Abstract
The pathways responsible for cell wall polysaccharide biosynthesis are vital in eukaryotic microorganisms. The corresponding synthases are potential targets of inhibitors such as fungicides. Despite their fundamental and economical importance, most polysaccharide synthases are not well characterized, and their molecular mechanisms are poorly understood. With the example of Saprolegnia monoica as a model organism, we show that chitin and (1→3)-β-d-glucan synthases are located in detergent-resistant membrane microdomains (DRMs) in oomycetes, a phylum that comprises some of the most devastating microorganisms in the agriculture and aquaculture industries. Interestingly, no cellulose synthase activity was detected in the DRMs. The purified DRMs exhibited similar biochemical features as lipid rafts from animal, plant, and yeast cells, although they contained some species-specific lipids. This report sheds light on the lipid environment of the (1→3)-β-d-glucan and chitin synthases, as well as on the sterol biosynthetic pathways in oomycetes. The results presented here are consistent with a function of lipid rafts in cell polarization and as platforms for sorting specific sets of proteins targeted to the plasma membrane, such as carbohydrate synthases. The involvement of DRMs in the biosynthesis of major cell wall polysaccharides in eukaryotic microorganisms suggests a function of lipid rafts in hyphal morphogenesis and tip growth.
A number of oomycete species are plant or animal pathogens responsible for severe environmental damage and economic loss (44). For instance, members of the genus Phytophthora can infect woody plants in natural ecosystems (32), as well as a wide range of agriculturally important plants such as potato, tomato, and soybean (19). Some of the most devastating fish pathogens also belong to the oomycete phylum (69). A typical example is Saprolegnia parasitica, a microorganism encountered worldwide in freshwater habitats, where it represents a serious threat to natural populations of salmonids (30, 69). Losses in the aquaculture industry due to oomycetes are estimated at tens of millions of dollars annually (27). In the southeastern United States, catfish farmers have reported losses due to S. parasitica to be as high as 50% during winter (12). Until recently, saprolegniosis, the group of diseases caused by Saprolegniales, was contained by using malachite green. However, this substance has been classified as a class II health hazard and banned for all food applications due to its mutagenic and carcinogenic effects and because of the persistence of one of its metabolites, leucomalachite green, in fish tissues (57). A recrudescence of saprolegniosis is observed worldwide as a consequence of the absence of other efficient inhibitors of proliferation of oomycetes. Thus, the development of alternative strategies to tackle pathogenic oomycetes has become a priority for the aquaculture industry.
Biochemical pathways that are not present in the host fishes and that play a central role in such vital processes as the morphogenesis and growth of oomycetes are potential targets for novel specific inhibitors. Typical examples are the biosynthetic pathways of cell wall polysaccharides and their corresponding enzymes. The potential of targeting such enzymes is illustrated by the current use of efficient herbicides directed toward plant cellulose synthases (61). However, the design of efficient inhibitors specific for polysaccharide synthases depends on the characterization of the molecular mechanisms of these enzymes. A crucial step that needs to be achieved is the identification of all of the protein components of the synthase complexes. Of particular importance is the characterization of the (1→3)-β-d-glucan and cellulose synthases, which catalyze the formation of the most abundant cell wall polysaccharides in oomycetes. The isolation of these enzymes has been particularly challenging because of their location in the plasma membrane and their inherent instability upon extraction with detergents. Even in the case of (1→3)-β-d-glucan synthase, which is the most stable of these two enzymes, a portion only of the activity originally present in total membranes can be solubilized efficiently (16). In addition, while detergent-insoluble membranes retain a high glucan synthase activity, the activity recovered in detergent extracts usually drops within a few hours, thus complicating enzyme purification (16). Altogether, these observations suggest that the (1→3)-β-d-glucan synthase is essentially located in membrane structures that are resistant to detergents. Consistent with this hypothesis, we report here the isolation and characterization of detergent-resistant microdomains (DRMs) from the plasma membrane of the oomycete Saprolegnia monoica and biochemical evidence that these structures contain (1→3)-β-d-glucan and chitin synthases but no detectable cellulose synthase activity. Our report provides insight on the lipid environment of the (1→3)-β-d-glucan and chitin synthases in oomycetes and sheds light on the reason why it is difficult to obtain a complete solubilization of these enzymes in a stable and active form. Interestingly, the DRMs from S. monoica exhibit similar biochemical features as lipid rafts from animal (66), plant (6, 39, 49, 56), and yeast (34) cells. The involvement of raft-like structures in cell wall polysaccharide biosynthesis is a novel concept that suggests a function of rafts in mycelial morphogenesis and growth.
MATERIALS AND METHODS
Reagents.
UDP-d-[U-14C]glucose (∼300 mCi mmol−1) and UDP-N-acetyl-d-[U-14C]glucosamine (288 mCi mmol−1) were obtained from Perkin-Elmer. [U-13C]glucose was from Cambridge Isotope Laboratories. The Bradford reagent (8) was from Bio-Rad, and Dextran T500 from Amersham Biosciences. The (1→3)-β-d-glucanase from Trichoderma sp. was purchased from Megazyme, and the recombinant cellulases Cel 6A and Cel 6B from Thermobifida fusca expressed in Escherichia coli were a generous gift from D. B. Wilson (Cornell University). All other reagents were purchased from Sigma-Aldrich.
Strain and growth conditions.
The strain S. monoica Pringsheim 53-967 Dick was obtained from the Centraal Bureau voor Schimmel Culture (Baarn, The Netherlands) and maintained on potato dextrose agar in 9-cm petri dishes. The mycelium used for all experiments was grown for 3 days at 24°C in 100 ml of liquid medium of Machlis (43) in 140-mm petri dishes. Each dish was inoculated with 30 agar plugs of ∼5 mm cut from cultures on potato dextrose agar.
Isolation of plasma membranes.
Cells grown in liquid medium from 12 petri dishes were harvested, washed with water, and dried under vacuum on filter paper. All of the following steps were performed at 4°C. The cells were homogenized in extraction buffer (10 mM Tris-HCl [pH 7.4]) using a Waring blender (total of four periods of 10 s at maximum speed separated by rest periods of 2 min to maintain the sample temperature in the range 4 to 8°C). Cell debris, cell walls, nuclear fractions, vacuoles, and other intracellular compartments of a high density were pelleted by low-speed centrifugation (10 min at 5,000 × g) and discarded. The supernatant was centrifuged at 50,000 × g for 1 h to sediment the total cell membranes, which were subsequently resuspended in 5 mM potassium phosphate buffer (pH 7.5) containing 0.33 M sucrose, 0.1 mM EDTA, and 0.5 mM dithiothreitol (DTT). The final protein concentration in the membrane suspension was adjusted to 5 mg ml−1 by addition of an adequate volume of resuspension buffer. Plasma membranes were then purified by two-phase partitioning essentially as described earlier (38), except for the following modifications. The two-phase system was composed of 6% (wt/wt) dextran T500, 6% (wt/wt) polyethylene glycol 3350, 5 mM KCl, 0.33 M sucrose, 0.1 mM EDTA, 0.5 mM DTT, and 5 mM potassium phosphate buffer (pH 7.5) (all of these are final concentrations). The plasma membranes obtained by repeating the phase partitioning three times were washed and resuspended in extraction buffer. The purity of the plasma membranes was assessed by measuring the activity of enzymatic markers of endoplasmic reticulum (cytochrome c reductase) (9), mitochondria (cytochrome c oxidase) (9), and plasma membrane [(1→3)-β-d-glucan synthase] (24).
Preparation of detergent-resistant membranes (DRMs).
Proteins in the plasma membranes were assayed and diluted in extraction buffer containing Triton X-100 in order to obtain a final concentration of 1% (wt/wt) detergent and a weight ratio of Triton to protein of 13. Membranes were solubilized at 4°C for 30 min and centrifuged at 120,000 × g for 35 min at the same temperature. The pellets containing the nonsolubilized membranes were then resuspended in extraction buffer and diluted with extraction buffer containing 80% (wt/wt) sucrose to reach a final concentration of 42% (wt/wt) sucrose. The sample was layered under a 26-ml linear sucrose gradient (20 to 40%) in extraction buffer and centrifuged at 250,000 × g for 20 h at 4°C. The protein concentration, carbohydrate synthase activities, and density were determined in 1-ml fractions collected from the bottom of the gradient tube.
Solubilization of the (1→3)-β-d-glucan synthase activity from DRMs.
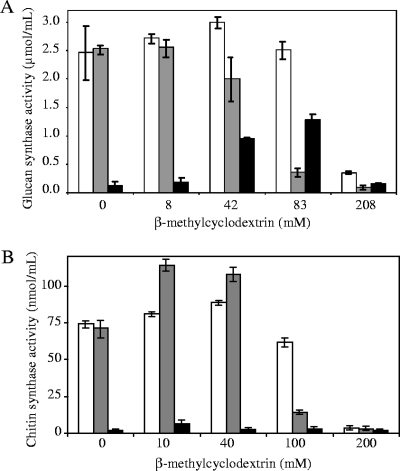
Purified DRMs were diluted twice in a solution of β-methylcyclodextrin in order to obtain the final concentrations indicated in Fig. 3. The samples were incubated at 25°C for 30 min and centrifuged at 120,000 × g for 35 min at 4°C. The supernatants were removed and the pellets were washed with 10 mM Tris-HCl buffer (pH 7.4) and resuspended in the same buffer. The (1→3)-β-d-glucan and chitin synthase activities were measured as described below in the fractions before centrifugation, as well as in the pellets and supernatants.
