Abstract
The microbial community structure of cork with marked musty-earthy aromas was analyzed using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Cork stoppers and discs were used for DNA extraction and were analyzed by using selective primers for bacteria and fungi. Stoppers clearly differed from discs harboring a different fungal community. Moreover, musty-earthy samples of both types were shown to have a specific microbiota. The fungi Penicillium glabrum and Neurospora spp. were present in all samples and were assumed to make only a small contribution to off-odor development. In contrast, Penicillium islandicum and Penicillium variabile were found almost exclusively in 2,4,6-trichloroanisole (TCA) tainted discs. Conversely, Rhodotorula minuta and Rhodotorula sloofiae were most common in cork stoppers, where only small amounts of TCA were detected. Alpha- and gammaproteobacteria were the most commonly found bacteria in either control or tainted cork stoppers. Specific Pseudomonas and Actinobacteria were detected in stoppers with low amounts of TCA and 2-methoxy-3,5-dimethylpyrazine. These results are discussed in terms of biological degradation of taint compounds by specific microorganisms. Reliable and straightforward microbial identification methods based on a molecular approach provided useful data to determine and evaluate the risk of taint formation in cork.
Cork stoppers provide the desired level of elasticity and strength to ensure an effective and safe closure of bottles of most wine types and are used as the preferred material for bottling of high-quality wines (45). However, the natural origin of cork and the long manufacturing process of stoppers facilitate a continuous contact of the raw material with different types of microorganisms, which may lead to the development of unpleasant off-odors as a consequence of their metabolism. The origins of undesirable sensory characteristics in wine are varied and may include contaminated grapes and wine cellars in addition to cork stoppers. Some musty-earthy (ME) aromas, such as must taint or mold taint, can occur for 0.5 to 6% of bottled wine (46), thus seriously affecting the reputation of cellars and causing significant economic losses. Such taints are often referred to as “cork-taint” regardless of whether or not the cork is the source.
The chemical analysis of tainted wines has revealed the presence of a variety of taint compounds. In an early work, Amon et al. (4) made a full list of chemical compounds detected both in wine and in cork stoppers that could be responsible for ME aromas. This list included 2,4,6-trichloroanisole (TCA), geosmin (GSM), 2-methyl-isoborneol (MIB), 1-octen-3-one, and 1-octen-3-ol, as the most relevant compounds. More recently, different methoxypyrazines, such as 2-methoxy-3-isopropylpyrazine (IPMP) and 2-methoxy-3,5-dimethylpyrazine (MDMP), together with 2,3,4,6-tetrachloroanisole or 2,4,6-tribromanisole, have also been described as causative agents of musty, earthy, or moldy aromas in wine and cork (9, 43). The main contributors to cork taint are considered to be TCA, MDMP, and TBA (43, 48, 49).
The biological synthesis and accumulation of chloroanisoles by the biomethylation of the corresponding chlorophenols is a well-documented process, which has been proven experimentally in cork spiked with 2,4,6-trichlorophenol as a substrate (14, 25, 31, 36, 39). Methylating reactions have been confirmed with a partially purified chlorophenol O-methyltransferase from Trichoderma longibrachiatum isolated from cork (3, 13). In addition, the participation of bacteria, mainly Streptomyces spp. and Bacillus spp., in the production of guaiacol (GUA) or GSM has been proven experimentally in cork (2, 46). In contrast, scientific knowledge about the biological synthesis and accumulation of methoxypyrazines is scarce and has been detected in only a few food and package products (6, 10, 16, 20, 42).
Cork is shown to constitute a true ecosystem, with a fairly complex and changing microbial community, as determined by cultivation-dependent methods (3, 15, 30, 37). Most authors have found a rather low richness at the final stages of the stopper manufacturing process, with Chrysonilia sytophila and Penicillium glabrum as the dominant species. The identification of Penicillium spp. isolates by using multilocus DNA sequence comparison revealed the presence of a high number of unusual species and some phylogenetically specific groups (44). Moreover, the diversity and the abundance of fungi in TCA tainted stoppers was found to be higher compared to control cork based on analyses of a limited number of isolates (39).
Molecular fingerprinting methods, such as denaturing gradient gel electrophoresis (DGGE) or temperature gradient gel electrophoresis, have been extensively applied to analyze microbial communities in a large variety of samples, including soils (5, 8), decaying wood (52), fermented products (23), or fresh grapes (17). Determination of yeast richness and relative abundances in wine fermentations has also been achieved by PCR-DGGE methods (12, 33). To our knowledge, common molecular fingerprinting methods have not been introduced in the cork industry as discriminatory methods to evaluate the risk of off-flavor development.
In the present study a PCR-DGGE method was applied to the study of the microbial composition of cork samples for the first time. The experimental design was aimed at establishing direct relationships between the presence of bacteria and fungi and the development of ME off-flavors in corks. The analysis was carried out using samples from the halfway and final production stages of cork stopper manufacturing.
MATERIALS AND METHODS
Cork samples.
The samples studied included two batches of cork discs and ready-to-use agglomerate cork stoppers for Spanish sparkling wine (D. O. Cava). Cork discs (35 mm in diameter, 6 mm thick) were obtained from direct chiseling of preboiled cork slabs and were representative of natural cork material at a middle stage of stopper manufacturing. Cork stoppers for sparkling wine (30 mm in diameter, 47 mm thick) consisted of a portion of agglomerated cork granules and two individual cork discs glued at one end of the cylinder.
