Abstract
Aphids are widespread agricultural pests that are capable of disseminating plant viral diseases; however, despite coming into frequent contact with epiphytic bacteria, aphids are considered to have no role in bacterial transmission. Here, we demonstrate the ability of pea aphids to vector the phytopathogen Pseudomonas syringae pv. syringae B728a (PsyB728a). While feeding on plants colonized by epiphytic bacteria, aphids acquire the bacteria, which colonize the digestive tract, multiply, and are excreted in the aphid honeydew, resulting in inoculation of the phyllosphere with up to 107 phytopathogenic bacteria per cm2. Within days of ingesting bacteria, aphids succumb to bacterial sepsis, indicating that aphids serve as an alternative, nonplant host for PsyB728a. The related strain Pseudomonas syringae pv. tomato DC3000 is >1,000-fold less virulent than PsyB728a in the pea aphid, suggesting that PsyB728a possesses strain-specific pathogenicity factors that allow it to exploit aphids as hosts. To identify these factors, we performed a mutagenesis screen and recovered PsyB728a mutants that were hypovirulent, including one defective in a gene required for flagellum formation and motility. These interactions illustrate that aphids can also vector bacterial pathogens and that even seemingly host-restricted pathogens can have alternative host specificities and lifestyles.
Aphids are prolific insect pests that impact numerous agricultural crop plants (8). Although direct parasitization of their hosts can reduce plant biomass, crop quality, and crop yield, aphids are of particular importance for their role in the dissemination of plant disease (24, 30). Phytopathogenic viruses can be acquired by aphids as they probe infected plant hosts with their stylet (29, 30), with the subsequent movement of infected aphids to healthy plants contributing significantly to the dissemination of plant disease (29). Surprisingly, however, aphids are considered strictly vectors of phytopathogenic viruses, despite their frequent physical contact with a variety of bacterial epiphytes (10).
Upon landing on a potential host, the aphid begins to explore the plant, probing the plant tissues with its stylet and puncturing through the epidermal layers to locate and access the phloem sieve tube elements to feed (3, 18, 24). As it traverses the plant surface, the aphid continually evaluates the suitability of the host, with some aphid species sampling plant fluids with droplets of saliva that are expelled and reingested (3). This saliva can contact the plant surface where many bacterial epiphytes reside, such as the ubiquitous phytopathogens Pantoea agglomerans and Pseudomonas syringae (4, 5, 10, 21, 22, 32).
The pea aphid, Acyrthosiphon pisum, feeds on a variety of agriculturally relevant crop plants in the Fabaceae, including soybean (Glycine max), fava/broad bean (Vicia faba), pea (Pisum sativum), and snap bean (Phaseolus vulgaris) (15, 28, 34-36), and has been shown to vector bean yellow mosaic virus between these hosts (15, 34-36). Several of these plants are also primary hosts for the highly aggressive bean pathogen Pseudomonas syringae pv. syringae B728a (PsyB728a), which establishes epiphytically and can invade the plant and cause disease (4, 5, 17). Although PsyB728a generally exhibits low survivorship on plant surfaces due to limitations of carbohydrates and water (23), it can form aggregates on plant surfaces, which can be dispersed further by rain splash (16, 17, 26, 37). Ultimately, under optimal environmental conditions, high titers of bacteria increase the likelihood of plant disease (17).
In this study, we describe the vectoring of the phytopathogenic bacterium P. syringae by the pea aphid. We illustrate infection of the aphid by epiphytic populations of PsyB728a, which multiply in the aphid and are deposited onto the plant surface in high titer via the aphid honeydew. In addition, we provide an account of P. syringae infecting a nonplant host and identify a PsyB728a mutant that is hypovirulent in A. pisum.
MATERIALS AND METHODS
Aphid lines, bacterial strains, and growth conditions.
