, 69, and 93).
The ability to scavenge iron is critical for survival in soils. All three genomes contained genes that should enable these isolates to take up iron from the environment, including the genes feoAB, which encode a high-affinity ferrous (Fe2+) iron transporter (38, 86). Iron is likely to exist mostly as ferrous ion only under reducing conditions caused by anaerobic and/or low-pH conditions, and the bacterial species for which this pathway has been characterized are all facultative anaerobes. The three sequenced acidobacterial strains were isolated under aerobic conditions, and their ability to grow under anaerobic conditions has not yet been tested. A different acidobacterial strain has been shown to oxidize ferrous iron at pH >3 (31), and A. capsulatum and related strains could catalyze dissimilatory ferric iron reduction under oxygen-limited conditions (14), suggesting that the acidobacteria may be capable of ferrous ion oxidation, as well as anaerobic or microaerophilic growth.
Ellin345 and A. capsulatum, but not Ellin6076, contained genes identified as encoding iron permease FTR1 and FTR1 family proteins, as well as sequences similar to that of fet3p, which encodes a multicopper oxidase involved in FTR1-mediated iron uptake in yeast (78). Experiments with Pseudomonas aeruginosa indicate that the multicopper oxidase mco is a ferroxidase that functions in Fe2+ uptake (34). The two A. capsulatum sequences (ORFA01590 and ORFA00877) have 49 and 48% amino acid identities to the P. aeruginosa mco gene (33), while the Ellin345 and Ellin6076 sequences had lower amino acid identities (22 to 39%) to P. aeruginosa mco. All of the acidobacterial sequences have domain structures similar to that of the P. aeruginosa gene product, suggesting that they too may be involved in Fe2+ uptake. The three genomes contain other iron transporter genes, including several genes that encode Mn2+ and Fe2+ transporters of the NRAMP family.
Mechanisms of dissimilatory ferric iron reduction have been investigated in Shewanella oneidensis and in Geobacter species. Searches for cytochrome c proteins shown to be involved in ferric iron reduction in S. oneidensis (OmcA, OmcB, MtrA, MtrB, CymA) identified mtrA homologs in all three acidobacteria, one omcA candidate in Ellin345, and five genes in Ellin6076 that could be candidates for either omcA or omcB. Geobacter sulfurreducens proteins (OmcB, OmcD, OmcE, FerA [= OmcB], PpcA, Fro-1) were also searched for but were not detected in the acidobacterial genomes. These results suggest that the acidobacteria do not use a pathway for ferric iron reduction similar to that of either Shewanella or Geobacter.
Bacteria that can scavenge iron via excreted siderophores have a potential advantage in soils where iron is tightly bound to soil colloids and the available concentration is growth limiting. BLAST searches of the A. capsulatum, Ellin345, and Ellin6076 genomes by using the sequences of proteins required for the synthesis of pyoverdine, mycobactin, enterobactin, bacillobactin, aerobactin, vibrioferrin, alcaligin, and other siderophores as queries revealed no highly similar sequences, leading to the initial conclusion that none of the three genomes contained any obvious genes for siderophore biosynthesis. However, there were candidates in each genome for some of the genes that make up the enterobactin biosynthetic pathway, namely, entA, entB, entD, entE, and entF, as well as the enterobactin exporter gene entS, and the three genomes contained genes that encode candidate siderophore receptors.
The identification and analysis of these genes provided several clues that these three acidobacteria may use siderophores produced by other microorganisms. (i) All three genomes contained multiple copies of the tonB, exbB, and exbD genes, which are necessary for siderophore transport (28, 50, 51, 65, 70). (ii) All three genomes contained many genes that encode TonB-dependent receptors, and the known siderophore receptors are TonB-dependent receptors (reviewed in references 13, 28, 50, and 65). (iii) With known siderophore receptor sequences as queries, the top BLAST hits (with an E-value cutoff of 1e-05) were to one or more of the TonB-dependent receptor genes in all three genomes. In addition to the candidate TonB-dependent siderophore receptor genes, we also found candidates for pvuE, which encodes the ATPase component of a vibrioferrin transporter, and fepB, which encodes the periplasmic solute-binding component of an enterobactin transporter. However, genes that encode the other components of these transporters were not found.
PKSs and NRPSs.
