Abstract
The atrazine chlorohydrolase AtzA has evolved within the past 50 years to catalyze the hydrolytic dechlorination of the herbicide atrazine. It is of wide research interest for two reasons: first, catalytic improvement of the enzyme would facilitate its application in bioremediation, and second, because of its recent evolution, it presents a rare opportunity to examine the early stages in the acquisition of new catalytic activities. Using a structural model of the AtzA-atrazine complex, a region of the substrate-binding pocket was targeted for combinatorial randomization. Identification of improved variants through this process informed the construction of a variant AtzA enzyme with 20-fold improvement in its kcat/Km value compared with that of the wild-type enzyme. The reduction in Km observed in the AtzA variants has allowed the full kinetic profile for the AtzA-catalyzed dechlorination of atrazine to be determined for the first time, revealing the hitherto-unreported substrate cooperativity in AtzA. Since substrate cooperativity is common among deaminases, which are the closest structural homologs of AtzA, it is possible that this phenomenon is a remnant of the catalytic activity of the evolutionary progenitor of AtzA. A catalytic mechanism that suggests a plausible mechanistic route for the evolution of dechlorinase activity in AtzA from an ancestral deaminase is proposed.
Current intensive farming practices are facilitated by the use of effective chemical weed control agents, such as the triazine herbicides. Atrazine (6-chloro-N2-ethyl-N4-isopropyl-1,3,5-triazine-2,4-diamine) is a highly effective pre- and postemergence triazine herbicide that has been used extensively for the control of broadleaf weed species since it was first introduced in 1958 (52). However, concentrations of atrazine found in the environment have been causally linked to endocrine dysfunction in vertebrate species; for example, demasculination of Xenopus laevis has been observed at atrazine concentrations as low as 1 ppb, ca. 4.5 nM (16-18). It has also been suggested that atrazine may be carcinogenic (21, 22). Due to their broad specificity, atrazine and related triazine herbicides can also be toxic to nontarget photosynthetic species, from phototropic bacteria and freshwater algae to mangrove trees (4, 31, 49). Atrazine is also environmentally persistent: the half-life of atrazine in soil has been estimated at between 4 and 57 weeks (5), and atrazine has been detected in both surface and ground waters in several countries (15, 51, 53) at concentrations up to 4.6 μM.
Several gene/enzyme systems that allow catabolism of the triazine herbicides as sources of carbon and nitrogen have evolved in prokaryotes. AtzA is the first enzyme of the most thoroughly characterized pathway, the atzABCDEF triazine-catabolic pathway, which was isolated in the early 1990s from Pseudomonas sp. strain ADP (12, 32) and is thought to have evolved recently in direct response to the use of chlorinated s-triazine herbicides (44, 55, 56). The entire catabolic pathway is encoded by the transmissible pADP1 plasmid (33). Atrazine and simazine (6-chloro-N2,N4-diethyl-1,3,5-triazine-2,4-diamine) (11) are successively dechlorinated and dealkylated by enzymes of the amidohydrolase family encoded by atzA, atzB, and atzC, yielding cyanuric acid (7, 11, 39). The latter is then converted to ammonia and carbon dioxide by the remaining hydrolases in the pathway, encoded by atzD, atzE, and atzF (10, 14, 45).
AtzA is an Fe(II)-dependent homohexamer of the amidohydrolase superfamily (20) and has been shown to catalyze the hydrolytic dechlorination of atrazine, yielding the nonherbicidal product 2-hydroxyatrazine (11). The closest known relative of AtzA is melamine deaminase (TriA from Pseudomonas sp. strain NRRL B-12227; 98% sequence identity) (41, 42). Despite their high sequence similarity, AtzA and TriA are catalytically distinct; TriA is a deaminase with low dechlorinase activity, while AtzA is a dechlorinase with no detectable deaminase activity. Previous work has shown that two of the nine amino acids that differ between the two proteins (Asn328 and Ser331 in AtzA) are largely responsible for the differences in catalytic specificity, although the mechanistic basis for this difference has not been elucidated (38).
In this work, a structural model of AtzA has been generated, providing insight into the catalytic mechanism and the evolution of catalytic activity toward atrazine. Using this model, five amino acids within the substrate-binding pocket were chosen for randomization based on their proximity to a docked substrate. Combinatorial limited site saturation mutagenesis was used to generate a library, and several AtzA variants that displayed improved catalytic efficiency were selected. Generation of the “consensus” mutant, containing the residues selected for at the highest frequency at each position, yielded a variant enzyme with 20-fold-greater catalytic efficiency than that of wild-type AtzA.
MATERIALS AND METHODS
Bacterial growth and DNA manipulation.
The plasmid encoding the atzA gene and promoter (pMD4) (11) was kindly provided by Lawrence Wackett (University of Minnesota). For these experiments, we constructed the pCS150 expression vector, in which the atzA promoter from pMD4 (12) was cloned between the AvaI and HindIII sites of pACYC184 using the primers CS284 (CAACCAATTATCTCGGGGAACTTCTTGAGCGCGGCCACAGCAG) and CS285 (CAACAAGCTTGGATCCTGCAGCTCAGCATGCGGCCGCCATATGATGTCTCCAATAGTGTGTTACAC). The reverse primer (CS285) contained a new multiple-cloning site that included an in-frame NdeI site and a BamHI site, which were used to clone the limited site saturation library discussed below using the primers CS203 (GAAGACATATGCAAACGCTCAGCATCCAGCACGGTA) and CS204 (GTTCTTGGATCCTAGAGGCTGCGCCAAGCTGGGTTAG).
We constructed a limited site saturation library (GeneArt) in which the identities of 5 amino acid residues predicted to lie in the binding pocket for the N-isopropyl side chain of atrazine were randomized to 1 of the 15 amino acids encoded by the degenerate codon BNS, where B is T, G, or C; N is any base; and S is C or G (Table 1). This generated a library containing approximately 8 × 106 different mutant genes, encoding 6 × 105 different peptides. The quality of the library was such that fewer than 1 copy in 20 encoded a mutation in addition to those included by design. DNA sequence determination from 20 random clones from the library showed that there was no bias in the incorporation of nucleotides during the synthesis of the library.
TABLE 1.
