Abstract
Searles Lake occupies a closed basin harboring salt-saturated, alkaline brines that have exceptionally high concentrations of arsenic oxyanions. Strain SLAS-1T was previously isolated from Searles Lake (R. S. Oremland, T. R. Kulp, J. Switzer Blum, S. E. Hoeft, S. Baesman, L. G. Miller, and J. F. Stolz, Science 308:1305-1308, 2005). We now describe this extremophile with regard to its substrate affinities, its unusual mode of motility, sequenced arrABD gene cluster, cell envelope lipids, and its phylogenetic alignment within the order Halanaerobacteriales, assigning it the name “Halarsenatibacter silvermanii” strain SLAS-1T. We also report on the substrate dynamics of an anaerobic enrichment culture obtained from Searles Lake that grows under conditions of salt saturation and whose members include a novel sulfate reducer of the order Desulfovibriales, the archaeon Halorhabdus utahensis, as well as a close homolog of strain SLAS-1T.
More than a decade has passed since the initial reports that documented the ability of two closely related freshwater members of the Epsilonproteobacteria, Sulfurospirillum arsenophilus and Sulfurospirillum barnesii, to grow using arsenate as a respiratory electron acceptor (1, 27, 37, 48). Since then, about 20 taxonomically diverse species from this bacterial domain that have the capacity to grow on this toxicant have been characterized in pure culture (see reviews in references 38, 47, and 49). Bacteria that respire arsenate can be readily cultured from pristine ecosystems (22), as well as from As-contaminated environments (1), suggesting that most species are opportunists. The majority of these described species are mesophiles, with only a few documented examples of moderate halophiles isolated from soda lakes (50) or moderate thermophiles isolated from terrestrial hot springs (10) and deep-sea hydrothermal vents (54). Indeed, until recently there were no reports of any true “extremophiles” from the domain Bacteria having the ability to respire arsenate at very high temperatures (≥90°C) or salinities (≥300 g/liter) or under highly alkaline (pH ≥ 11) or acidic conditions (pH ≤ 2). With regard to the Archaea, there is but one report of arsenate respiration within the entire domain. Two members of the order Thermoproteales, namely, Pyrobaculum arsenaticum and Pyrobaculum aerophilum, exhibited arsenate-dependent anaerobic growth at 95°C (16). They remain the only documented examples of arsenate-respiring extremophiles.
The motivation for the work reported herein came from the absence of any described species of extreme halophiles, either from the bacterial or archaeal domains, that have the capacity to conserve energy for growth by respiring arsenate. This work is also relevant to the question of whether microbes could have existed within the dense, caustic brines thought to have been present on the surface of ancient Mars (57). Because dissolved arsenic is often abundant in the waters of hot springs (e.g., see references 10, 17, and 25), as well as in closed-basin desert soda lakes that receive hydrothermal inflows (39), we opted to examine the latter as a source to enrich for extremely halophilic prokaryotes having the capacity for arsenate respiration. We chose Searles Lake because its brine represents an exceptionally harsh physiological challenge to life, being salt saturated (salinity, ∼346 g/liter), alkaline (pH 9.8), and high in arsenate and borate content (∼3.9 mM and 0.46 molal, respectively [24]). Our discovery that active arsenate respiration could be readily established with the sediments underlying this brine led to the isolation of strain SLAS-1 (40). Subsequent studies of 16S rRNA gene amplicons of DNA extracted from Searles Lake sediments have noted the persistent presence of close homologs of this strain, such as SLAS-3 (95% 16S rRNA gene sequence similarity) (23, 24). These observations underscored the importance of strain SLAS-1 as representative of a portion of the flora within this ecosystem that carries out arsenate respiration.
Herein we describe strain SLAS-1 and propose the name Halarsenatibacter silvermanii strain SLAS-1T as the first reported example of an arsenate-respiring extreme halophile, taxonomically classified within the order Halanaerobacteriales of the domain Bacteria. To learn more about the potential substrate interactions of strain SLAS-1T with other diverse extremophiles that live in the Searles Lake brine, we established an antibiotic-fed enrichment culture to encourage the growth of anaerobic prokaryotes, especially members of the Archaea. We also describe a stable, mixed enrichment culture composed of an extremely halophilic sulfate reducer, an archaeon not previously noted for any ability to respire arsenate, as well as a close relative of strain SLAS-1T.
MATERIALS AND METHODS
Enrichment, isolation, and substrate affinities of strain SLAS-1.
An artificial brine (see below) was prepared and supplemented with 10 ml of a vitamin mix (37), 5 mM As(V), 10 mM Na lactate, with a small amount of yeast extract (0.2 g/liter) added as a nutrient supplement and 0.025% (wt/vol) cysteine-HCl to serve as a reducing agent. The medium (10 ml) was dispensed into 25-ml Balch tubes and sealed under N2, using standard procedures for cultivation of strict anaerobes (49, 50). About 1 g of Searles Lake sediment was added to a medium-containing tube, and after a few weeks time it was noted that As(V) had been entirely replaced by As(III), with the concomitant oxidation of lactate to acetate. A 1-ml portion of the slurry was injected into fresh medium and incubated for an additional 3 weeks. After several months of such a transfer/incubation regimen, a stable enrichment culture was established. It was purified by serial dilution, with the highest-dilution tube (10−6) showing a microbial culture having uniform morphology. We were not able to achieve colony growth on solid medium of various ionic strengths using a variety of solidifying agents. The purity of the culture was confirmed by the presence of only two close eluant bands on denaturing gradient gel electrophoresis gels (35) that were found to have identical 16S rRNA gene sequences, indicating initial overloading of the gel rather than the presence of a separate species (see below). Initial growth experiments were conducted with heterotrophic (lactate) and autotrophic (sulfide) electron donors (40). Electron donors tested for growth in this report (with 7 mM arsenate as the electron acceptor) include (10 mM, unless indicated otherwise) lactate, glutamate, formate, galactose, serine, aspartate, propionate, succinate, citrate, malate, glycine, glucose, acetate plus H2 gas phase, acetate, fructose, pyruvate, glycerol, sulfide (2 mM), methanol (7 mM), ethanol (7 mM), and butyrate (7 mM). Electron acceptors tested for growth (with 10 mM lactate as the electron donor) include (10 mM, unless indicated otherwise) oxygen (5% or 10% headspace volume), nitrate, nitrite, sulfate, thiosulfate, fumarate, tungstate, selenate, selenite, trimethylamine oxide, Fe(III)-nitrilotriacetic acid, elemental sulfur (∼100 mmol/liter), and manganese dioxide (10 to 20 mmol/liter).
Artificial brine composition.
Artificial brine composition (g/liter of deionized water are shown in parentheses) was NaCl (180), Na2SO4 (100), K2SO4 (30), Na2CO3 (27), NaHCO3 (5.0), H3BO3 (4.0), (NH4)2SO4 (0.05), KH2PO4 (0.08), K2HPO4 (0.15), MgSO4·7H2O (0.025), Na2WO4 (0.075), Na2SeO4 (0.00001), and 3.0 ml of SL10 trace element solution (62). The pH was adjusted to 9.5 with NaOH.
TEM.
For negative stains, cells were harvested from culture and layered on a 200-mesh, carbon- and Formvar-coated transmission electron microscopy (TEM) grid that was then floated on a drop of distilled water to reduce the concentration of interfering salts before being negatively stained with 2% (wt/vol) uranyl acetate. For thin sections, a growing broth culture was made 2% (vol/vol) with glutaraldehyde cells to initiate fixation. Since high salt inhibits the polymerization of the embedding plastic, the salt concentration of the growth medium was gradually reduced from 100% to 0% in 25% steps via washing in deionized water and centrifugation. The cells were then postfixed with 2% (wt/vol) osmium tetroxide, washed with deionized water, and embedded in LR White embedding medium (London Resins, Inc., United Kingdom). The embeddings were thin sectioned and stained with 2% uranyl acetate, followed by lead citrate (2). All electron microscopy was done using a Philips CM10 operating at 80 kV.
16S rRNA gene sequencing.