Isolates of Pseudomonas syringae were grown overnight with continuous shaking in 5 ml LB medium at 25°C. Escherichia coli DH5a and RK600 were grown in 5 ml LB medium at 37°C. When applicable, antibiotics were supplemented at the following concentrations: chloramphenicol, 64 μg/ml; kanamycin, 50 μg/ml; rifampin, 50 μg/ml; and ampicillin, 150 μg/ml. The pea aphid, Acyrthosiphon pisum strain 5A, was maintained at 20°C on fava bean plants. For screening and growth assays, a modified aphid artificial diet containing essential amino acids, vitamins, and sucrose was used (11). Fava bean plants were grown by sowing fava bean seeds in a 1:1 ratio of Sunshine Mix to vermiculite. Approximately 11-day-old seedlings were used in all assays.
Transposon mutagenesis and screening.
Transposon mutagenesis of PsyB728a was performed via triparental mating, whereby a helper strain containing a self-transmissible plasmid assists in mobilizing a plasmid from a donor to the recipient strain. The mini-Tn5 vector pBSL118 in E. coli VPE42 (carrying kanamycin and ampicillin resistance) was used as the donor, E. coli RK600 (chloramphenicol resistance) was used as the helper, and PsyB728a was the intended recipient. The three parental strains were combined in a 1:1:1 ratio, spotted onto LB medium without antibiotics, and incubated overnight at 28°C to 30°C. Bacterial lawns were resuspended in 10 mM MgSO4 and serial dilutions plated onto LB medium containing rifampin and kanamycin. Single transconjugants were dipped into individual wells of a 96-well microtiter plate, each of which contained 350 μl of artificial aphid diet. Positive (wild-type PsyB728a) and negative (no bacteria) controls were included on each plate. Plates were sealed with parafilm, allowing the aphid diet in each chamber to contact the parafilm. These plates were each placed on top of a second plate that contained a single second or third aphid instar in each well so that the wells aligned, allowing the aphids to feed on diet containing a single transconjugant. Plates were kept at ambient temperature and scored after 3 to 4 days. Transconjugants were recovered from those wells in which the aphid survived longer than the wild-type control. Positives were then confirmed to be P. syringae via both blue/white screening and PCR and retested eight times. Approximately 1,500 mutants were screened.
Mutated genes were identified by inverse PCR (31). Briefly, genomic DNA from each mutant was extracted using the Gentra Puregene kit (Qiagen, CA) according to manufacturer's instructions. Between 1 and 2 μg of genomic DNA was digested using HincII and EcoRI in a 20-μl volume. After heat inactivation, 10 μl of each digest was used in a unimolecular ligation, consisting of 15 units of T4 DNA ligase (Fermentas, MD) in a final volume of 200 μl. After overnight incubation at 16°C, the ligation was purified using the Qiagen PCR purification kit (Qiagen, CA), eluting in a final volume of 30 μl. Individual PCRs were performed on 2, 5, and 10 μl of the purified product, using primers npt-41 (5′-AGCCGAATAGCCTCTCCACCCAAG-3′) and npt+772 (5′-TTCGCAGCGCATCGCCTTCTATC-3′).
Site-directed mutagenesis and homologous integration.
Genes targeted for disruption included YP_237273.1 (Psy4205), a 7,509-bp gene that has homology to the TcdA1 toxin complex gene of Photorhabdus luminescens (AAF05542.1) but which lacks any major protein domains, and YP_237102.1 (Psy4034), a 2,946-bp gene with homology to the TccA3 toxin complex gene of Photorhabdus luminescens (NP_928150.1), which also lacks any informative protein folds or domains. A trp terminator was introduced into pBluescript (Stratagene, CA) by the ligation of oligonucleotides 5′-AATTGAGCCCGCCTAATGAGCGGGCTTTTTTTTG-3′ and 5′-AATTCAAAAAAAAGCCCGCTCATTAGGCGGGCTC-3′ into the EcoRI restriction site, creating pBluescript-trp. A 500-bp portion of Psy4034 was amplified from P. syringae pv. syringae B728a by using primers Psy4034+1 (5′-ATGACCGAGCAACCCTTCTCCC-3′) and Psy4034-568 (5′-GATTGACGATGTGCAGCGTAGGC-3′) and was subsequently ligated into pBluescript-trp. A fragment of Psy4205 was amplified using Psy4205+64 (5′-GCTGCACAGGCGTTGATATCCC-3′) and Psy4205-549 (5′-CGTTACATCCAGCTTCAGCGACTCTC-3′) and was also ligated into pBluescript-trp. Both constructs were introduced into P. syringae pv. syringae B728a independently by electroporation in 0.2-cm cuvettes, using a MicroPulser electroporator (Bio-Rad) with a setting of 2.5 kV.
