Abstract
The dense hyphal network directly underneath the fruiting bodies of ectomycorrhizal fungi might exert strong influences on the bacterial community of soil. Such fruiting bodies might serve as hot spots for bacterial activity, for instance by providing nutrients and colonization sites in soil. Here, we assessed the putative selection of specific members of the Sphingomonadaceae family at the bases of the fruiting bodies of the ectomycorrhizal fungi Laccaria proxima and Russula exalbicans in comparison to the adjacent bulk soil. To do so, we used a previously designed Sphingomonadaceae-specific PCR-denaturing gradient gel electrophoresis (DGGE) system and complemented this with analyses of sequences from a Sphingomonadaceae-specific clone library. The analyses showed clear selective effects of the fruiting bodies of both fungi on the Sphingomonadaceae community structures. The effect was especially prevalent with R. exalbicans. Strikingly, similar fungi sampled approximately 100 m apart showed similar DGGE patterns, while corresponding bulk soil-derived patterns differed from each other. However, the mycospheres of L. proxima and R. exalbicans still revealed divergent community structures, indicating that different fungi select for different members of the Sphingomonadaceae family. Excision of specific bands from the DGGE patterns, as well as analyses of the clone libraries generated from both habitats, revealed fruiting body-specific Sphingomonadaceae types. It further showed that major groups from the mycospheres of R. exalbicans and L. proxima did not cluster with known bacteria from the database, indicating new groups within the family of Sphingomonadaceae present in these environments.
Soil is generally regarded a carbon-limited environment for its inhabitants (35, 41). There are, however, so-called hot spots for bacterial activity in soil, in which carbonaceous compounds become increasingly available for the soil microbiota. In addition to the well-known rhizosphere, in which root exudates that contain easily available carbonaceous compounds are provided by plant roots (6, 22, 23, 34), the feet of fruiting bodies of ectomycorrhizal fungi (the mycosphere) may constitute another hot spot in which bacterial activity is stimulated (8, 9, 16) by the provision of such compounds. This has been called the mycosphere effect (43). In many cases, the interactions between soil bacteria and fungi may have evolved toward mutual benefit for both the bacteria and the fungal partners (12, 14); however, we still understand very little about the selective processes exerted on the bacteria in the mycosphere.
Leveau and Preston (19) recently described three ways by which soil bacteria can access the carbon present in ectomycorrhizal fungi: (i) extracellular necrotrophy (living off compounds of dead/dying fungal tissue), (ii) extracellular biotrophy (living outside of fungal tissue on the basis of released compounds), and (iii) endocellular biotrophy (becoming endomycotic, thus utilizing cytoplasmic compounds). The compounds captured and metabolized by bacteria in the mycosphere, as well as the potential recognition and signaling between the two partners in the interaction, remain, however, largely unknown. This in spite of the fact that the availability of carbon substrates is thought to be key to the ecological success of the soil bacteria involved. For instance, Frey et al. (11) reported that specific Pseudomonas fluorescens types in the Laccaria laccata hyphosphere were capable of degrading fungus-derived trehalose, while Sahin (28) found that Methylobacterium spp. in soil were able to use oxalate or oxalic acid—both of which are often derived from fungi—as a carbon source.
Various members of the Sphingomonadaceae family (previously subdivided into the genera Sphingomonas, Sphingopyxis, Novosphingobium, and others) are known for their capability to utilize a wide variety of carbon sources, and in fact, several are renowned degraders of recalcitrant (xenobiotic) molecules (4, 17, 20, 32). Members of the family are ubiquitous, as they are found in different soils (1, 7, 20), sediments (10), and pelagic aquatic environments (7, 36, 38). Recent studies show that particular sphingomonads may also play important roles in the mycorrhizosphere, defined as the zone in the rhizosphere that is affected by mycorrhizal hyphae (40; P. Lemanceau, unpublished data; S. Moulin, unpublished data). However, the putative selection of members of the Sphingomonadaceae in the mycosphere of ectomycorrhizal fungi has not yet been described in detail.
During a recent study in our lab on the selective effect of the Laccaria proxima mycosphere on the soil bacterial community (43, 44), the selection of a particular group of (culturable) sphingomonads was observed (J. A. Warmink, unpublished data). Furthermore, Uroz et al. (40) recently described a Sphingomonas type that was capable of inciting mineral weathering in the hyphosphere of Scleroderma citrinum. These findings hint at the selection of, and a role for, particular sphingomonads in the mycosphere. We here tested this hypothesis and report the selection of specific sphingomonads in the mycospheres of different fungi, with emphasis on two important ectomycorrhizal fungi of hazelnut and coniferous trees, Laccaria proxima and Russula exalbicans. Cultivation-independent methods, consisting of mycosphere- and soil DNA-based Sphingomonadaceae-specific PCR-denaturing gradient gel electrophoresis (DGGE) and clone library sequence analysis, were used.
MATERIALS AND METHODS
Sampling of ectomycorrhizal fruiting bodies (fungi) and soil.
Triplicate samples of fungi belonging to Laccaria proxima were obtained in October 2006 from an area near hazel trees in Gieterveen, Drenthe, The Netherlands. The fungi were dug out as a whole, including the shallow (4-cm) soil layer that adhered tightly to the fungus feet. For control samples, three corresponding bulk soil samples were taken at approximately 1-m distance from each fungus. Sampling, fungal identification, and the characteristics of the Gieterveen soil (G soil) were as described by Warmink and van Elsas (43).
In addition, 16 fungi and corresponding bulk soil samples were sampled in a forest in Noordlaren, Drenthe, The Netherlands, in November 2006. The Noordlaren soil (N soil) and its vegetation were previously characterized by Warmink et al. (44). The 16 fungi were collected at distances of approximately 50 to 100 m apart. For controls, corresponding bulk soil samples were taken at 1-m distances from each fungus. Soil characteristics were comparable at each sampling location. Identification of the fungi was as described by Warmink et al. (44).
Following sampling, all fungi were taken to the laboratory, where they were processed immediately by the method of Warmink and van Elsas (43). Briefly, the fruiting bodies were cut from the fungus feet, and excess soil was removed by tapping and shaking from the dense hyphal network of the fungus feet. This yielded the fungus base (trunk), containing hyphae intruding into the adjacent soil, which was thus presumably directly influenced by the hyphae. The adhering soil, denoted mycosphere soil, was sampled by cutting and scratching, and used for subsequent DNA isolation.
DNA isolation from fungus feet and bulk soil.
Mycosphere and bulk soil DNA was obtained by the method of Warmink and van Elsas (43), using the Mo Bio Ultraclean soil DNA isolation kit (MoBio Laboratories, Carlsbad, CA), followed by subsequent removal of humic acids using the Wizard DNA Clean-Up System kit (Promega, Leiden, The Netherlands). The DNA was finally taken up in the buffer prescribed by the manufacturer.