Amino acids encoded by the degenerate BNS codon used in this study
| First base (B) | Amino acid with indicated second base (N)
|
Third base (S) | |||
|---|---|---|---|---|---|
| T | C | A | G | ||
| T | Phe | Ser | Tyr | Cys | C |
| Leu | Ser | Stop | Trp | G | |
| C | Leu | Pro | His | Arg | C |
| Leu | Pro | Gln | Arg | G | |
| G | Val | Ala | Asp | Gly | C |
| Val | Ala | Glu | Gly | G | |
DNA sequencing of library variants was conducted by Micromon (Department of Microbiology, Monash University, Melbourne, Australia) using the vector-specific primers pCS150For (GACGTGCGGGATGACCACCCAGTTGCGGTGC) and pCS150Rev (GAGATTACGAGAAGACCAAAACGATCTCAAGAAGATCATC) and the atzA-specific primer atzA529F (CAAGTCGAACTGTGCTCGAT). The pCS150 derivatives containing the libraries were used to transform chemically competent Escherichia coli JM109 (Promega). Site-directed mutagenesis, using the PCR-based method of Higiuchi et al. (19), was used to generate the consensus improved AtzA variant (see below).
Library screening.
Variant enzymes with potentially improved atrazine dechlorinase activities were selected from the library on the basis of their capacity to produce zones of clearance on LB agar containing 1 mg·ml−1 atrazine and supplemented with 34 μg·ml−1 chloramphenicol. E. coli JM109 was used as the screening strain. Wild-type atzA was used as a control to establish the extent of wild-type AtzA activity in this assay, which formed visible zones of clearance after 48 h of growth at 37°C.
Protein purification.
Both TriA and the AtzA variants were expressed from pCS150, grown in 2-liter shake flasks (shaken at 250 rpm) in LB medium supplemented with chloramphenicol (34 μg·ml−1) at 37°C for 48 h. Cultures were sedimented by centrifugation and resuspended in AtzA reaction buffer (11). Cells were lysed by freeze-thawing, followed by sonication (Branson sonifier, model 250), and the soluble fraction was obtained by centrifugation at 14,000 × g for 30 min. TriA, AtzA, and the AtzA variants constituted greater than 50% of the total protein in the enriched sample by the ammonium-sulfate precipitation method described by de Souza et al. (11) and quantified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis densitometry. Samples of bovine serum albumin (5, 10, and 20 μg) were loaded onto each gel analyzed by densitometry to provide an internal standard. Protein determinations were verified using the Bio-Rad protein assay dye (Bio-Rad). AtzA recovered from pCS150 containing wild-type atzA had approximately the same specific activity as the published value (11).
Enzyme kinetics.
The colorimetric chloro-s-triazine detection method of Tawfik and coworkers (50) was adapted to a 96-well microtiter plate format to allow moderate-throughput screening of specific activity and determination of the kinetic parameters of the variants. Briefly, pyridine is used to detect atrazine, as it reacts with chloro-s-triazines to produce colored products that can be quantified by measuring their absorbance at 436 nm. Determination of atrazine concentration from samples and standards was performed using a SpectroMAX 190 spectrophotometer (Molecular Devices). Specific activity assays were performed with 34.6 nM enzyme and 23 μM atrazine, equivalent to approximately 16% of the published Km of AtzA for atrazine (11). Initial velocities were determined using samples in which 10% or less of the substrate had been dechlorinated.
Steady-state kinetic analysis of wild-type AtzA and variants utilized the colorimetric protocol, using atrazine concentrations between 4.6 and 153 μM and enzyme concentrations of 10 nM. Where possible, the Km, the maximal reaction rate (Vmax), and the Hill coefficient for each reaction was calculated using a curve-fitting program (CurveExpert 1.3; http://curveexpert.webhop.biz/) that uses the Hill equation (1), V = Vmax([S]n/[KA]n + [S]n, where V is the reaction rate, [s] is the substrate concentration, KA is the substrate concentration at half the Vmax, and n is the Hill coefficient. The second-order rate constant (kcat/Km) for wild-type AtzA was calculated under the assumption that when [S] is much less than Km, V is equal to kcat/Km[E][S], where [E] is the enzyme concentration.
Liquid chromatography-mass spectrometry (LC-MS) was used to validate the colorimetric method used in the moderate-throughput specific-activity assay and to determine the kinetic properties of the TriA-mediated deamination of melamine. The substrates and products were first separated using an SB-C18 column (Zorbax) with a 0-to-100% methanol gradient. Samples were run in a 0.1% formate solution (high-pressure liquid chromatography grade). Atrazine, 2-hydroxyatrazine, melamine, and 2-hydroxymelamine were then detected using an Agilent G1969 LC-MS time of flight instrument. Initial velocities were determined using samples in which 10% or less of the substrate had been dechlorinated or deaminated. PESTANAL analytical standards of atrazine, hydroxyatrazine, and hydroxymelamine were obtained from Sigma-Aldrich. Melamine (>99% purity) was also obtained from Sigma-Aldrich.
Computational procedures.
The closest structural relatives of AtzA were identified by submitting the AtzA sequence to the FUGUE server (46; http://www-cryst.bioc.cam.ac.uk/fugue/). The closest relatives with known structures were a protein of unknown function from Thermotoga maritima (1J6P, 25% identity [unpublished]), the imidazolone propionase from Agrobacterium tumefaciens (2GOK, 17% identity [unpublished]), and the cytosine deaminase from E. coli (1K6W, 17% identity [24]). The sequence of AtzA was initially aligned with these templates using the T-COFFEE server. 1J6P was used as the primary template, and with a T-COFFEE pairwise alignment (36), the AtzA sequence was threaded onto the template using the program DEEPVIEW (25). The preliminary model was then optimized using the SWISS-MODEL workspace (http://swissmodel.expasy.org/workspace) (3), which successfully ligated all loops and minimized the structure. The robustness of the structure was monitored through analysis of plots of the Anolea mean force potential (34) and GROMOS (6) empirical force field energy, as implemented in the workspace. The structure was of good quality, consistent with the quality of the alignments.