Batch-cultured lactate-grown SLAS-1T cells (1 liter) were harvested by centrifugation and stored at −20°C until DNA extraction. DNA was extracted by using the modifications of the FastPrep kit (QBiogene, United States) described by Yeates and Gillings (64). Bacterial 16S rRNA genes were amplified with primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGYTACCTTGTTACGACTT-3′) (26), and archaeal 16S rRNA genes were amplified with primers A21F (5′-TTCCGGTTGATCCYGCCGGA-3′) and A958R (5′-YCCGGCGTTGAMTCCAATT-3′) (42). To minimize variation and PCR bias, five replicate 16S rRNA gene PCR products were combined prior to cloning. PCR products were cloned by using a TOPO TA cloning kit (Invitrogen, CA). Twenty randomly selected clones were sequenced. Taxonomic assignments were done by comparing the clone sequences with the nonredundant nucleotide database in the RDP database by using RDP classifier (4). All positions containing gaps and missing data were eliminated from the data set. A total of 500 positions in the final data set were used for phylogenetic analysis. The neighbor-joining tree was constructed by using MEGA4 (55).
Sequencing of SLAS-1 arr operon.
The full-length arrA gene from SLAS-1T was sequenced by using an inverse PCR (iPCR) approach (44). The partial SLAS-1 arrA sequence reported in Kulp et al. (24) was used to design primers for iPCR (SLAS-PW-R1, 5′-ATT TAC AGC AGG CGA AGA AG-3′, and SLAS-PW-F1, 5′-GGG GTA GTA AAA TGG TGG AA-3′). Genomic DNA was purified from a pellet of SLAS-1T cells by using a Qiagen-tip 500. The DNA was digested with PstI or SalI, followed by intramolecular ligation and iPCR. The PCR products were cloned by using Invitrogen's TA TOPO cloning kit, and the resulting plasmids were sequenced by using the vector-specific primers. Primer walking and sequencing were done on the cloned iPCR product, as well as on PCR products amplified from the uncut genomic DNA. The sequence data was assembled by using AssemblyLIGN and analyzed for open reading frames by using MacVector. The final arrA-containing sequence was ∼4 kb.
Phylogenetic analysis of SLAS-1 ArrA.
The predicted amino acid translation of the SLAS-1T ArrA was compared to ArrAs of other arsenate-respiring bacteria or to ArrA-like sequences identified in the genomes of recently sequenced microorganisms. Sequences were aligned with ClustalW 1.82 (56). Phylogenetic analysis was performed by using PAUP*, version 4.0b10 (52). The distance criterion was used to construct an unrooted neighbor-joining tree. One other dimethyl sulfoxide reductase family enzyme (32) was also included in the analysis (Wolinella Psr). After ignoring gaps in the multisequence alignment, the final number of amino acids included in the phylogeny was 542.
Determination of RubisCO activity.
Washed whole-cell preparations of strain SLAS-1T grown on As(V) either as a heterotroph with lactate as the electron donor or as a chemoautotroph (CO2 as the sole carbon source) with sulfide as the electron donor were stored at −80°C until use. Frozen cells were thawed on ice, resuspended in 0.5 ml of ice-cold lysis buffer [50 mM N,N-bis(2-hydroxyethyl)glycine (bicine)-NaOH, pH 8.0, 1 mM EDTA, 10 mM MgCl2, 100 mM NaCl, 10 mM NaHCO3, 1 mM dithiothreitol], and sonicated for 2.5 min in 5-s cycles. The resulting lysate was centrifuged at 4°C (15 min at 12,000 × g) to remove unbroken cells and cell debris. The cleared lysate was then immediately assayed for RubisCO activity according to established procedures (46, 61). Aliquots of 5 μl or 20 μl of lysate were assayed in a total volume of 250 μl containing 50 mM bicine-NaOH at pH 8.3 and NaCl concentrations of 100 mM. All assays were performed at 30°C and were done in triplicate. Additionally, each set of assays included negative controls (omitting ribulose biphosphate) and positive controls (cleared lysate from photoautotrophically grown Rhodobacter capsulatus).
Determination of cellular lipids.
Freeze-dried cells (145 mg) of SLAS-1 cultures were extracted using methanol-chloroform-water (10:5:4). The solid cellular residue was recovered by centrifugation, and the solvent phase partitioned by the addition of chloroform and water to a final ratio of 10:10:9. The lower chloroform layer containing the total lipid extract was removed and dried under N2. Fatty acid methyl esters (FAME) were prepared by treatment of the total lipid extract by transesterification with 0.1 N methanolic NaOH for 60 min at 37°C (60). FAME and accompanying lipids were recovered by the extraction procedure described above and separated by thin-layer chromatography using silica gel G plates developed with methylene chloride to 15 cm twice. Standards for hydrocarbon, FAME, and hydroxy-FAME were used to identify appropriate zones for compound recovery. Gas chromatography-mass spectrometry analysis of the hydrocarbon fraction and a trimethylsilyl derivative of the hydroxy-FAME fraction were negative. FAME were identified by gas chromatography-mass spectrometry as described previously (18). The double-bond positions of FAME were determined by preparing dimethyl disulfide adducts by heating at 35°C for 35 min (63). FAME structure was confirmed by comparison to the relative retention time and spectra of standard FAME (Matreya, Ultra Scientific).
Analytical methods.
Arsenic speciation and organic acids were determined by using high-performance liquid chromatography (13), and sulfide by the method of Cline (3). Cell abundances were determined by acridine orange direct counts (12).
Searles Lake antibiotic-fed enrichment culture.
A stable enrichment culture of microbes was established using a lactate-arsenate medium (see above) but supplemented (after autoclaving) with an antibiotic mix to favor the development of anaerobic archaea. The antibiotic mix was composed of vancomycin, kanamycin, penicillin, and tetracycline, each at a final concentration of 20 mg/liter, and was always present in the medium. The enrichment culture was maintained with bimonthly transfers and incubated in the dark at 38°C. Sulfate reduction was assayed by injecting [35S]sulfate (∼4.0 × 109 Bq/30 ml culture). After 57 days of incubation, the [35S]sulfide produced was trapped and quantified by liquid scintillation spectroscopy as outlined elsewhere (40). PCR techniques were used to amplify, separate by denaturing gradient gel electrophoresis (23), and sequence the 16S rRNA genes of a 500-ml culture of lactate-grown cells (see above). We also used PCR to detect the presence of the arsenate respiratory reductase gene, arrA, within the microbial community of the antibiotic enrichment cultures. A degenerate primer set (30) modified for extreme halophiles by using Halobacterium archaeal codon-biased arrA primers (HAArrA-D1F, 5′-CCG CTA CTA CAC CGA GGG CWW YTG GGR NTA-3′, and HAArrA-G2R, 5′-CGT GCG GTC CTT GAG CTC NWD RTT CCA CC-3′) (23, 24), and SLAS-1-specific primers (eSLAS1-F2, 5′-TTG CGG GAT ATT CTC TAC GG-3′, and eSLAS1-R2, 5′-GGT TGG ATT GTA TGG GAT CG-3′) were used to detect the presence of arrA in the extracted DNA. The HAarrA and eSLAS1 PCR products were cloned and sequenced using previously reported methods (24). Of the 20 HAarrA clones sequenced, there was only one major HAarrA phylotype.
Nucleotide sequence accession numbers.
The SLAS-1T 16S rRNA gene sequences were deposited in the NCBI database under GenBank accession numbers EU855125 to EU855129. The final SLAS-1T arrA-containing sequence was submitted to GenBank and assigned the accession number EU723191. The single major HAArrA phylotype sequence from the enrichment culture was submitted to GenBank and assigned the accession number EU723192.
RESULTS
Cell morphology, physiology, and substrate affinities of strain SLAS-1T.