FIG. 3.
Solubilization of (1→3)-β-d-glucan and chitin synthase activities from DRMs. DRMs were incubated with various concentrations of β-methylcyclodextrin, and the samples were centrifuged at 120,000 × g for 35 min at 4°C. The (1→3)-β-d-glucan (A) and chitin (B) synthase activities were measured in the pellets (░⃞) and supernatants (▪), as well as in the samples before centrifugation (□).
Assay of carbohydrate synthase activities.
(1→3)-β-d-Glucan synthase activity was measured by mixing 1 volume of enzyme fraction with 2.7 volumes of the reaction mixture in order to obtain final concentrations of 8 mM PIPES-Tris (pH 6.0), 10 mM cellobiose, 1.3 mM DTT, 1 mM UDP-glucose, and 0.16 μM UDP-d-[U-14C]glucose. The samples were incubated at 25°C for 1 h, and the reaction was terminated by adding 2 volumes of ethanol. After an overnight precipitation at −20°C, insoluble polysaccharides were recovered on Whatman GF/C glass-fiber filters and successively washed with water and 66% ethanol. The radioactivity retained on the filters was detected by liquid scintillation (Wallac WinSpectral 1414 counter). Chitin synthase was measured as described earlier (23) by mixing 1 volume of enzyme fraction with 3 volumes of reaction mixture. The final concentrations were 10 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 20 mM N-acetylglucosamine, 1.25 μg of trypsin ml−1, 0.5 mM UDP-N-acetylglucosamine, and 434 nM UDP-N-acetyl-d-[U-14C]glucosamine. The reaction was stopped by adding 5 volumes of ethanol. Precipitation of the newly synthesized polysaccharides and liquid scintillation counting were performed as for the assay of (1→3)-β-d-glucan synthase.
Enzymatic hydrolysis of the polysaccharides synthesized in vitro.
After in vitro synthesis of polysaccharides under the conditions described above, the radioactive insoluble polymers were pelleted by centrifugation at 10,000 × g for 15 min at 4°C, washed in hydrolysis buffer (50 mM sodium acetate buffer [pH 5.5]), and resuspended in 100 μl of the same buffer. Hydrolyses were performed for 20 h in the presence of 0.5 U of chitinase ml−1 from Serratia marcescens (Sigma) at 25°C (1 U releases 1.0 mg of N-acetyl-d-glucosamine from chitin per h at pH 6.0 at 25°C) or 0.4 U of (1→3)-β-d-glucanase ml−1 at 40°C (1 U releases 1 μmol of Glc from laminarin per min at pH 4 at 40°C) or 75.5 mU of cellulase Cel 6A ml−1 and 30 μU of cellulase Cel 6B ml−1 at 50°C (1 U releases 1 μmol of cellobiose from cellulose swollen in phosphoric acid per min at pH 5 at 50°C). Controls were performed in the same conditions but in the absence of hydrolytic enzyme. The reactions were stopped by adding 2 volumes of ethanol to the mixtures. After 1 h at −20°C the insoluble nonhydrolyzed polysaccharides were recovered on glass-fiber filters, and the corresponding radioactivity was measured by liquid scintillation as described above. The specificity of each hydrolytic enzyme preparation was verified on well-characterized polysaccharides (cellulose from Gluconacetobacter xylinus, curdlan [(1→3)-β-d-glucan] from Agrobacterium spp., and chitin from crab), and each hydrolase was found to be active only on its specific substrate (not shown).
13C-NMR spectroscopy.
13C-enriched polysaccharides were synthesized in vitro by the DRMs obtained from one sucrose gradient. The conditions were as described for the assay of (1→3)-β-d-glucan synthase activity except for the use of 13C-enriched UDP-glucose (UDP-d-[U-13C]glucose, 1 mM) that was synthesized enzymatically (42) and used as described earlier for polysaccharide characterization (20). The insoluble products were collected by centrifugation at 5,000 × g for 10 min at 4°C, purified (54), freeze-dried, and dissolved in (CD3)2SO. 13C-nuclear magnetic resonance (NMR) spectra were recorded by using a Brucker spectrometer operated at 300 K and 75.5 MHz. The central peak of the (CD3)2SO multiplet (39.5 ppm) was used as a reference.
TEM.
The (1→3)-β-d-glucan and chitin synthesized in vitro by DRMs were purified (54) and observed by transmission electron microscopy (TEM) on carbon-coated copper grids. TEM observations were performed at the Centre Technologique des Microstructures (University of Lyon, Lyon, France) and at the Research Institute for Sustainable Humanosphere (University of Kyoto, Japan) using a Philips CM120 and a JEOL-2000EXII microscope, respectively. Specimens were negatively stained with a solution containing 4% uranyl acetate and 1% trehalose and observed under an accelerating voltage of 100 kV.
Preparation of lipid fractions.
Lipids were extracted as described elsewhere (5). Plasma membranes or DRMs were first heated at 100°C for 5 min in order to denature lipases and preserve the lipids. Volumes (3.75) of chloroform-methanol (1:2) were added to 1 volume of sample. After homogenization and incubation of the mixture at room temperature for 20 min, the insoluble material was pelleted at 450 × g for 10 min at 17°C and used for a second extraction performed in the same conditions. The supernatants from the two extractions were pooled and mixed with 2.5 volumes of water and 2.5 volumes of chloroform. After a vigorous homogenization, the mixture was centrifuged at 450 × g for 10 min at 17°C and the chloroformic phase containing the lipids was recovered and dried under nitrogen. Phospholipids were then solubilized in chloroform and assayed (67). Glycerophospholipids were methanolyzed by incubation of the total lipids at 70°C for 30 min in 1.5 M KOH in methanol. The hydrophobic products were recovered in the chloroform phase after acidification by addition of HCl and phase separation in chloroform/methanol (final conditions: chloroform-methanol-water [1:1:0.9]).
Lipid analysis by TLC.
Lipids were analyzed by one-dimensional thin-layer chromatography (1D-TLC) on silica gel 60 plates using a two-solvent system. Development was performed first to half the height of the plate in ethyl acetate-propanol-chloroform-methanol-0.25% (wt/vol) aqueous KCl (25:25:25:10:9) and then to three-quarters the height of the plate in hexane-diethyl ether-acetic acid (75:21:4). For detailed identification, lipids were separated by 2D high-performance TLC (2D-HPTLC) using a CAMAG horizontal developing chamber. The developing solvent used for the first dimension was chloroform-methanol-NH4OH (65:25:5), whereas the second dimension was developed in chloroform-acetone-methanol-acetic acid-water (30:40:10:10:5). For quantitative analysis, the lipids separated by 1D-TLC were stained by rapidly immersing the plate in a solution containing 10% (wt/vol) CuSO4 and 8% (vol/vol) H3PO4 and subsequently heating it at 180°C. The plate was then scanned by using a CAMAG II TLC scanning densitometer. Quantification was performed by comparing the intensities of the spots present in the samples to standard curves obtained from the migration of known quantities of lipid standards on the same plate. For the detection of the lipids containing amino groups, the plates were sprayed with a solution containing 0.25% ninhydrin in acetone-dimethylpyridine (9:1) and heated at 100°C.
Fatty acid analysis by GC.
Total lipids were separated by 1D-TLC as described above and stained by spraying the plate with a solution of 0.02% (wt/vol) dichlorofluorescein in 95% methanol. The spots corresponding to the poorly resolved glycerophospholipids (phosphatidylcholine [PC], phosphatidylserine [PS], phosphatidylinositol [PI], lysophosphatidylethanolamine [LPE], and lysophosphatidylcholine [LPC]) on one hand and the spot corresponding to phosphatidylethanolamine (PE) on the other hand were scraped and transesterified at 100°C for 90 min in 250 μl of toluene-methanol (40:60) and 250 μl of BF3 in 14% methanol. The reaction was stopped in ice by adding 1.5 ml of 10% K2CO3, and fatty acid methyl esters were extracted with 2 ml of isooctan. The derivatives were analyzed by gas chromatography (GC) using an Agilent Technologies chromatograph (model 6890) fitted with a BPX 70 fused silica capillary column (60 mm by 0.25 mm [inner diameter], 0.25-μm film thickness; SGE Europe, Ltd., France). The oven temperature was set at 80°C for 1.5 min and increased to 150°C at 20°C min−1 and then to 250°C at 2°C min−1. The temperature was maintained at 250°C for 10 min before returning to the initial conditions. Helium was used as the carrier gas at 1 ml min−1. The temperatures of the split/splitless injector and the flame ionization detector were set at 230 and 280°C, respectively.
GC-mass spectrometry (MS) analysis of sterols.