Polyketide synthase (PKS) and nonribosomal peptide synthase (NRPS) enzymes are well known for their roles in the synthesis of siderophores, as well as other natural products such as antibiotics, antifungals, antivirals, antitumor agents, and antinematodal agents (reviewed in references 9, 15, 30, 47, and 58). NRPSs also make biosurfactants (79). The A. capsulatum genome contained three clustered genes that encode NRPSs, several genes that encode PKSs, and one hybrid NRPS-PKS gene. The NRPS genes contained domains present in EntE and EntF. These are subunits of the enterobactin synthase multienzyme complex (identified by SEARCHPKS [92] and CD-Search [54]). Several PKS genes were found in the two soil strains, although they were not localized in one region as in the A. capsulatum genome.
Cellulose synthesis.
The A. capsulatum genome contained an operon with all of the genes identified as necessary for cellulose synthesis and is one of very few non-proteobacterial genomes reported to contain this operon. The two genomes from soil strains contained clear orthologs of all but one of the genes in this operon. An ortholog of the final cellulose synthesis gene (celAB) was not identified. Ellin345 and Ellin6076 both contained a glycosyltransferase family 2 gene with some sequence similarity to a UDP-forming cellulose synthase from Nitrosomonas eutropha C71 (29 and 32% amino acid identities, respectively). Their celAB genes may be too novel to define by similarity to genes of other organisms. Cellulose microfibrils have not been microscopically observed in cultures of the sequenced strains, but filaments have been observed microscopically associated with acidobacteria in soils (C. R. Kuske, unpublished data) and the two soil strains form visible “clumps” or “flocs” in liquid culture (72).
Bacterial cellulose has been implicated in several functions related to survival in soil, including biofilm production, enhanced survival under stressful conditions by retaining hydration under dry conditions, and promoting aeration and water-holding capacity, that contribute to soil aggregate formation (85, 89). Johnson et al. (36) suggested that cellulose synthesis by acidobacteria could promote a biofilm that enhances their abilities to adhere to ferric iron-rich substrates and to produce biofilm-forming, ferric iron-reducing “bioshrouds” to protect sulfidic mine tailings. Acidobacteria may also synthesize a network of cellulose that traps air and provides a loose network for water and nutrient percolation through the soil. Future experimental characterization of cellulose production could include testing of the ability to form microfibrils, which would enhance putative network formation.
Stress and starvation response.
The genomes of all three acidobacteria contained genes that encode putative addiction modules, consisting of a toxin-and-antitoxin pair and originally described as mechanisms for maintenance of plasmids. The addiction modules may operate rapidly to inhibit the synthesis of DNA and protein in response to stress or starvation, as has been previously demonstrated by gene expression profiling of Escherichia coli growth transitions (10). Such a response would allow the acidobacteria to make the best use of scarce resources in an environment with recurring periods of carbon and energy limitation.
Acid tolerance.
The phylum Acidobacteria was named after the first cultured isolate, A. capsulatum, which was isolated from sediments in an acidic mine drainage. The phylum name implies acid tolerance or preference, and the three sequenced acidobacterial strains are moderately acidophilic. A. capsulatum grows best at a pH of 3.0 to 6.0, Ellin345 at 4.0 to 6.5, and Ellin6076 at 3.5 to 6.5, and none of the strains grow at a pH below 3 or above 6.5. All three sequenced genomes contained candidate genes in the AR3 system that may suggest a mechanism for tolerance of acidic conditions. However, genes involved in the AR1, AR2, or AR4 inducible acid resistance system were not found. We did not find evidence of adaptation to acidic conditions by comparisons of individual amino acid frequencies.
Preference for an acidic pH has been found to be a trait of other subdivision 1 acidobacteria (72) but is probably not characteristic of the entire phylum. For example, acidobacterial sequences in subdivision 4 have been found to be abundant in soils of neutral or alkaline pH (in 12 different soils) but their presence was variable in soils with pHs below 5.4, and they were not detected in soils with pHs of less than 4.0 (3). In addition, multiple members of the phylum Acidobacteria have been found to be abundant in alkaline soils (18, 19).
Novel genome traits.
The acidobacterial genomes include novel features that are abundant and that set the members of this phylum apart from those of other bacterial phyla. In most cases, the functions of these genes are not known or completely described and further investigation of protein structure and function is needed to link them to physiology and environmentally relevant functions.