To assess molecular size, quantity, and purity, the DNA was electrophoresed on 1% agarose gels and visualized using a UV transilluminator after staining with ethidium bromide. Average molecular fragment size, yield, and purity were estimated by comparison with a commercially available marker, i.e., the Smart ladder (Eurogentec, Maastricht, The Netherlands).
Sphingomonadaceae-specific PCR.
To examine putative community shifts in the mycosphere, Sphingomonadaceae-specific 16S rRNA gene-based PCR was performed using forward primer Sphingo108f (5′-GCGTAACGCGTGGGAATCTG-3′) and reverse primer Sphingo420r (5′-TTACAACCCTAAGGCCTTC-3′) by the method of Leys et al. (20). To facilitate this Sphingomonadaceae-specific PCR, an initial 20-cycle bacterial PCR was performed on bulk soil and mycosphere DNA using primers B8f (5′-AGAGTTTGATCMTGGCTCAG-3′) and U1492r (5′-GGTTACCTTGTTACGACTT-3′) (18). For this, 50-μl mixes containing 5 μl of PCR buffer (Roche, Basel, Switzerland), 200 μM of each deoxynucleoside triphosphate, 2.5 μM MgCl2, 200 nM of each primer, 0.1 μl Taq DNA polymerase (5 U/μl) (Roche, Basel, Switzerland), 44.7 μl H2O, and 10 to 50 ng of mycosphere or bulk soil DNA (1 μl) were used. The PCR program started with a 5-min denaturing step at 94°C, followed by 25 cycles where 1 cycle consisted of 45 s at 94°C, 45 s at 58°C, and 30 s at 74°C, and finished by a final extension step at 74°C for 10 min. To yield amplicons for subsequent DGGE analysis, an amplification system consisting of the Sphingomonadaceae forward and reverse primers (the latter equipped with a 40-nucleotide GC clamp [5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG-3′] at its 5′ end) was used on a 1-μl subsample added to a fresh 50-μl PCR mix (same as above) (24). The PCR program of Leys et al. (20) was used in this second run (25 cycles). Thus, 321-bp amplicons (361 bp with GC clamp) were produced on the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA).
Preparation of clone library and sequencing.
The 321-bp amplicons generated from pooled samples of the mycospheres of Russula exalbicans and Laccaria proxima, as well as the corresponding bulk soils, were cloned into the pGEM-T vector following the manufacturer's protocol (Invitrogen, Breda, The Netherlands) and subsequently introduced into Escherichia coli MM294 via transformation. Following transformant growth, selected colonies were checked for the presence of inserts of the expected size using PCR based on primers T7f and SP6r, which recognize the insert flanks. A random subselection of 96 clones with insert per habitat was then subjected to direct sequencing. Prior to sequencing, the PCR products were cleaned using the polyethylene glycol-sodium acetate method (43). Sequencing was performed on an ABI3130 DNA sequencer, using primer T7f (Applied Biosystems, Foster City, CA). The sequences were checked and manually improved, where needed, using the Chromas (http://www.technelysium.com.au/chromas.html) software. Sequences are deposited in the NCBI database under accession numbers FJ685775 to FJ685963.
Sphingomonadaceae community DGGE fingerprinting.
Sphingomonadaceae-specific 16S rRNA gene-based PCR products (approximately 30 ng per lane) were loaded onto gels in a PhorU2 system (Ingeny International, Goes, The Netherlands) according to the manufacturer's protocol. We used a gradient of 40 to 70% denaturants (urea and formamide) (100% denaturants is 8 M urea plus 40% formamide), a half-strength Tris-acetate-EDTA (TAE) buffer at pH 8.0 and a temperature of 60°C, at 110 V for 18 h. After the gels were run, they were removed from the apparatus and stained using silver nitrate (15). When bands had to be excised, Sybr gold (final concentration, 0.5 μg/liter TAE buffer) (Invitrogen, Breda, The Netherlands) was used to stain the gels.
Excision of bands from DGGE and subsequent sequencing.
After Sybr gold staining, the gels were inspected for the emergence of putative mycosphere-specific bands. Selected bands were then excised using a scalpel. To obtain band DNA in solution, the bands were “crushed” in 10 μl H2O and then kept for 24 h at 4°C. Following this extraction, PCR reamplification was performed using the aforementioned reaction mixture for primers Sphingo108f and Sphingo420r with the addition of 2 μl of 50% acetamide. Products were checked for their migratory behavior on DGGs. Of the initial 19 bands, 6 did not run their expected migration distance, leaving a total of 13 relevant bands. The six “nonrelevant” bands were identified as being homologous to the dominant band in the same lane; they were not considered in further analyses. The 13 products that ran according to expectation and that were pure were then subjected to sequencing directly from the PCR product using primer Sphingo108f. Five of the 13 sequences were retrieved from bands with migration behavior equal to those of bands from different lanes. These five sequences indeed confirmed the nature of their counterparts. The total diversity thus analyzed consisted of nine sequences. These sequences were deposited in the NCBI database under accession numbers FJ685964 to FJ685972.
Statistics.
DGGE patterns were analyzed and compared using GelCompar II (Applied Maths, Belgium). Using the GelCompar II program, bands were identified and quantified using the normalized relative band intensity (area under the curve) to allow comparison of the different samples. Clustering was performed using the unweighted-pair group method with mathematical averages (UPMGA). A table containing the band positions and relative intensity values was exported and used for multivariate analysis using Canoco (Canoco 4.5; Biometris, Wageningen, The Netherlands). In the multivariate analysis, the correlation between DGGE bands and environmental variables was determined. The table exported from GelCompar II was used as the species input, while habitat (mycosphere versus bulk soil) and fungal species were considered environmental factors. In addition, the Shannon-Weaver indices of diversity (31) were calculated using the band intensity values extracted from the GelCompar program.
Classification of clones obtained from the libraries was done using the Naive Bayesian rRNA Classifier (version 1.0; Ribosomal Database Project [RDP]). Phylogenetic trees were built with the Mega4 program (37) using the neighbor-joining method (29) for calculations of evolutionary history. Rarefaction analysis was performed to examine the coverage of the four clone libraries. Rarefaction diagrams were made by plotting the number of operational taxonomic units (OTUs) (defined as sequences that showed >97% similarity) as a function of the number of individual clones sampled from the different clone library. Rarefaction analysis was performed by making use of the DOTUR software (http://www.plantpath.wisc.edu/fac/joh/dotur.html) (30). Library comparisons were performed using LIBSHUFF (33) and UniFrac (21, 37). The two programs were used, as both use different algorithms for comparing the libraries. Similarity matrices, needed as input for LIBSHUFF, were generated using DOTUR. Phylogenetic trees of OTUs, which were needed as input for the program UniFrac, were generated using Mega4.