The ab initio conformation of atrazine was calculated in the gas phase at the B3-LYP/6-to-31G(d) level of theory, as implemented in the SPARTAN software package (Wavefunction, Inc.) (59). Docking of atrazine at the active site of the AtzA model was undertaken first through manual positioning, using the program COOT (13), based on the positions of 4-imidazolone-5-propanoate and 4-hydroxy-3,4-dihydro-1H-pyrimidin-2-one in the templates 2GOK and 1K6W, respectively. The enzyme-substrate system was then minimized with the nonredundant basis set and the CHARMm (8) force field over 1,000 steps with a root mean square gradient of 0.1 kcal/(mol × Å), as implemented in the Discovery Studio software suite (Accelrys). Unless stated otherwise, default settings were used. Images were produced using PyMol Viewer v0.99rc6 (DeLano Scientific LLC.).
RESULTS
Structural model.
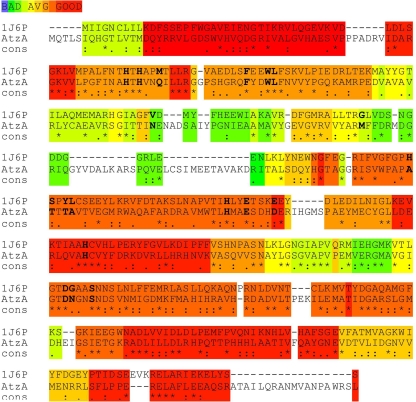
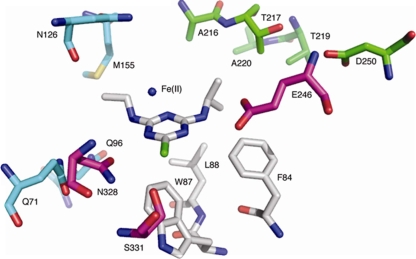
AtzA is known to be a metallohydrolase from the urease/amidohydrolase superfamily (11), although its atomic structure has yet to be elucidated. To construct a homology model of AtzA, its closest homolog with a known structure (a protein of unknown function from T. maritima) was identified using the FUGUE server and used as a template for model building (Fig. 1). Atrazine was docked into the active site of the AtzA model, and the CHARMm force field was then used in an energy minimization step to optimize the orientation. Based on the modeled enzyme-substrate complex, the active site and substrate-binding pocket could be divided into five sections (Fig. 2; section i has been omitted from this figure for clarity): (i) residues His66, His68, His243, His276, and Asp327, which are essential for coordination of the active-site metal ion (43); (ii) residues Glu246, Asn328, and Ser331, which are involved in coordination of the substrate; (iii) residues Phe84, Trp87, and Leu88, which form the hydrophobic “base” of the active site and are likely to be essential for π-π interaction with the aromatic ring of the substrate; (iv) the N-isopropyl side chain pocket (Ala216, Thr217, Thr219, Ala220, and Asp250); and (v) the N-ethyl side chain pocket (Gln71, Gln96, Asn126, and Met155).
FIG. 1.
Alignment of the amino acid sequences of 1J6P and AtzA. The homology between AtzA and 1J6P is indicated. Conserved active-site- and substrate-binding pocket residues are in boldface.
FIG. 2.
Model of atrazine (center) docked in the active site of AtzA. The residues forming the hydrophobic base (Phe84, Trp87, and Leu88 [white]), the residues involved in coordination of the substrate (Glu246, Asn328, and Ser331 [purple]), the N-isopropyl binding pocket (Ala216, Thr217, Thr219, Ala220, and Asp250 [green]), and the N-ethyl binding pocket (Gln71, Gln96, Asn126, and Met155 [blue]) are shown. The residues that coordinate the active-site metal (His66, His68, His243, His276, and Asp327) have been omitted for clarity.
As seen in Fig. 1, sections i to iv are all in regions of average-to-good homology, whereas region v is in a region of average-to-poor homology. The overall identity between AtzA and 1J6P is relatively low (25%), which is largely due to several insertions that are located in surface-exposed regions and are remote from the active site and substrate-binding pocket. Indeed, the regions comprising the active site and substrate-binding pocket, which are the only regions for which we require the model, have considerably higher identity (51%). Furthermore, if the predicted N-ethyl side chain pocket (Gln71, Gln96, Asn126, and Met155) is excluded, as was done for the protein engineering described later, the sequence identity increases to 67%. Thus, for the purposes of this work, namely, redesigning the substrate-binding pocket and investigating the catalytic mechanism, the model is sufficiently accurate.
Combinatorial mutagenesis.
Examination of the 1J6P-AtzA alignment (Fig. 1) reveals that the N-isopropyl side chain-binding pocket is in a region of good homology and that the model is likely to be relatively accurate. This was not the case for the pocket predicted to accommodate the N-ethyl group of the substrate. Thus, only the residues of the N-isopropyl side chain-binding pocket were selected to randomize in an attempt to improve the catalytic efficiency of AtzA. Since the amino acid residues that comprise the N-isopropyl pocket (Ala216, Thr217, Thr219, Ala220, and Asp250) did not appear to form a complementary pocket for the N-isopropyl side chain of atrazine, they were selected for randomization, with the rationale that by altering the composition of this pocket, it might be possible to reduce the Km of AtzA for atrazine by allowing more-productive contact with the substrate via their substitution by alternative amino acids. A combinatorial, limited site saturation library in which these amino acid residues were each replaced with 1 of the 15 amino acids encoded by the degenerate BNS codon was therefore made (Table 1).
The variant enzymes encoded by this library were screened for their ability to clear a “halo” in LB agar containing atrazine precipitate. The DNA sequences of variants that demonstrated more-rapid clearing than wild-type AtzA were determined, and variants with mutations outside the targeted region were discarded. From approximately 106 clones (12.5% of the possible diversity of the library at the DNA level; twofold screening coverage at the amino acid level), we selected 126 that formed zones of clearance more rapidly than wild-type AtzA and lacked additional mutations.
A secondary screen was then used to identify AtzA variants with high catalytic efficiency at low substrate concentrations, i.e., displaying low Km constants. Therefore, the 126 variants selected from the plate-clearing assay were assayed at a concentration of 23 μM atrazine (approximately 16% of the published Km of wild-type AtzA) (11). Sixty-three of these variants had specific activities at least twice that of the wild-type enzyme under these conditions, and 46 of them were at least five times more active than the wild-type enzyme (Fig. 3).