One family of duplicated orphan permeases is mostly restricted to the acidobacteria, with a particular expansion in Ellin6076. These permeases are transport proteins that contain two efflux ABC transporter permease domains (PF02687) and lack the typical ATP-binding subunit. The orphan permeases are found, in every case, as members of a gene pair also containing a member of a PadR transcriptional regulator subfamily unique to the acidobacteria. The unusual configuration of this gene pair suggests a role in transport (either passive or powered by a mechanism not requiring an ATP-binding cassette) and/or signaling.
The Ellin6076 genome contained at least 45 high-molecular-weight proteins of unknown function that were predicted to be extracellular or cell surface proteins. Abundant genes in this same class (n = 74) have been found in Cytophaga hutchinsonii, where it was suggested that they may be part of the cell-bound cellulolytic machinery (91). Ellin6076 may also have specialized protein-targeting capabilities.
The Ellin6076 genome also contained the highest number of PEP-CTERM proteins (n = 61) yet reported in any microbial genome. PEP-CTERM appears to be a transpeptidase-mediated protein export signal associated with EPS biosynthesis (29). The gene for the predicted PEP-CTERM sorting enzyme EpsH typically is found within EPS loci, and there are three copies in the genome of Ellin6076. The PEP-CTERM proteins themselves are postulated to add a proteinaceous component to extracellular material (29). In 63 bacterial species across 47 genera, an individual genome encodes either cellulose biosynthesis (29 species) or the PEP-CTERM system (34 species), but not both. The acidobacteria, with cellulose biosynthesis in A. capsulatum but EpsH/PEP-CTERM in Ellin6076, now join the alphaproteobacteria, betaproteobacteria, and gammaproteobacteria as subdivisions with at least one example of each. The disjointed distribution of these two systems supports the hypothesis that they have chemically distinct but functionally similar roles.
Summary and future directions.
The three acidobacterial genomes are only distantly related to other organisms in the domain Bacteria. For this reason, annotating acidobacterial genes based on similarity to species in other bacterial phyla was challenging due to the often low percent similarities. Physiological studies on the acidobacteria and members of other diverse phyla are sorely needed to enable more accurate annotation of genes by function. Without functional studies, the wealth of genomic data becoming available is of diminished use.
This comparative genomic study of members of the phylum Acidobacteria has demonstrated the monophyly and phylogenetic position of the phylum and has identified distinguishing genomic features that may predict their metabolic capabilities and lifestyle in soils. They share traits with the Proteobacteria but also with the Cyanobacteria and Fungi. The bacterial species falling within the phylum Acidobacteria are very diverse, and the acidobacteria are abundant in soils that differ greatly in physical and chemical characteristics (7, 18, 19). It is therefore difficult to generalize metabolic traits across the phylum based on only these three genomes.
The three sequenced species are aerobic heterotrophs typified by a low rRNA copy number and slow growth in culture. They have only been culturable in low-nutrient media. The presence of copious extracellular slime in cultures and the potential ability to synthesize extracellular cellulose suggest an ability to withstand cycles of drying and rehydration. These strains potentially have the ability to use a wide variety of carbon substrates that include complex substrates, and they rely on low-specificity and high-affinity sugar transporters. These culture and genomic traits are all consistent with a broad metabolic capability and a lifestyle suited to slow growth in nutrient-limited environments. Such a lifestyle would be characterized by the use of substrates at low concentrations, catalytic enzymes with high substrate affinities and low Vmax characteristics, and slow cell turnover rates (25). In this lifestyle, individual cells might remain metabolically active or survive in soil for long periods of time, with individual bacterial cells dividing only a few times per year in soil (31). Based on the combination of physiological and genomic evidence, we postulate that cells of these isolates are long-lived, divide slowly, exhibit slow metabolic rates under low-nutrient conditions, and are well equipped to tolerate fluctuations in hydration. Experimental validation of these predictions will reveal how acidobacterial activities affect soil quality through organic matter decomposition, nutrient cycling, and the formation of soil aggregates.
The genome results have not identified a single major factor that would allow simpler or more rapid cultivation of members of this phylum. Indeed, our difficulty in culturing these bacteria may be due more to our lack of patience and the inability of acidobacteria to compete in culture with other species than to a lack of appropriate nutritional conditions. A wider variety of cultured representatives may be difficult to achieve but, together with continued comparative genomic and gene expression analyses, would contribute significantly to our understanding of this diverse and environmentally abundant phylum.