Bacterial growth assays.
Second- and third-instar aphids were allowed to feed for 14 h on aphid artificial diet containing 107 CFU/ml of either P. syringae pv. syringae B728a or P. syringae pv. syringae DC3000, after which aphids were transferred to fresh diet. Three aphids were sampled at 12-hour intervals and macerated in 100 μl of 10 mM MgSO4, and dilutions were plated onto LB containing rifampin. Growth assays were performed three times.
Vectoring experiment.
Second and third instars were allowed to feed for 14 h on artificial diet containing 107 CFU/ml of P. syringae pv. syringae B728a. Up to 20 aphids were transferred to fava bean plants and allowed to incubate for 7 days at 24°C under ambient light. After 7 days, leaves were harvested and washed in 10 mM MgSO4, and the wash was then plated onto LB containing rifampin. Colony counts were performed after 24 to 48 h. Leaf surface area was calculated by tracing the leaves on grid paper. Experiments were performed twice, with at least five plants used per experiment.
Aphid infection from epiphytic bacterial populations.
Fava bean leaves and stems were brushed with PsyB728a and resuspended in 10 mM MgSO4 (optical density at 600 nm of 1.8), and second and third instars of A. pisum 5A were introduced at the base of the plant. After 24 h under ambient light, all aphids on the plant were collected, and each was placed into an individual 1.5-ml Microfuge tube containing 100 μl of 10 mM MgSO4. Surface bacteria were washed from the aphid by agitation, and 10 μl of the wash was plated onto LB plates containing rifampin. Each aphid was then macerated with a pestle in the remaining 90 μl, and 10 μl of the homogenate was plated onto LB containing rifampin. Bacterial growth was examined after 24 to 36 h. Aphids were scored as being infected if bacteria were completely absent from the wash, but at least 10 CFU were present on plates containing the homogenate. Experiments were repeated twice, each with 30 to 80 aphids screened from each of three plants.
Swimming and swarming assays.
Swimming and swarming assays of P. syringae strains were conducted by inoculating 5 μl of an overnight culture (optical density at 600 nm of >1) into the center of 0.2% to 0.6% LB agar plates and incubating at room temperature (25°C) overnight.
RESULTS
Infected aphid honeydew inoculates plant surfaces.
To determine if aphids become infected from plants colonized epiphytically with PsyB728a, healthy aphids were introduced onto fava bean plants that were surface inoculated with PsyB728a. Recovered aphids surveyed for the presence of ingested bacteria revealed an infection rate of 15%, with infection rates of up to 29% in some experiments. As a result of these studies, we found that aphid honeydew can contain high titers of bacterial inoculum, as assayed by a plating method. Because of this, we postulated that infected aphids contribute to the inoculation of plants with PsyB728a. To quantify the relative size of the PsyB728a inoculum that can be introduced through aphid honeydew onto the leaf surfaces, infected aphids were transferred to healthy fava bean plants, and leaves were assayed for the presence of bacteria. Leaves contained bacterial titers averaging 3.6 × 108 CFU per leaf (4.5 × 107 CFU/cm2), with some leaves harboring titers of up to 1.6 × 109 CFU (1.6 × 108 CFU/cm2). Each aphid was responsible for depositing, on average, 2 × 107 CFU on leaves over the duration of the experiment.
PsyB728a, but not PtoDC3000, is pathogenic to the pea aphid.