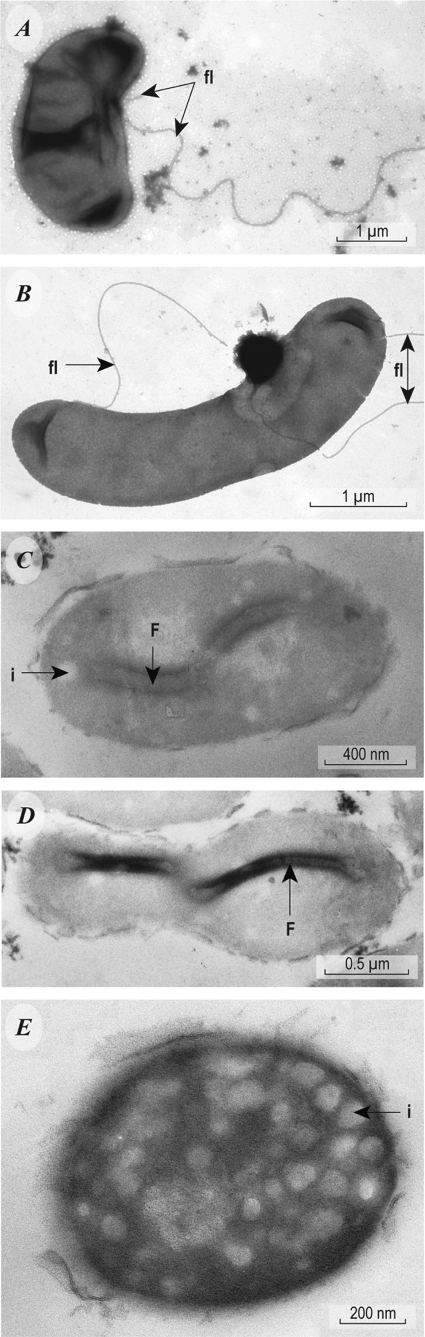
Cells were motile, gram-negative curved rods (3.0 by 0.5 μm). TEMs revealed the presence of paired flagella inserted approximately a third to halfway down the side of the organism rather than being located at either pole (Fig. 1A and B). Thin-section TEMs also revealed the presence of an unusual, linear internal structure that spanned the cytoplasm and appeared to be anchored to each side of the internal portion of the cell envelope (Fig. 1C and D). Strain SLAS-1T displays an unusual mode of locomotion characterized by both helical and sinuous aspects; the latter is best described as an eel-like, full-body-length undulation (see film clip in the supplemental material). TEM images also revealed the presence of what appear to be numerous cytoplasmic inclusions, perhaps vesicular intracytoplasmic membranes (Fig. 1E).
FIG. 1.
TEM images of cells of strain SLAS-1T. (A and B) Images showing locations of two flagella (fl) located approximately one-third of the way down the side of the organism. (C and D) Thin sections revealing the presence of longitudinal internal filaments (f). i, see description for panel E. (E) Transverse thin section showing the presence of numerous cytoplasmic inclusions, possibly vesicular intracytoplasmic membranes (i).
Growth was observed over a relatively narrow pH range (8.7 to 9.8), with an optimum at 9.4 (μ = 0.015 h−1). While no growth occurred at salinities below 200 g/liter, roughly equivalent growth rates (0.015 to 0.018 h−1) were observed at higher salinities, including at salt saturation (∼340 g/liter) (40). Growth occurred over a temperature range of 28 to 55°C, with an optimum at 44°C (see Fig. S1 in the supplemental material). In addition to lactate or sulfide, strain SLAS-1T could also use pyruvate and some sugars (e.g., glucose) as electron donors for growth but, notably, not acetate or H2 or acetate plus H2 (Table 1). Fermentative growth in the absence of arsenate was not observed with either glucose or lactate. Growth was observed with arsenate, Fe(III), or elemental sulfur as respiratory electron acceptors, but strain SLAS-1T was unable to use nitrate, sulfate, thiosulfate, fumarate, selenate, or Mn(IV) as oxidants. Strain SLAS-1T is an obligate anaerobe, being unable to grow under either fully aerobic or microaerophilic conditions (Table 1). The pure culture of strain SLAS-1T did not grow when the antibiotic mix of vancomycin, kanamycin, penicillin, and tetracycline was included in the medium.
TABLE 1.
Electron donors and acceptors tested for growth with strain SLAS-1T
| Electron donor or acceptora | Growthb |
|---|---|
| Donorsc | |
| Lactate | + |
| Galactose | + |
| Malate | + |
| Glucose | + |
| Fructose | + |
| Pyruvate | + |
| Glycine | − |
| Glutamate | − |
| Aspartate | − |
| Succinate | − |
| Citrate | − |
| Glycerol | − |
| Acetate | − |
| Acetate + H2 | − |
| H2 | − |
| Propionate | − |
| Sulfide | + |
| Acceptorsd | |
| Arsenate | + |
| Fe(III)-NTA | + |
| Sulfur | + |
| Oxygen (10%) | − |
| Oxygen (5%) | − |
| Nitrate | − |
| Nitrite | − |
| Sulfate | − |
| Thiosulfate | − |
| Fumarate | − |
| Tungstate | − |
| Selenate | − |
| Selenite | − |
| Mn(IV) | − |
| TMAO | − |
NTA, nitrilotriacetic acid; TMAO, trimethylamine oxide.
+, growth occurred; −, no growth occurred.
As(V) was the electron acceptor.
Lactate was the electron donor.
Cell membrane lipids.
The SLAS-1T isolate was harvested in late exponential phase, when cells express their unique motility. The fatty acids from this analysis represent both the cytoplasmic and outer membranes and are a complex mixture of 39 individual molecular structures, some representing only 0.1% of the total (Table 2). Four major fatty acids (iso-15:0 > n-18:0 > iso-17:0 > n-16:0) accounted for 62.8%, while another 21% contained one double bond (monounsaturated) occurring at distinct multiple carbon positions in both branched- and straight-chain acids (Table 2). Two fatty acids, 16:1Δ6 and 18:1Δ8, present at low levels, have not been identified previously in Halanaerobiaceae (7). The C-2 elongation pattern (e.g., 16:1Δ7 + C2 to 18:1Δ9 + C2 to 20:1Δ11) is observed in both straight- and branched-chain acids and is characteristic of the anaerobic mechanism for the biosynthesis of unsaturated fatty acids as established for Proteobacteria (5, 6). The iso-branched fatty acids were more abundant than the anteiso, with a respective ratio of 8.8.
TABLE 2.
Fatty acid composition of strain SLAS-1T
| Fatty acida | % |
|---|---|
| Branched | |
| Saturated | |
| i14:0 | 0.6 |
| i15:0 | 21.3 |
| a15:0 | 2.9 |
| i16:0 | 1.4 |
| i17:0 | 13.1 |
| a17:0 | 1.6 |
| i18:0 | 0.6 |
| i19:0 | 3.9 |
| a19:0 | <0.1 |
| i20:0 | 0.4 |
| i21:0 | 0.1 |
| a21:0 | <0.1 |
| Total | 46.0 |
| Unsaturated | |
| i15:1Δ5 | 0.3 |
| i15:1Δ7 | 2.1 |
| a15:1Δ7 | 0.4 |
| i16:1Δ7 | 0.3 |
| i16:1Δ9 | 0.5 |
| i17:1Δ7 | 2.5 |
| i17:1Δ9 | 1.2 |
| a17:1Δ7 | 0.6 |
| i18:1Δ9 | 0.5 |
| i18:1Δ11 | 0.3 |
| i19:1Δ7 | 0.4 |
| i19:1Δ9 | 0.8 |
| Total | 10.0 |
| Normal | |
| Saturated | |
| 14:0 | 0.7 |
| 15:0 | 0.6 |
| 16:0 | 11.7 |
| 17:0 | 1.0 |
| 18:0 | 16.7 |
| 19:0 | 0.6 |
| 20:0 | 0.6 |
| 21:0 | 0.6 |
| Total | 32.5 |
| Unsaturated | |
| 16:1Δ6 | 0.4 |
| 16:1Δ7 | 0.8 |
| 16:1Δ9 | 0.7 |
| 18:1Δ8 | 0.1 |
| 18:1Δ9 | 5.3 |
| 18:1Δ11 | 1.6 |
| 20:1Δ11 | 0.6 |
| 20:1Δ13 | 0.2 |
| 22:1Δ13 | 1.5 |
| Total | 11.2 |
Carbon number for double-bond position (Δ) relative to the carboxyl carbon; “i” indicates iso and “a” indicates anteiso branched fatty acids.
RubisCO activity.
Although our prior work indicated CO2 fixation activity in autotrophically grown SLAS-1 strains (40), no RubisCO activity was detected in lysates derived from cells grown either under autotrophic (sulfide as electron donor) or heterotrophic (lactate as electron donor) conditions (data not shown). Thus, alternate C fixation pathways must be present in this strain.
Sequence analysis of the SLAS-1T arrA gene cluster.