Supplementary Material
Acknowledgments
This work was supported by the U.S. DOE Microbial Genome Program (under U.S. DOE contract W-7405-ENG-36), the NSF Microbial Genome Sequencing Program (under NSF award 0237365), the U.S. DOE Joint Genome Institute, and The Institute for Genomic Research. Support for N.L.W. was also provided under NSF award EPS-0447681.
Footnotes
Published ahead of print on 5 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barabote, R. D., and M. H. Saier, Jr. 2005. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69:608-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barns, S. M., E. C. Cain, L. Sommerville, and C. R. Kuske. 2007. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 73:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blöthe, M., D. M. Akob, J. E. Kostka, K. Göschel, H. L. Drake, and K. Küsel. 2008. pH gradient-induced heterogeneity of Fe(III)-reducing microorganisms in coal mining-associated lake sediments. Appl. Environ. Microbiol. 74:1019-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branco, R., A. P. Chung, A. Verissimo, and P. V. Morais. 2005. Impact of chromium-contaminated wastewaters on the microbial community of a river. FEMS Microbiol. Ecol. 54:35-46. [DOI] [PubMed] [Google Scholar]
- 6.Bryant, D. A., A. M. Costas, J. A. Maresca, A. G. Chew, C. G. Klatt, M. M. Bateson, L. J. Tallon, J. Hostetler, W. C. Nelson, J. F. Heidelberg, and D. M. Ward. 2007. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317:523-526. [DOI] [PubMed] [Google Scholar]
- 7.Buckley, D. H., and T. M. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 8.Cava, F., O. Zafra, M. S. da Costa, and J. Berenguer. 2008. The role of the nitrate respiration element of Thermus thermophilus in the control and activity of the denitrification apparatus. Environ. Microbiol. 10:522-533. [DOI] [PubMed] [Google Scholar]
- 9.Challis, G. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6:601-611. [DOI] [PubMed] [Google Scholar]
- 10.Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 11.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283-1287. [DOI] [PubMed] [Google Scholar]
- 12.Coates, J. D., D. J. Ellis, C. V. Gaw, and D. R. Lovley. 1999. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Bacteriol. 49(Pt. 4):1615-1622. [DOI] [PubMed] [Google Scholar]
- 13.Cornelis, P., and S. Matthijs. 2002. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4:787-798. [DOI] [PubMed] [Google Scholar]
- 14.Coutinho, P. M., M. Stam, E. Blanc, and B. Henrissat. 2003. Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci. 8:563-565. [DOI] [PubMed] [Google Scholar]
- 15.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis, K. E., S. J. Joseph, and P. H. Janssen. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dedysh, S. N., T. A. Pankratov, S. E. Belova, I. S. Kulichevskaya, and W. Liesack. 2006. Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eichorst, S. A., J. A. Breznak, and T. M. Schmidt. 2007. Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 73:2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisen, J. A. 1998. A phylogenomic study of the MutS family of proteins. Nucleic Acids Res. 26:4291-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisen, J. A., and C. M. Fraser. 2003. Phylogenomics: intersection of evolution and genomics. Science 300:1706-1707. [DOI] [PubMed] [Google Scholar]
- 24.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fierer, N., M. A. Bradford, and R. B. Jackson. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354-1364. [DOI] [PubMed] [Google Scholar]
- 26.Fracchia, L., A. B. Dohrmann, M. G. Martinotti, and C. C. Tebbe. 2006. Bacterial diversity in a finished compost and vermicompost: differences revealed by cultivation-independent analyses of PCR-amplified 16S rRNA genes. Appl. Microbiol. Biotechnol. 71:942-952. [DOI] [PubMed] [Google Scholar]