RESULTS
Analysis of Sphingomonadaceae communities at the feet of ectomycorrhizal fruiting bodies in N soil samples.
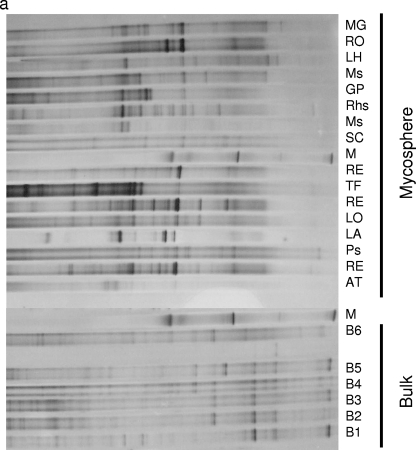
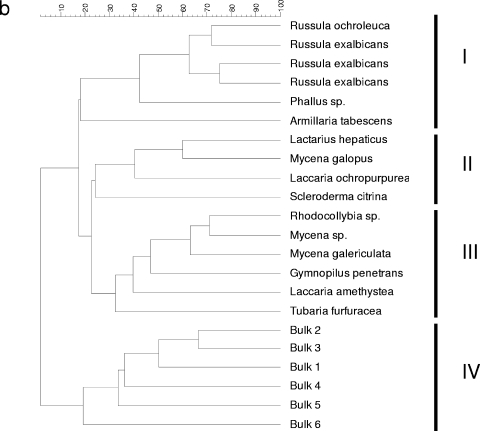
To analyze whether natural mushroom-forming fungi select for specific members of the Sphingomonadaceae at their feet, Sphingomonadaceae-specific PCR-DGGE was performed on DNA extracted from the mycospheres of 16 different fungi sampled in the N soil. DNA of N bulk soil was used as the comparator. Internal transcribed spacer sequencing (44) showed that the sampled fungi represented diverse saprophytic, ectomycorrhizal, and pathogenic fungi. Typically, four fungi belonged to the family Russulaceae, with three being affiliated with Russula exalbicans and the fourth a bit more distant. Thus, DNA of about 10 to 20 kb was successfully obtained from all samples in quantities of approximately 2 to 5 μg per g (dry weight) mycosphere or bulk soil (43). Subsequent Sphingomonadaceae-specific PCR consistently yielded amplicons of the expected 361-bp size, as evidenced by electrophoresis in agarose gels (not shown). These amplicons were then separated out on DGGEs. Comparison of the DGGE banding patterns for the mycospheres and the corresponding bulk soils clearly showed that specific members of the Sphingomonadaceae family were selected in the different mycospheres (Fig. 1a and b). Specifically, four clusters (I to IV) could be distinguished, three clusters (I to III) representing all mycosphere samples and one cluster (IV) representing all bulk soil samples. The patterns derived from Russulaceae mycospheres clustered tightly together in cluster I, indicating the selection of similar members of the Sphingomonadaceae by these mycospheres. In contrast, the remaining two mycosphere clusters contained diverse fungal species.
FIG. 1.
DGGE (a) and UPMGA clustering analysis (b) of the Sphingomonadaceae community of the mycospheres of 16 fungi and six corresponding bulk soil samples isolated from N soil. Only six bulk soil samples (B1 to B6) are shown, as the patterns of all bulk soil samples were found to be highly similar. Abbreviations: MG, Mycena galopus; RO, Russula ochroleuca; LH, Lactarius hepaticus; Ms, Mycena sp.; GP, Gymnopilus penetrans; Rhs, Rhodocollybia sp.; SC, Scleroderma citrina; RE, Russula exalbicans; TF, Tubaria furfuracea; LO, Laccaria ochropurpurea; LA, Laccaria amethystea; Ps, Phallus sp.; AT, Armillaria tabescens; M, markers. (b) Clusters I to IV are shown. The scale at the top shows percent similarity.
To achieve statistical significance with the key fungal species, we selected R. exalbicans and Laccaria proxima (isolated from G soil; Warmink, unpublished) as the target fungi for further study. Thus, four putative R. exalbicans mycospheres, in addition to three L. proxima mycospheres taken from the field, were analyzed.
Molecular analysis of Sphingomonadaceae communities in Russula exalbicans and Laccaria proxima mycospheres and corresponding bulk soil samples.
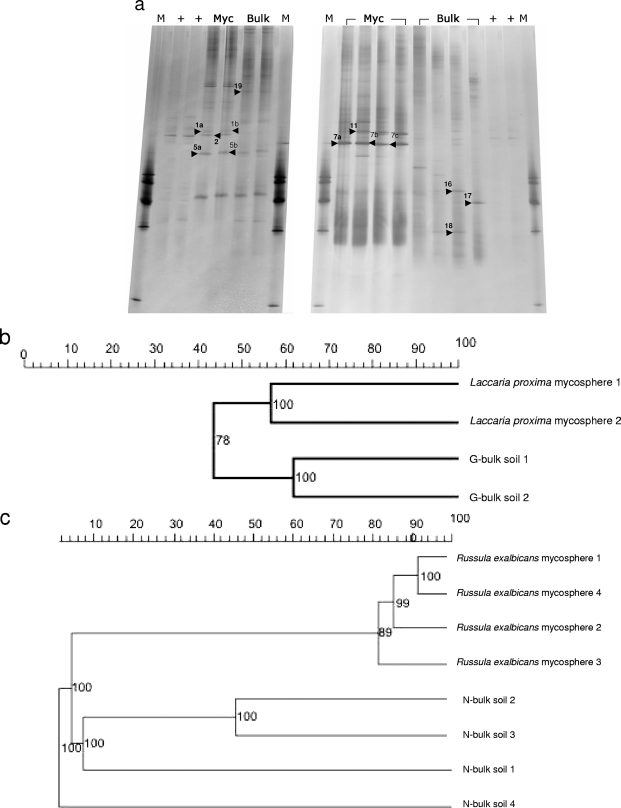
To assess the Sphingomonadaceae communities in the mycosphere soil of L. proxima and R. exalbicans compared to corresponding bulk soil samples, additional PCR-DGGE analyses were performed. Figure 2a shows the results of these analyses. Unfortunately, in one L. proxima sample, the Sphingomonadaceae-specific 361-bp fragment could not be amplified. Overall, the patterns showed semicomplex communities represented by roughly 10 bands. Specifically, averages of 10 (±0) and 10.25 (±0.71) bands were found for the mycospheres of L. proxima and R. exalbicans, respectively. The corresponding bulk soil samples showed 9.5 (±1.71) and 11.5 (±2.38) bands, respectively. Thus, no significant difference in the number of bands was detected between bulk soil and any of the two mycospheres, which was corroborated by calculating the Shannon-Weaver index of diversity (H′). Thus, we found H′ = 2.0 ± 0.02 and 1.9 ± 0.3 for the L. proxima mycosphere and corresponding bulk soil, respectively, and H′ = 1.8 ± 0.3 and 1.7 ± 0.3 for the R. exalbicans mycosphere and its corresponding bulk soil, respectively. No significant difference was detected in these values for the two mycospheres. There were, however, differences in the relative dominance of particular bands between mycosphere- and bulk soil-derived DGGE patterns. Specifically, the L. proxima mycosphere patterns revealed 4 or 5 dominant bands among the 10 bands. At least three bands (Fig. 2a, mycosphere, bands 1 and 2, and bulk soil, band 19) from each pattern appeared to be habitat specific. For the R. exalbicans mycosphere patterns, the differences were even more striking, as two or three dominating bands were found (e.g., bands 7 and 11), but these were absent from the bulk soil patterns. Conversely, three bulk soil-derived bands, i.e., bands 16, 17, and 18, could not be detected in the corresponding mycosphere patterns.