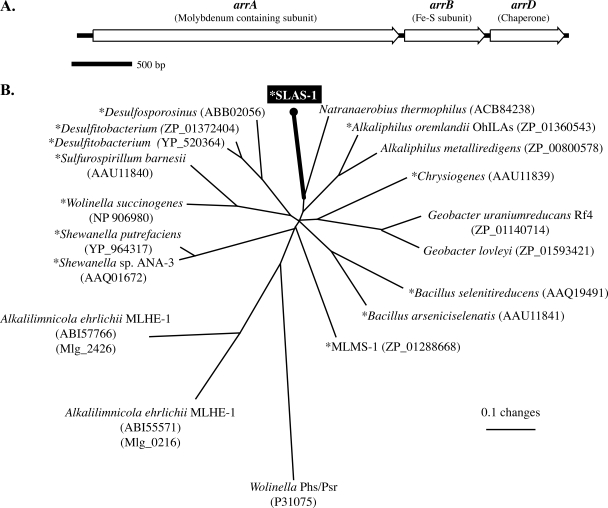
In our previous work (23), we generated a 250-bp sequence of the arrA gene using PCR with degenerate primers (30). Using this known sequence, we used iPCR and primer walking to sequence the flanking DNA of the original 250-bp SLAS-1T partial arrA gene. The final sequence was assembled into one ∼4.5-kb contig. Analysis of this DNA sequence revealed three open reading frames with high degrees of similarity to arsenate respiratory reductases: arrA; an iron sulfur cluster-containing subunit, arrB; and a putative molybdenum cofactor chaperone, arrD (Fig. 2A).
FIG. 2.
Genomic map (A) and phylogenetic analysis (B) of the arsenate respiratory reductase of SLAS-1T. (A) The arr gene cluster contains at least three genes arranged as arrABD, with likely additional arr genes downstream of arrD. (B) The phylogenetic analysis was performed by using previously established computation methods and representative members of the arsenate respiratory reductase clade of the dimethyl sulfoxide reductase family of molybdenum-containing enzymes (32). An asterisk indicates that the organism is known to respire arsenate. GenBank accession numbers are indicated in parentheses. The second numbers in parentheses for A. ehrlichii are gene identification numbers within the genome sequence.
The phylogenetic analysis of the predicted ArrA amino acid sequence of SLAS-1T revealed that it was most similar to the ArrA-like sequence from Natranaerobius thermophilus (64% identity and 81% similarity), a halalkalithermophilic, low-G+C, gram-positive bacterium recently isolated from a saline lake in Egypt (34) (Fig. 2B). Although SLAS-1T ArrA was also similar to ArrA-like sequences of Alkaliphilus (55% identity and 72% similarity), the phylogenetic tree showed SLAS-1T ArrA clustered with the Natranaerobius ArrA-like sequence. It is also clear that the SLAS-1T ArrA sequence is very distant from the two respiratory ArrA gene sequences annotated in the genome of Alkalilimnicola ehrlichii (located at locus tags Mlg_0216 and Mlg_2426), an arsenite-oxidizing haloalkaliphile. The amino acid identities and similarities of SLAS-1T ArrA to the translated products of these arrA-like genes are low: 32 to 33% and 47 to 49%, respectively. The Alkalilimnicola ehrlichii strain MLHE-1 enzymes are thought to function in vivo as arsenite oxidases (14). The SLAS-1T iron sulfur subunit for ArrA, ArrB, was most similar to an ArrB-like sequence in Natranaerobius thermophilus (58% identity and 71% similarity) and the putative ArrBs of Alkalilimnicola ehrlichii and Alkaliphilus oremlandii (55% identity and 69% similarity to both species' ArrBs). The analysis of the predicted proteins encoded by arrAB provides bioinformatic evidence that these genes may encode an arsenate respiratory reductase.
The TorD-like molybdenum cofactor chaperone, ArrD, was less conserved than ArrA and ArrB when compared to putative ArrD sequences from other organisms. Although still very similar to Alkaliphilus oremlandii OhILAs (37% and 59% identity and similarity, respectively), the amino acid similarity of SLAS-1T ArrD to the Alkaliphilus metalliredigens strain QYMF putative ArrD was even lower (32% and 50% identity and similarity, respectively). It is not known what functional role ArrD has in dissimilatory arsenate reduction.
Growth and substrate dynamics of the antibiotic-fed enrichment culture.
In the presence of arsenate, the initial oxidation of lactate to acetate coincided with logarithmic growth and reduction of As(V) to As(III) (Fig. 3A). After 25 days of incubation, the supply of As(V) was exhausted and stationary phase was reached. However, by 40 days a modest increase in cell number was noted which coincided with increased rates of lactate oxidation to acetate, the appearance of sulfide, and a decline in the abundance of As(III), the latter probably due to the formation of thioarsenicals (8). When the enrichment culture was incubated without As(V), growth was initially slower but reached significantly higher cell densities than before, and sulfide concentrations were also higher (about 2.2-fold) (Fig. 3B). Sulfate reduction was confirmed by the reduction of [35S]sulfate in cultures as follows ([35S]sulfide counts are given as percentage of [35S]sulfate initially added ± 1 standard deviation; n = 3): lactate plus sulfate (4.9 ± 1.6), lactate plus sulfate plus arsenate (2.7 ± 0.8), heat-killed lactate plus sulfate control (0.08 ± 0.02). Growth and As(V) reduction to As(III) were also noted when sulfide served as an electron donor in lieu of lactate (Fig. 3C). There was a 4:1 ratio of sulfide consumed to As(V) reduced; however, growth was far slower and less extensive than that achieved in the lactate-amended conditions with either As(V) plus sulfate (Fig. 3A) or with only sulfate (Fig. 3B) present as electron acceptors. In the control without any added electron donor (lactate or sulfide) there was still some detectable As(V) reduction to As(III), as well as limited growth (Fig. 3D), presumably caused by carryover of some of the electron donor from the inoculum.
FIG. 3.
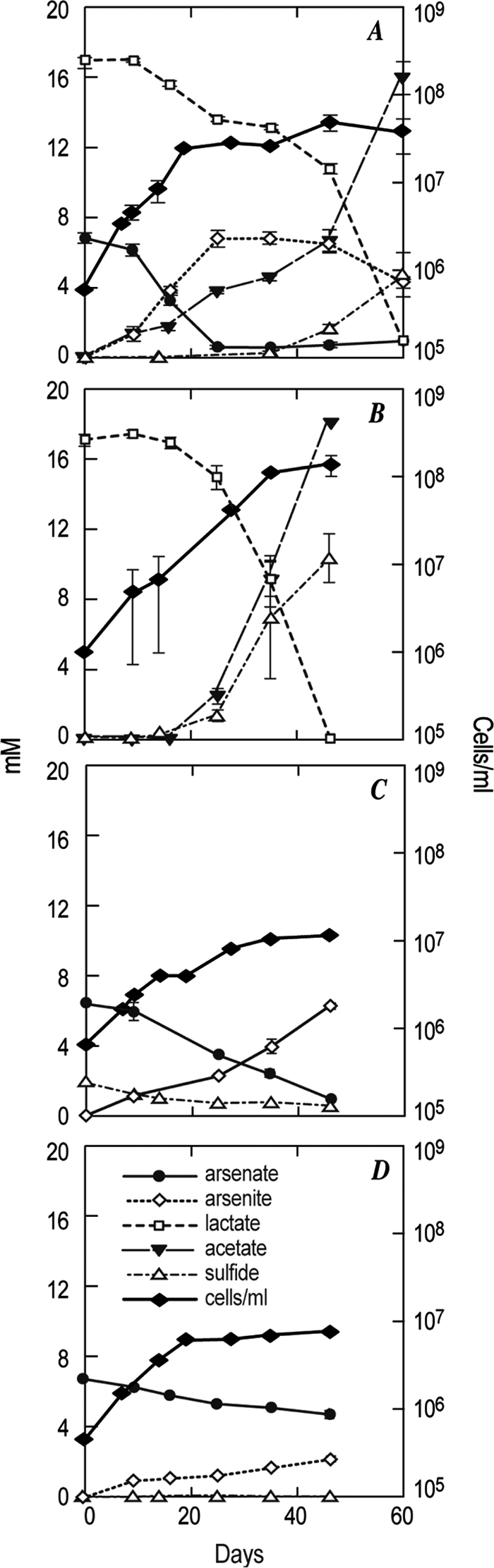
Time course of metabolic kinetics and growth of the antibiotic enrichment culture under the following incubation conditions: lactate as the electron donor and sulfate plus arsenate as the available electron acceptors (A), lactate and sulfate only (B), sulfide as the electron donor with arsenate plus sulfate (C), and without any added electron donor but with arsenate plus sulfate (D). All incubation conditions contained abundant sulfate ions (>700 mM). Error bars show standard deviations of the mean (n = 3 cultures).