Olfactory determination of ME off-flavors.
Cork samples were selected, tested, and classified according to the presence or absence of undesirable sensory characteristics. Cork samples were macerated for 24 h in double-distilled water according to the method proposed by the AENOR regulation UNE 56928 (50). Sensory evaluation was done by three independent trained judges at the providing industries. Two specific 1-h training sessions were carried out before the panel evaluated the macerates. In the first training session, judges generated descriptive terms for the sensory deviations present in different cork samples. In the second session, different aroma standards were presented and discussed by the panel. Three terms were selected through these discussions and used for further descriptive analysis: no undesirable aromas (samples designated as controls [C]), ME taints, and ME samples exhibiting a recognizable TCA (ME-TCA). Despite the sampling effort, all tainted cork discs found exhibited a strong TCA aroma, and no representatives of the ME class were found. All three classes were found with the cork stopper samples.
Analysis of off-flavor compounds by HS-SPME-GC/MS.
Headspace solid-phase microextraction (HS-SPME) was chosen as the extraction and preconcentration method for the screening of TCA, IPMP, GSM, MIB, and GUA. Although GUA has a smoky aroma, it was included in the analysis since its presence in cork and wine has been previously reported. Samples consisted of single cork stoppers (9 g on average) or 2 U of cork discs (∼2 g each). Macerates for chemical analyses were obtained after soaking cork in 100 ml of distilled water for 24 h without agitation at room temperature. Pyrex glass bottles of appropriate volume to ensure a complete submersion of cork were used. SPME analyses of 5-ml aliquots of cork macerates were performed with a manual fiber holder with a 50/30-μm divinylbenzene-carboxen-polydimethylsiloxane fiber exactly as previously described (40). Gas chromatography with mass spectrometry (GC/MS) detection was used for the analysis of extracted compounds. The selected m/z ratios used for quantification were 210 and 212 for TCA, 137 for IPMP, 112 for GSM, 95 for MIB, and 124 for GUA. SPME calibration graphs for TCA, IPMP, GSM, MIB, and GUA were obtained by using d5-TCA as internal standard.
Determination of MDMP.
In order to quantify the presence in the macerates of MDMP at lower levels than achieved previously (47), an improved analytical procedure with a limit of detection of 1 ng/liter for this analyte was developed. The new method utilized the weakly basic properties of MDMP to allow this compound to be concentrated in aqueous solution by extraction into pentane and then back extraction into mineral acid, which was neutralized and analyzed by HS-SPME-GC/MS. The SPME method was optimized for fiber type, ionic strength, incubation temperature, and fiber extraction time. The optimal sample preparation and analytical conditions were as follows: 2H3-MDMP, prepared as described by Simpson et al. (47) was added, as an internal standard, as an ethanol solution (10 ng/ml, 125 μl) to cork macerate (25 ml) in a volumetric flask. The macerate was then extracted with pentane (three times with 5 ml each time). The combined pentane extract was back extracted with 1 M HCl (5 ml), the acidic portion was collected into an SPME vial and then blown down with a stream of nitrogen to remove any traces of pentane. The solution was neutralized with sodium carbonate (typically, 50 mg), and sodium chloride (2 g) was added prior to capping. GC/MS was carried out as described previously (47) with the following modifications. The Gerstel MPS2 was operated in solid-phase microextraction injection mode with a 65-μm-pore-size polydimethylsiloxane-divinylbenzene (blue; Supelco part 57311) fiber fitted. The gas chromatograph was fitted with an approximately 30-m-by-0.25-mm J&W fused silica capillary column DB-1701 (0.25-μm film thickness). The carrier gas (helium) flow rate was 1.8 ml/min. The SPME method was set in prep-ahead mode, i.e., each vial was agitated at 500 rpm for 10 min at 30°C prior to fiber exposure. The fiber was exposed to each sample for 40 min at 50°C. The fiber was then desorbed into the injector at 240°C for 15 min and held at this temperature throughout the run. The splitter, at 42:1, was opened after 36 s. The injection was done in pulse splitless mode with an inlet pressure of 25.0 lb/in2 maintained until splitting. The oven temperature was started at 50°C, held at this temperature for 1 min, increased to 130°C at 5°C/min, and then increased to 260°C at 50°C/min and held at this upper temperature for 10 min. The mass spectrometer was operated in selected ion monitoring mode. The analytical method was validated by a series of standard additions of unlabeled MDMP (0, 0.5 to 100 ng/liter, n = 8 for the analyte) to a dry white wine (9.5% ethanol [pH 3.2]) and a dry red wine (12.5% ethanol [pH 3.4]). The standard addition curves obtained were linear throughout the concentration range, with a coefficient of determination (r2) of 1.000 and linear regression equation of y = 1.98x - 0.0042. To determine the precision of the analysis, seven replicate white wine samples were spiked at two different concentrations: 50 and 5 ng/liter. The standard deviations for each were 0.6 and 0.1 ng/liter, respectively.
Nucleic acid extraction.