- 27.Gascuel, O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685-695. [DOI] [PubMed] [Google Scholar]
- 28.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 29.Haft, D. H., I. T. Paulsen, N. Ward, and J. D. Selengut. 2006. Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol. 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallberg, K., and D. Johnson. 2003. Novel acidophiles isolated from moderately acidic mine drainage waters. Hydrometallurgy 71:139-148. [Google Scholar]
- 31.Harris, D., and E. A. Paul. 1994. Measurement of bacterial growth rates in soil. Appl. Soil Ecol. 1:277-290. [Google Scholar]
- 32.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huston, W. M., M. P. Jennings, and A. G. McEwan. 2002. The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 45:1741-1750. [DOI] [PubMed] [Google Scholar]
- 34.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, D. B., B. Stallwood, S. Kimura, and K. B. Hallberg. 2006. Isolation and characterization of Acidicaldus organivorus, gen. nov., sp. nov.: a novel sulfur-oxidizing, ferric iron-reducing thermo-acidophilic heterotrophic Proteobacterium. Arch. Microbiol. 185:212-221. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, D. B., L. Yajie, and N. Okibe. 2008. “Bioshrouding”: a novel approach for securing reactive mineral tailings. Biotechnol. Lett. 30:445-449. [DOI] [PubMed] [Google Scholar]
- 37.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanokratana, P., S. Chanapan, K. Pootanakit, and L. Eurwilaichitr. 2004. Diversity and abundance of Bacteria and Archaea in the Bor Khlueng Hot Spring in Thailand. J. Basic Microbiol. 44:430-444. [DOI] [PubMed] [Google Scholar]
- 40.King, G. M., and C. F. Weber. 2007. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5:107-118. [DOI] [PubMed] [Google Scholar]
- 41.Kishimoto, N., Y. Kosako, and T. Tano. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinsteuber, S., F. D. Muller, A. Chatzinotas, K. Wendt-Potthoff, and H. Harms. 2008. Diversity and in situ quantification of Acidobacteria subdivision 1 in an acidic mining lake. FEMS Microbiol. Ecol. 63:107-117. [DOI] [PubMed] [Google Scholar]
- 44.Koch, I. H., F. Gich, P. F. Dunfield, and J. Overmann. 2008. Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., acidobacteria isolated from alpine and forest soils. Int. J. Syst. Evol. Microbiol. 58:1114-1122. [DOI] [PubMed] [Google Scholar]
- 45.Kuske, C. R., K. L. Banton, D. L. Adorada, P. C. Stark, K. K. Hill, and P. J. Jackson. 1998. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 64:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lal, R., R. Kumari, H. Kaur, R. Khanna, N. Dhingra, and D. Tuteja. 2000. Regulation and manipulation of the gene clusters encoding type-I PKSs. Trends Biotechnol. 18:264-274. [DOI] [PubMed] [Google Scholar]
- 48.Lee, S. H., J. O. Ka, and J. C. Cho. 2008. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 285:263-269. [DOI] [PubMed] [Google Scholar]
- 49.Liesack, W., F. Bak, J. U. Kreft, and E. Stackebrandt. 1994. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162:85-90. [DOI] [PubMed] [Google Scholar]
- 50.Llamas, M. A., and W. Bitter. 2006. Iron gate: the translocation system. J. Bacteriol. 188:3172-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Llamas, M. A., M. Sparrius, R. Kloet, C. R. Jimenez, C. Vandenbroucke-Grauls, and W. Bitter. 2006. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol. 188:1882-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.López-García, P., S. Duperron, P. Philippot, J. Foriel, J. Susini, and D. Moreira. 2003. Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ. Microbiol. 5:961-976. [DOI] [PubMed] [Google Scholar]
- 53.Ludwig, W., and H. P. Klenk. 2005. Overview: a phylogenetic backbone and taxonomic framework for prokaryotic systematics. Springer, New York, NY.