The sterol spots were scraped from dichlorofluorescein-stained TLC plates. The sterols were eluted from the silica powder by incubation in 0.5 ml of chloroform-methanol (2:1). The supernatant was recovered, the solvent was evaporated, and the products were silylated in 100 μl of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) for 30 min at 60°C before analysis by GC-MS in a Hewlett-Packard quadrupole mass spectrometer interfaced with a Hewlett-Packard gas chromatograph (Les Ullis, France). The gas chromatograph was equipped with a DB-17MS fused-silica capillary column (60 m by 0.25 mm [inner diameter], 0.25-μm film thickness; Agilent Technologies), which was held at 57°C. The oven temperature program consisted of 5 min at 57°C, followed by an increase of temperature to 200°C at 40°C min−1, an increase to 310°C at 10°C min−1, and a final step at 310°C for 20 min. The interface, injector, and ion source were kept at 280, 280, and 150°C, respectively. The electron energy was set at 70 eV and helium was used as the carrier gas. The electron multiplier voltage was set at 1,400 V. Mass spectra were acquired from 100 to 600 Da using the electron impact ionization mode. Sterols were identified by comparison of their retention times and mass spectra to those of commercial standard sterols silylated in the conditions described above and quantified using their total ion current signals.
2D-PAGE analysis and protein identification by MS.
The protein profile of the DRM fraction was analyzed by 2D polyacrylamide gel electrophoresis (PAGE) as detailed below. Some of the 2D gels used for protein identification, and all MS analyses were performed by the company Innova Proteomics (Rennes, France) as described elsewhere (17). Before performing 2D PAGE analyses, sucrose was eliminated from the gradient fractions by dialysis against 10 mM Tris-HCl buffer (pH 7.4). Proteins were precipitated at −20°C for 15 min in the presence of 2 volumes of absolute ethanol and washed once with 66% ethanol. The resulting pellet containing a theoretical amount of 100 μg of total protein was dissolved in denaturing buffer consisting of 5 M urea, 2 M thiourea, 2% Zwittergent 3-10, 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 20 mM DTT. The samples were then loaded onto Ready Strip IPG (Bio-Rad) strips (7 or 11 cm, linear or nonlinear pH gradient in the range 3 to 10 or 5 to 8), and the gels were actively rehydrated in the sample at 50 V and 20°C for 10 h. Isoelectric focalization was performed using the Protean IEF cell from Bio-Rad and the following steps: 250 V for 15 min, from 250 to 6,000 V in 150 min, and 6,000 V for 6 h. Strips were successively incubated for 20 min in a reducing buffer (6 M urea, 2% sodium dodecyl sulfate [SDS], 0.375 M Tris-HCl [pH 6.8], 20% glycerol, 130 mM DTT) and in an alkylating buffer which had the same composition as the reducing buffer, except for the replacement of DTT by 135 mM iodoacetamide, and the addition of 0.01% bromophenol blue. Equilibrated strips were placed on top of SDS-10% PAGE gels and overlaid with 0.5% agarose in SDS running buffer. After electrophoresis, 2D gels were silver stained (48). Individual spots were then excised from the gel and subjected to fingerprint analysis by MS after hydrolysis with trypsin (17). For several proteins of interest, de novo sequencing of some of the trypsic peptides was performed by tandem MS analysis (17). Peptide fingerprints and sequences were used for protein identification through searches in all available public sequence databases, including oomycete EST databases (22; http://www.oomycete.org; http://www.pfgd.org/) and draft genome databases of Phytophthora species (68; http://genome.jgi-psf.org/), using the Mascot (55; http://www.matrixscience.com/) and BLAST (1; http://www.ncbi.nlm.nih.gov/BLAST) search tools.
RESULTS
(1→3)-β-d-Glucan and chitin synthases are located in DRMs.
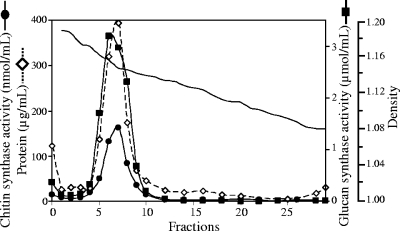
Plasma membranes were prepared by two-phase partitioning in an aqueous system consisting of polyethylene glycol and dextran as detailed in Materials and Methods. The upper phase contained 70% of the (1→3)-β-d-glucan synthase activity that was originally present in the total cell membranes (Table 1) as opposed to only 4.5 and 0.9% for cytochrome c oxidase and cytochrome c reductase, respectively. These data indicate a clear enrichment of plasma membranes in the upper phase, as expected. In addition, a significant increase in the (1→3)-β-d-glucan synthase specific activity was observed in the upper phase compared to the total cell membranes, whereas the specific activities of the mitochondrial and endoplasmic reticulum markers decreased (Table 1). No better enrichment of plasma membranes could be obtained with any of the other conditions tested for two-phase partitioning. Thus, upper phases prepared as described in Materials and Methods were used as a starting material for the isolation of DRMs. For this purpose, the enriched plasma membranes were solubilized by 1% Triton X-100, and the resulting DRMs were purified by flotation in a linear sucrose gradient in which they were visible as a translucent band coinciding with the major peak of protein at a density of 1.14 to 1.16 (Fig. 1). The DRMs were clearly separated from a second peak of protein corresponding to the pellet of the gradient (Fig. 1). Interestingly, the detectable chitin and (1→3)-β-glucan synthase activities obtained after flotation of the Triton-treated plasma membranes were almost entirely present in the peak of protein corresponding to the DRMs (Fig. 1). This represented averages of 72% (71.9% ± 2.7%) and 60% (59.6% ± 5.3%), respectively, of the total chitin and (1→3)-β-glucan synthase activities originally present in the plasma membranes. These data demonstrate a clear specific enrichment of both carbohydrate synthases in DRMs since an average of only ∼14% (13.9% ± 3.1%) of the total protein originally present in the plasma membranes was recovered in the DRM peak. The proportions of carbohydrate synthases recovered in the DRMs are most likely underestimated since these enzymes, particularly the (1→3)-β-glucan synthase activity, is partially inhibited upon incubation in the presence of Triton X-100 (data not shown).
TABLE 1.
Purification of plasma membranes by two-phase partitioning
| Enzymatic marker(s) | Sp act (nmol min−1 mg of protein−1) ± SD in:
|
% in plasma membranes ± SD | |
|---|---|---|---|
| Total cell membranes | Plasma membranes | ||
| Cytochrome c reductase (endoplasmic reticulum) | 340 ± 21 | 24.2 ± 3.9 | 0.9 ± 0.6 |
| Cytochrome c oxidase (mitochondria) | 137 ± 23 | 19.2 ± 8.6 | 4.5 ± 2.7 |
| Glucan synthase (plasma membrane) | 1.9 ± 0.47 | 14.4 ± 2.5 | 70.5 ± 6.7 |
| Proteins | 11.8 ± 3.2 | ||
FIG. 1.
Purification of DRMs by flotation in sucrose gradient. After solubilization of plasma membranes by Triton X-100, the insoluble material was layered under a linear 20 to 40% sucrose gradient and ultracentrifuged (see Materials and Methods). Two peaks of protein were observed, one corresponding to DRMs (fractions 5 to 8) and the other corresponding to a pellet containing high-density structures not solubilized by Triton X-100 (fraction “0”). Chitin and (1→3)-β-d-glucan synthase activities were assayed in each fraction of the gradient. They are expressed as nmol of GlcNAc incorporated into chitin per ml of fraction in 1 h and as μmol of Glc incorporated into (1→3)-β-d-glucan per ml of fraction in 1 h, respectively.
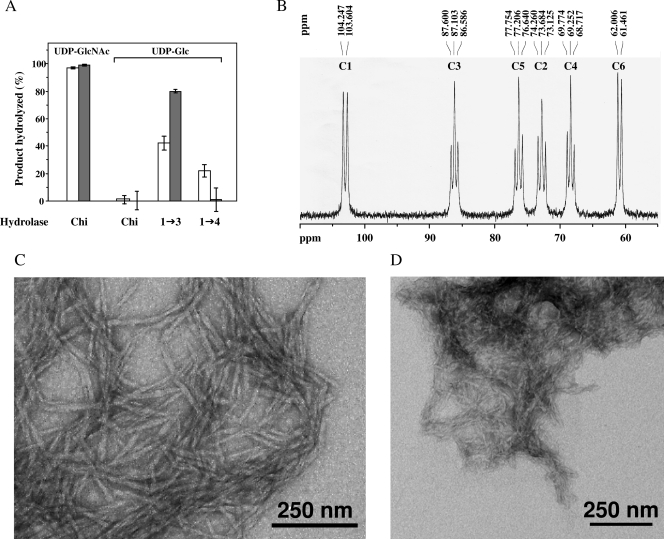
The polysaccharides synthesized in vitro by the plasma membranes and DRMs were first characterized by using glycoside hydrolases, as detailed in Materials and Methods. Nearly 100% of the polysaccharides synthesized in vitro by the plasma membranes and DRMs using radioactive UDP-N-acetylglucosamine were hydrolyzed by chitinase, showing that the newly synthesized carbohydrates corresponded to chitin (Fig. 2A). This result not only confirms the previous finding that oomycetes contain chitin synthase activity (15, 23) but it also demonstrates the occurrence of chitin synthase in DRMs. When the in vitro synthesis was performed in the presence of 14C-labeled UDP-glucose, no chitin was synthesized by the plasma membrane and DRMs fractions, as expected (Fig. 2A). The action of the (1→3)-β-d-glucanase showed that the polysaccharides synthesized in these conditions were (1→3)-β-d-glucans. However, the polysaccharides were not completely hydrolyzed: the percentage of hydrolysis reached up to 45% in the case of the polysaccharides synthesized by the plasma membranes and up to 80% for the (1→3)-β-d-glucan synthesized by the DRMs (Fig. 2A). This can be explained by the fact that the action of (1→3)-β-d-glucanase is dependent on the crystallinity of the polysaccharides, the more crystalline polysaccharides being more resistant to the action of hydrolytic enzymes (54). Thus, the results presented in Fig. 2A suggest that the degree of crystallinity of the (1→3)-β-d-glucan synthesized by the DRMs is lower than that of the polysaccharide synthesized by plasma membranes. The structure of the (1→3)-β-d-glucan synthesized in vitro by the DRMs was confirmed by 13C-NMR spectroscopy (Fig. 2B). The substrate used for in vitro synthesis was uniformly enriched in 13C (UDP-[U-13C]glucose) to facilitate product characterization by 13C-NMR spectroscopy (20). The resonance signals appeared as doublets or triplets due to homonuclear C-C coupling in the fully 13C-enriched polysaccharide (Fig. 2B). The spectrum is characteristic of a strictly linear (1→3)-β-d-glucan for which resonance peaks are expected at 61.0, 68.5, 72.9, 76.4, 86.3, and 103.1 ppm for C6, C4, C2, C5, C3, and C1, respectively (33, 54, 62). It is comparable to those obtained for well-characterized (1→3)-β-d-glucans (33, 62) and for in vitro (1→3)-β-d-glucans synthesized by enzymes from S. monoica (54) and plants (14, 36, 37).