FIG. 2.
DGGE (a) and UPMGA clustering analysis (b and c) of the Sphingomonadaceae community of the G-soil-derived mycosphere of Laccaria proxima and its corresponding bulk soil (left gel in panel a) and the N-soil-derived mycosphere of Russula exalbicans and its corresponding bulk soil (right gel in panel a) based on 361-bp Sphingomonadaceae-specific 16S rRNA gene fragments. The arrowheads point to the excised bands. Lanes: M, markers: +, 16S rRNA gene fragment of a Sphingomonas echinoides isolate denoted HB44; Myc, mycosphere; Bulk, bulk soil. The scales at the top of panels b and c show percent similarity. The numbers at the nodes indicate the bootstrap value as a percentage (1,000 replications).
Thus, the DGGE patterns showed clear habitat-specific groupings and considerable similarities among the replicates of each habitat (Fig. 2b and c). However, the N bulk soil patterns were internally divergent.
The analysis of the R. exalbicans mycosphere and bulk soil patterns (Fig. 2c) showed a similarity of 80% among all mycosphere patterns and a similarity of 55% among the corresponding bulk soil patterns. In contrast, the similarity between the mycosphere and bulk soil patterns was a mere 5%.
For the L. proxima/G-soil comparison, the mycosphere patterns grouped together at 55% similarity and the bulk soil patterns grouped together at 65% similarity, respectively (Fig. 2b), whereas the similarity level between the two clusters was only 40%.
CCA.
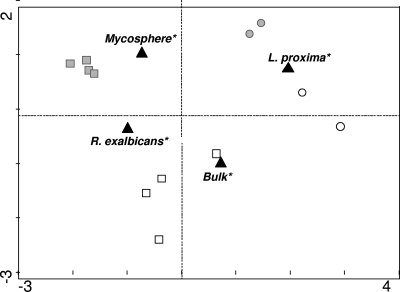
To more precisely analyze the (dis)similarity between the different DGGE patterns, we performed canonical correspondence analysis (CCA) (Fig. 3). The expected difference between the mycosphere- and bulk soil-derived patterns was clearly illustrated by the two-dimensional location of the patterns. Furthermore, the Sphingomonadaceae communities in replicate mycospheres of each of the two ectomycorrhizal fungi were more closely related to each other than those of the bulk soil samples were to each other. Also, the patterns derived from each of the mycospheres were widely separated by CCA, and this CCA did not show a close relationship between the bulk soil samples.
FIG. 3.
CCA of the Sphingomonadaceae-specific DGGE from Fig. 2a. G-soil-derived samples (circles), N-soil-derived samples (squares), mycosphere samples (gray symbols), bulk soil samples (white symbols), and nominal environmental samples (black triangles) are indicated.
Analysis of Sphingomonadaceae community members by identification of bands from DGGE patterns.
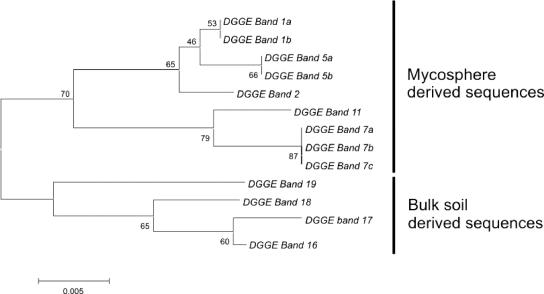
The aforementioned DGGE patterns revealed that several bands were clearly selected in the L. proxima and R. exalbicans mycospheres, indicating ectomycorrhizal fungus-specific selection of particular Sphingomonadaceae types. Nine bands, which presumably represented members of the Sphingomonadaceae abundant in the L. proxima mycospheres (Fig. 2a, bands 1a/b, 2, and 5a/b) and R. exalbicans mycospheres (Fig. 2a, 7a/b/c and 11) were excised from the gel (indicated by black arrowheads in Fig. 2a). Four other conspicuous bands were selected from bulk soils (Fig. 2a, G soil, band 19, and N soil, bands 16, 17, and 18). All bands were purified and subjected to sequence analysis. Figure 4 shows the internal clustering of these DGGE band sequences. Interestingly, all sequences grouped into three distinct groups which were defined by habitat, i.e., two mycosphere clusters (one L. proxima and the other R. exalbicans) and one bulk soil cluster became apparent. The two mycosphere-specific groups together made up a larger cluster which thus encompassed all mycosphere-derived sequences.
FIG. 4.
Dendrogram showing the phylogenetic relationship of the sequences of the excised bands depicted by the black arrowheads in Fig. 2a. Clustering was performed using the neighbor-joining method. Bands 1a/b, 2, and 5a/b were excised from the pattern of the G-soil-derived mycosphere of Laccaria proxima. Bands 7a/b/c and 11a/b were excised from the patterns of the N-soil-derived mycosphere of Russula exalbicans. Bands 16 to 18 were excised from the patterns from the N-soil-derived bulk soil, while band 19 was excised from the G-soil-derived bulk soil. The numbers at the nodes indicate the bootstrap values as percentages (1,000 replications). The scale bar shows 0.005 alterations per 1,000 nucleotides.