Cork stoppers were treated as individual samples. For the analysis of cork discs, two units were combined to facilitate the detection of microorganisms. Either cork discs or stoppers were placed in 250-ml Erlenmeyer flasks and washed twice in 50 ml of a sterile isotonic solution for 1 h at 30°C using a rotary shaker HT Infors Unitron (InforsAG, Bottmingen, Switzerland) at 250 rpm. The two washing solutions were combined into one 100-ml sample and centrifuged at 10,000 rpm for 30 min at 4°C using a RC 5BPlus Sorvall centrifuge (Sorvall/Du Pont Co., Wilmington, DE). DNA was extracted from pellets by using a CTAB (cetyltrimethylammonium bromide)-based method suitable for decaying plant material modified from Pla et al. (38). Briefly, cell pellets were resuspended in 600 μl of preheated CTAB extraction buffer (100 mM Tris-HCl [pH 8.0], 20 mM EDTA, 1.4 M NaCl, 2% CTAB [wt/vol]), and the cells were disrupted twice in a glass bead mill Mini-Bead-Beater (BioSpec Products, Bartlesville, OH) operating at maximum speed for 45 s. Polyvinylpolypyrrolidone and β-mercaptoethanol (2% [wt/vol], final concentration of each) were added to the extraction buffer, and samples were incubated at 65°C for 30 min. Tubes were cooled down to room temperature for at least 10 min. Chloroform-isoamyl alcohol (25:1, 700 μl) was added, and the tubes were gently mixed by inversion and centrifuged at 6,000 rpm for 15 min. The aqueous phase was transferred to a clean sterile tube, and the last rinsing step was repeated. Nucleic acid precipitation was done by using a standard ammonium acetate-ethanol method at 4°C for at least 12 h at 4°C. Nucleic acids were pelleted by centrifugation at 10,000 rpm for 10 min. The supernatant was discarded, and the pellet was washed with cold (−20°C) 70% (vol/vol) ethanol. The ethanol was evaporated in a speed vacuum centrifuge (Vac VR1; Heto, Denmark), and the pellets were finally rehydrated in 50 to 100 μl of distilled water. The amount of DNA was quantified spectrophotometrically using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE) and stored at −80°C.
PCR amplification.
All chemicals and Taq polymerase used for the amplification of nucleic acids of both bacteria and fungi were provided by Applied Biosystems (Foster City, CA). Amplifications were done in a 9700 GeneAmp thermal cycler (PE/Applied Biosystems). Partial 16S rRNA gene fragments (∼620 bp) were obtained by PCR amplification using the Bacteria universal primers 357F and 907R. Reaction mixtures and PCR amplification conditions and programs were applied as previously described (29).
Partial sequences of the internal transcribed gene spacer ITS1 from fungi were obtained by using a seminested PCR amplification approach. Primers ITS-1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) (19) and ITS-4 (5′-TCCTCCGCTTATTGATATGC-3′) (53) were used for the first amplification step. Nested PCRs were conducted by using the primers ITS-1F and ITS-2 (5′-GCTGCGTTCTTCATCGATGC-3′) (53). PCR mixtures for both steps contained 5 μl of 10× PCR buffer, 2 mM MgCl2, 200 μM concentrations of each deoxynucleotide triphosphate, a 0.4 mM concentration of each primer, and 400 ng of bovine serum albumin/μl in a total volume of 50 μl.
A 44-bp GC-rich clamp sequence was added to the 5′ ends of primers 357F and ITS-1F for separation of the PCR products in DGGE (35). PCR conditions using primers 357F-GC or ITS-1F-GC were identical to those stated above.
DGGE.
PCR products were concentrated when necessary to an approximate concentration of 100 ng of DNA/μl. Concentrated samples (30 μl) were loaded on DGGE gels. Appropriate standards containing a mixture of PCR products of five known microorganisms were loaded on both sides and the center of acrylamide gels and used for comparison. DGGE standards consisted of PCR-amplified 16S rRNA gene fragments of Micrococcus luteus, Pseudomonas aeruginosa, and Sulfolobus solfataricus and the ITS1 fragments of Mucor plumbeus and Saccharomyces cerevisiae. Analyses were carried out using an INGENY PhorU system (Ingeny, Goes, The Netherlands). Six percent (wt/vol) acrylamide-bisacrylamide (37.5:1) gels were prepared with a 20 to 70% urea-formamide vertical gradient according to the instructions of the manufacturer. One hundred percent denaturing solutions contained forty percent formamide and 7 M urea in 1× Tris-acetate-EDTA buffer. Electrophoresis was performed in 1× Tris-acetate-EDTA buffer at 120V for 17 h at a constant temperature of 60°C. Gels were stained for 45 min with SybrGold (1:10,000; Invitrogen Molecular Probes, Eugene, OR) and visualized under UV excitation.
Bands of interest were chosen after detailed analyses of images and excised from gels for further reamplification and sequencing. DNA was rehydrated in 50 μl of 10 mM Tris-HCl (pH 7.4) and eluted after incubation at 65°C for 30 min.
Gel image analysis.
Digital images of acrylamide gels were analyzed by using the GELCompar II v.4.0 software package (Applied Maths BVBA, Sint-Martens-Latem, Belgium). Lanes were manually defined, and band positions were identified from corrected intensity plots. Comparison between samples loaded on different DGGE gels was completed using normalized values derived from known standards. A binary matrix showing the presence or absence of identified bands was made for all gel lanes. Further, a similarity matrix based on the Dice coefficient was calculated, and samples were clustered according to the UPGMA (unweighted pair-group method with arithmetic averages) algorithm using a tolerance position value of 1.5%.
DNA sequencing and data analysis.