FIG. 2.
Analysis of the polysaccharides synthesized in vitro by membrane fractions. (A) Polysaccharides synthesized by plasma membranes (□) or DRMs (░⃞) in the presence of either UDP-N-acetyl-[14C]glucosamine (UDP-GlcNAc) or UDP-[14C]glucose (UDP-Glc) were hydrolyzed by specific glycoside hydrolases. Chi, chitinase; 1→3, (1→3)-β-d-glucanase; 1→4, cellulase mixture. The percentage of hydrolysis was determined by liquid scintillation counting as described in Materials and Methods. (B) 13C-NMR analysis in (CD3)2SO of the polysaccharides synthesized by DRMs in the presence of 13C-enriched UDP-glucose. The spectrum is characteristic of a linear (1→3)-β-d-glucan. (C) Transmission electron micrograph of the (1→3)-β-d-glucan synthesized in vitro by DRMs incubated in the presence of UDP-glucose. The sample was negatively stained with 4% uranyl acetate. (D) Same as in panel C, but in the case of chitin synthesized in vitro in the presence of UDP-GlcNAc.
The use of specific cellulases showed that some cellulose was synthesized in vitro by the plasma membranes, but no incorporation of radioactive glucose into cellulose was observed in the case of the purified DRMs (Fig. 2A). This observation suggests that the cellulose synthase complex is not located in DRMs, as opposed to (1→3)-β-d-glucan and chitin synthases. Alternatively, the absence of detectable cellulose synthase activity in DRMs might be due to a spontaneous inactivation of the highly unstable cellulose synthase machinery (18, 36) or to its inhibition by Triton X-100 during the preparation of DRMs.
The (1→3)-β-d-glucan synthesized by the DRMs exhibits a microfibrilar morphology typical of (1→3)-β-d-glucans of a high-molecular-weight (Fig. 2C). In particular its morphology is similar to that of in vitro synthesized (1→3)-β-d-glucans of a degree of polymerization of about 20,000 (54). The chitin synthesized in vitro also consisted of microfibrils (Fig. 2D). These results suggest that the purified DRMs contain all of the proteins required for polymerization and assembly of (1→3)-β-d-glucan and chitin into microfibrillar structures. Thus, DRMs represent an ideal starting material for the identification of all of the components of the carbohydrate synthase complexes.
The use of β-methylcyclodextrin, which specifically interacts with sterols (47), showed that it is possible to release in an active and soluble form the (1→3)-β-d-glucan synthase from purified DRMs. Increasing levels of activity were recovered in the supernatants obtained after incubation of the purified DRMs with increasing concentrations of β-methylcyclodextrin in the range from 0 to 80 mM (Fig. 3A). Higher concentrations of β-methylcyclodextrin inactivated the enzyme (Fig. 3A). Interestingly, chitin synthase remained in an insoluble form after incubation of the DRMs with 0 to 100 mM β-methylcyclodextrin (Fig. 3B). As for (1→3)-β-d-glucan synthase, inhibition of chitin synthase was observed when using β-methylcyclodextrin at concentrations higher than 100 mM (Fig. 3B). Altogether, these results suggest that (1→3)-β-d-glucan synthase interacts with sterols within DRMs, while chitin synthase occurs in a different lipid environment either within the same population of DRMs or within a different one.
Lipid analyses of the purified DRMs.
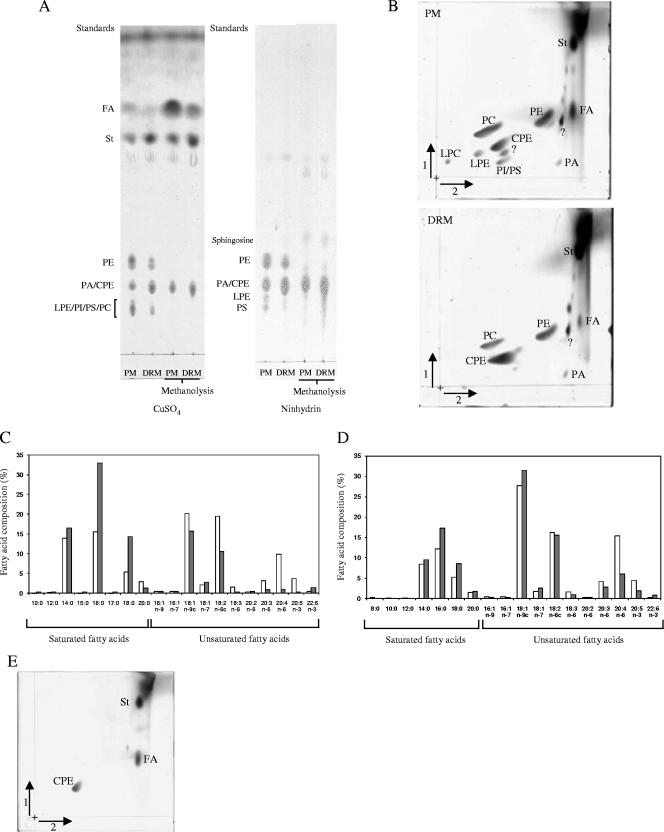
Qualitative and semiquantitative lipid analyses revealed that the glycerophospholipid profile of the DRMs was simpler than that of the plasma membranes (Fig. 4). In particular, 2D-HPTLC analyses of total lipids showed that the purified DRMs contained hardly any PI, PS, LPE, and LPC as opposed to plasma membranes (Fig. 4B). In addition, the amount of PC and, to a lower extent, of PE was significantly lower in the DRMs compared to the plasma membranes (Fig. 4B). Most saturated fatty acids such as myristic and palmitic acids were predominant in the glycerophospholipids isolated from the DRMs, whereas the glycerophospholipids from the plasma membranes were composed of a higher proportion of unsaturated fatty acids such as, for example, oleic, linoleic, and arachidonic acids (Fig. 4C and D). The percentage of total saturated fatty acids in the glycerophospholipids from the plasma membranes was of 35%, as opposed to 65% in the DRMs.
FIG. 4.
Lipid analyses of plasma membranes and DRMs. (A) The lipids from plasma membranes (PM) and DRMs were analyzed by 1D-TLC before or after alkaline methanolysis. All lipids were stained with CuSO4, whereas ninhydrin was used to detect specifically lipids containing free amino groups. The position of standards of a known identity is indicated at the left of each chromatogram. PA, phosphatidic acid; PI, phosphatidylinositol; PS, phosphatidylserine; FA, fatty acids; St, sterols; CPE, ceramide-phosphorylethanolamine. (B) Analysis of the lipids from plasma membranes (PM) and DRMs by 2D-HPTLC (staining with CuSO4). All lipids are abbreviated as in panel A or as defined in the text. Question marks indicate unidentified lipids. (C) Analysis of the fatty acids in the glycerophospholipid fraction (pool of PC, PI, PS, LPE, and LPC) of plasma membranes (□) and DRMs (░⃞). The results are expressed as a percentage of the total fatty acids identified in glycerophospholipids from plasma membranes and DRMs. The data from a typical experiment of five independent analyses are shown. In the case of fatty acids representing <2% of the total fatty acids, the standard deviation did not exceed 50%; a maximum of 15% standard deviation was obtained for fatty acids occurring in proportions higher than 2%. c, double bond with a cis configuration. The percentage of total saturated fatty acids in the glycerophospholipids from the plasma membrane was 35% as opposed to 65% in the DRMs. (D) Same as in panel C, but in the case of the fatty acids present in the PE fraction. (E) 2D-HPTLC of methanolyzed lipids from DRMs stained with CuSO4; identical results were obtained with the lipids from plasma membranes (not shown). Lipid abbreviations are as in panel A.
The relative proportion of sphingolipids and sterols was clearly higher in DRMs than in the plasma membranes compared to total glycerophospholipids (Fig. 4A and B and Fig. 5). Sphingolipids were unequivocally distinguished from phospholipids by their resistance to mild alkaline methanolysis (50) (Fig. 4A, and compare Fig. 4B, lower panel, with Fig. 4E). The DRMs were found to be greatly enriched in a sphingolipid identified as ceramide phosphorylethanolamine by comparing its chromatographic mobility with a commercial standard and by staining with ninhydrin, which specifically detects free amino groups (Fig. 4A, B, and E and data not shown). To our knowledge, this unusual sphingolipid has been reported in total cellular lipid extracts only in a few eukaryotic microorganisms, which are all oomycete plant pathogens, but its subcellular location has never been determined (50).