Microbial diversity of the antibiotic-fed enrichment culture.
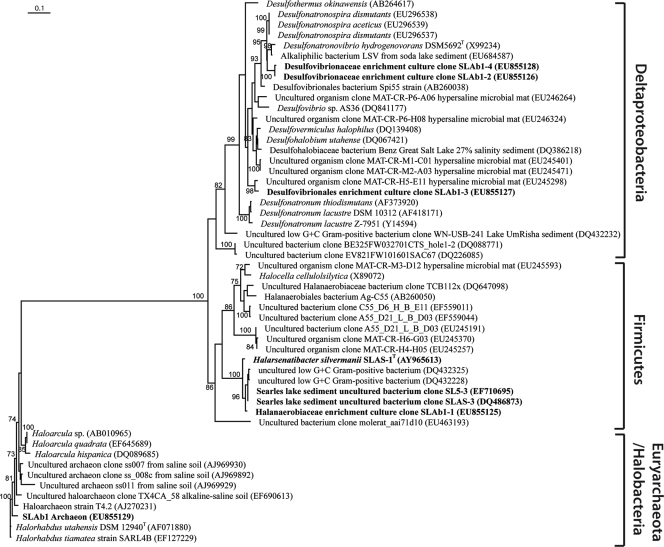
The 16S rRNA gene clone library analysis revealed four bacterial and one archaeal sequence group (based on 97% sequence similarity; Fig. 4). The first sequence group, labeled SLAb1-1, was the most abundant in the clone library and was 97% similar to strain SLAS-1T (see below). SLAb1-2 and SLAb1-4 were affiliated with sulfate-reducing members of the Deltaproteobacteria, with the highest similarity (88% each) to Desulfonatronovibrio hydrogenovorans (X99234), a member of the Desulfovibrionaceae. SLAb-3 was also affiliated with the Desulfovibrionaceae; however, it is a deep-branching affiliate unrelated to any specific member of this family of sulfate-reducing bacteria. Only a single archaeal sequence was revealed from clone library analysis. The SLAb1 archaeon was affiliated with the Halobacteriales in the Euryarchaeota, showing high similarity (94%) to Halorhabdus utahensis, an extremely halophilic archaeon isolated from Great Salt Lake, UT (59).
FIG. 4.
Phylogenetic analysis of Halarsenatibacter silvermanii strain SLAS-1T and the antibiotic enrichment culture, showing the relationship of the 16S rRNA genes of isolates and enrichment culture to known species and environmental clones. Sequences from this study are shown in boldface, and the GenBank accession numbers for sequences obtained from the NCBI database are shown in parentheses. All sequences were automatically aligned according to the SILVA SSU reference alignment (SILVA Incremental Aligner [43]). All aligned sequences were imported and analyzed using the ARB software package (version 08.08.13) (28). Manual refinement of the alignment was carried out in ARB editor4, taking into account the secondary structure information of the rRNA. Tree reconstruction was performed with 52 sequences using maximum likelihood (Rhyml version 2.4.5) (11). The final tree was calculated with 503 positions (all ambiguous bases and nonaligned positions were omitted from the analysis) using PHYML (model, GTR and GAMMA). Bootstrap values were generated by 100 replications, and bootstrap values above 70 are shown next to the branches. The bar in the upper left corner refers to 0.1 fixed nucleotide substitutions per site.
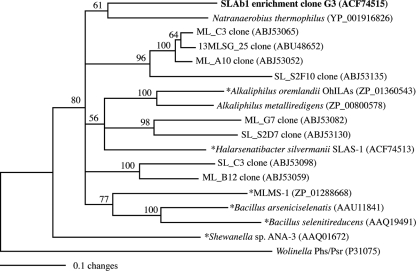
The functional gene for respiratory arsenate reductase (arrA) was also detected in the antibiotic enrichment culture by using the Halobacterium archaeal codon-biased arrA primer set (HAArrA primer set) that had previously been shown to be effective at amplifying these genes in Searles Lake sediments (23, 24). Twenty randomly picked clones had either identical sequences or one or two nucleotide differences within the 359-bp PCR product. Sequence analysis of the amino acid sequence translation of the dominant arrA phylotype (SLAb1 ArrA clone G3) revealed that it was ∼84% similar to the amino acid translations of a variety of partial arrA sequences previously retrieved from Mono Lake and Searles Lake sediments. In comparison to pure strains, SLAb1 ArrA was most similar to a predicted ArrA-like protein sequence within the genome of Natranaerobius thermophilus (84% similarity to sequence accession number Nther_0643). There are no reports demonstrating arsenate respiration in N. thermophilus. In addition to identifying the SLAb1 arrA phylotype, we also detected the SLAS-1T-specific arrA gene in the antibiotic enrichment culture. This was done by using primers designed specifically to target the SLAS-1T arrA. These results demonstrate that SLAS-1T and possibly a novel arr-containing prokaryotic strain were present in the antibiotic enrichment culture.
DISCUSSION
Strain SLAS-1T is classified in the order Halanaerobacteriales, members of which are all halophilic bacteria with fermentative types of metabolism. Within the order it is most closely aligned (based on 16S rRNA sequences) with Halothermothrix orenii (83.9%) and Halocella cellulosilytica (83.5%) (40). The only other member of this order demonstrating a respiratory metabolism is Selenihalanaerobacter shriftii, which can respire with either selenate or nitrate with concomitant oxidation of glucose or glycerol but lacks any capacity for respiration of arsenate (51). S. shriftii however, is only distantly related to strain SLAS-1T based on its 16S rRNA gene sequences (81%). Strain SLAS-1T is more versatile metabolically than S. shriftii, having a broader range of electron donors and a different array of electron acceptors available that support its growth, but it conversely lacks the ability to respire nitrate or selenate (Table 1). Heterotrophic growth on lactate results in the formation of acetate, bicarbonate, and As(III) that closely aligns with the following stoichiometry (40):
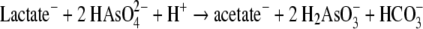 |
In addition, strain SLAS-1T has a capacity for chemoautotrophic growth using sulfide as its electron donor and arsenate as its electron acceptor, a mode of lithotrophic metabolism first identified for the obligate chemoautotroph strain MLMS-1 (13), as well as other isolates from Mono Lake (15). Although strain SLAS-1T can incorporate [14C]bicarbonate into biomass when grown as an autotroph (40), we did not detect any RubisCO activity in the efforts reported herein, a situation similar to that of strain MLMS-1 (13), which in a preliminary annotation of its genome lacked any homologous cbb genes encoding RubisCO (J. F. Stolz and R. S. Oremland, unpublished data). Although RubisCO proteins are widely distributed in the microbial world and the Calvin-Benson-Bassham pathway represents the major means of biological CO2 fixation on Earth, it is not the only biochemical means available for incorporation of CO2 into organic matter (53).
Members of the Halanaerobiaceae synthesize a diverse group of branched and monounsaturated fatty acids (7). C15 to C19 branched-chain fatty acids with both iso- and anteiso-methyl groups have been identified, together with normal (n-) chain fatty acids. The monounsaturated n-fatty acids of the Halanaerobiaceae characteristically contain double bonds (Δ) at multiple carbon positions (e.g., Δ7, Δ9, and Δ11 C16:1). Strain SLAS-1T fatty acids display similar attributes, containing 31 individual branched-chain and/or multipositional monounsaturated isomers in the range of C14 to C22, and additionally include a novel isomeric suite of unsaturated iso- and anteiso-branched fatty acids (Table 2). These fatty acids comprise 67% of the membrane lipids.