PCR-amplified DNA products obtained from multiple DGGE bands at the same relative position were sequenced using forward ITS-1F and reverse 907R primers for fungi and bacteria, respectively (Macrogen, Seoul, South Korea). Sequences were checked for chimera detection, manually refined by using the BioEdit package (22), and aligned using CLUSTAL W (51). Aligned sequences were analyzed with the Blast2 advanced program at the National Center for Biotechnology Information and fungal or bacterial species identified as closer similarities to known sequences in the database. Nucleotide sequences were used for construction of phylogenetic trees with the distance (neighbor-joining [NJ]) and maximum-parsimony methods, respectively. NJ and maximum-parsimony trees were obtained by using the MEGA version 4.0 software with 1,000 bootstrap replicates (28).
Statistical analyses.
Differences in the concentrations of taint compounds were tested for the effects of sample type (cork discs and stoppers) and sensory deviations (C, ME, and ME-TCA samples) with a Mann-Whitney test for unequal variances using SPSS software (SPSS, Inc.). Differences between mean concentrations of taint compounds of sensory classes of corks were further analyzed using Tukey's HSD post hoc test at P ≤ 0.05 with the general linear model procedure of the SPSS software.
Nucleotide sequence accession numbers.
Partial sequences of the bacterial 16S rRNA and fungal ITS1 regions of the dominant bands obtained in the DGGE gels have been deposited in the GenBank database under accession numbers FJ217223 to FJ217256 and FJ217257 to FJ217328, respectively.
RESULTS
Chemical characterization of selected cork samples.
Quantitative analyses of aqueous cork macerates by HS-SPME-GC/MS revealed the presence of GSM in only three of the tainted cork discs, with concentrations ranging from 2.0 to 19.9 ng/liter. IPMP was not detected in any of the samples used. As a result, GSM and IPMP were considered to contribute little to the unpleasant odors detected in the sensorial analyses. GUA was invariably found at the μg/liter level in all samples, but no significant differences (P > 0.5) were found in any of the comparisons made (Table 1). The main differences between sample types were found for TCA and MDMP. ME-TCA samples of both discs and finished corks exhibited a high concentration of releasable TCA compared to the corresponding control or ME samples. Highly significant differences were recorded for cork discs (P < 0.001). Moreover, the TCA content was significantly higher in stoppers than in cork discs in either control or ME-TCA sensory classes. Despite the high variability found between replicates of the same sample type, the content of MDMP was found to be significantly different in the three sensory classes analyzed (P < 0.05). A more detailed analysis using a Tukey's HSD test indicated that the MDMP content of ME samples differed significantly from ME-TCA samples (P = 0.03) but not from control, untainted samples (P = 0.13).
TABLE 1.
Mean concentration values for TCA, GUA, and MDMP found in cork macerates
| Source (no. of samples)a | Mean concn (SE)
|
||
|---|---|---|---|
| TCA (ng/liter) | GUA (μg/liter) | MDMP (ng/liter) | |
| Cork stoppers | |||
| C (6) | 4.6 (0.9) | 1.4 (0.3) | 3.4 (1.0) |
| ME (10) | 6.1 (1.1) | 1.2 (0.1) | 11.2 (3.6) |
| ME-TCA (8) | 85.6 (22.1) | 1.4 (0.1) | 1.3 (0.2) |
| Cork discs | |||
| C (7) | 0.4 (0.2) | 1.3 (0.1) | NDb |
| ME-TCA (18) | 35.8 (8.8) | 2.9 (1.4) | ND |
C, nontainted cork (control); ME, ME taint; ME-TCA, ME-TCA taint.
ND, not determined.
DGGE fingerprints of cork samples.
The recovered nucleic acids varied between 3.5 to 260 ng of total DNA/μl and was less in the case of cork discs. The observed differences were not related to previous sensory classification of corks and seemed to relate to other factors such as the heterogeneity in microbial colonization or the presence of cork dust. Positive PCR amplifications of fungal internal transcribed spacer 1 (ITS1) fragments of the expected size could be obtained for most of the samples. Unfortunately, no PCR products were obtained for any of the ME-TCA stoppers provided. This negative result may be due to the presence of phenolic compounds, which will affect DNA extraction and may act as PCR inhibitors.