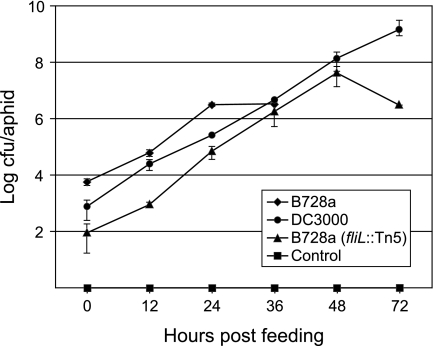
We conducted pathogenicity assays with the bean pathogen PsyB728a and the tomato pathogen Pseudomonas syringae pv. tomato DC3000 (PtoDC3000) to determine if entomopathogenicity is either strain or species specific. After feeding on an artificial diet supplemented with bacteria, the PsyB728a titer in aphids was approximately 6 × 103 CFU/aphid, increasing to 3 × 106 CFU/aphid after 24 h (Fig. 1). Titers remained at this level for 36 h, after which mortality set in, with only 5 to 10% of aphids surviving 48 h. By comparison, titers of PtoDC3000 at transfer were 8 × 102 CFU/aphid, which increased exponentially to 1 × 109 CFU/aphid by 72 h, exceeding the lethal PsyB728a concentration of 3 × 106 CFU/aphid (Fig. 1).
FIG. 1.
Growth of bacteria in the aphid. Titers of aphid-pathogenic P. syringae pv. syringae B728a, aphid-nonpathogenic P. syringae pv. tomato DC3000, and the fliL mutant in the pea aphid following an initial feeding for 14 h on artificial diet containing bacteria. All aphids feeding on wild-type P. syringae pv. syringae B728a were dead within 36 h. Control aphids were fed 10 mM MgSO4.
Putative insecticidal genes of PsyB728a are not responsible for virulence in pea aphids.
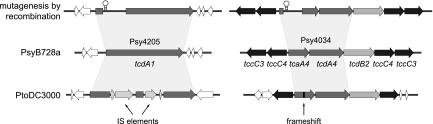
Interspecific genomic analysis of PsyB728a revealed the presence of several conserved genes implicated in entomotoxicity in other systems. These include homologs of the Photorhabdus luminescens toxin complex genes, of which PsyB728a retains a cluster containing at least one homolog of tcaA, tccC, tcdA, and tcdB (Fig. 2). A genomic comparison of PsyB728a and PtoDC3000 exposed disruptions or deletions of some of these genes in PtoDC3000 that remained intact in PsyB728a (Fig. 2), possibly accounting for the observed differences in entomopathogenicity in the aphid pathosystem. To test the involvement of these genes in pathogenicity, we disrupted the open reading frames of Psy4034 and Psy4205 in PsyB728a independently by single-crossover homologous recombination (Fig. 2); however, each mutant retained full virulence, as determined by a qualitative lethality assay (data not shown).
FIG. 2.
Genomic comparison of toxin complex genes between aphid-pathogenic P. syringae pv. syringae B728a and aphid-nonpathogenic P. syringae pv. tomato DC3000. In P. syringae pv. tomato DC3000, the Psy4205 homolog is disrupted by two IS elements, whereas the Psy4034 homolog contains a frameshift mutation. Gene knockouts of Psy4205 and Psy4034 in P. syringae pv. syringae B728a were achieved through homologous recombination of a gene fragment with a trp terminator (stem-loop structure). The corresponding Photorhabdus luminescens W14 homologs are indicated below each gene.
A PsyB728a fliL mutant is attenuated in virulence.
A transposon-mutagenesis screen designed to recover genetic loci contributing to PsyB728a pathogenicity in the pea aphid identified one hypovirulent mutant that was disrupted in the flagellar basal body-associated protein, FliL. A qualitative pathogenicity assay of this mutant indicated significantly reduced virulence, with aphids surviving for up to 120 h (data not shown), unlike wild-type PsyB728a, which caused almost complete aphid mortality in 36 h.