- 54.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Markowitz, V. M., F. Korzeniewski, K. Palaniappan, E. Szeto, G. Werner, A. Padki, X. Zhao, I. Dubchak, P. Hugenholtz, I. Anderson, A. Lykidis, K. Mavromatis, N. Ivanova, and N. C. Kyrpides. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34:D344-D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martín-Cuadrado, A. B., P. Lopez-Garcia, J. C. Alba, D. Moreira, L. Monticelli, A. Strittmatter, G. Gottschalk, and F. Rodriguez-Valera. 2007. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS ONE 2:e914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazón, G., S. Campoy, I. Erill, and J. Barbe. 2006. Identification of the Acidobacterium capsulatum LexA box reveals a lateral acquisition of the Alphaproteobacteria lexA gene. Microbiology 152:1109-1118. [DOI] [PubMed] [Google Scholar]
- 58.Mootz, H. D., D. Schwarzer, and M. A. Marahiel. 2002. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. Chembiochem 3:490-504. [DOI] [PubMed] [Google Scholar]
- 59.Moran, M. A., A. Buchan, J. M. Gonzalez, J. F. Heidelberg, W. B. Whitman, R. P. Kiene, J. R. Henriksen, G. M. King, R. Belas, C. Fuqua, L. Brinkac, M. Lewis, S. Johri, B. Weaver, G. Pai, J. A. Eisen, E. Rahe, W. M. Sheldon, W. Ye, T. R. Miller, J. Carlton, D. A. Rasko, I. T. Paulsen, Q. Ren, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, M. J. Rosovitz, D. H. Haft, J. Selengut, and N. Ward. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913. [DOI] [PubMed] [Google Scholar]
- 60.Myers, E. W., G. G. Sutton, A. L. Delcher, I. M. Dew, D. P. Fasulo, M. J. Flanigan, S. A. Kravitz, C. M. Mobarry, K. H. Reinert, K. A. Remington, E. L. Anson, R. A. Bolanos, H. H. Chou, C. M. Jordan, A. L. Halpern, S. Lonardi, E. M. Beasley, R. C. Brandon, L. Chen, P. J. Dunn, Z. Lai, Y. Liang, D. R. Nusskern, M. Zhan, Q. Zhang, X. Zheng, G. M. Rubin, M. D. Adams, and J. C. Venter. 2000. A whole-genome assembly of Drosophila. Science 287:2196-2204. [DOI] [PubMed] [Google Scholar]
- 61.Pankratov, T. A., Y. M. Serkebaeva, I. S. Kulichevskaya, W. Liesack, and S. N. Dedysh. 2008. Substrate-induced growth and isolation of Acidobacteria from acidic Sphagnum peat. ISME J. 2:551-560. [DOI] [PubMed] [Google Scholar]
- 62.Park, D. S., H. W. Oh, W. J. Jeong, H. Kim, H. Y. Park, and K. S. Bae. 2007. A culture-based study of the bacterial communities within the guts of nine longicorn beetle species and their exo-enzyme producing properties for degrading xylan and pectin. J. Microbiol. 45:394-401. [PubMed] [Google Scholar]
- 63.Paulsen, I. T., M. K. Sliwinski, and M. H. Saier, Jr. 1998. Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J. Mol. Biol. 277:573-592. [DOI] [PubMed] [Google Scholar]
- 64.Penn, K., D. Wu, J. A. Eisen, and N. Ward. 2006. Characterization of bacterial communities associated with deep-sea corals on Gulf of Alaska seamounts. Appl. Environ. Microbiol. 72:1680-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 66.Quaiser, A., T. Ochsenreiter, C. Lanz, S. C. Schuster, A. H. Treusch, J. Eck, and C. Schleper. 2003. Acidobacteria form a coherent but highly diverse group within the bacterial domain: evidence from environmental genomics. Mol. Microbiol. 50:563-575. [DOI] [PubMed] [Google Scholar]
- 67.Riaz, K., C. Elmerich, D. Moreira, A. Raffoux, Y. Dessaux, and D. Faure. 2008. A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ. Microbiol. 10:560-570. [DOI] [PubMed] [Google Scholar]
- 68.Richardson, D. J., B. C. Berks, D. A. Russell, S. Spiro, and C. J. Taylor. 2001. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci. 58:165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rowe, O. F., J. Sanchez-Espana, K. B. Hallberg, and D. B. Johnson. 2007. Microbial communities and geochemical dynamics in an extremely acidic, metal-rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ. Microbiol. 9:1761-1771. [DOI] [PubMed] [Google Scholar]
- 70.Rutz, J. M., J. Liu, J. A. Lyons, J. Goranson, S. K. Armstrong, M. A. McIntosh, J. B. Feix, and P. E. Klebba. 1992. Formation of a gated channel by a ligand-specific transport protein in the bacterial outer membrane. Science 258:471-475. [DOI] [PubMed] [Google Scholar]
- 71.Sait, M. 2008. Isolation and characterization of soil acidobacteria. Ph.D. thesis. Department of Microbiology and Immunology, University of Melbourne, Melbourne, Australia.