FIG. 3.
Distribution of the improvements (n-fold) in the specific activities of variants selected from the limited site saturation library. Specific activities were determined using a 23 μM initial concentration of atrazine and 34.6 nM enzyme.
Sequence data from the top 46 variants were used to analyze the amino acid preference at each of the five randomized sites (Table 2). Residues 216 and 217 appear to have the greatest influence of the five targeted residues upon AtzA activity. Tyrosine and aspartic acid were, respectively, identified 8.3 and 10.4 times more frequently at these positions than was expected, given the content of the degenerate BNS codon. There was less, but still significant, selective pressure at positions 220 and 250: histidine was 5.2-fold overrepresented at position 220, and glutamine was 5.2-fold overrepresented at 250. Finally, there was no clear preference at position 219 (glycine was 2.6-fold overrepresented). The consensus of the amino acid preferences in the 46 variants with at least a fivefold increase in specific activities was therefore Ala216Tyr, Thr217Asp, Ala220His, and Asp250Glu, with no clear preference at residue 219 (Table 2). Because the consensus sequence was not isolated from the library, either due to insufficient sampling of the library or unfavorable enzymatic properties of the consensus variant, it was generated by site-directed mutagenesis for further analysis and found to confer a fast-clearing phenotype upon E. coli JM109 (data not shown).
TABLE 2.
Distribution of amino acids among the AtzA variants with specific activities greater than fivefold more than that of the wild type
| Amino acid | Fold overrepresentation at indicated position (consensus sequence; wild-type amino acid)a
|
||||
|---|---|---|---|---|---|
| 216 (YAS; A) | 217 (DGA; T) | 219 (GYE; T) | 220 (HAS; A) | 250 (EDGYW; D) | |
| A | 12 (3.1) | 10 (2.6) | 2 (0.5) | 12 (3.1) | 2 (0.5) |
| L | ND | ND | 4 (0.7) | ND | 2 (0.3) |
| G | 6 (1.6) | 12 (3.1) | 10 (2.6) | 2 (0.5) | 8 (2.1) |
| V | ND | ND | 4 (1.0) | 2 (0.5) | 2 (0.5) |
| S | 8 (2.1) | 4 (1.0) | 2 (0.5) | 12 (3.1) | 4 (1.0) |
| Q | ND | ND | ND | 2 (1.0) | ND |
| Y | 16 (8.3) | ND | 4 (2.1) | ND | 4 (2.1) |
| P | ND | ND | 6 (1.5) | 2 (0.5) | 2 (0.5) |
| D | ND | 20 (10.4) | 6 (1.5) | ND | 4 (2.1) |
| E | ND | ND | 4 (2.1) | ND | 10 (5.2) |
| R | ND | ND | 2 (0.5) | ND | ND |
| C | ND | ND | ND | ND | 2 (1.0) |
| H | 4 (2.1) | ND | 2 (1.0) | 10 (5.2) | 2 (1.0) |
| W | ND | ND | ND | 2 (1.0) | 4 (2.1) |
| F | ND | ND | ND | 2 (1.0) | ND |
Specific activities were determined with an initial concentration of 23 μM atrazine to select for variants with reduced Km values. The values in parentheses indicate the selective pressure at each position (the frequency of each amino acid compared with that expected from the frequency of each base in the degenerate BNS codon). Where an amino acid has been selected greater than fivefold more frequently than expected, it is in boldface in the consensus sequence. ND, not detected.
Kinetic analysis of AtzA variants.
Kinetic parameters were obtained for wild-type AtzA, the 10 most improved variants recovered from the library, and the consensus variant (Table 3). The 10 variants isolated from the library with the highest specific activities had Km values between 49 and 105 μM (Table 3), significantly lower than the published wild-type value (11), while the consensus variant had a Km of 93 μM. The highest kcat from a variant isolated from the library was 15.1 s−1 (variant 734), while the kcat observed for the consensus variant was 27 s−1 (Table 3). The kcat/Km values for the 10 library variants were distributed within a sevenfold range (3.4 × 104 to 2.4 × 105 s−1·M−1), with that of the consensus variant being somewhat higher again (3.0 × 105 s−1·M−1). Based on the rate of spontaneous, noncatalyzed dechlorination of atrazine (knon) estimated by Lei and coworkers (29), the rate of enhancement (kcat/knon) of the consensus N-isopropyl pocket variant generated in this study was equivalent to approximately 1.3 × 108. Thus, an >20-fold improvement of the catalytic power of AtzA was achieved through a single round of combinatorial randomization of the substrate-binding pocket, followed by construction of the consensus mutant.
TABLE 3.
Characterization of wild-type TriA, AtzA, and the most improved AtzA variants derived by limited site saturation mutagenesisa
| Variant or position of variation in AtzA | Amino acid at position:
|
Km (μM) | kcat (s−1) | kcat/Km (s−1·M−1) | Hill coefficientb | ||||
|---|---|---|---|---|---|---|---|---|---|
| 216 | 217 | 219 | 220 | 250 | |||||
| TriA | A | I | P | A | D | 305 | 4.1 | 1.3 × 104 | 3.8 |
| AtzAc | A | T | T | A | D | >153 | 2.2 | 1.5 × 104 | ND |
| 357 | S | A | P | F | G | 67 | 2.3 | 3.4 × 104 | 3.6 |
| 288 | A | D | E | A | D | 49 | 4.3 | 8.8 × 104 | 2.8 |
| 305 | G | D | A | V | W | 95 | 6.2 | 6.5 × 104 | 2.5 |
| 422 | Y | D | Y | H | V | 92 | 6.5 | 7.1 × 104 | 3.5 |
| 297 | S | D | V | H | G | 92 | 6.8 | 7.4 × 104 | 3.3 |
| 662 | H | A | E | S | S | 100 | 7.4 | 7.4 × 104 | 3.4 |
| 431 | A | S | H | G | Y | 76 | 8.0 | 1.1 × 105 | 3.8 |
| 841 | S | D | G | S | D | 105 | 8.5 | 8.1 × 104 | 3.8 |
| 430 | G | D | G | H | G | 90 | 12.7 | 1.4 × 105 | 3.8 |
| 734 | G | D | G | H | D | 62 | 15.1 | 2.4 × 105 | 3.6 |
| Consensus | Y | D | T | H | E | 93 | 27.9 | 3.0 × 105 | 3.2 |
Kinetic data for atrazine dechlorination by AtzA and the AtzA variants were obtained using 4.6 to 153 μM atrazine and 10 nM enzyme. Kinetic data for melamine deamination by TriA were obtained using 80 to 560 μM melamine and 10 nM TriA. Values for the Km, the kcat, and the Hill coefficient for each enzyme varied by less than 10% in replicate experiments.