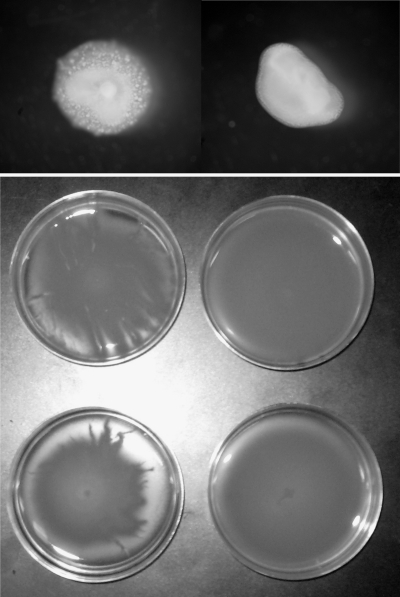
To determine the relative growth rate of this mutant during aphid colonization, we assayed bacterial titers over a 72-hour period. The PsyB728a (fliL::Tn5) mutant began at lower titers at transfer (<102 CFU/aphid) but grew exponentially, reaching titers of 4 × 107 CFU/aphid by 48 h, which fell to 3 × 106 CFU/aphid by 72 h. The fliL mutant was able to reach and surpass the lethal dose of the wild-type PsyB728a concentration of 3 × 106 CFU/aphid, closely resembling the growth curve of the avirulent PtoDC3000 (Fig. 1). Given that the fliL mutant bacteria were able to achieve even higher densities than wild-type PsyB728a and given the prominent role of fliL in bacterial motility, we decided to evaluate swimming and swarming proficiencies. Swarming assays of PtoDC3000, PsyB728a, and PsyB728a (fliL::Tn5) revealed that mutation in the fliL gene reduced swimming ability (Fig. 3, top panel) and eliminated swarming ability (Fig. 3, bottom panel). Wild-type PtoDC3000, which has been shown to be capable of swarming (7, 9), also exhibited swarming proficiency in our experiments (data not shown).
FIG. 3.
Motility assays of fliL mutant. Colony morphology of wild-type P. syringae pv. syringae B728a and fliL mutant on 0.3% LB agar (top panel). Comparison of swarming abilities of wild-type and fliL mutant on 0.4% LB agar (bottom panel). Tentacle-like extensions projecting outward from the growing edge of colonies were seen only on plates containing wild-type B728a.
DISCUSSION
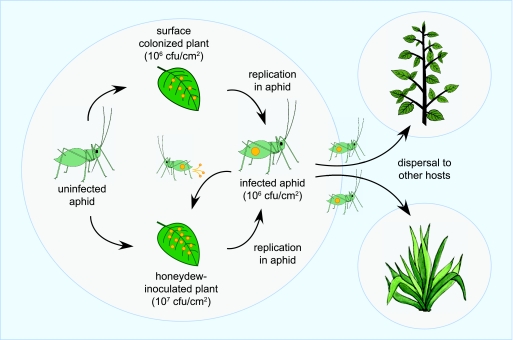
The current model of P. syringae dissemination onto and between host plants involves rain splash-mediated inoculation from infected to uninfected plants, facilitated largely by the aggressive epiphytic and aggregation capabilities of P. syringae (17, 25, 27). We have extended this model by showing that aphids contribute directly to pathogen amplification and inoculation of the plant phyllosphere, following aphid infection through contact with epiphytic populations of PsyB728a (Fig. 4). Transient ingestion of surface bacteria by aphids likely occurs during stylet-mediated plant host probing, which is consistent with the observation that aphids probe candidate host plants repeatedly during initial colonization (3, 18). Ingested bacteria migrate through the aphid digestive tract, establish in the gut (our unpublished data), multiply, and are eventually deposited onto leaves in the aphid honeydew. As the bacterial load in the aphid approaches lethal titers, aphids stop feeding and begin to wander, continually depositing infected honeydew over plant surfaces in the process. Moreover, because aphid honeydew is rich in carbohydrate, deposited bacteria are effectively suspended in a nutrient-laden droplet that may enhance their survival on the leaf surface. Epiphytic bacterial populations typically have high mortality in the phyllosphere, with 75% of the epiphytic population becoming nonviable after 80 h (38, 39); thus, the deposition in a carbohydrate-rich droplet suspension likely contributes to the persistence of the bacteria on the leaf surface and is also consistent with previous findings of sugar availability in the phyllosphere enhancing bacterial colonization of host plants (23).