Table 1 shows an overview of the identities assigned to the different band sequences. Seven of the 9 different 16S rRNA gene sequences obtained from the bands were closely related to those of particular members of the Sphingomonadaceae, whereas the remaining 2 sequences were more remotely similar to the Sphingomonadaceae (>90%). Specifically, the sequences of bands 1, 2, and 5 (bands 1 and 5 extracted from two lanes, thus encompassing a and b forms), from the mycosphere of L. proxima, affiliated with (at 98 to 99% identity) uncultured Sphingomonadaceae types (closest database hit clone Amb_16S_608 [GenBank accession number EF018252] from aspen rhizosphere). Type strain matching revealed that bands 1 and 2 were closely related to Sphingosinicella microcystinivorans (97%), while band 5 had 98% similarity with Sphingomonas sp. strain T5-04. Band 19, obtained from the corresponding bulk soil, was affiliated (99%) with an uncultured forest soil bacterium (GenBank accession number AY913534) belonging to the alphaproteobacteria. Type strain matching showed this band to be remotely (95% similarity) related to Pleomorphomonas oryzea, another alphaproteobacterium. This band also showed 94% similarity with Sphingobium sp. strain RL-2005. Bands 16, 17, and 18 from the N bulk soil were also affiliated with an uncultured forest soil bacterium (GenBank accession number AY913608) isolated in the aforementioned study, and similarities ranged from 96 and 98%. Type strain matching revealed bands 16 to 18 to be most related to Sphingomonas yunnanensis, at 91% (band 18) and 95% similarity (bands 16 and 17). Bands 7 and 11, which stood out in the Russula exalbicans mycosphere patterns, both matched (95% for band 11; 96% for band 7) the sequence of an uncultured unidentified eubacterium (GenBank accession number AJ292593). Type strain matching of these sequences for bands 7 and 11 revealed only remote affiliation with Sphingomonas sp. strain T5-04 (7 to 90% and 11 to 92%, respectively). On the other hand, band 7 was 96% similar to a sequence of Magnetospirillum magnetotacticum (GenBank accession number Y10110).
TABLE 1.
Identification of the excised bands depicted by the black arrowheads in Fig. 2a
| Banda | Match total/match type strainb | % Similarity to match total/match type strain |
|---|---|---|
| 1a/b | Uncultured Sphingomonadaceae (EF072445)/Sphingosinicella microcystinivorans (AB084247) | 99/98 |
| 2 | Uncultured Sphingomonadaceae (EF072445)/Sphingosinicella microcystinivorans (AB084247) | 98/97 |
| 5a/b | Uncultured Sphingomonadaceae (EF072445)/Sphingomonas sp. strain T5-04 (AB166883) | 98/98 |
| 7a/b/c | Uncultured eubacterium (AJ292593)/Magnetospirillum magnetotacticum (Y10110) | 96/96 |
| 11 | Uncultured eubacterium (AJ292593)/Sphingomonas sp. strain T5-04 (AB166883) | 95/92 |
| 16 | Uncultured forest soil bacterium (AY913608)/Sphingomonas yunnanensis (AY251818) | 98/95 |
| 17 | Uncultured forest soil bacterium (AY913608)/Sphingomonas yunnanensis (AY894691) | 97/95 |
| 18 | Uncultured forest soil bacterium (AY913608)/Sphingomonas yunnanensis (AY894691) | 96/91 |
| 19 | Uncultured forest soil bacterium (AY913534)/Pleomorphomonas oryzae (AB159680) | 99/95 |
Excised bands depicted by the black arrowheads in Fig. 2a.
The closest match in the GenBank database is indicated by the accession number within parentheses.
Analysis of sequences in the Sphingomonadaceae-specific clone library.
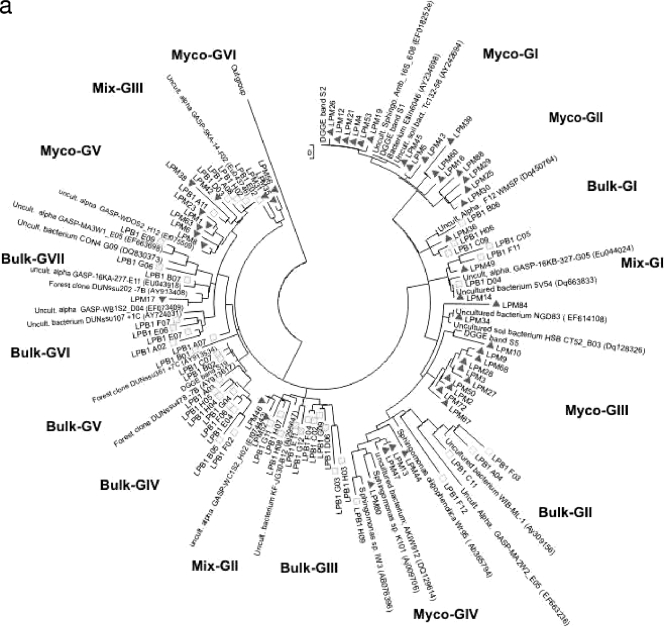
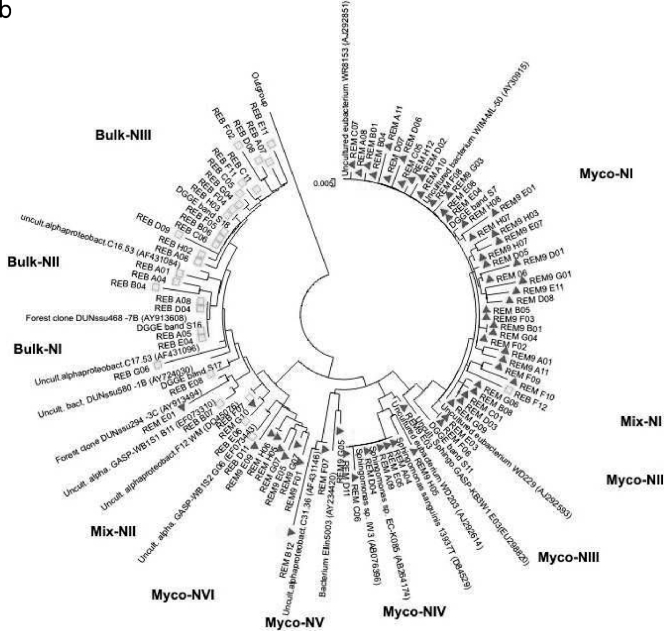
To allow an analysis of the Sphingomonadaceae community structure and diversity in the selected mycospheres, four clone libraries of pooled samples (per habitat) were constructed. The libraries thus obtained consisted of 49 clones from the L. proxima mycosphere, 48 clones from the corresponding G soil, 67 clones from the R. exalbicans mycosphere and 31 clones from the corresponding N soil. One hundred ninety of the 195 sequences fell within the Sphingomonadales group of the alphaproteobacteria or presumably belonged to this group but could be shown only to be affiliated with as-yet-uncharacterized (alpha)proteobacteria (a criterion of at least 89% similarity with the Sphingomonadales was used).