Membrane lipid homeostasis in bacteria depends on an ability to adjust lipid composition in response to environmental conditions (65). Gram-negative bacteria of the Proteobacteria group generally maintain a relatively fluid membrane by synthesizing an increased proportion of cis-unsaturated fatty acids (45), while gram-positive bacteria increase the proportion of branched-chain fatty acids (20). A correlation of an increased proportion of unsaturated and branched-chain fatty acids with the presence of motility in Cytophaga has been observed (33). A relatively fluid outer membrane in Spirochaetes is thought to play an important role in the unique motility possessed by this group (58). The highly branched and unsaturated fatty acid composition of SLAS-1T suggests a highly fluid membrane of potential importance to the unique style of motility and necessary flexibility of the cell envelope.
Just how strain SLAS-1T achieves its unusual locomotion is not clear. We speculate that the odd positioning of its two external flagella imparts a vector-generated moment that causes an overall helical motion (Fig. 1A and B), while the sinuous aspect may be a consequence of contractions and/or undulations of its curious internal filament structure (Fig. 1C and D). Actin-like filaments have been noted in Bacillus subtilis (19) and Caulobacter crescentus (31) and are thought to have a structural “cytoskeletal” role. Involvement in locomotion by actin proteins would presumably also require myosin for actual contraction. In any case, the unusual motility of strain SLAS-1T allowed us to clearly identify its presence in the antibiotic-fed enrichment culture, a fact that was subsequently confirmed by analysis of 16S rRNA gene sequences (see below).
Gene sequences for respiratory arsenate reductase are employed as a means of determining the diversity of this functional gene in environmental samples (23, 24, 30). This gives impetus to more fully characterize the arsenate reductases from novel bacteria isolated from the environment, so as to better interpret those in situ diversity patterns. The SLAS-1T arr gene cluster contains at least three genes (Fig. 2): arrA, encoding a molybdenum oxidoreductase with strong homology to other arsenate respiratory reductases; arrB, encoding an iron sulfur subunit; and a coding sequence for a TorD-like molybdenum cofactor chaperone, arrD. The arrangement of the core arrAB gene is consistent with that of the arr operons of Shewanella species. Moreover, the genome sequences of other haloalkaliphilic arsenate-respiring bacteria, such as Alkaliphilus oremlandii OhILAs, Bacillus selenitireducens MLS10, Bacillus arseniciselenatis, and the deltaproteobacterium species strain MLMS-1 have revealed the presence of arr operons with the core set of arrAB. Genome-sequencing projects with nonextremophiles have also revealed the presence of numerous arr-like gene clusters. Some of these gene clusters contain arrD, which in addition to strain SLAS-1T, is present in Alkaliphilus oremlandii OhILAs, Bacillus selenitireducens strain MLS10, strain MLMS-1, Geobacter lovleyi, and Desulfitobacterium hafniense. In addition to arr genes, the DNA sequence upstream and on the minus strand contains an incomplete reading frame with a translated amino acid sequence similar to an ACR3-like arsenite efflux pump. This observation suggests that SLAS-1T likely contains arsenic resistance genes, which is commonly observed in other genomes of arsenate-respiring bacteria, e.g., Shewanella sp. strain ANA-3 (44). Additional sequencing will be necessary to further define the genetic composition of the arr and possible arsenic resistance operons in SLAS-1T.
The presence of an SLAS-1T-like microorganism in the enrichment culture was unexpected because the original isolate had proved sensitive to the antibiotic mixture that was routinely included in the milieu in an effort to screen out bacteria and enrich for archaea. Nonetheless, its presence as a component of the enrichment was first suspected by noting its characteristic motility pattern upon microscopic examination and was further confirmed by 16S rRNA gene sequence analysis (Fig. 4). Presumably, either this organism, which is apparently highly similar in other features to SLAS-1T, was resistant to the antibiotics or the presence of other microbes in the enrichment somehow ameliorated their inhibitory effects. Nonetheless, respiratory arsenate reduction linked to lactate oxidation was noted in the primary enrichment (Fig. 3A), as was chemoautotrophic arsenate reduction linked to sulfide oxidation (Fig. 3A and C), both of which are consistent with the basic metabolic capabilities of strain SLAS-1T (Table 1).
Sulfate reduction was clearly detected in the enrichment, as determined by the appearance of sulfide after all the arsenate was consumed (Fig. 3A), by the much larger amount of sulfide formed in the absence of arsenate (Fig. 3B) and by the observed reduction of [35S]sulfate to [35S]sulfide (see Results). Again, the presence of sulfate reducers was confirmed by 16S rRNA gene sequence analysis (Fig. 4), which indicated a population of relatives of Desulfonatronovibrio and deeply branching members of the Desulfovibrionaceae, a family of sulfate-reducing bacteria. Note that higher diversity was seen in representatives from the Bacteria than from the Archaea in this enrichment; therefore, it illustrates the pitfalls of routinely employing cell wall antibiotics as a strategic means to selectively screen for members of the Archaea.
The measured sulfate reduction activity and presence of 16S rRNA gene sequences closely related to those of sulfate reducers within the salt-saturated enrichment culture poses another paradox, namely, that sulfate reducers are thought to be excluded from extremely hypersaline systems as a consequence of energy conservation considerations (41). Indeed, we were unable to detect [35S]sulfate reduction in sediments from Searles Lake, even when we took a number of steps to conjure its activity by adding electron donors and/or by lowering the sulfate content to increase the specific activity of the added [35S]sulfate radiotracer (23). Nonetheless, high rates of sediment sulfate reduction have been reported in extremely hypersaline soda lakes in Russia (9). Although sulfate reducers have been detected in and cultured from the highly saline (∼270 g/liter) northern arm of Great Salt Lake, they were operating therein at their physiological limits with regard to salt tolerance (21). In the case of mineral-rich Searles Lake, sulfate reduction appears to be constrained in part by its high salinity (∼345 g/liter) but also by the high levels of borate present in the brine (24). Nonetheless, intact anoxic sediment cores from Searles Lake contain discernible levels of free sulfide (0.1 to 0.2 mM) (23, 40), suggesting that some level of either sulfate reduction or reduction of other intermediates of the sulfur cycle (e.g., thiosulfate or elemental sulfur) occurs therein. These sulfidogenic processes would open niches for the observed sulfide-linked chemoautotrophic growth of strain SLAS-1T. Indeed, we have recently isolated a sulfate reducer from this enrichment culture (strain SLSR-1) that can grow under conditions of salt saturation (J. Switzer Blum, unpublished data), which suggests that in situ sulfate reduction in Searles Lake is probably constrained by a multiplicity of factors in addition to salinity. It is also possible that these sulfate reducers have the capacity to respire arsenate, a stronger oxidant than sulfate, as has been shown with examples from both gram-positive and gram-negative sulfate reducers cultured from freshwater environments (29, 36). This could potentially put them in competition with strain SLAS-1T for available arsenate.
Clearly, extremely halophilic bacteria like strain SLAS-1T fill a niche in Searles Lake and are readily detectable as 16S rRNA gene amplicons at all depths sampled in sediment cores (23) and incubated sediment slurries (24), as well as in the enrichment culture described in this report. Some additional circumstantial evidence supports the possible occurrence of halophilic arsenate-respiring archaea, including the recovery of diverse arrA amplicons from the sediments of both Mono and Searles Lakes from the use of a degenerate primer set having codons of halophilic archaea (HAArrA primer set) (23). One dominant phylotype that is likely an HAarrA amplicon was retrieved in the enrichment culture (Fig. 5), and this is distinct from a partial arrA gene sequence from the enrichment culture that was identical to that from SLAS-1T. The HAarrA amplicons recovered from the enrichment culture may have been linked to the Archaea rather than the Bacteria, as their sequences are significantly different than that obtained for strain SLAS-1T. The presence in the enrichment of a halophilic archaeon closely related to Halorhabdus utahensis was observed in the 16S rRNA gene analysis (Fig. 4). However, while H. utahensis is capable of either aerobic or fermentative growth, it has no reported ability to grow via anaerobic respiration (59). Indeed, we were unable to achieve anaerobic, arsenate-dependent respiratory growth of this organism when we examined the type strain (J. Switzer Blum, unpublished data). Hence, either our detection of this archaeon in the enrichment was merely fortuitous and unrelated to the presence of arsenate or the possibility exists that a yet-to-be-described close relative of the genus Halorhabdus exists that can grow under conditions of salt saturation by respiring arsenate.
FIG. 5.