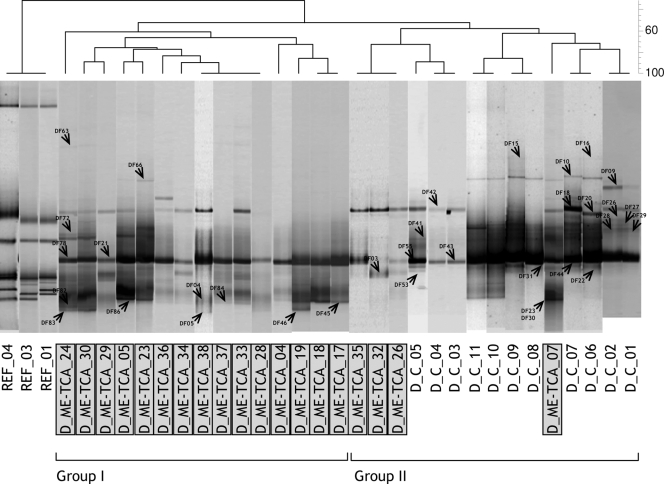
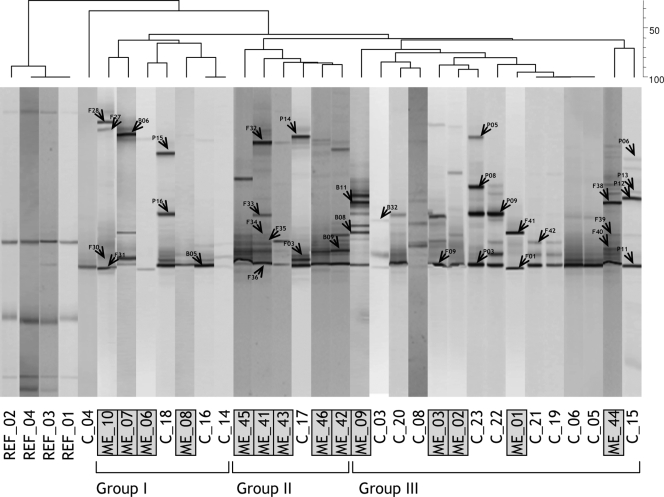
DGGE profiles revealed the presence of a fairly simple fungal community with no more than 10 different bands in a single sample. Cork discs, either control or TCA tainted, showed similar levels of fingerprint richness (i.e., the number of DGGE bands). The numbers of bands in DGGE ranged from eight to two in tainted and from seven to four in control discs (Fig. 1). Samples clustered in two distinct groups at a 50% similarity level. Group I contained all control samples, together with three ME-TCA cork discs. Group II contained the rest of ME-TCA samples. The PCR-DGGE approach was less discriminatory between replicates where high ME tainted cork stoppers were compared to untainted stoppers, and the differences between replicates were shown to be high (Fig. 2). Although different groups could be distinguished at a 60% similarity level, no consistency was found in clusters according to sensory deviations.
FIG. 1.
Negative image composition of SybrGold-stained DGGE gels of environmental ITS1 genes of fungi obtained with the specific primer set ITS-1F-ITS2. The dendrogram on top is based on a similarity matrix (Dice index) calculated from a presence-absence binary matrix of bands. Grouping was based on the UPGMA method. Bands indicated with an arrow head were excised and sequenced. REF, reference position markers for gel comparison; D_ME-TCA, TCA tainted cork discs; D_C, control untainted cork discs.
FIG. 2.
Negative image composition of SybrGold-stained DGGE gels of environmental ITS1 genes of fungi obtained with the specific primer set ITS-1F-ITS2 from cork stoppers. The dendrogram on top is based on a calculated similarity matrix (Dice index) from a presence-absence binary matrix of bands. Grouping has been made by using a UPGMA method. Bands indicated with an arrow head were excised and sequenced. REF, reference position markers for gel comparison; ME, ME tainted cork stoppers; C, control untainted cork stoppers.
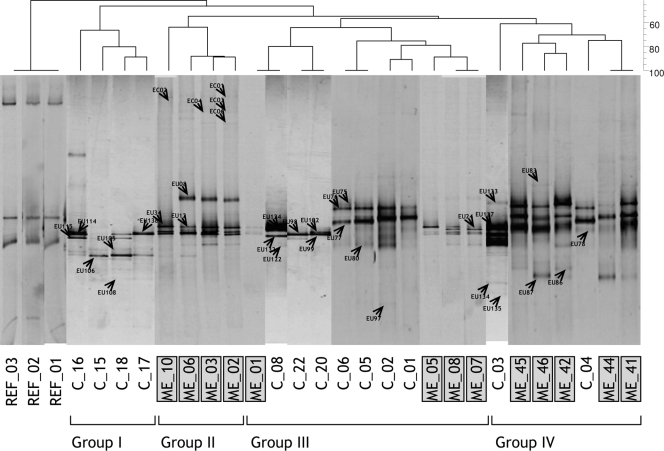
The analysis of the fungal community structure of finished cork stoppers was complemented with the molecular fingerprint of bacteria in order to increase the differentiation capacity of the sensorial groups. We failed in the amplification of some ME and control samples, and the number was reduced to 13 each (Fig. 3). Fingerprint richness was slightly higher in tainted samples (the number of bands ranging from 10 to 2) than in control samples (from 8 to 2 bands). Similar to what was observed for fungi, not all samples clustered consistently with classes defined after sensorial analysis. Samples distributed in four different groups at a 60% similarity level. Groups I and II contained exclusively control and ME cork stoppers, respectively, and were clearly separate from the rest. Groups III and IV contained both control and tainted samples.
FIG. 3.
Negative image composition of SybrGold-stained DGGE gels of environmental bacterial 16S rRNA genes obtained with the specific primer set 357F-907R from cork stoppers. The dendrogram on top is based on a calculated similarity matrix (Dice index) from a presence-absence binary matrix of bands. Grouping has been made by using a UPGMA method. Bands indicated with an arrow head were excised and sequenced. REF, reference position markers for gel comparison; ME, ME tainted cork stoppers; C, control untainted cork stoppers.
Identification of microorganisms and sequence analysis.
Ninety-two bands were excised and treated for further PCR reamplification and sequencing. From these, 69 bands produced useful sequences with no ambiguous positions that matched with the ITS1 region from fungi in databases and were used for identification. Most sequences yielded very high similarity values (98 to 100%) with previously published sequences. Moreover, some bands present at different positions in the gel resulted in identical sequences (i.e., bands DF26, DF27, DF28, and DF29 in Fig. 1), which may be due to variable melting behaviors or the presence of multiple ribosomal gene copies in a single organism. The result of unspecific amplifications due to the use of a seminested protocol was discarded as an explanation for band duplication since similar results have been previously reported with completely different samples and PCR protocols and seem to be common in the analysis of fungal ITSs (5).