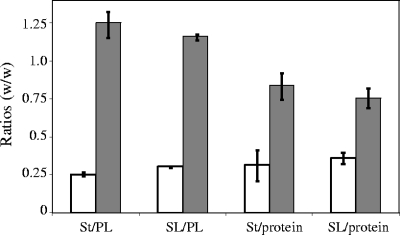
FIG. 5.
Lipid/lipid and lipid/protein ratios in plasma membranes (□) and DRMs (░⃞) (mean values of three independent experiments). St, sterols; PL, glycerophospholipids; SL, sphingolipids.
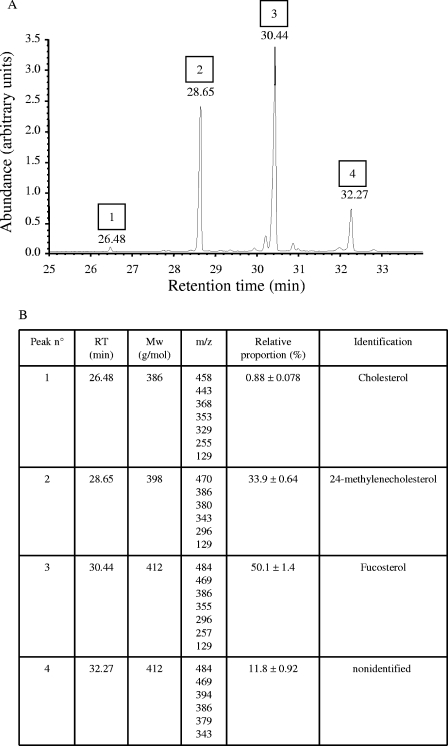
The ratios of total sterols to glycerophospholipids and proteins were higher in the DRMs than in the plasma membranes (Fig. 5). Both types of membranes contained at least four different sterols. Three of them—cholesterol, 24-methylenecholesterol, and fucosterol—were unequivocally identified by GC-MS (Fig. 6). A fourth sterol corresponding to peak 4 in Fig. 6A could not be identified. The relative proportion of each sterol within the total sterol fraction from DRMs is presented in Fig. 6B. Fucosterol was by far occurring in the highest proportion, representing 50% of the total sterols in DRMs (Fig. 6B). Identical relative proportions as shown in Fig. 6B were measured in the case of the plasma membranes (not shown).
FIG. 6.
GC-MS analysis of sterols from the purified DRMs. (A) Gas chromatogram of silylated sterols. (B) Identification of the sterol derivatives separated in the chromatogram shown in A by electron impact mass spectrometry. The table presents the retention times (RT) and peak numbers highlighted in A, as well as the m/z ratios of the characteristic ion fragments obtained by fragmentation of each sterol derivative.
Identification of proteins in DRMs.
Proteins from the purified DRMs were separated by 2D PAGE, stained with silver, and subjected to de novo sequencing by tandem MS and peptide fingerprint after trypsin digestion. Searches in the oomycete EST databases (22) and in the draft genome databases of Phytophthora species (68) were performed. This allowed the identification of some of the most abundant proteins in the preparation.
The most abundant proteins in the purified DRMs were located in three different areas of the 2D gels, corresponding to apparent molecular masses of 30 to 35, 55, and 66 kDa (see Fig. S1 in the supplemental material). When a higher amount of total protein was loaded on the gel (250 μg instead of 100 μg), numerous additional spots were easily distinguishable while the abundant proteins in the 30- to 35-, 55-, and 66-kDa areas were detected as horizontal smears (see Fig. S1B in the supplemental material). Thus, the gel presented in Fig. S1A in the supplemental material was used for identification of the major proteins, whereas the one shown in Fig. S1B in the supplemental material was used for analysis of some of the less-abundant proteins. None of the analyses performed allowed the identification of peptides arising from (1→3)-β-d-glucan and chitin synthases. This is most likely due to the fact that these proteins contain multiple transmembrane domains and exhibit high molecular masses, which seriously challenges their recovery in 2D gels [200 kDa for all known (1→3)-β-d-glucan synthases (see for instance reference 11) and 98.5 kDa for the chitin synthase from S. monoica (52)]. Indeed, the proteins detected on 2D gels in amounts allowing peptide fingerprint analyses or de novo sequencing exhibited a maximum apparent molecular mass of ∼70 kDa (see Fig. S1 in the supplemental material).
Proteins exhibiting heterogeneous apparent isoelectric points and corresponding to the A and B subunits of V-ATPase were clearly identified by peptide fingerprint with 26 to 36% sequence coverage (see Table S1 in the supplemental material). Two other well-conserved proteins, actin and a protein belonging to the heat shock protein 70 (Hsp70) family, were identified in the DRMs with 38 and 24% sequence coverage, respectively (see Table S1 in the supplemental material). The protein profile of the 30- to 35-kDa area of the 2D gels was rather complex (see Fig. S1 in the supplemental material). Peptide fingerprint analysis clearly showed that three of the most abundant proteins in this area correspond to isoforms of a 35-kDa annexin (see Fig. S1A and Table S1 in the supplemental material), which is an activator of the (1→3)-β-d-glucan synthase from S. monoica (accession no. DQ323662) (7). In addition, the following peptides arising from the trypsic digest of each isoform showed 100% identity with segments of the 35-kDa annexin: GIGTDEYGLSAAIVR, KLLQLLAQPLEDAEALIVR, SLFSEIRGETSGDYGK, QFQNDLVVVLADDLSGDLK, and FYLAIVNQMAQPYNPAIHTQA. Attempts to identify additional proteins using MS approaches were unsuccessful essentially because the sequences obtained were tentative or contained tags that were too short to be exploited for protein identification. In several instances, especially for some of the minor silver-stained spots, the amount of protein recovered from the 2D gels was a limiting factor. The analyses based on peptide fingerprint and de novo sequencing were complicated by the fact that the only databases available for Saprolegniales are partial EST databases (22). Protein identification using the Phytophthora genome databases (68) was possible for proteins exhibiting well-conserved sequences only (see Table S1 in the supplemental material).
DISCUSSION
Oomycetes have long been considered as a separate class within the kingdom Fungi, but they are in fact taxonomically unrelated to true fungi and belong to the stramenopile eukaryotic kingdom, which includes heterokont algae and water molds (4, 35, 44, 53). We demonstrate here for the first time the existence of DRMs in the plasma membrane of a stramenopile species, the oomycete S. monoica, and the occurrence of important carbohydrate synthases in these detergent-insoluble membrane structures. In addition, the isolated DRMs exhibit similar biochemical properties as lipid rafts in animal cells. The relationship between DRMs and physiological lipid rafts is currently debated (25, 41, 73). In particular, it is argued that the formation of detergent-resistant structures may be artificially induced by the nonphysiological experimental conditions used for their preparation, i.e., the use of detergents at 4°C, and that populations of DRMs do not fully represent the original in vivo lipid raft structures (41). It remains nonetheless that the experimental isolation of DRMs based on the treatment of membranes with Triton X-100 at 4°C reflects differential affinities of specific sets of membranes proteins to various lipid environments (41). Thus, the isolation and composition analysis of DRMs is a valuable tool for understanding the interaction and function of membrane proteins that segregate in membrane microdomains enriched in sterols and sphingolipids (41).
The DRMs isolated from S. monoica are enriched in sphingolipids and sterols compared to total plasma membranes, and their glycerophospholipids contain a higher proportion (∼65%) of saturated fatty acids. Thus, they exhibit the same general lipid characteristics as DRMs from animal (66), plant (6, 39, 49, 56), and yeast (34) cells but with differences in the nature and relative proportions of specific fatty acids and lipids from the sphingolipid and sterol families.
Consistent with the early suggestion that oomycetes should be grouped with brown algae rather than with true fungi on the basis of their sterol composition (13), the plasma membranes and DRMs from S. monoica were devoid of ergosterol and the corresponding unsaturated derivatives, which are the major sterols of true fungi and products of the lanosterol pathway (46, 72). None of the Δ5-sterols that are major sterols in DRMs from higher plants, such as sitosterol and stigmasterol (6, 39, 49), were present in detectable amounts in the purified DRMs from S. monoica. Instead, fucosterol, 24-methylenecholesterol, and cholesterol, all products of the cycloartenol pathway (72), were unequivocally identified as components of the oomycete DRMs, together with a fourth unidentified sterol (Fig. 6). The proportions between these four molecular species were identical to those determined in the plasma membrane fraction, suggesting that no selective sorting of the different sterols occurs during the assembly of DRMs. Fucosterol was the major sterol (∼50%), followed by 24-methylenecholesterol (∼34%), the unidentified sterol (∼12%), and cholesterol (∼1%). An early study on various oomycete species demonstrated the occurrence of comparable proportions of fucosterol, 24-methylenecholesterol, and cholesterol in the total mycelium of the Leptomitales Apodachlyella completa (46). In addition, similarly to S. monoica, A. completa did not contain any detectable amount of desmosterol (46), which is the direct unsaturated precursor of cholesterol. This is somewhat expected, given the low proportion of cholesterol in both species. Interestingly, none of the Saprolegniales analyzed in the pioneering work of McCorkindale et al. (46) exhibited a sterol composition similar to that of S. monoica. In particular, the mycelium of other Saprolegniales contained up to nearly 75% of either 24-methylenecholesterol or cholesterol (46). These significant differences might be the result of a spontaneous adaptation of different oomycete species to different environments and/or reflect phylogenetic evolution. However, it must be kept in mind that variations in the composition of the culture media used to grow the mycelia of the different species might have an effect on the relative expression of the enzymes involved in the biosynthetic sterol pathways and thus on the observed sterol composition. In the case of our investigations on S. monoica, the mycelium was grown on the synthetic medium of Machlis (43), which is devoid of exogenous sterols. Thus, unlike parasitic Peronosporales such as Pythium and Phytophthora species, which are unable to epoxidize squalene and consequently synthesize sterols (72), S. monoica is able to produce its own pool of sterols without requiring any exogenous sterol. This observation and the occurrence of fucosterol in S. monoica are consistent with the cyclization of 2,3-oxidosqualene to cycloartenol and thus with the biosynthesis of sterols through the cycloartenol pathway in Saprolegniales (72).