Phylogenetic analysis of the amino acid translations of the dominant HAarrA fragment (SLAb1 clone G3) retrieved from the Searles Lake antibiotic enrichment culture. Several representative environmental arrA clones, as well as arrAs of a subset of bacteria, were included in the analysis. Only nodes with bootstrap values greater than 50% from 1,000 replications were retained in the final phylogenetic tree. An asterisk indicates that the eSLAS1 arrA sequence is 100% identical to positions 1024 to 1204 of the SLAS-1T arrA gene.
Description of Halarsenatibacter Switzer Blum et al., gen. nov.
Halarsenatibacter (Hal.ar.se.na.ti.bac′ter. Gr. n. hals, halos, salt. N.L. arsenas, -atis, arsenate, N.L. masc. n. bacter rod, N.L. Halarsenatibacter halophilic arsenate-utilizing rod). Gram-negative, motile, strictly anaerobic, slightly curved rods (3.0 by 0.5 μm). Motility achieved by a pair of flagella located along the side of the organism, but motion has both helical and sinuous components. Extreme halophile that grows between 200 and 350 g/liter salinity with an optimum at salt saturation (∼350 g/liter) and no growth below 200 g/liter. Alkaliphilic with a pH range from 8.7 to 9.8 with an optimum at 9.4. Temperature range of 28 to 55°C with an optimum at 44°C. A limited number of organic substrates support growth, including a few sugars and organic acids but not fatty acids or amino acids. Fermentative growth or microaerophilic growth not observed. Growth is by dissimilatory (respiratory) reduction of arsenate to arsenite, Fe(III) to Fe(II), and elemental sulfur to sulfide. Chemoautotrophic growth occurs with sulfide as the electron donor and arsenate as the electron acceptor. Catalase positive. Sensitive to antibiotic mixture of vancomycin, kanamycin, penicillin, and tetracycline. G+C content is 45.2%. Major cell membrane lipids (62.8% of total) are comprised of four fatty acid (iso-15:0 > n-18:0 > iso-17:0 > n-16:0) triglycerides.
Description of Halarsenatibacter silvermanii Switzer Blum et al., sp. nov.
Halarsenatibacter silvermanii (sil.ver.ma′ni.i. N.L. gen. n. silvermanii, of/from Silverman, to honor the late American microbiologist Melvin P. Silverman of NASA [Ames Research Center] for his contributions to the field of geomicrobiology, especially with regard to the metabolism of inorganic iron and sulfur compounds by extremophilic bacteria). The description of Halarsenatibacter silvermanii is identical to that of the genus description given above with the following additions. The following organic substrates served as electron donors: glucose, malate, fructose, galactose, lactate, and pyruvate with arsenate as the electron acceptor. The G+C content of the DNA of the only strain known in the species is 45.2%. The type strain, strain SLAS-1T, isolated from Searles Lake, CA, has been deposited in the American Type Culture Collection (Manassas, VA) as strain BAA-1651.
Supplementary Material
Acknowledgments
This work was supported by the USGS National Research Program and by a NASA Exobiology grant.
We are indebted to H. Trüper with his help with the epithet. We are grateful to J. Stolz, C. Pearce, and R. Sierra for constructive criticism of earlier drafts of the manuscript. The TEMs were conducted by S.L. and T.J.B. at the University of Guelph.
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahmann, D., A. L. Roberts, L. R. Krumholtz, and F. M. M. Morel. 1994. Microbe grows by reducing arsenic. Nature 371:750. [DOI] [PubMed] [Google Scholar]
- 2.Beveridge, T. J., D. Moyles, and R. Harris. 2007. Electron microscopy.In C. A. Reddy, T. J. Beveridge, J. A. Breznak, L. Snyder, T. M. Schmidt, and G. A. Marzluf (ed.), Methods for general and molecular microbiology, 3rd ed. ASM Press, Washington, DC.
- 3.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-459. [Google Scholar]
- 4.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronan, J. E., Jr., and C. O. Rock. 1987. Biosynthesis of membrane lipids, p. 474-497. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, American Society for Microbiology, Washington, DC.
- 6.Cronan, J. E. 2003. Bacterial membrane lipids: where do we stand? Annu. Rev. Microbiol. 57:203-224. [DOI] [PubMed] [Google Scholar]
- 7.Eder, W., L. L. Jahnke, M. Schmidt, and R. Huber. 2001. Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl. Environ. Microbiol. 67:3077-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher, J. C., D. Wallschläger, B. Planer-Friedrich, and J. T. Hollibaugh. 2008. A new role for sulfur in arsenic cycling. Environ. Sci. Technol. 42:81-85. [DOI] [PubMed] [Google Scholar]
- 9.Foti, M. J., D. Y. Sorokin, E. E. Zacharova, N. V. Pimenov, J. G. Kuenen, and G. Muyzer. 2008. Bacterial diversity and activity along a salinity gradient in soda lakes of the Kulunda Steppe (Altai, Russia). Extremophiles 12:133-145. [DOI] [PubMed] [Google Scholar]
- 10.Gihring, T. M., and J. F. Banfield. 2001. Arsenite oxidation and arsenate respiration by a new Thermus isolate. FEMS Microbiol. Lett. 204:335-340. [DOI] [PubMed] [Google Scholar]
- 11.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 12.Hobbie, J. E., R. L. Daley, and S. Jaspar. 1977. Use of Nuclepore filters for counting bacteria for fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoeft, S. E., T. R. Kulp, J. F. Stolz, J. T. Hollibaugh, and R. S. Oremland. 2004. Dissimilatory arsenate reduction with sulfide as the electron donor: experiments with Mono Lake water and isolation of strain MLMS-1, a chemoautotrophic arsenate respirer. Appl. Environ. Microbiol. 70:2741-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeft, S. E., J. S. Blum, J. F. Stolz, F. R. Tabita, B. Witte, G. M. King, J. M. Santini, and R. S. Oremland. 2007. Alkalilimnicola ehrlichii, sp. nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int. J. Syst. Evol. Microbiol. 57:504-512. [DOI] [PubMed] [Google Scholar]
- 15.Hollibaugh, J. T., C. Budinoff, R. A. Hollibaugh, B. Ranson, and N. Bano. 2006. Sulfide oxidation coupled to arsenate reduction by a diverse microbial community in a soda lake. Appl. Environ. Microbiol. 72:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber, R., M. Sacher, A. Vollmann, H. Huber, and D. Rose. 2000. Respiration of arsenate and selenate by hyperthermophilic Archaea. Syst. Appl. Microbiol. 23:305-314. [DOI] [PubMed] [Google Scholar]
- 17.Inskeep, W. P., R. E. Macur, N. Hamamura, T. P. Warelow, S. A. Ward, and J. M. Santini. 2007. Detection, diversity, and expression of aerobic bacterial arsenite oxidase genes. Environ. Microbiol. 9:934-943. [DOI] [PubMed] [Google Scholar]
- 18.Jahnke, L. L., T. Embaye, J. Hope, K. A. Turk, M. Van Zuilen, D. J. Des Marais, J. D. Farmer, and R. E. Summons. 2004. Lipid biomarker and carbon isotopic signatures for stromatolite-forming, microbial mat communities and Phormidium cultures from Yellowstone National Park. Geobiology 2:31-47. [Google Scholar]
- 19.Jones, L. J. F., R. Carballido-López, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 20.Kaneda, T. 1991. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjeldsen, K. U., A. Loy, T. F. Jakobsen, T. R. Thomsen, M. Wagner, and K. Ingvorsen. 2007. Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah). FEMS Microbiol. Ecol. 60:287-298. [DOI] [PubMed] [Google Scholar]
- 22.Kulp, T. R., S. E. Hoeft, and R. S. Oremland. 2004. Redox transformation of arsenic oxyanions in periphyton communities. Appl. Environ. Microbiol. 70:6428-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulp, T. R., S. E. Hoeft, L. G. Miller, C. Saltikov, J. N. Murphy, S. Han, B. Lanoil, and R. S. Oremland. 2006. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl. Environ. Microbiol. 72:6514-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulp, T. R., S. Han, C. W. Saltikov, B. D. Lanoil, K. Zargar, and R. S. Oremland. 2007. Effects of imposed salinity gradients on dissimilatory arsenate reduction and other microbial processes in sediments from two California soda lakes. Appl. Environ. Microbiol. 73:5130-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulp, T. R., S. E. Hoeft, M. Asao, M. T. Madigan, J. C. Fisher, J. F. Stolz, C. W. Culbertson, L. G. Miller, and R. S. Oremland. 2008. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science 321:967-970. [DOI] [PubMed] [Google Scholar]
- 26.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, London, United Kingdom.