ND, not determined.
As the Km of wild-type AtzA for atrazine exceeded the water solubility of atrazine, it was not possible to directly determine the Km, kcat, or Hill coefficient for wild-type AtzA; the maximum rate of wild-type AtzA, normalized for enzyme concentration, was used in place of kcat.
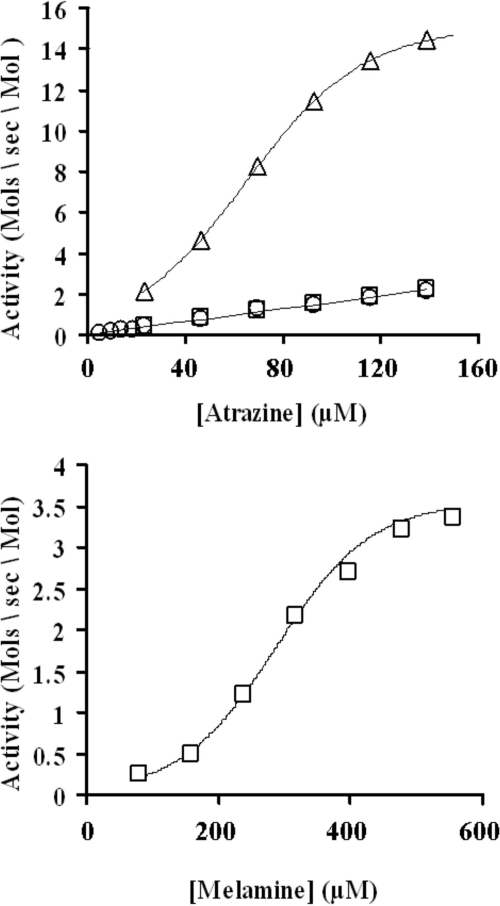
The relationship between the reaction rate of wild-type AtzA and substrate concentration was linear up to the aqueous solubility limit of atrazine (153 μM) (Fig. 4) (52). This finding was confirmed by LC-MS analysis of hydroxyatrazine production. Thus, it was not possible to corroborate the previous determination of the Km of wild-type AtzA for atrazine (149 μM) (11). Indeed, the data presented here suggest that the Km of AtzA for atrazine exceeds the water solubility of atrazine.
FIG. 4.
(Top) Kinetic profiles for dechlorination of atrazine by wild-type AtzA (squares) and the AtzA 734 variant (triangles) as determined by colorimetric assay and by wild-type AtzA as determined by LC-MS (circles); (bottom) deamination of melamine by TriA as determined by LC-MS. Data varied by less than 10% in replicate experiments.
Surprisingly, unlike wild-type AtzA, the improved variants displayed a sigmoidal response to increasing substrate concentration (with r values of 0.996 to 0.999) (Fig. 4). Such a relationship suggests that AtzA may be subject to positive substrate-dependent cooperative behavior (with Hill coefficients of 2.5 to 3.8) (Table 3). Substrate cooperativity (with a Hill coefficient of 3.8) was also observed in TriA-catalyzed melamine deamination (Fig. 4; Table 3). TriA catalyzed the reaction with a kcat value of 4.1 s−1 and a Km value of 305 μM for melamine (kcat/Km = 1.3 × 104) (Table 3).
Structural effects of the mutations.
The potential effects of the mutations found in the consensus variant were examined in the structural model. As seen in Fig. 5 and Table 2, there is a clear relationship between the selective pressure at each position and the proximity of the residues to the substrate. The positions under the strongest selective pressure (216 and 217) were closest to the substrate, followed by those positions under moderate selection pressure (220 and 250), while the position that was under no significant selective pressure was the most remote (219). Additionally, larger residues were substituted for the wild-type amino acids at the four closer sites (i.e., A216Y, T217D, A220H, and D250E). These changes are almost certain to result in closer contacts between the enzyme and the substrate, which is consistent with the large reduction in Km observed in the kinetic analysis.
FIG. 5.
Model of atrazine docked in the active site of the wild-type AtzA N-isopropyl pocket (A) and the consensus N-isopropyl pocket variant of AtzA (B). Residues in the N-isopropyl pocket that were randomized are shown, demonstrating the closer contacts between the enzyme and substrate in the consensus variant.
DISCUSSION
Mechanism and evolution of atrazine dechlorination by AtzA.
Using the structural model described above, and with reference to the well-characterized catalytic mechanism of the closely related Saccharomyces cerevisiae cytosine deaminase (23), a plausible catalytic mechanism can be proposed (Fig. 6). We suggest that substrate binding will involve π-π stacking between the aromatic ring of the substrate and the hydrophobic base of the cavity formed by Phe84, Trp87, and Leu88, in addition to hydrogen bonding with the conserved glutamate (Glu246). The active-site metal ion (Fe2+) and Asp327 are positioned to activate a solvent molecule, resulting in the formation of a nucleophilic hydroxide. Nucleophilic attack at the C-4 carbon of atrazine would result in the formation of a tetrahedral intermediate. Breakdown of the intermediate and departure of the leaving group would be stabilized through the interaction between the halide and Ser331, which is in turn stabilized by hydrogen bonding to Asn328. The importance of positions 331 and 328 to catalysis has been verified elsewhere by mutagenesis (38).
FIG. 6.
Proposed catalytic mechanism for AtzA-mediated dechlorination of atrazine. The reaction proceeds by nucleophilic attack at C-4 of atrazine by the hydroxyl nucleophile (A), leading to the formation of the tetrahedral intermediate (B), followed by breakdown of the intermediate and departure of the chloride leaving group (C). Other residues that coordinate the active-site metal (His66, His68, His243, His276) have been omitted for clarity.