Interestingly, our analyses revealed the occurrence of ceramide phosphorylethanolamine in the DRMs from S. monoica. This type of sphingolipid has been previously reported only in a limited number of organisms, including Phytophthora and Pythium species (50), the freshwater snail Sinotaia histrica (29), and three genera of anaerobic Bacteroides (31). In the case of oomycetes, two types of ceramide phosphorylethanolamine containing either a common 16-carbon 1,3-diOH sphingoid or an unusual 19-carbon branched triunsaturated sphingoid were identified in total cellular lipid extracts (50). However, their subcellular location was not determined, although it was hypothesized that they are likely to occur in the plasma membrane like most of the sphingolipids present in other species (50). With the example of S. monoica, we demonstrate that ceramide phosphorylethanolamines indeed occur in the plasma membrane, in which they are more specifically concentrated in microdomains that are resistant to detergents (DRMs). Even though the function and physiological significance of ceramide phosphorylethanolamine in DRMs remain to be determined, it may be expected that such sphingolipids contribute to the stabilization and physical properties of DRMs and/or that they specifically interact and stabilize proteins or protein complexes carrying a DRM-specific biological activity.
The protein composition of DRMs reflects at least partially the composition of physiological microdomains or lipid rafts. Hence, the identification of the proteins in DRMs represents a valuable approach toward the identification of the physiological functions of lipid rafts. With this goal in mind, we have undertaken the identification of the proteins in the purified DRMs from S. monoica using an MS approach. This was, however, hindered by the fact that there is currently no sequence database available for the species S. monoica. Thus, only well-conserved proteins could be identified by screening the partial EST databases currently available for other Saprolegniales (22), as well as the draft genomes of Phytophthora ramorum and Phytophthora sojae (68). A more thorough proteomic investigation on DRMs from Saprolegniales is thus dependent on the future sequencing of the genomes of some of the most representative and/or economically important species such as, for instance, S. parasitica, Saprolegnia ferax, and S. monoica.
Our analyses show that V-type ATPase subunits are present in the DRMs from S. monoica. V-type ATPases are known constituents of animal DRMs (21, 40) and, even though they are primarily vacuolar in plants, there is evidence that they are also present in the plasma membranes of plant cells (58, 60). In addition, several recent reports have shown an enrichment of V-type ATPases in DRMs from Arabidopsis thaliana (6), Nicotiana tabacum (51), and Medicago truncatula (39). Thus, the occurrence of V-ATPases in DRMs seems to be a common feature of animals, plants, and Saprolegniales. It remains, however, to determine the function of this type of ATPases in DRMs. Another well-conserved protein, actin, was identified in the DRMs from S. monoica. Since actin filaments are involved in a wide range of cellular processes in which lipid rafts are also presumably involved, such as for instance cell polarization and cell surface processes including secretion and endocytosis, it is somehow expected that actin copurifies with DRMs. This is further supported by the occurrence of this protein in plant DRMs (51). In the latter case, it has been proposed that lipid rafts are involved in cytoskeleton organization (49). A protein similar to Hsp70 is present in the purified DRMs from S. monoica. The possibility that this protein arises from the cytoplasm as a free cytosoluble protein contaminating the DRM fraction cannot be completely ruled out. However, the rather complex fractionation procedure used to purify the DRMs, which is well adapted for the selective isolation of membrane proteins, coupled with our extensive lipid and structural analyses and the occurrence of other known DRM-associated proteins in the preparation, strongly suggest that the DRM fraction used is highly enriched in DRM-specific proteins and devoid of free cytosoluble proteins. Hsps act as molecular chaperones in various intracellular compartments and, although these proteins do not exhibit transmembrane domains and lack secretory signal sequences, accumulating evidence in mammalian cells support the occurrence of some members of the Hsp superfamily in the plasma membrane (10, 26). Lipid rafts have been shown to be involved in plasma membrane delivery of Hsp, thus representing an alternative to the classical exocytic pathway (10). In addition, membrane-bound and secreted Hsp70 seems to be involved in processes such as macrophage phagocytosis, immune response, and signal transduction, in some cases through their binding and interaction with lipid rafts or raft-associated proteins (70, 71). However, their detailed function and mode of action in plasma membrane microdomains remain poorly understood. Interestingly, Hsp70 proteins have also been recently identified in plant DRMs (39, 51).
The purified DRMs from S. monoica contain several isoforms of a 35-kDa annexin. Recent work in our group has demonstrated that one of the 35-kDa annexins from S. monoica positively regulates (1→3)-β-d-glucan synthase activity (7). Thus, the presence of annexin isoforms in our DRM fraction is also consistent with our novel finding that (1→3)-β-d-glucan synthase is associated with DRMs. Interestingly, annexins have also been shown to occur in DRMs from plant cells (39). It is noteworthy that peptides arising from a putative (1→3)-β-d-glucan (callose) synthase have been reported in DRMs from tobacco cells; however, the percentage of sequence coverage does not exceed 10% (51), and the function of the corresponding protein was not demonstrated. Altogether, these results and observations support the novel concept that DRMs similar to lipid rafts are involved in cell wall polysaccharide biosynthesis and that at least one isoform of an annexin is involved in the regulation of this process in S. monoica.
Enzymatic assays performed on the purified DRMs from S. monoica revealed that these structures contain carbohydrate synthase activities. In particular, (1→3)-β-d-glucan and chitin synthases were clearly enriched in the DRMs. Indeed, 72 and 60%, respectively, of the chitin and (1→3)-β-d-glucan synthase activities originally present in the plasma membranes were recovered in the purified DRMs, while only 14% of the total proteins from the plasma membranes partitioned in the DRMs. The polysaccharides synthesized in vitro by the isolated DRMs were characterized and unequivocally shown to be microfibrillar (1→3)-β-d-glucan and chitin, respectively. The occurrence of cell wall polysaccharide synthases in DRMs is consistent with a role of lipid rafts in cell polarization and tip growth. Interestingly, the involvement of lipid rafts in these cellular processes has been demonstrated in other eukaryotic microorganisms such as Saccharomyces cerevisiae and Candida albicans (2, 3, 45). By analogy with the situation in yeast (2, 3, 45), we propose here that lipid rafts in S. monoica are involved in protein targeting, tip growth, and hyphal polarization. The involvement of lipid raft in apical elongation is consistent with the occurrence of cell wall carbohydrate synthases, namely, (1→3)-β-d-glucan and chitin synthases, in the DRMs from S. monoica. It is noteworthy that beyond the case of eukaryotic microorganisms, animal rafts have also been proposed to be involved in cell polarization processes and to function as sorting platforms for plasma membrane proteins (28, 59, 63, 66).
Cellulose synthase assays in DRMs revealed the absence of detectable activity. The most direct explanation for this result is that cellulose synthase is not located in DRMs. In this case, and given the fact that lipid rafts are most likely involved in cell polarization and tip growth, cellulose synthase would not be delivered to the apex by lipid rafts. As a consequence, it can be expected that it would not significantly contribute to the biosynthesis of cell wall polysaccharides in the fast-growing hyphal tip, unless the enzyme is delivered to this active zone of the mycelium by the classical secretion pathway that does not involve lipid rafts. The hypothesis that cellulose synthase activity is either low or absent at the apex of the growing mycelium is supported by the earlier observation in the oomycete Achlya bisexualis (a species closely related to S. monoica) that the newly synthesized cell wall at the elongating apex of hyphae lacks microfibrillar cellulose (65). In addition, since (1→3)-β-d-glucan is present at the apex of the hyphae of A. bisexualis (64), it has been proposed that this polysaccharide is synthesized before cellulose in the elongating apical zone (65). This is in good agreement with our finding that (1→3)-β-d-glucan synthase activity is present in membrane microdomains, which are most likely polarized to the apical zone where cell wall polysaccharide biosynthesis is quantitatively more important than in the rest of the hyphae. Interestingly, even though chitin synthase activity is present in the mycelium of S. monoica, the total amount of chitin in the walls of the mycelium does not exceed 1% of the total cell wall carbohydrate content (15). This raises the question of the role of chitin biosynthesis in Saprolegniales. A possible answer to this question is provided by our discovery that chitin synthase activity occurs in DRMs and thus that it is most likely involved in cell wall polysaccharide biosynthesis at the tip of the mycelium where lipid rafts are proposed to be polarized. The delivery of active chitin synthase to the apical zone by polarized lipid rafts might transiently compensate for the absence of cellulose synthesis in this part of the elongating cell. Although the above hypothesis is quite attractive in terms of mechanisms of hyphal growth and morphogenesis, we cannot rule out at this stage the possibility that cellulose synthase, which is known to be a highly unstable complex (18, 36), is in fact located in lipid rafts, together with (1→3)-β-d-glucan and chitin synthases, but that it is inactivated during the preparation of DRMs.