Based on the number of identified fungal species, the community structure in finished cork stoppers showed the highest complexity (Table 2). Some fungi, mainly Neurospora spp. and Penicillium glabrum, were found in almost all samples, confirming their broad distribution in cork. Stoppers showed a higher presence of Basidiomycota, which are rarely present in cork discs. Specific organisms only found in finished stoppers included Fusarium poae and Fusarium sambucinum, Cladosporium cladosporioides, Eurotium repens, Cryptococcus carnescens, Udeniomyces pannonicus, Rhodotorula minuta, Rhodotorula sloofiae, and Rhodotorula glutinis. Specific sequences found only in cork discs included Nectria mauritiicola, Sporotrichum laxum, and two species of Penicillium.
TABLE 2.
Most probable sequence identification and occurrence of DGGE bands in control and ME and ME-TCA tainted cork stoppers and discs: closest fungal speciesa
| Group and band no.b | Closest fungal species | % Presence in:
|
|||
|---|---|---|---|---|---|
| Cork discs
|
Cork stoppers
|
||||
| C (n = 11) | ME-TCA (n = 19) | C (n = 14) | ME (n = 16) | ||
| Group 1 | Ascomycota (Sordariomycetes) | ||||
| F42, F41 | Fusarium poae bba65499 (AF414968) | 28.6 | 50.0 | ||
| B09 | Fusarium sambucinum NRRL20765 (X65482) | 14.3 | |||
| DF03 | Nectria mauritiicola RNHRCFC042 (AJ558114) | 31.6 | |||
| DF16, DF18, DF42, P09, P16, B08, F33 | Neurospora sitophila FGSC1135 (AF388926) | 63.6 | 84.2 | 18.8 | |
| F10 | Neurospora intermedia (EF197071) | 18.2 | 10.5 | 71.4 | 25.0 |
| Group 2 | Ascomycota (Dothideomycetes) | ||||
| P01 | Cladosporium cladosporioides STE U 3683 (AY251074) | 28.6 | |||
| Group 3 | Ascomycota (Eurotiomycetes) | ||||
| F01 | Eurotium repens ATCC 66457 (AY373890) | 37.5 | |||
| DF53 | Penicillium diversum NRRL2122 (DQ308554) | 9.0 | |||
| 24 bands | Penicillium glabrum FRR835 (AY373915) | 100.0 | 100.0 | 100.0 | 100.0 |
| DF45, DF46 | Penicillium islandicum FRR2239 (AY373919) | 5.3 | |||
| DF23, DF30 | Penicillium islandicum Wb156 (AF455543) | 27.3 | 26.3 | ||
| DF04, DF05, DF72, DF83 to DF86 | Penicillium variabile FRR1290 (AY373936) | 18.2 | 84.2 | ||
| Group 4 | Basidiomycota (Tremellomycetes) | ||||
| F03, F29, F30, F35 | Cryptococcus carnescens IAM14506 (AB105438) | 7.1 | 6.3 | ||
| DF09 | Sporotrichum laxum (AJ428045) | 18.2 | 10.5 | ||
| P08 | Udeniomyces pannonicus JCM11149 (AB072232) | 7.1 | |||
| Group 5 | Basidiomycota (Cystobasidiomycetes) | ||||
| P05, P15 | Rhodotorula minuta SY87 (AB025997) | 7.1 | 12.5 | ||
| F07, F27, F28, F32, B06, P04, P14 | Rhodotorula sloofiae CBS5706 (AM901759) | 28.6 | 68.8 | ||
| Group 6 | Basidiomicota (Microbotryomycetes) | ||||
| F38, P06, P12, P13 | Rhodotorula glutinis HB1215 (AM160643) | 28.5 | 18.8 | ||
Control (C) and ME and ME-TCA tainted cork stoppers and discs were examined. The closest matches (>98% similarity) to sequences of cultured fungi in the GenBank database are indicated. Accession numbers appear in parentheses in column 2. n, number of samples.
Bands are grouped based on the division or phylum categorizations in column 2.
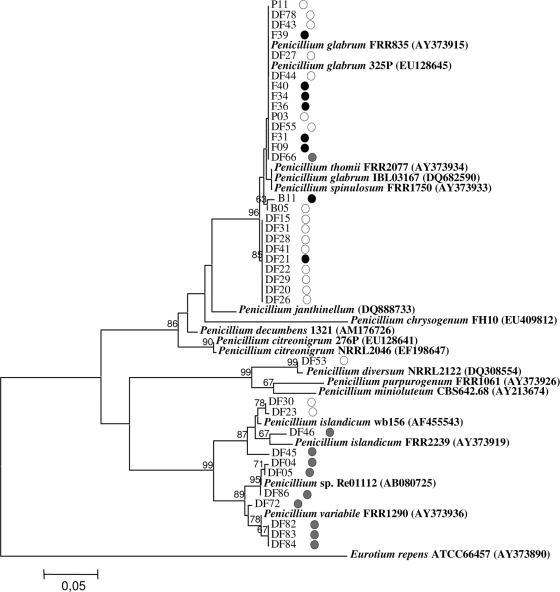
The unexpected diversity of Penicillium related sequences in cork discs was further investigated. All 37 DGGE bands showing high homologies with Penicillium species clustered separately according to the sample origin. Most of the sequences clustered with P. glabrum FRR835 (>99% homology) and were found in control and tainted samples of cork discs and stoppers (Fig. 4). Other Penicillium species found in untainted samples included Penicillium diversum NRRL2122 (sequences DF53 and DF30) and Penicillium islandicum Wb156 (sequence DF23). Finally, nine sequences obtained from bands present exclusively in ME-TCA discs clustered separately and showed high homologies (>98%) with P. islandicum FRR2239, Penicillium variabile FRR1290, and the unidentified Penicillium strain Re01112.