- 72.Sait, M., K. E. Davis, and P. H. Janssen. 2006. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl. Environ. Microbiol. 72:1852-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 74.Salzberg, S. L., O. White, J. Peterson, and J. A. Eisen. 2001. Microbial genes in the human genome: lateral transfer or gene loss? Science 292:1903-1906. [DOI] [PubMed] [Google Scholar]
- 75.Schoenborn, L., P. S. Yates, B. E. Grinton, P. Hugenholtz, and P. H. Janssen. 2004. Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl. Environ. Microbiol. 70:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Selenska-Pobell, S., G. Kampf, K. Hemming, G. Radeva, and G. Satchanska. 2001. Bacterial diversity in soil samples from two uranium waste piles as determined by rep-APD, RISA and 16S rDNA retrieval. Antonie van Leeuwenhoek 79:149-161. [DOI] [PubMed] [Google Scholar]
- 77.Sievert, S. M., J. Kuever, and G. Muyzer. 2000. Identification of 16S ribosomal DNA-defined bacterial populations at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 66:3102-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 79.Stein, T., and J. Vater. 1996. Amino acid activation and polymerization at modular multienzymes in nonribosomal peptide biosynthesis. Amino Acids (Vienna) 10:201-227. [DOI] [PubMed] [Google Scholar]
- 80.Stevenson, B. S., and T. M. Schmidt. 2004. Life history implications of rRNA gene copy number in Escherichia coli. Appl. Environ. Microbiol. 70:6670-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stott, M. B., M. A. Crowe, B. W. Mountain, A. V. Smirnova, S. Hou, M. Alam, and P. F. Dunfield. 2008. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ. Microbiol. 10:2030-2041. [DOI] [PubMed] [Google Scholar]
- 82.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 83.Tettelin, H., D. Radune, S. Kasif, H. Khouri, and S. L. Salzberg. 1999. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics 62:500-507. [DOI] [PubMed] [Google Scholar]
- 84.Tringe, S. G., C. von Mering, A. Kobayashi, A. A. Salamov, K. Chen, H. W. Chang, M. Podar, J. M. Short, E. J. Mathur, J. C. Detter, P. Bork, P. Hugenholtz, and E. M. Rubin. 2005. Comparative metagenomics of microbial communities. Science 308:554-557. [DOI] [PubMed] [Google Scholar]
- 85.Ude, S., D. L. Arnold, C. D. Moon, T. Timms-Wilson, and A. J. Spiers. 2006. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ. Microbiol. 8:1997-2011. [DOI] [PubMed] [Google Scholar]
- 86.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 87.Venter, J. C., H. O. Smith, and L. Hood. 1996. A new strategy for genome sequencing. Nature 381:364-366. [DOI] [PubMed] [Google Scholar]
- 88.Volfovsky, N., B. J. Haas, and S. L. Salzberg. 2001. A clustering method for repeat analysis in DNA sequences. Genome Biol. 2:Research0027.1-0027.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.White, A. P., D. L. Gibson, W. Kim, W. W. Kay, and M. G. Surette. 2006. Thin aggregative fibriae and cellulose enhance the long-term survival and persistence of Salmonella. J. Bacteriol. 188:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wicker, N., G. R. Perrin, J. C. Thierry, and O. Poch. 2001. Secator: a program for inferring protein subfamilies from phylogenetic trees. Mol. Biol. Evol. 18:1435-1441. [DOI] [PubMed] [Google Scholar]
- 91.Xie, G., D. C. Bruce, J. F. Challacombe, O. Chertkov, J. C. Detter, P. Gilna, C. S. Han, S. Lucas, M. Misra, G. L. Myers, P. Richardson, R. Tapia, N. Thayer, L. S. Thompson, T. S. Brettin, B. Henrissat, D. B. Wilson, and M. J. McBride. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 73:3536-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yadav, G., R. S. Gokhale, and D. Mohanty. 2003. SEARCHPKS: a program for detection and analysis of polyketide synthase domains. Nucleic Acids Res. 31:3654-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yin, H., L. Cao, M. Xie, Q. Chen, G. Qiu, J. Zhou, L. Wu, D. Wang, and X. Liu. 2008. Bacterial diversity based on 16S rRNA and gyrB genes at Yinshan mine, China. Syst. Appl. Microbiol. 31:302-311. [DOI] [PubMed] [Google Scholar]
- 94.Zimmermann, J., J. M. Gonzalez, C. Saiz-Jimenez, and W. Ludwig. 2005. Detection and phylogenetic relationships of highly diverse uncultured acidobacterial communities in Altamira Cave using 23S rRNA sequence analysis. Geomicrobiol. J. 22:379-388. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.