- 27.Laverman, A. M., J. S. Blum, J. K. Schaeffer, E. J. Philips, D. R. Lovley, and R. S. Oremland. 1995. Growth of strain SES-3 with arsenate and other diverse electron acceptors. Appl. Environ. Microbiol. 61:3556-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macy, J. M., J. M. Santini, B. V. Pauling, A. H. O'Neil, and L. I. Sly. 2000. Two new arsenate/sulfate-reducing bacteria: mechanisms of arsenate reduction. Arch. Microbiol. 173:49-57. [DOI] [PubMed] [Google Scholar]
- 30.Malasarn, D., C. W. Saltikov, K. M. Campbell, J. M. Santini, J. G. Hering, and D. K. Newman. 2004. arrA is a reliable marker for As(V) respiration. Science 306:455. [DOI] [PubMed] [Google Scholar]
- 31.Margolin, W. 2004. Bacterial shape: concave coiled coils curve Caulobacter. Curr. Biol. 14:R242-R244. [DOI] [PubMed] [Google Scholar]
- 32.McEwan, A. G., J. P. Ridge, C. A. Mcdevitt, and P. Hugenholtz. 2002. The DMSO reductase family of microbial molybdenum enzymes; molecular properties and role in the dissimilatory reduction of toxic elements. Geomicrobiol. J. 19:3-21. [Google Scholar]
- 33.McGrath, C. F., C. W. Moss, and R. P. Burchard. 1990. Effect of temperature shifts on gliding motility, adhesion, and fatty acid composition of Cytophaga sp. strain U67. J. Bacteriol. 172:1978-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mesbah, N. M., D. B. Hedrick, A. D. Peacock, M. Rohde, and J. Wiegel. 2007. Natranaerobius thermophilus gen. nov., sp. nov., a halophilic, alkalithermophilic bacterium from soda lakes of the Wadi An Natrun, Egypt, and proposal of Natrananaerobiacea fam. nov. and Natrananaerobiales ord. nov. Int. J. Syst. Evol. Microbiol. 57:2507-2512. [DOI] [PubMed] [Google Scholar]
- 35.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman, D. K., T. J. Beveridge, and F. M. M. Morel. 1997. Precipitation of arsenic trisulfide by Desulfotomaculum auripigmentum. Appl. Environ. Microbiol. 63:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oremland, R. S., J. Switzer Blum, C. W. Culbertson, P. T. Visscher, L. G. Miller, P. Dowdle, and F. E. Strohmaier. 1994. Isolation, growth and metabolism of an obligately anaerobic, selenate-respiring bacterium, strain SES-3. Appl. Environ. Microbiol. 60:3011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oremland, R. S., and J. F. Stolz. 2003. The ecology of arsenic. Science 300:939-944. [DOI] [PubMed] [Google Scholar]
- 39.Oremland, R. S., J. F. Stolz, and J. T. Hollibaugh. 2004. The microbial arsenic cycle in Mono Lake, California. FEMS Microbiol. Ecol. 48:15-27. [DOI] [PubMed] [Google Scholar]
- 40.Oremland, R. S., T. R. Kulp, J. Switzer Blum, S. E. Hoeft, S. Baesman, L. G. Miller, and J. F. Stolz. 2005. A microbial arsenic cycle in a salt-saturated, extreme environment. Science 308:1305-1308. [DOI] [PubMed] [Google Scholar]
- 41.Oren, A. 1999. Bioenergetic aspects of halophism. Microbiol. Mol. Biol. Rev. 63:334-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Øvreås, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Sælenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruesse, E., C. Quast, K. Knittel, B. Fuchs, W. Ludwig, J. Peplies, and F. O. Glöckner. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saltikov, C. W., and D. K. Newman. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. USA 100:10983-10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinensky, M. 1974. Homeoviscous adaptation: a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. USA 71:522-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, S. A., and F. R. Tabita. 2004. Glycine 176 affects catalytic properties and stability of Synecocccus sp. strain PCC6301 ribulose-1,5 biphosphate carboxylase/oxygenase. J. Biol. Chem. Biophys. 426:43-54. [DOI] [PubMed] [Google Scholar]
- 47.Stolz, J. F., and R. S. Oremland. 1999. Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23:615-627. [DOI] [PubMed] [Google Scholar]
- 48.Stolz, J. F., D. J. Ellis, J. S. Blum, D. Ahmann, D. R. Lovley, and R. S. Oremland. 1999. Sulfurospirillum barnesii sp. nov., Sulfurospirillum arsenophilus sp. nov., and the Sulfurospirillum clade in the epsilon proteobacteria. Int. J. Syst. Bacteriol. 49:1177-1180. [DOI] [PubMed] [Google Scholar]
- 49.Stolz, J. F., P. Basu, J. M. Santini, and R. S. Oremland. 2006. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60:107-130. [DOI] [PubMed] [Google Scholar]
- 50.Switzer Blum, J., A. Burns Bindi, J. Buzelli, J. F. Stolz, and R. S. Oremland. 1998. Bacillus arsenicoselenatis sp. nov., and Bacillus selenitireducens sp. nov.: two haloalkaliphiles from Mono Lake, California, which respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19-30. [DOI] [PubMed] [Google Scholar]
- 51.Switzer Blum, J., J. F. Stolz, A. Oren, and R. S. Oremland. 2001. Selenihalanaerobacter shriftii gen. nov., sp. nov.: a halophilic anaerobe from Dead Sea sediments that respires selenate. Arch. Microbiol. 175:208-219. [DOI] [PubMed] [Google Scholar]
- 52.Swofford, D. L. 1999. Phylogenetic analysis using parsimony (and other methods), version 4.0b10b. Sinauer Associates, Sunderland, MA.
- 53.Tabita, F. R., T. E. Hanson, H. Li, S. Satagopan, J. Singh, and S. Chan. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubsiCO homologs. Microbiol. Mol. Biol. Rev. 71:576-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takai, K., H. Kobayashi, K. H. Nealson, and K. Horikoshi. 2003. Deferribacter desulfuricans sp. nov., a novel sulfur-, nitrate-, and arsenate-reducing thermophile isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 53:839-846. [DOI] [PubMed] [Google Scholar]
- 55.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tosca, N. J., A. H. Knoll, and S. M. McLennan. 2008. Water activity and the challenge for life on early Mars. Science 320:1204-1207. [DOI] [PubMed] [Google Scholar]
- 58.Vinogradov, E., C. J. Paul, J. Li, Y. Zhou, F. A. Lyle, R. I. Tapping, A. M. Kropinski, and M. B. Perry. 2004. The structure and biological characteristics of the Spirochaeta aurantia outer membrane glycolipid LGLB. Eur. J. Biochem. 271:4685-4695. [DOI] [PubMed] [Google Scholar]
- 59.Wainø, M., B. J. Tindall, and K. Ingvorsen. 2000. Halorahbdus utahensis gen. nov., sp. nov., an aerobic, extremely halophilic member of the Archaea from Great Salt Lake, Utah. Int. J. Syst. Evol. Microbiol. 50:183-190. [DOI] [PubMed] [Google Scholar]
- 60.White, D. C., W. M. Davis, J. S. Nickels, J. D. King, and R. J. Bobbie. 1979. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 40:51-62. [DOI] [PubMed] [Google Scholar]
- 61.Whitman, W., and F. R. Tabita. 1976. Inhibition of d-ribulose 1,5-bisphosphate carboxylase by pyridoxal 5′-phosphate. Biochem. Biophys. Res. Commun. 71:1034-1039. [DOI] [PubMed] [Google Scholar]
- 62.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
- 63.Yamamoto, K., A. Shibahara, T. Nakayama, and G. Kajimoto. 1991. Determination of double-bond positions in methylene-interrupted dienoic fatty acids by GC-MS as their dimethyl disulfide adducts. Chem. Phys. Lipids 60:39-50. [Google Scholar]
- 64.Yeates, C., and M. R. Gillings. 1998. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 27:49-53. [Google Scholar]
- 65.Zhang, Y.-M., and C. O. Rock. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222-233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.