It has been proposed that the chlorohydrolase atzA gene evolved from the deaminase triA gene in response to the anthropogenic selection pressure induced by the use of the synthetic triazine herbicide atrazine (41, 44, 54), conferring a selective advantage on its host by allowing the exploitation of atrazine as a carbon and nitrogen source (42). This complete change in activity has resulted from only nine amino acid substitutions and particularly through a change in the Asp328-Cys331 dyad to an Asn328-Ser331 dyad, as described above (38). Thus, a simple mechanistic route from deaminase activity to chlorohydrolase activity must exist.
The chemical differences between the leaving groups in the chlorohydrolase and deaminase reactions and the chemical properties of these distinct catalytic dyads (Asp-Cys/Asn-Ser) provide a mechanistic explanation for the rapid change in activity. As chloride leaving groups are considerably more labile than the amine leaving groups, it is reasonable to assume that activation of the amine group of melamine is important for efficient catalysis but that the chloride leaving group of atrazine is not. The catalytic dyads of the two enzymes appear perfectly suited to these distinct requirements. In the Asn328-Ser331 dyad of AtzA, the hydroxyl group of Ser331 may hydrogen bond to the leaving group and stabilize the developing negative charge in the transition state to lower the activation energy (Ser331 would be stabilized through its hydrogen bonding interaction with Asn328 but not strongly polarized). In contrast, in the Asp328-Cys331 dyad of TriA, the aspartic acid may abstract a proton from the cysteine and generate a thiolate that may activate the amine leaving group, thereby making it more electron withdrawing and amenable to departure.
This model predicts that the TriA dyad, or an intermediate dyad containing Asp328-Ser331, would be able to catalyze both reactions, since Ser331, polarized by Asp328, may substitute for Cys331 in activating the amine leaving group. In contrast, the second alternative dyad of Asn328-Cys331 would be catalytically inactive, since Asn328 would not be able to substitute for Asp328 in abstracting a proton from Cys331. Indeed, this is what is observed in the data of Raillard and coworkers (38). Thus, a clear evolutionary pathway from deaminase activity to exclusive chlorohydrolase activity, via a latent promiscuous activity (28), can involve as few as two amino acid substitutions.
Substrate cooperativity: an evolutionary remnant?
Although substrate cooperativity has not been observed in wild-type AtzA, its observation in the AtzA variants and TriA suggests that it is highly likely to exist in the wild-type enzyme and is not seen only because the Km for atrazine exceeds the solubility limit of the substrate. Thus, this kinetic phenomenon is most probably a remnant of an evolutionary progenitor with a far more soluble substrate. Indeed, amine-substituted aromatics have much higher solubility limits than their chlorine analogs, e.g., melamine (25 mM) versus atrazine (153 μM) (52). Melamine itself is a man-made compound and has been in the environment for only ca. 70 years, and it would be surprising for such a trait to have evolved so rapidly. It is therefore likely that AtzA and TriA share a common deaminase ancestor that displays substrate cooperativity, especially considering the observation that substrate cooperativity is a relatively common trait in deaminases (9, 35). Efforts are under way to probe the early stages of divergence in more detail and to identify the ancestor of TriA and AtzA in order to map the mechanism of AtzA's evolution more completely.
Evolutionary limits of AtzA.
Because of the recent emergence of AtzA, it is unlikely that the enzyme has fulfilled its evolutionary potential for the catalysis of atrazine hydrolysis. Indeed, the data presented here (Table 3) demonstrate that a 20-fold improvement in catalytic efficiency can be obtained by combinatorial randomization of only 5 residues, largely through a reduction in Km. Additionally, the high Hill coefficient values obtained from the variant AtzA enzymes (2.5 to 3.8) suggest that the positive cooperativity of the enzyme is highly optimized but for a much more soluble substrate.
Moreover, while the consensus AtzA variant has an improved turnover value (kcat = 27.9 s−1) compared with wild-type AtzA, the structurally and mechanistically related E. coli cytosine deaminase has a kcat value of 185 s−1 (37), a sevenfold increase compared to the kcat value of the consensus AtzA variant. These relative deaminase/dechlorinase rates are inconsistent with the chemical stability of cytosine and atrazine: the half-life of cytosine in water at pH 7.0 is estimated to be ∼340 years (30), whereas the half-life of atrazine is estimated to be <3 years (29). Considering that the cytosine deaminase has had many millions of years to evolve toward efficient deaminase activity, and AtzA has had less than a century, these relative rates are not entirely surprising and suggest that there is much potential for further evolution of dechlorinases.
Biotechnological significance of a catalytically improved AtzA.
AtzA has potential utility in both the generation of herbicide-resistant crops (57) and the bioremediation of atrazine contamination. Indeed, Kawahigashi and coworkers have proposed expressing mammalian cytochrome P450 in transgenic plants to phytoremediate atrazine (26, 27). Transgenic AtzA-expressing E. coli has also been employed in field trials to remove residual atrazine contamination in situ (47). Free-enzyme bioremediation is another option for the development of bioremediants which do not require the release of genetically modified organisms and also has many additional advantages (discussed in depth elsewhere) (2, 40, 48). Irrespective of the bioremediation strategy used, the demands on an enzyme for cost-effective use as a commercial bioremediant are somewhat stringent (2, 48) and include a requirement for a low Km to allow removal of contaminants to concentrations below the maximum residue limits prescribed by regulatory bodies.
The need to improve AtzA and functionally related enzymes has been noted by other investigators. There is a report of attempts to improve the catalytic characteristics of AtzA by creating chimeras of AtzA and TriA, yielding enzymes with broadened substrate specificities (38). Chimeras of TrzN, a triazine chlorohydrolase functionally related to AtzA, have been generated from naturally occurring variants, revealing the identities of residues important in determining substrate specificity (58). We envisage that catalytically improved atrazine dechlorinase enzymes, such as those described here and elsewhere, will play an important role in future bioremediation strategies.
Acknowledgments
We thank Lawrence Wackett for kindly providing plasmid pMD4.