FIG. 4.
Model of aphid-mediated dissemination of P. syringae. Aphids may become infected after feeding on plant tissues colonized with epiphytic populations of P. syringae (yellow dots). As bacterial titers in infected aphids increase, droplets of honeydew laden with bacteria inoculate the plant surface, with movement of aphids to other hosts resulting in the dissemination of P. syringae and plant disease.
The dispersal of P. syringae by an insect vector is consistent not only with its epiphytic life stages and likelihood of encountering insects in a foliar ecosystem but also with the presence of many genetic loci commonly attributed to insect association and entomopathogenicity in P. syringae genomes (20). Although many of these attributes are present in other pseudomonads, only one other documented case of insect-mediated dispersal of a Pseudomonas species has been documented. The rhizobacterium Pseudomonas chlororaphis is vectored by the spotted cucumber beetle, Diabrotica undecimpunctata, following infection through feeding on infected roots or foliage (33). Other reports highlight indirect dispersal of pseudomonads, in which the insect does not serve as host but merely carries the bacteria externally (17, 19). In the case of the P. syringae-aphid interaction, the bacteria not only are internalized but also can replicate in their insect vector prior to dispersal.
Although typically considered a dedicated plant pathogen, PsyB728a clearly exploits the aphid as a secondary host, since it can support pathogen replication, escape, and dispersal. The drastic difference between the pathogenic abilities of PtoDC3000 and PsyB728a in this pathosystem suggests strain-specific pathogenicity determinants that enable PsyB728a to colonize the pea aphid. This difference is not due to variability in survival or growth rates between PsyB728a and PtoDC3000 in artificial diet or to a greater propensity for wild-type PsyB728a to be ingested during the initial feeding. In fact, similar initial titers and growth rates were observed for both strains, yet aphids were unaffected by titers of PtoDC3000 that were lethal with PsyB728a.
We had predicted based on genomic comparisons that the toxin complex genes, whose homologs are known to contribute to Photorhabdus luminescens entomopathogenicity, might be at least partly responsible for the observed differences in virulence phenotype (12); however, these genes appear to have no role in this pathosystem. Instead, the isolation and characterization of a hypovirulent fliL mutant suggest an active virulence mechanism that is linked to bacterial motility and, more specifically, bacterial swarming. Because fliL is the leading gene of the FliL operon, transposon disruption may have had polar effects, impacting the proper assembly of the flagellum. The PsyB728a fliL mutant was slightly defective in swimming ability and completely defective in swarming ability, consistent with observations of both Salmonella and E. coli fliL mutants (2); however, swarming alone was not sufficient for virulence, as wild-type PtoDC3000, which is swarming proficient, is avirulent in the aphid. This suggests that fliL and the associated swarming phenotype have a role in the regulation of other PsyB728a-specific virulence factors. In Proteus mirabilis, fliL is responsible for inducing swarmer cell differentiation and regulating virulence gene expression, including hemolysin, urease, and protease (1, 6), whereas swarming in Vibrio cholerae is linked to the induction of a variety of virulence-associated factors, including pilus formation, and the production of hemagglutinin, hemolysins, adhesins, and cholera toxin (13). Thus, in P. syringae, the differentiation of vegetative cells into swarm cells may induce virulence gene expression that contributes to aphid colonization and death. The pathogenicity of Dickeya dadantii toward the pea aphid is mediated by Cyt-like toxin genes (14); however, homologs of these genes are not present in P. syringae, suggesting the involvement of other, as-yet-unidentified, strain-specific virulence factors. The identity of such factors could prove useful for the development of effective pest control strategies.