FIG. 4.
NJ tree generated from the alignment of Penicillium spp. ITS1 sequences (188 to 201 bp). Sequences and accession numbers of selected organisms retrieved from GenBank appear in boldface. Samples numbering corresponds to DGGE bands in Fig. 1 and 2. Bootstraps values >60% are indicated at branch nodes. ITS1 sequence from Eurotium repens (AY373890) was used as outgroup. Origin of sequences is depicted by shaded dots on the right side of each name: white, control discs; gray, TCA tainted discs; black, cork stoppers.
Alpha- and gammaproteobacteria were particularly diverse in cork stoppers, including eight and five different phylotypes, respectively. Sequences with a high homology (>99%) to Sphingobium yanoikuyae DD109 were the only ones found in all stoppers (Table 3). Only small differences corresponding to the presence of certain species of bacteria were found between control and tainted samples. Species belonging to Actinobacteria (three sequences), Pseudomonas putida, and P. aeruginosa were specifically detected in control stoppers. In contrast, Acinetobacter spp., Pseudomonas strain pl-72 and Flavobacterium spp. were particularly frequent in ME tainted samples.
TABLE 3.
Most probable sequence identification and occurrence of DGGE bands in control and ME tainted cork samples: closest bacterial speciesa
| Group and band no.b | Closest bacterial species | % Presence in cork samples
|
|
|---|---|---|---|
| C (n = 13) | ME (n = 13) | ||
| Group 1 | Alphaproteobacteria | ||
| EU105, EU106 | Methylobacterium B25 (EU240206) | 38.5 | 22.1 |
| EU83, EU129 | Novosphingobium subarcticum LH128 (AY151394) | 46.2 | |
| EU125, EU86 | Paracoccus marcusii (Y12703) | 15.4 | 30.8 |
| EU126 | Paracoccus sp. strain B61Y (EU368762) | 15.4 | |
| EU117 | Sphingobium yanoikuyae BF-18 (EU307932) | 7.7 | |
| EU77, EU98, EU99, EU102, EU114, EU128, EU137 | Sphingobium yanoikuyae DD109 (AY574367) | 100.0 | 100.0 |
| EU87 | Sphingobium sp. strain 2F5-2 (EU286535) | 30.8 | |
| EU115 | Sphingomonas sp. strain D31C2 (AY162145) | 30.8 | 15.4 |
| Group 2 | Betaproteobacteria | ||
| EU132 | Zooglea sp. strain AI-20 (AY437629) | 7.7 | |
| Group 3 | Gammaproteobacteria | ||
| EU12, EU34, EU75, EU76, EU124 | Acinetobacter sp. strain SH825131.2 (AJ633636) | 53.8 | 100.0 |
| EU24 | Acinetobacter lwofii PTA-152 (AM293676) | 15.4 | |
| EU78, EU80, EU138 | Pseudomonas putida NABI (AY395005) | 46.2 | |
| EU122 | Pseudomonas aeruginosa SG-1 (EU391389) | 15.4 | |
| EC01, EC02, EC03, EC04, EC05, EC06 | Pseudomonas sp. strain pl-72 (EU375376) | 46.2 | |
| Group 4 | Actinobacteria | ||
| EU108 | Brachybacterium paraconglomeratum GN0406 (DQ890508) | 7.7 | |
| EU97, EU134 | Microbacterium oxydans H190 (EF204430) | 30.8 | |
| EU135 | Microbacterium sp. strain Atl-19 (EF028128) | 7.7 | |
| Group 5 | Bacteroidetes | ||
| EU08, EU133 | Flavobacterium sp. strain THWCSN41 (AM888190) | 7.7 | 46.2 |
Control (C) and ME tainted cork samples were examined. The closest matches (>98% similarity) to sequences of cultured bacteria in GenBank database are indicated. Accession numbers appear in parentheses in column 2. n, number of samples.
Bands are grouped based on the division or phylum categorizations in column 2.
DISCUSSION
The measurement of releasable TCA in aqueous cork macerates has been used as a common means of assessment of cork quality by manufacturing industries (7, 9, 24, 26, 32). Although chlorinated phenols and anisoles have attracted most of the attention in the field, other molecules, such as IPMP and MDMP, have recently been incriminated in off-odor production in corks (43, 47, 48). Anisoles are biologically produced by the methylation of the corresponding phenolic substrate. This reaction is frequent in many fungi isolated from cork (3, 31, 39). Pyrazines derive from temperature-dependent chemical reactions (11) or biological transformations of amino acids catalyzed by different bacteria and fungi (1, 16, 42). It is generally assumed that anisoles and other sensory active molecules are produced at initial stages of cork manufacturing and even before harvesting of cork slabs. However, we were not able to find any report of a conclusive experiment that demonstrated that off-odor development during the cork manufacturing process is not possible. More interestingly, Jäger et al. (25) showed that many bacterial and yeast isolated from cork were able to develop unpleasant aromas when inoculated in test models including cork granules in wine (11.5% [vol/vol] ethanol), suggesting that off-odors can develop even after bottling.