Supplementary Material
Acknowledgments
We thank Chiori Ito and Junji Sugiyama (Kyoto University, Kyoto, Japan) for help with electron microscopy imaging and David B. Wilson (Cornell University) for the generous gift of recombinant cellulases. Fatty acid and sterol analyses were performed using the facilities of the Lipidomic Platform associated with the Institut Multidisciplinaire de Biochimie des Lipides (Lyon, France).
This study was supported by the Swedish Centre for Biomimetic Fiber Engineering and by grants from the EU (contract QLK5-CT-2001-00443), the French Ministry of Research, and the Japanese Society for the Promotion of Science to V.B.
Footnotes
Published ahead of print on 5 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnat, M., and K. Simons. 2002. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA 99:14183-14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagnat, M., and K. Simons. 2002. Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol. Chem. 383:1475-1480. [DOI] [PubMed] [Google Scholar]
- 4.Baldauf, S. L., A. J. Roger, I. Wenk-Siefert, and W. F. Doolittle. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972-977. [DOI] [PubMed] [Google Scholar]
- 5.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 6.Borner, G. H. H., D. J. Sherrier, T. Weimar, L. V. Michaelson, N. D. Hawkins, A. MacAskill, J. A. Napier, M. H. Beale, K. S. Lilley, and P. Dupree. 2005. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137:104-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouzenzana, J., L. Pelosi, A. Briolay, J. Briolay, and V. Bulone. 2006. Identification of the first oomycete annexin as a (1→3)-β-d-glucan synthase activator. Mol. Microbiol. 62:552-565. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Briskin, D. P., R. T. Leonard, and T. K. Hodges. 1987. Isolation of the plasma-membrane: membrane markers and general principles. Methods Enzymol. 148:542-558. [Google Scholar]
- 10.Broquet, A. H., G. Thomas, J. Masliah, G. Trugnan, and M. Bachelet. 2003. Expression of the molecular chaperone Hsp70 in detergent-resistant microdomains correlates with its membrane delivery and release. J. Biol. Chem. 278:21601-21606. [DOI] [PubMed] [Google Scholar]
- 11.Brownfield, L., K. Ford, M. S. Doblin, E. Newbigin, S. Read, and A. Bacic. 2007. Proteomic and biochemical evidence links the callose synthase in Nicotiana alata pollen tubes to the product of the NaGSL1 gene. Plant J. 52:147-156. [DOI] [PubMed] [Google Scholar]
- 12.Bruno, D. W., and B. P. Wood. 1999. Saprolegnia and other oomycetes, p. 599-659. In P. T. K Woo and D. W. Bruno (ed.), Fish diseases and disorders, vol. 3. CABI Publishing, Wallingford, Oxon, United Kingdom. [Google Scholar]
- 13.Bu'Lock, J. D., and A. U. Osagie. 1976. Sterol biosynthesis via cycloartenol in Saprolegnia. Phytochemistry 15:1249-1251. [Google Scholar]
- 14.Bulone, V., G. B. Fincher, and B. A. Stone. 1995. In vitro synthesis of a microfibrillar (1→3)-β-glucan by a ryegrass (Lolium multiflorum) endosperm (1→3)-β-glucan synthase enriched by product entrapment. Plant J. 8:213-225. [Google Scholar]
- 15.Bulone, V., H. Chanzy, L. Gay, V. Girard, and M. Fèvre. 1992. Characterization of chitin and chitin synthase from the cellulosic cell wall fungus Saprolegnia monoica. Exp. Mycol. 16:8-21. [Google Scholar]
- 16.Bulone, V., V. Girard, and M. Fèvre. 1990. Separation and partial purification of (1→3)-β-glucan and (1→4)-β-glucan synthases from Saprolegnia. Plant Physiol. 94:1748-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Com, E., B. Evrard, P. Roepstorff, F. Aubry, and C. Pineau. 2003. New insights into the rat spermatogonial proteome. Mol. Cell. Proteomics 2:248-261. [DOI] [PubMed] [Google Scholar]
- 18.Delmer, D. P. 1999. Cellulose biosynthesis: exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:245-276. [DOI] [PubMed] [Google Scholar]
- 19.Erwin, D. C., and O. K. Ribeiro. 1996. Phytophthora diseases worldwide. APS Press, St. Paul, MN.
- 20.Fairweather, J. K., J. Lai Kee Him, L. Heux, H. Driguez, and V. Bulone. 2004. Structural characterization by 13C-NMR spectroscopy of products synthesized in vitro by polysaccharide synthases using 13C-enriched glycosyl donors. Application to a UDP-glucose:(1→3)-β-d-glucan synthase from blackberry (Rubus fruticosus). Glycobiology 14:775-781. [DOI] [PubMed] [Google Scholar]
- 21.Foster, L. J., C. L. De Hoog, and M. Mann. 2003. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. USA 100:5813-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gajendran, K., M. D. Gonzales, A. Farmer, E. Archuleta, J. Win, M. E. Waugh, and S. Kamoun. 2006. Phytophthora functional genomics database (PFGD): functional genomics of Phytophthora-plant interactions. Nucleic Acids Res. 34:D465-D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gay, L., H. Chanzy, V. Bulone, V. Girard, and M. Fèvre. 1993. Synthesis in vitro of crystalline chitin by a solubilized enzyme from the cellulosic fungus Saprolegnia monoica. J. Gen. Microbiol. 139:2117-2122. [Google Scholar]
- 24.Girard, V., and M. Fèvre. 1984. β-1,4 and β-1,3 glucan synthases are associated with the plasma membrane of the fungus Saprolegnia. Planta 160:400-406. [DOI] [PubMed] [Google Scholar]
- 25.Hancock, J. F. 2006. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 7:456-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirai, I., N. Sato, W. Qi, S. Ohtani, T. Torigoe, and K. Kikuchi. 1998. Localization of pNT22 70-kDa heat shock cognate-like protein in the plasma membrane. Cell. Struct. Funct. 23:153-158. [DOI] [PubMed] [Google Scholar]
- 27.Hussein, M. M. A., and K. Hatai. 2002. Pathogenicity of Saprolegnia species associated with outbreaks of salmonid saprolegniosis in Japan. Fish. Sci. 68:1067-1072. [Google Scholar]
- 28.Ikonen, E. 2001. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 13:470-477. [DOI] [PubMed] [Google Scholar]
- 29.Itaska, O., T. Hori, K. Sasahara, Y. Wakabayashi, F. Takahashi, and H.-I. Rhee. 1984. Analysis of phospho- and phosphonosphingolipids by high-performance liquid chromatography. J. Biochem. 95:1671-1675. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, T. W., Jr., R. L. Seymour, and D. E. Padgett. 2002. Biology and systematics of the Saprolegniaceae. National Science Digital Library. http://dl.uncw.edu/digilib/biology/fungi/taxonomy%20and%20systematics/padgett%20book/.
- 31.Kato, M., Y. Muto, K. Tanakabandoh, K. Watanabe, and K. Ueno. 1995. Sphingolipid composition in Bacteroides species. Anaerobe 1:135-139. [DOI] [PubMed] [Google Scholar]
- 32.Knight, J. 2002. Fears mount as oak blight infects redwoods. Nature 415:251. [DOI] [PubMed] [Google Scholar]
- 33.Kogan, G., J. Alföldi, and L. Masler. 1988. Carbon-13 NMR spectroscopic investigation of two yeast cell wall β-d-glucans. Biopolymers 27:1055-1063. [Google Scholar]
- 34.Kübler, E., H. G. Dohlman, and M. P. Lisanti. 1996. Identification of Triton X-100 insoluble membrane domains in the yeast Saccharomyces cerevisiae: lipid requirements for targeting of heterotrimeric G protein subunits. J. Biol. Chem. 271:32975-32980. [DOI] [PubMed] [Google Scholar]
- 35.Kumar, C., and A. Rzhetsky. 1996. Evolutionary relationships of eukaryotic kingdoms. J. Mol. Evol. 42:183-193. [DOI] [PubMed] [Google Scholar]
- 36.Lai Kee Him, J., H. Chanzy, M. Müller, J.-L. Putaux, T. Imai, and V. Bulone. 2002. In vitro versus in vivo cellulose microfibrils from plant primary wall synthases: structural differences. J. Biol. Chem. 277:36931-36939. [DOI] [PubMed] [Google Scholar]
- 37.Lai Kee Him, J., L. Pelosi, H. Chanzy, J.-L. Putaux, and V. Bulone. 2001. Biosynthesis of (1→3)-β-d-glucan (callose) by detergent extracts of a microsomal fraction from Arabidopsis thaliana. Eur. J. Biochem. 268:4628-4638. [DOI] [PubMed] [Google Scholar]
- 38.Larsson, C., M. Sommarin, and S. Widell. 1994. Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles, p. 451-459. In H. Walter and G. Johansson (ed.), Aqueous two-phase systems, vol. 228. Academic Press, Inc., San Diego, CA. [Google Scholar]
- 39.Lefebvre, B., F. Furt, M.-A. Hartmann, L. V. Michaelson, J.-P. Carde, F. Sargueil-Boiron, M. Rossignol, J. A. Napier, J. Cullimore, J.-J. Bessoule, and S. Mongrand. 2007. Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 144:402-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, N., A. Mak, D. P. Richards, C. Naber, B. O. Keller, L. Li, and A. R. Shaw. 2003. Monocyte lipid rafts contain proteins implicated in vesicular trafficking and phagosome formation. Proteomics 3:536-548. [DOI] [PubMed] [Google Scholar]
- 41.Lichtenberg, D., F. M. Goñi, and H. Heerklotz. 2005. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem. Sci. 30:430-436. [DOI] [PubMed] [Google Scholar]
- 42.Ma, X., and J. Stöckigt. 2001. High yielding one-pot enzyme-catalyzed synthesis of UDP-glucose in gram scales. Carbohydr. Res. 333:159-163. [DOI] [PubMed] [Google Scholar]
- 43.Machlis, L. 1953. Growth and nutrition of watermolds in the subgenus Euallomyces. II. Optimal composition of the minimal medium. Am. J. Bot. 40:449-460. [Google Scholar]
- 44.Margulis, L., and K. V. Schwartz. 2000. Five kingdoms: an illustrated guide to the phyla of life on Earth. W. H. Freeman and Co., New York, NY.