Pathogens often reach a virulence optimum that maximizes both their aggressiveness and transmission potential. Because a highly aggressive pathogen may kill its host before it has an opportunity to disperse, natural selection will favor a reduction in aggressiveness to increase host survivorship and enhance pathogen dissemination (40). In the case of the interaction between P. syringae and the pea aphid, pathogen aggressiveness appears to be extremely high, such that infection results in rapid aphid mortality; however, because P. syringae has a direct and continual escape route from its host, it can exploit the aphid maximally by replicating rapidly without the tradeoff of host survivorship. Thus, the interaction between the phytopathogenic/entomopathogenic PsyB728a and the pea aphid represents a novel host-pathogen relationship in which the pathogen has achieved a virulence optimum that is less dependent on host survivorship and which reduces competition for resources while simultaneously enhancing pathogen dispersal and fitness in a foliar environment.
Acknowledgments
We thank Alexander No for technical assistance, Steven Lindow for providing the sequenced isolate of Pseudomonas syringae pv. syringae B728a, and Lori Burrows for advice on swimming and swarming assays. We thank Nancy Moran for support and advice throughout this work and Becky Nankivell for technical assistance.
This work was supported by NIH grant GM56120 to Howard Ochman. John Stavrinides is supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Allison, C., H. C. Lai, and C. Hughes. 1992. Coordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6:1583-1591. [DOI] [PubMed] [Google Scholar]
- 2.Attmannspacher, U., B. E. Scharf, and R. M. Harshey. 2008. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol. Microbiol. 68:328-341. [DOI] [PubMed] [Google Scholar]
- 3.Auclair, J. L. 1963. Aphid feeding and nutrition. Annu. Rev. Entomol. 8:439-490. [Google Scholar]
- 4.Beattie, G. A., and S. E. Lindow. 1999. Bacterial colonization of leaves: a spectrum of strategies. Phytopathology 89:353-359. [DOI] [PubMed] [Google Scholar]
- 5.Beattie, G. A., and S. E. Lindow. 1995. The secret life of foliar bacterial pathogens on leaves. Annu. Rev. Phytopathol. 33:145-172. [DOI] [PubMed] [Google Scholar]
- 6.Belas, R., and R. Suvanasuthi. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J. Bacteriol. 187:6789-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berti, A. D., N. J. Greve, Q. H. Christensen, and M. G. Thomas. 2007. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 189:6312-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackman, R. L., and V. F. Eastop. 2000. Aphids on the world's crops: an identification and information guide, 2nd ed. J. Wiley, London, England.
- 9.Chatterjee, A., Y. Y. Cui, H. L. Yang, A. Collmer, J. R. Alfano, and A. K. Chatterjee. 2003. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant-Microbe Interact. 16:1106-1117. [DOI] [PubMed] [Google Scholar]
- 10.Danhorn, T., and C. Fuqua. 2007. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61:401-422. [DOI] [PubMed] [Google Scholar]
- 11.Febvay, G., B. Delobel, and Y. Rhabe. 1988. Influence of amino acid balance on the improvement of an artificial diet for a biotype of Acyrthosiphon pisum (Homoptera: Aphididae). Can. J. Zool. 66:2449-2453. [Google Scholar]
- 12.ffrench-Constant, R. H., and N. R. Waterfield. 2006. Ground control for insect pests. Nat. Biotechnol. 24:660-661. [DOI] [PubMed] [Google Scholar]
- 13.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grenier, A. M., G. Duport, S. Pages, G. Condemine, and Y. Rahbe. 2006. The phytopathogen Dickeya dadantii (Erwinia chrysanthemi 3937) is a pathogen of the pea aphid. Appl. Environ. Microbiol. 72:1956-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hampton, R. O., A. Jensen, and G. T. Hagel. 2005. Attributes of bean yellow mosaic potyvirus transmission from clover to snap beans by four species of aphids (Homoptera: Aphididae). J. Econ. Entomol. 98:1816-1823. [DOI] [PubMed] [Google Scholar]