The levels of uncharacterized (alpha)proteobacteria were 4.5% and 20% in the mycospheres of L. proxima and R. exalbicans, respectively, whereas this level was higher for the bulk soils, i.e., 55% (N soil) and 65% (G soil). Figure 5a and b show the phylogenies of the Sphingomonadaceae-related sequences obtained from the L. proxima and R. exalbicans mycospheres and the corresponding bulk soils. In both trees, a clear separation between clones isolated from the mycosphere and clones isolated from bulk soil was visible. The separation was clearest in the tree containing the R. exalbicans and N bulk soil sequences and less clear in the L. proxima samples, which corroborates the DGGE results. Specifically, the phylogenetic tree constructed on the basis of the N-soil sequences (Fig. 5b) contained 11 different clusters consisting of two or more sequences, whereas five sequences formed singletons. Nine clusters were habitat specific, with six clusters specific for the mycosphere and three for the bulk soil. The remaining clusters (Mix-NI and Mix-NII) contained mixed (soil- and mycosphere-derived) sequences. Of the total sequences retrieved from the N soil, most (95%) grouped in the habitat-specific clusters (Fig. 5b), whereas the “mixed” clusters encompassed only a minority (three and two sequences, respectively) of sequences (5% of the total). Of the five mycosphere-specific clusters, three contained five or more sequences, indicating clonal selection in the mycosphere. The largest mycosphere-specific cluster (Myco-NI) encompassed 33 clones. Interestingly, sequences in this cluster did not closely group with any known database sequence. The closest match was an as-yet-uncultured eubacterium at 96% similarity (GenBank accession number AJ292593), while the cluster grouped, at 90% similarity, with Sphingomonas sp. strain DP524 (GenBank accession number AY227693). Strikingly, this cluster matched DGGE band 7, the band of greatest intensity in the R. exalbicans mycosphere patterns. As this band matched the closest database sequence (GenBank accession number AJ292593) at only 96% (Table 1), the cluster may represent a new Sphingomonadaceae group.
FIG. 5.
Phylogenetic analysis of the clones from the mycosphere of Laccaria proxima and its corresponding G bulk soil (a) and the clones from the mycosphere of Russula exalbicans and its corresponding N bulk soil (b). The clustering was performed using the neighbor-joining method for calculations of evolutionary history. Clones derived from mycosphere samples (triangles) and clones derived from bulk soil samples (squares) are indicated. Abbreviations: Uncult. alpha, uncultured alphaproteobacterium; bact., bacterium; Sphingo, Sphingomonadaceae; alphaproteobact., alphaproteobacterium.
Moreover, R. exalbicans mycosphere-derived band 11 grouped closely with the mycosphere-specific clusters, but not within a specific cluster. Two other large clusters, both encompassing seven sequences (Fig. 5b), were Myco-NIV (matching [100% similarity] Sphingomonas sp. strain EC-K085 [GenBank accession number AB264174] and Sphingomonas sp. strain IW3 [GenBank accession number AB076396]) and Myco-NVI (matching an uncultured alphaproteobacterium [GenBank accession number EF073443] at 97% similarity). Among the bulk soil clusters, Bulk-NIII was the largest, containing 15 sequences. It included bulk soil DGGE band 18, matching an uncultured forest soil bacterium sequence with GenBank accession number AY913608 (97% similarity). Furthermore, cluster Bulk-NI, encompassing four sequences, including DGGE band 16, matched uncultured forest soil clone (GenBank accession number AY913608) at 97% similarity. Finally, singleton REB_E08 matched band 17 and thus is in the bulk soil cluster.
The tree of the G-soil sequences (Fig. 5a) encompassed 16 clusters, whereas five sequences were present as singletons. The largest mycosphere-specific group, Myco-GI (12 sequences), matched an uncultured Sphingomonadaceae species with GenBank accession number EF018252at 99% similarity. The group also matched DGGE bands 1 and 2, which were closely related. DGGE band 5 clustered near the large mycosphere cluster Myco-GIII; however, its closest neighbor was the singleton sequence LPM10 (related at 96% similarity to Sphingomonadaceae bacterium Ellin7076 [GenBank accession number AY673242]). Cluster Myco-GIII, containing nine clones, had as its closest match an uncultured Sphingomonadacae type (GenBank accession number EF018252) at 96% similarity, possibly indicating a new group related to the Sphingomonadacae. The two largest bulk soil clusters, Bulk-GIII and Bulk-GIV, both containing eight clones, were closely affiliated, at 97% and 98% similarity, with as-yet-uncultured German forest-derived alphaproteobacterial clones (GenBank accession numbers AY913617and AY913735). The latter sequences also provided the closest matches to DGGE bands 16 to 18 isolated from N bulk soil. A cluster of two sequences matched DGGE band 19 and closely resembled another German forest clone (GenBank accession number AY913451).
Rarefaction analyses.
Rarefaction analyses were performed to examine the coverage of our four clone libraries (see Fig. S1 in the supplemental material). The diversity of Sphingomonadacae sequences among G-soil-derived bulk and mycosphere samples was higher than that among the N-soil ones. Although all four rarefaction curves showed a decline in the rate of OTU (sequence having >97% similarity) detection, no clone library fully covered the diversity in its habitat. However, both mycospheres appeared to “saturate” quicker than the corresponding bulk soils, hinting at a selective (“bottleneck”) effect of the mycosphere on the Sphingomonadaceae communities in soil.
To determine whether the four clone libraries, and therefore the four habitats, were significantly different, S-LIBSHUFF and UniFrac analyses were performed. The S-LIBSHUFF analysis showed all libraries to be significantly different (P < 0.05), except for the library constructed from the G soil, which was overlapped by that from the N soil (P = 0.064). UniFrac analyses reinforced the contention that all four libraries had different compositions. Here, the highest similarity was between the libraries from the bulk soil of Laccaria proxima and the mycosphere of Russula exalbicans. In the latter comparison, major differences between the libraries were still present, as a difference of 87% was measured. Thus, only a small fraction (13%) of the Sphingomonadaceae diversity was shared between these two environments.
DISCUSSION
So far, most studies on bacterial-fungal relationships in soil describe the interplay between plants, fungi, and bacteria, i.e., they describe interactions that occur in the mycorrhizosphere (8, 12). Some studies have addressed the role of soil bacteria in the establishment of the symbiotic relationship between plants and mycorrhizal fungi (2, 12, 42). In contrast, not much is known about the putative interactions of bacteria with the dense hyphal network underneath fungal fruiting bodies that constitutes the mycosphere. Moreover, the selection of bacteria by fungi in soil is being elaborately addressed only recently (19), and most studies have focused on the culturable bacterial fraction, thus overlooking the as-yet-unculturable organisms of soil (3, 27). Warmink and van Elsas (43) described a clear shift in the soil bacterial community in the Laccaria proxima mycosphere, indicating the selection of specific bacteria by this ectomycorrhizal fungus. Warmink et al. (44) extended this work to the mycospheres of other basidiomycetous fungi and found that the mycospheres of these fungi indeed exerted a selective effect on both the total bacterial community and the Pseudomonas community.