This work was supported in part by Orica Australia Ltd.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Adair, G. S. 1925. The hemoglobin system. VI. The oxygen dissociation curve of hemoglobin. J. Biol. Chem. 63:529-545. [Google Scholar]
- 2.Alcalde, M., M. Ferrer, F. J. Plou, and A. Ballesteros. 2006. Environmental biocatalysis: from remediation with enzymes to novel green processes. Trends Biotechnol. 24:281-287. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 4.Bell, A. M., and N. C. Duke. 2005. Effects of Photosystem II inhibiting herbicides on mangroves—preliminary toxicology trials. Mar. Pollut. Bull. 51:297-307. [DOI] [PubMed] [Google Scholar]
- 5.Belluck, D. A., S. L. Benjamin, and T. Dawson. 1991. Groundwater contamination by atrazine and its metabolites—risk assessment, policy, and legal implications. ACS Symp. Ser. 459:254-273. [Google Scholar]
- 6.Billeter, S. R., and W. F. van Gunsteren. 1997. A modular molecular dynamics quantum dynamics program for non-adiabatic proton transfers in solution. Comput. Physics Commun. 107:61-91. [Google Scholar]
- 7.Boundy-Mills, K. L., M. L. deSouza, R. T. Mandelbaum, L. P. Wackett, and M. J. Sadowsky. 1997. The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl. Environ. Microbiol. 63:916-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy minimization and dynamics calculations. J. Comp. Chem. 4:187-217. [Google Scholar]
- 9.Bustos-Jaimes, I., A. Sosa-Peinado, E. Rudino-Pinera, E. Horjales, and M. L. Calcagno. 2002. On the role of the conformational flexibility of the active-site lid on the allosteric kinetics of glucosamine-6-phosphate deaminase. J. Mol. Biol. 319:183-189. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, G., N. Shapir, M. J. Sadowsky, and L. P. Wackett. 2005. Allophanate hydrolase, not urease, functions in bacterial cyanuric acid metabolism. Appl. Environ. Microbiol. 71:4437-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Souza, M. L., M. J. Sadowsky, and L. P. Wackett. 1996. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization. J. Bacteriol. 178:4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Souza, M. L., L. P. Wackett, K. L. Boundymills, R. T. Mandelbaum, and M. J. Sadowsky. 1995. Cloning, characterization, and expression of a gene region from Pseudomonas sp. strain ADP involved in the dechlorination of atrazine. Appl. Environ. Microbiol. 61:3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 14.Fruchey, I., N. Shapir, M. J. Sadowsky, and L. P. Wackett. 2003. On the origins of cyanuric acid hydrolase: purification, substrates, and prevalence of AtzD from Pseudomonas sp. strain ADP. Appl. Environ. Microbiol. 69:3653-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavrilescu, M. 2005. Fate of pesticides in the environment and its bioremediation. Eng. Life Sci. 5:497-526. [Google Scholar]
- 16.Hayes, T., K. Haston, M. Tsui, A. Hoang, C. Haeffele, and A. Vonk. 2003. Atrazine-induced hermaphroditism at 0.1 ppb in American leopard frogs (Rana pipiens): laboratory and field evidence. Environ. Health Perspect. 111:568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes, T. B., A. Collins, M. Lee, M. Mendoza, N. Noriega, A. A. Stuart, and A. Vonk. 2002. Hermaphroditic, demasculinized frogs after exposure to the herbicide atrazine at low ecologically relevant doses. Proc. Natl. Acad. Sci. USA 99:5476-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes, T. B., A. A. Stuart, M. Mendoza, A. Collins, N. Noriega, A. Vonk, G. Johnston, R. Liu, and D. Kpodzo. 2006. Characterization of atrazine-induced gonadal malformations in African clawed frogs (Xenopus laevis) and comparisons with effects of an androgen antagonist (cyproterone acetate) and exogenous estrogen (17 beta-estradiol): support for the demasculinization/feminization hypothesis. Environ. Health Perspect. 114:134-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higiuchi, R., B. Krummel, and R. Saiki. 1988. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm, L., and C. Sander. 1997. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins 28:72-82. [PubMed] [Google Scholar]
- 21.Huff, J. 2002. IARC monographs, industry influence, and upgrading, downgrading, and under-grading chemicals—a personal point of view. Int. J. Occup. Environ. Health 8:249-270. [DOI] [PubMed] [Google Scholar]
- 22.Huff, J., and J. Sass. 2007. Atrazine—a likely human carcinogen? Int. J. Occup. Environ. Health 13:356-358.17915550 [Google Scholar]
- 23.Ireton, G. C., M. E. Black, and B. L. Stoddard. 2001. Crystallization and preliminary X-ray analysis of bacterial cytosine deaminase. Acta Crystallogr. D 57:1643-1645. [DOI] [PubMed] [Google Scholar]
- 24.Ireton, G. C., G. McDermott, M. E. Black, and B. L. Stoddard. 2002. The structure of Escherichia coli cytosine deaminase. J. Mol. Biol. 315:687-697. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan, W., and T. G. Littlejohn. 2001. Swiss-PDB Viewer (Deep View). Brief. Bioinform. 2:195-197. [DOI] [PubMed] [Google Scholar]
- 26.Kawahigashi, H., S. Hirose, H. Ohkawa, and Y. Ohkawa. 2007. Herbicide resistance of transgenic rice plants expressing human CYP1A1. Biotechnol. Adv. 25:75-84. [DOI] [PubMed] [Google Scholar]
- 27.Kawahigashi, H., S. Hirose, H. Ohkawa, and Y. Ohkawa. 2006. Phytoremediation of the herbicides atrazine and metolachlor by transgenic rice plants expressing human CYP1A1, CYP2B6, and CYP2C19. J. Agric. Food Chem. 54:2985-2991. [DOI] [PubMed] [Google Scholar]
- 28.Khersonsky, O., C. Roodveldt, and D. S. Tawfik. 2006. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10:498-508. [DOI] [PubMed] [Google Scholar]
- 29.Lei, Z. F., C. M. Ye, and X. J. Wang. 2001. Hydrolysis kinetics of atrazine and influence factors. J. Environ. Sci. (China) 13:99-103. [PubMed] [Google Scholar]