- 16.Hirano, S. S., L. S. Baker, and C. D. Upper. 1996. Raindrop momentum triggers growth of leaf-associated populations of Pseudomonas syringae on field-grown snap bean plants. Appl. Environ. Microbiol. 62:2560-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringaep—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy, J. S., and H. L. G. Stroyan. 1959. Biology of aphids. Annu. Rev. Entomol. 4:139-160. [Google Scholar]
- 19.Lilley, A. K., R. S. Hails, J. S. Cory, and M. J. Bailey. 1997. The dispersal and establishment of pseudomonad populations in the phyllosphere of sugar beet by phytophagous caterpillars. FEMS Microbiol. Ecol. 24:151-157. [Google Scholar]
- 20.Lindeberg, M., C. R. Myers, A. Collmer, and D. J. Schneider. 2008. Roadmap to new virulence determinants in Pseudomonas syringae: insights from comparative genomics and genome organization. Mol. Plant-Microbe Interact. 21:685-700. [DOI] [PubMed] [Google Scholar]
- 21.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindow, S. E., and J. H. Leveau. 2002. Phyllosphere microbiology. Curr. Opin. Biotechnol. 13:238-243. [DOI] [PubMed] [Google Scholar]
- 23.Mercier, J., and S. E. Lindow. 2000. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl. Environ. Microbiol. 66:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miles, P. W. 1999. Aphid saliva. Biol. Rev. 74:41-85. [Google Scholar]
- 25.Monier, J. M., and S. E. Lindow. 2003. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. USA 100:15977-15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monier, J. M., and S. E. Lindow. 2004. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 70:346-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monier, J. M., and S. E. Lindow. 2005. Spatial organization of dual-species bacterial aggregates on leaf surfaces. Appl. Environ. Microbiol. 71:5484-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nault, B. A., D. A. Shah, H. R. Dillard, and A. C. McFaul. 2004. Seasonal and spatial dynamics of alate aphid dispersal in snap bean fields in proximity to alfalfa and implications for virus management. Environ. Entomol. 33:1593-1601. [Google Scholar]
- 29.Ng, J. C. K., and B. W. Falk. 2006. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 44:183-212. [DOI] [PubMed] [Google Scholar]
- 30.Ng, J. C. K., and K. L. Perry. 2004. Transmission of plant viruses by aphid vectors. Mol. Plant Pathol. 5:505-511. [DOI] [PubMed] [Google Scholar]
- 31.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabaratnam, S., and G. A. Beattie. 2003. Differences between Pseudomonas syringae pv. syringae B728a and Pantoea agglomerans BRT98 in epiphytic and endophytic colonization of leaves. Appl. Environ. Microbiol. 69:1220-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder, W. E., D. W. Tonkyn, and D. Kluepfel. 1998. Insect-mediated dispersal of the rhizobacterium Pseudomonas chlororaphis. Phytopathology 88:1248-1254. [DOI] [PubMed] [Google Scholar]
- 34.Sohi, S. S., and K. G. Swenson. 1964. Pea aphid biotypes differing in bean yellow mosaic virus transmission. Entomol. Exp. Appl. 7:9-14. [Google Scholar]
- 35.Swenson, K. G. 1954. Aphid transmission of a bean yellow mosaic virus. J. Econ. Entomol. 47:1121-1123. [Google Scholar]
- 36.Swenson, K. G. 1957. Transmission of bean yellow mosaic virus by aphids. J. Econ. Entomol. 50:727-731. [Google Scholar]
- 37.Upper, C. D., S. S. Hirano, K. K. Dodd, and M. K. Clayton. 2003. Factors that affect spread of Pseudomonas syringae in the phyllosphere. Phytopathology 93:1082-1092. [DOI] [PubMed] [Google Scholar]
- 38.Wilson, M., and S. E. Lindow. 1994. Inoculum density-dependent mortality and colonization of the phyllosphere by Pseudomonas syringae. Appl. Environ. Microbiol. 60:2232-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, M., and S. E. Lindow. 1992. Relationship of total viable and culturable cells in epiphytic populations of Pseudomonas syringae. Appl. Environ. Microbiol. 58:3908-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woolhouse, M. E. J., L. H. Taylor, and D. T. Haydon. 2001. Population biology of multihost pathogens. Science 292:1109-1112. [DOI] [PubMed] [Google Scholar]