- 45.Martin, S. W., and J. B. Konopka. 2004. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot. Cell 3:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCorkindale, N. J., S. A. Hutchinson, B. A. Pursey, W. T. Scott, and R. Wheeler. 1969. A comparison of the types of sterol found in species of the Saprolegniales and Leptomitales with those found in some other Phycomycetes. Phytochemistry 8:861-867. [Google Scholar]
- 47.Mitter, D., C. Reisinger, B. Hinz, S. Hollmann, S. V. Yelamanchili, S. Treiber-Held, T. G. Ohm, A. Herrmann, and G. Ahnert-Hilger. 2003. The synaptophysin/synaptobrevin interaction critically depends on the cholesterol content. J. Neurochem. 84:35-42. [DOI] [PubMed] [Google Scholar]
- 48.Moertz, E., T. N. Krogh, H. Vorum, and A. Gorg. 2001. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 1:1359-1363. [DOI] [PubMed] [Google Scholar]
- 49.Mongrand, S., J. Morel, J. Laroche, S. Claverol, J. P. Carde, M. A. Hartmann, M. Bonneu, F. Simon-Plas, R. Lessire, and J.-J. Bessoule. 2004. Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279:36277-36286. [DOI] [PubMed] [Google Scholar]
- 50.Moreau, R. A., D. H. Young, P. O. Danis, M. J. Powell, C. J. Quinn, K. Beshah, R. A. Slawecki, and R. L. Dilliplane. 1998. Identification of ceramide-phosphorylethanolamine in oomycete plant pathogens: Pythium ultimum, Phytophthora infestans, and Phytophthora capsici. Lipids 33:307-317. [DOI] [PubMed] [Google Scholar]
- 51.Morel, J., S. Claverol, S. Mongrand, F. Furt, J. Fromentin, J.-J. Bessoule, J.-P. Blein, and F. Simon-Plas. 2006. Proteomics of plant detergent-resistant membranes. Mol. Cell. Proteomics 5:1396-1411. [DOI] [PubMed] [Google Scholar]
- 52.Mort-Bontemps, M., L. Gay, and M. Fèvre. 1997. CHS2, a chitin synthase gene from the oomycete Saprolegnia monoica. Microbiology 143:2009-2020. [DOI] [PubMed] [Google Scholar]
- 53.Paquin, B., M. J. Laforest, L. Forget, I. Roewer, Z. Wang, J. Longcore, and B. F. Lang. 1997. The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr. Genet. 31:380-395. [DOI] [PubMed] [Google Scholar]
- 54.Pelosi, L., T. Imai, H. Chanzy, L. Heux, E. Buhler, and V. Bulone. 2003. Structural and morphological diversity of (1→3)-β-d-glucans synthesized in vitro by enzymes from Saprolegnia monoica: comparison with a corresponding in vitro product from blackberry (Rubus fruticosus). Biochemistry 42:6264-6274. [DOI] [PubMed] [Google Scholar]
- 55.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 56.Peskan, T., M. Westermann, and R. Oelmüller. 2000. Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur. J. Biochem. 267:6989-6995. [DOI] [PubMed] [Google Scholar]
- 57.Plakas, S. M., K. R. El Said, G. R. Stehly, W. H. Gingerich, and J. L. Allen. 1996. Uptake, tissue distribution, and metabolism of malachite green in the channel catfish (Ictalurus punctatus). Can. J. Fish. Aquat. Sci. 53:1427-1433. [Google Scholar]
- 58.Prime, T. A., D. J. Sherrier, P. Mahon, L. C. Packman, and P. Dupree. 2000. A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21:3488-3499. [DOI] [PubMed] [Google Scholar]
- 59.Rajendran, L., and K. Simons. 2005. Lipid rafts and membrane dynamics. J. Cell Sci. 118:1099-1102. [DOI] [PubMed] [Google Scholar]
- 60.Robinson, D. G., H. P. Haschke, G. Hinz, B. Hoh, M. Maeshima, and F. Marty. 1996. Immunological detection of tonoplast polypeptides in the plasma membrane of pea cotyledons. Planta 198:95-103. [Google Scholar]
- 61.Sabba, R. P., and K. C. Vaughn. 1999. Herbicides that inhibit cellulose biosynthesis. Weed Sci. 47:757-763. [Google Scholar]
- 62.Saito, H., T. Ohki, and T. Sasaki. 1979. A 13C-nuclear magnetic resonance study of polysaccharide gels. Molecular architecture in the gels consisting of fungal, branched (1→3)-β-d-glucans (lentinan and schizophyllan) as manifested by conformational changes induced by sodium hydroxide. Carbohydr. Res. 74:227-240. [Google Scholar]
- 63.Schuck, S., and K. Simons. 2004. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 117:5955-5964. [DOI] [PubMed] [Google Scholar]
- 64.Shapiro, A., and J. T. Mullins. 2002. Distribution of 1,3-β-glucans in elongating and non-elongating regions of the wall. Mycologia 94:267-272. [PubMed] [Google Scholar]
- 65.Shapiro, A., and J. T. Mullins. 2002. II. Distribution of cellulose in elongating and non-elongating regions of the wall. Mycologia 94:273-279. [PubMed] [Google Scholar]
- 66.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 67.Stewart, J. C. 1980. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 104:10-14. [DOI] [PubMed] [Google Scholar]
- 68.Tyler, B. M., S. Tripathy, X. M. Zhang, P. Dehal, R. H. Y. Jiang, A. Aerts, F. D. Arredondo, L. Baxter, D. Bensasson, J. L. Beynon, J. Chapman, C. M. B. Damasceno, A. E. Dorrance, D. L. Dou, A. W. Dickerman, I. L. Dubchak, M. Garbelotto, M. Gijzen, S. G. Gordon, F. Govers, N. J. Grunwald, W. Huang, K. L. Ivors, R. W. Jones, S. Kamoun, K. Krampis, K. H. Lamour, M. K. Lee, W. H. McDonald, M. Medina, H. J. G. Meijer, E. K. Nordberg, D. J. Maclean, M. D. Ospina-Giraldo, P. F. Morris, V. Phuntumart, N. H. Putnam, S. Rash, J. K. C. Rose, Y. Sakihama, A. A. Salamov, A. Savidor, C. F. Scheuring, B. M. Smith, B. W. S. Sobral, A. Terry, T. A. Torto-Alalibo, J. Win, Z. Y. Xu, H. B. Zhang, I. V. Grigoriev, D. S. Rokhsar, and J. L. Boore. 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313:1261-1266. [DOI] [PubMed] [Google Scholar]
- 69.van West, P. 2006. Saprolegnia parasitica, an oomycete pathogen with a fishy appetite: new challenges for an old problem. Mycologist 20:99-104. [Google Scholar]
- 70.Wang, R., J. T. Kovalchin, P. Muhlenkamp, and R. Y. Chandawarkar. 2006. Exogenous heat shock protein 70 binds macrophage lipid raft microdomain and stimulates phagocytosis, processing, and MHC-II presentation of antigens. Blood 107:1636-1642. [DOI] [PubMed] [Google Scholar]
- 71.Wang, R., T. Town, V. Gokarn, R. A. Flavel, and R. Y. Chandawarkar. 2006. HSP70 enhances macrophage phagocytosis by interaction with lipid raft-associated TLR-7 and upregulating p38 MAPK and PI3K pathways. J. Surg. Res. 136:58-69. [DOI] [PubMed] [Google Scholar]
- 72.Warner, S. A., D. F. Eierman, G. W. Sovocool, and A. J. Domnas. 1982. Cycloartenol-derived sterol biosynthesis in the Peronosporales. Proc. Natl. Acad. Sci. USA 79:3769-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zappel, N. F., and R. Panstruga. 2008. Heterogeneity and lateral compartmentalization of plant plasma membranes. Curr. Opin. Plant Biol. 11:632-640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.