We show here, on the basis of the results of a molecular analysis, that specific members of the family Sphingomonadaceae are selected in the mycospheres of various fungi, indicating a clear mycosphere effect. Of the two fungal species chosen for further analysis, L. proxima and R. exalbicans, the mycosphere effect was most prominent in the latter. Presumably, the hyphae of R. exalbicans influence the surrounding soil more strongly than those of L. proxima in terms of providing favorable conditions for specific members of the Sphingomonadaceae.
The DGGE community analyses showed that replicate fungi largely select for similar Sphingomonadaceae types, as the patterns of replicate mycospheres were very similar. This was especially striking for the N-soil samples. Not only were the fungal fruiting bodies in this soil sampled at least 50 m apart, but the bulk soil also revealed various patterns, indicating different Sphingomonadaceae communities. Thus, even though the Sphingomonadaceae in the bulk soil varied, R. exalbicans still selected the same or similar types from this family. Moreover, L. proxima also selected similar Sphingomonadaceae types, in this case in a soil with similar Sphingomonadaceae community structures. These findings indicate a possibly specific bacterial capability that allows their successful establishment in the specific niche. Mycorrhizal fungi may even select sphingomonads with particular capabilities, such as mineral weathering, as Uroz et al. (40) recently described the selection of such a Sphingomonas type by Scleroderma citrinum. The fungus-responsive members of the Sphingomonadaceae presumably use particular fungus-exuded compounds as carbon sources (19) by, for instance, extracellular biotrophy or necrotrophy, and the organisms identified by us likely utilize either strategy. Multiple studies have described the breadth of carbon source utilization by several Sphingomonadaceae types (5, 26), and it is very likely that some of the compounds used occur in the fungal species studied here.
Strikingly, sequences of DGGE bands from patterns from similar habitats consistently showed the same top hit in the RDP database (Table 1), even though different migration in DGGE was observed. Minor variation in the retrieved sequences may explain the different band positions. The DGGE band results corroborated the clone library analysis results, as sequences from the same habitat clustered closely together. As in the DGGE analysis, separation between bulk soil and mycosphere was clearest in the clone library analysis of the N-soil samples with R. exalbicans, again indicating strong selection in the R. exalbicans mycosphere. Mycosphere cluster Myco-NI, which included DGGE band 7, even contained 33 sequences, suggesting clonal selection and possibly outgrowth and an important role for the underlying organism in the R. exalbicans mycosphere. The next largest group, Myco-NIII, containing seven clones, clustered with Sphingomonas sp. strain EC-K085, which was recently reported (25) to promote the growth of rhizosphere-inhabiting Frateuria spp. Strikingly, sequences of bands from bulk soil and most clones from the G-soil and N-soil (bulk soil) clone libraries all cluster with sequences from the same German forest, hinting that Sphingomonadaceae communities from bulk soils might be similar in different locations.
Interestingly, the rarefaction curves showed a typical selective effect of the mycospheres of L. proxima and R. exalbicans for Sphingomonadaceae in that reduced richness of Sphingomonadaceae was noted in both mycospheres compared to the corresponding bulk soils. However, from the curves, the diversity of the Sphingomonadaceae was predicted to be considerably higher than described here, a common observation in soil microbial diversity assessments (13, 39).
In summary, in this report, a clear selective process on specific Sphingomonadaceae in the mycospheres of both R. exalbicans and L. proxima was described. Furthermore, different fungi were found to select for different members of the Sphingomonadaceae, but within fungal species, similar types were selected even in different bulk soil backgrounds.
In the future, the specific role of the Sphingomonadaceae community in the mycosphere should be better characterized, clarifying whether specific members of the Sphingomonadaceae capture particular nutrients, take possession of colonization sites, or can even establish a mutualistic relationship with the fungal partner. Our group is currently actively involved in such studies, which are partially performed in soil microcosms with model fungi.
Supplementary Material
Acknowledgments
Fernando Dini Andreote received a grant from the Soil Biotechnology Foundation.
Footnotes
Published ahead of print on 30 January 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adkins, A. 1999. Degradation of the phenoxy acid herbicide diclofop-methyl by Sphingomonas paucimobilis isolated from a Canadian prairie soil. J. Ind. Microbiol. Biotechnol. 23:332-335. [DOI] [PubMed] [Google Scholar]
- 2.Babana, A. H., and H. Antoun. 2006. Effect of Tilemsi phosphate rock-solubilizing microorganisms on phosphorus uptake and yield of field-grown wheat (Triticum aestivum L.) in Mali. Plant Soil 287:51-58. [Google Scholar]
- 3.Bending, G. D., E. J. Poole, J. M. Whipps, and D. J. Read. 2002. Characterisation of bacteria from Pinus sylvestris-Suillus luteus mycorrhizas and their effects on root-fungus interactions and plant growth. FEMS Microbiol. Ecol. 39:219-227. [DOI] [PubMed] [Google Scholar]
- 4.Boersma, F. G., W. C. McRoberts, S. L. Cobb, and C. D. Murphy. 2004. A 19F NMR study of fluorobenzoate biodegradation by Sphingomonas sp. HB-1. FEMS Microbiol. Lett. 237:355-361. [DOI] [PubMed] [Google Scholar]
- 5.Corvini, P. F., A. Schaffer, and D. Schlosser. 2006. Microbial degradation of nonylphenol and other alkylphenols—our evolving view. Appl. Microbiol. Biotechnol. 72:223-243. [DOI] [PubMed] [Google Scholar]
- 6.Costa, R., N. C. M. Gomes, R. S. Peixoto, N. Rumjanek, G. Berg, L. C. S. Mendonca-Hagler, and K. Smalla. 2006. Diversity and antagonistic potential of Pseudomonas spp. associated to the rhizosphere of maize grown in a subtropical organic farm. Soil Biol. Biochem. 38:2434-2447. [Google Scholar]
- 7.Daane, L. L., I. Harjono, G. J. Zylstra, and M. M. Häggblom. 2001. Isolation and characterization of polycyclic aromatic hydrocarbon-degrading bacteria associated with the rhizosphere of salt marsh plants. Appl. Environ. Microbiol. 67:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Boer, W., L. B. Folman, R. C. Summerbell, and L. Boddy. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29:795-811. [DOI] [PubMed] [Google Scholar]
- 9.Finlay, R. D. 2004. Mycorrhizal fungi and their multifunctional roles. Mycologist 18:91-96. [Google Scholar]
- 10.Fredrickson, J. K., D. L. Balkwill, M. F. Romine, and T. Shi. 1999. Ecology, physiology, and phylogeny of deep subsurface Sphingomonas sp. J. Ind. Microbiol. Biotechnol. 23:273-283. [DOI] [PubMed] [Google Scholar]
- 11.Frey, P., P. Frey-Klett, J. Garbaye, O. Berge, and T. Heulin. 1997. Metabolic and genotypic fingerprinting of fluorescent pseudomonads associated with the Douglas fir-Laccaria bicolor mycorrhizosphere. Appl. Environ. Microbiol. 63:1852-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey-Klett, P., J. Garbaye, and M. Tarkka. 2007. The mycorrhiza helper bacteria revisited. New Phytol. 176:22-36. [DOI] [PubMed] [Google Scholar]
- 13.Gans, J., M. Wolinsky, and J. Dunbar. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387-1390. [DOI] [PubMed] [Google Scholar]
- 14.Garbaye, J. 1994. Helper bacteria—a new dimension to the mycorrhizal symbiosis. New Phytol. 128:197-210. [DOI] [PubMed] [Google Scholar]
- 15.Heuer, H., J. Wieland, J. Schönfeld, A. Schönwälder, N. C. M. Gomes, and K. Smalla. 2001. Bacterial community profiling using DGGE or TGGE analysis, p. 177-190. In P. Rouchelle (ed.), Environmental molecular microbiology: protocols and applications. Horizon Scientific Press, Wymondham, United Kingdom.