- 30.Levy, M., and S. L. Miller. 1998. The stability of the RNA bases: implications for the origin of life. Proc. Natl. Acad. Sci. USA 95:7933-7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lockert, C. K., K. D. Hoagland, and B. D. Siegfried. 2006. Comparative sensitivity of freshwater algae to atrazine. Bull. Environ. Contam. Toxicol. 76:73-79. [DOI] [PubMed] [Google Scholar]
- 32.Mandelbaum, R. T., D. L. Allan, and L. P. Wackett. 1995. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol. 61:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melo, F., and E. Feytmans. 1998. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 277:1141-1152. [DOI] [PubMed] [Google Scholar]
- 35.Merkler, D. J., and V. L. Schramm. 1990. Catalytic and regulatory site composition of yeast amp deaminase by comparative binding and rate studies—resolution of the cooperative mechanism. J. Biol. Chem. 265:4420-4426. [PubMed] [Google Scholar]
- 36.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 37.Porter, D. J. T., and E. A. Austin. 1993. Cytosine deaminase—the roles of divalent metal-ions in catalysis. J. Biol. Chem. 268:24005-24011. [PubMed] [Google Scholar]
- 38.Raillard, S., A. Krebber, Y. C. Chen, J. E. Ness, E. Bermudez, R. Trinidad, R. Fullem, C. Davis, M. Welch, J. Seffernick, L. P. Wackett, W. P. C. Stemmer, and J. Minshull. 2001. Novel enzyme activities and functional plasticity revealed by recombining highly homologous enzymes. Chem. Biol. 8:891-898. [DOI] [PubMed] [Google Scholar]
- 39.Sadowsky, M. J., Z. K. Tong, M. de Souza, and L. P. Wackett. 1998. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J. Bacteriol. 180:152-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott, C., G. Pandey, C. J. Hartley, C. J. Jackson, M. J. Cheesman, M. C. Taylor, R. Pandey, J. L. Khurana, M. Teese, C. W. Coppin, K. M. Weir, R. K. Jain, R. Lal, R. J. Russell, and J. G. Oakeshott. 2008. The enzymatic basis for bioremediation. Indian J. Microbiol. 48:65-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seffernick, J. L., M. L. de Souza, M. J. Sadowsky, and L. P. Wackett. 2001. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J. Bacteriol. 183:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seffernick, J. L., and L. P. Wackett. 2001. Rapid evolution of bacterial catabolic enzymes: a case study with atrazine chlorohydrolase. Biochemistry 40:12747-12753. [DOI] [PubMed] [Google Scholar]
- 43.Seibert, C. M., and F. M. Raushel. 2005. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44:6383-6391. [DOI] [PubMed] [Google Scholar]
- 44.Shapir, N., E. F. Mongodin, M. J. Sadowsky, S. C. Daugherty, K. E. Nelson, and L. P. Wackett. 2007. Evolution of catabolic pathways: genomic insights into microbial s-triazine metabolism. J. Bacteriol. 189:674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapir, N., M. J. Sadowsky, and L. P. Wackett. 2005. Purification and characterization of allophanate hydrolase (AtzF) from Pseudomonas sp. strain ADP. J. Bacteriol. 187:3731-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, J., T. L. Blundell, and K. Mizuguchi. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310:243-257. [DOI] [PubMed] [Google Scholar]
- 47.Strong, L. C., H. McTavish, M. J. Sadowsky, and L. P. Wackett. 2000. Field-scale remediation of atrazine-contaminated soil using recombinant Escherichia coli expressing atrazine chlorohydrolase. Environ. Microbiol. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 48.Sutherland, T. D., I. Horne, K. M. Weir, C. W. Coppin, M. R. Williams, M. Selleck, R. J. Russell, and J. G. Oakeshott. 2004. Enzymatic bioremediation: from enzyme discovery to applications. Clin. Exp. Pharmacol. Physiol. 31:817-821. [DOI] [PubMed] [Google Scholar]
- 49.Sutton, W. F., A. E. Brown, and B. Truelove. 1984. Atrazine-resistant and diuron-resistant strains of Rhodopseudomonas sphaeroides. Weed Sci. 32:664-669. [Google Scholar]
- 50.Tawfik, M., H. Ragab, and J. P. McCollum. 1968. Colorimetric methods for the determination of simazime and related chloro-s-triazines. J. Agric. Food Chem. 16:284-289. [Google Scholar]
- 51.Thurman, E. M., and M. T. Meyer. 1996. Herbicide metabolites in surface water and groundwater: introduction and overview. ACS Symp. Ser. 630:1-15. [Google Scholar]
- 52.Tomlin, C. D. S. 2006. The pesticide manual, 14th ed. British Crop Protection Council, Alton, United Kingdom.
- 53.van der Meer, J. R. 2006. Environmental pollution promotes selection of microbial degradation pathways. Front. Ecol. Environ. 4:35-42. [Google Scholar]
- 54.Wackett, L. P. 1998. Directed evolution of new enzymes and pathways for environmental biocatalysis. Enzyme Eng. 864:142-152. [DOI] [PubMed] [Google Scholar]
- 55.Wackett, L. P. 2004. Evolution of enzymes for the metabolism of new chemical inputs into the environment. J. Biol. Chem. 279:41259-41262. [DOI] [PubMed] [Google Scholar]
- 56.Wackett, L. P. 2004. Evolution of new enzymes and pathways: soil microbes adapt to s-triazine herbicides. Pestic. Decontam. Detox. 863:37-48. [Google Scholar]
- 57.Wang, L., D. A. Samac, N. Shapir, L. P. Wackett, C. P. Vance, N. E. Olszewski, and M. J. Sadowsky. 2005. Biodegradation of atrazine in transgenic plants expressing a modified bacterial atrazine chlorohydrolase (atzA) gene. Plant Biotechnol. J. 3:475-486. [DOI] [PubMed] [Google Scholar]
- 58.Yamazaki, K., K. Fujii, A. Iwasaki, K. Takagi, K. Satsuma, N. Harada, and T. Uchimura. 2008. Different substrate specificities of two triazine hydrolases (TrzNs) from Nocardioides species. FEMS Microbiol. Lett. 286:171-177. [DOI] [PubMed] [Google Scholar]
- 59.Young, D. 2001. SPARTAN, p. 330. In Computational chemistry. Wiley-Interscience, New York, NY.