- 16.Johansson, J. F., L. R. Paul, and R. D. Finlay. 2004. Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol. Ecol. 48:1-13. [DOI] [PubMed] [Google Scholar]
- 17.Keck, A., D. Conradt, A. Mahler, A. Stolz, R. Mattes, and J. Klein. 2006. Identification and functional analysis of the genes for naphthalenesulfonate catabolism by Sphingomonas xenophaga BN6. Microbiology 152:1929-1940. [DOI] [PubMed] [Google Scholar]
- 18.Lane, J. D. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 19.Leveau, J. H., and G. M. Preston. 2008. Bacterial mycophagy: definition and diagnosis of a unique bacterial-fungal interaction. New Phytol. 177:859-876. [DOI] [PubMed] [Google Scholar]
- 20.Leys, N. M., A. Ryngaert, L. Bastiaens, W. Verstraete, E. M. Top, and D. Springael. 2004. Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:1944-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matilla, M. A., M. Espinosa-Urgel, J. J. Rodriguez-Herva, J. L. Ramos, and M. I. Ramos-Gonzalez. 2007. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 8:R179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan, J. A., G. D. Bending, and P. J. White. 2005. Biological costs and benefits to plant-microbe interactions in the rhizosphere. J. Exp. Bot. 56:1729-1739. [DOI] [PubMed] [Google Scholar]
- 24.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogita, N., Y. Hashidoko, S. H. Limin, and S. Tahara. 2006. Linear 3-hydroxybutyrate tetramer (HB4) produced by Sphingomonas sp. is characterized as a growth promoting factor for some rhizomicrofloral composers. Biosci. Biotechnol. Biochem. 70:2325-2329. [DOI] [PubMed] [Google Scholar]
- 26.Pinyakong, O., H. Habe, and T. Omori. 2003. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs). J. Gen. Appl. Microbiol. 49:1-19. [DOI] [PubMed] [Google Scholar]
- 27.Rangel-Castro, J. I., J. J. Levenfors, and E. Danell. 2002. Physiological and genetic characterization of fluorescent Pseudomonas associated with Cantharellus cibarius. Can. J. Microbiol. 48:739-748. [DOI] [PubMed] [Google Scholar]
- 28.Sahin, N. 2003. Oxalotrophic bacteria. Res. Microbiol. 154:399-407. [DOI] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon, C. E. 1997. The mathematical theory of communication. 1963. MD Comput. 14:306-317. [PubMed] [Google Scholar]
- 32.Shintani, M., M. Urata, K. Inoue, K. Eto, H. Habe, T. Omori, H. Yamane, and H. Nojiri. 2007. The Sphingomonas plasmid pCAR3 is involved in complete mineralization of carbazole. J. Bacteriol. 189:2007-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorensen, J. 1997. Rhizosphere as a habitat for soil microorganisms, p. 21-45. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, Inc., New York, NY.
- 35.Standing, D., and K. Killham. 2007. The soil environment, p. 1-22. In J. D. van Elsas, J. K. Jansson, and J. T. Trevors (ed.), Modern soil microbiology. CRC Press, New York, NY.
- 36.Tabata, K., K.-I. Kasuya, H. Abe, K. Masuda, and Y. Doi. 1999. Poly(aspartic acid) degradation by a Sphingomonas sp. isolated from freshwater. Appl. Environ. Microbiol. 65:4268-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 38.Tiirola, M. A., M. K. Mannisto, J. A. Puhakka, and M. S. Kulomaa. 2002. Isolation and characterization of Novosphingobium sp. strain MT1, a dominant polychlorophenol-degrading strain in a groundwater bioremediation system. Appl. Environ. Microbiol. 68:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torsvik, V., L. Ovreas, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 40.Uroz, S., C. Calvaruso, M. P. Turpault, J. C. Pierrat, C. Mustin, and P. Frey-Klett. 2007. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl. Environ. Microbiol. 73:3019-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Elsas, J. D., V. Torsvik, A. Hartmann, L. Ovreas, and J. K. Jansson. 2007. The Bacteria and Archaea in soil, p. 83-104. In J. D. van Elsas, J. K. Jansson, and J. T. Trevors (ed.), Modern soil microbiology. CRC Press, New York, NY.
- 42.Vivas, A., J. M. Barea, and R. Azcon. 2005. Interactive effect of Brevibacillus brevis and Glomus mosseae, both isolated from Cd contaminated soil, on plant growth, physiological mycorrhizal fungal characteristics and soil enzymatic activities in Cd polluted soil. Environ. Pollut. 134:257-266. [DOI] [PubMed] [Google Scholar]
- 43.Warmink, J., and J. D. van Elsas. 2008. Selection of bacterial populations in the mycosphere of Laccaria proxima: is type III secretion involved? ISME J. 2:887-900. [DOI] [PubMed] [Google Scholar]
- 44.Warmink, J. A., R. Nazir, and J. D. van Elsas. 2009. Universal and species-specific bacterial ‘fungiphiles’ in the mycospheres of different basidiomycetous fungi. Environ. Microbiol. 11:300-312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.