Filamentous fungi are relatively abundant in most stages of cork manufacturing, with values ranging from 104 to 107 CFU/g of cork (3) and diminishing to levels below 102 microorganisms/g of cork when cork stoppers are treated with SO2. However, significantly higher densities of microorganisms (>102 microorganisms/g of cork) have been measured in ready-to-use stoppers (39). Storage conditions of finished stoppers may be of critical importance to prevent growth of molds and bacteria and potential off-odor development. Therefore, knowing the diversity of microorganisms at final cork manufacturing stages becomes crucial (15, 17, 46).
TCA tainted discs were clearly distinguishable from the control samples using two experimental approaches, an HS-SPME-GC/MS analysis of cork macerates and a PCR-DGGE fingerprint of the fungal community. Specific ITS1 sequences retrieved from tainted discs showed high similarities with P. islandicum and P. variabile. The latter species has recently being described in cork (44). The closest species based on ITS1 gene sequence comparison described as a moderate TCA producer is Penicillium purpurogenum (3).
The relationship between the presence of taint compounds other than TCA and specific microorganisms was not evident from our data. There could be several reasons for this. First, the chemical composition of ME cork macerates appeared to be more complex, and the presence of MDMP co-occurred with TCA and GUA. No significant differences were found between control and ME corks when MDMP concentration was analyzed, indicating that combined effects of different compounds may occur. Second, cork stoppers are composed of cork granules of different origins, cork discs, and casein-derived glues, which may increase the intrinsic microbial variability within samples. Biosynthesis of MDMP or related dialkyl methoxypyrazines have been proven in Pseudomonas perolens, several Serratia strains, and Chondromyces crocatus among other isolated microorganisms when incubated in the presence of available amino acids (10, 16, 18). The fingerprinting analysis revealed that microorganisms restricted to ME tainted stoppers were the fungi Neurospora sitophila and Eurotium repens and the bacteria Novosphingobium subarcticum, Sphingobium spp., Acinetobacter lwofii, and Pseudomonas spp., all found at relatively low frequencies (<47% of samples). Unfortunately, no experimental data reporting methoxypyrazine production exists for the detected or phylogenetically similar organisms except for Pseudomonas species (10, 34). Another interesting aspect to consider when trying to explain MDMP accumulation in cork is the selective detection of P. putida and P. aeruginosa, together with some other Actinobacteria in untainted cork stoppers. The presence of specific bacteria in control stoppers suggest that these bacteria may potentially be implicated in the degradation of taint compounds. Pyrazine degradation by microbial activity has been previously documented in several bacterial isolates, including Pseudomonas, Rhodococcus, and Mycobacterium strains, although no specific data exist for MDMP (27, 41). In addition, chloroanisole degradation has been proven in P. putida INBP1 and Acinetobacter radioresistens INBS1 (21), suggesting that Pseudomonas species may have a complex interaction with taint compounds. A further investigation with the use of selected producing or degrading isolates is still needed to fully understand pyrazine accumulation in cork matrices.
In conclusion, the use of PCR-DGGE fingerprinting as a selective technique has allowed us to characterize the relationship between microbial community composition and some of the most common taints in cork. The detection of TCA in TCA tainted cork discs appeared concomitantly to sequences related to P. variabile in almost all samples analyzed. Non-TCA-related ME taint was assigned to the presence of MDMP, although an additive effect of various chemical compounds was not discounted. The biological accumulation of MDMP in cork appeared to be a more complex process, and the contribution of different microorganisms was assumed.
Acknowledgments
C.P. is the recipient of a Ph.D. grant from the University of Girona and the Council of Palafrugell (BRAE 04/03). O.R.-R. is the recipient of a Ph.D grant from the Generalitat de Catalunya.
We thank Elena Bosch, Jordi Deulofeu, and Anna Martínez for help with the sensory determinations and valuable discussions of the results. Cork samples were generously provided by TESA and Geyru SA (Palafrugell, Girona, Spain).
Footnotes
Published ahead of print on 5 February 2009.
REFERENCES
- 1.Adams, A., and N. De Kimpe. 2007. Formation of pyrazines and 2-acetyl-1-pyrroline by Bacillus cereus. Food Chem. 101:1230-1238. [Google Scholar]
- 2.Álvarez-Rodríguez, M. L., C. Belloch, M. Villa, F. Uruburu, G. Larriba, and J. J. R. Coque. 2003. Degradation of vanillic acid and production of guaiacol by microorganisms isolated from cork samples. FEMS Microbiol. Lett. 220:49-55. [DOI] [PubMed] [Google Scholar]
- 3.Álvarez-Rodríguez, M. L., L. López-Ocaña, J. M. López-Coronado, E. Rodríguez, M. J. Martínez, G. Larriba, and J. J. R. Coque. 2002. Cork taint of wines: role of the filamentous fungi isolated from cork in the formation of 2,4,6-trichloroanisole by O methylation of 2,4,6-trichlorophenol. Appl. Environ. Microbiol. 68:5860-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amon, J. M., J. M. Vandepeer, and R. F. Simpson. 1989. Compounds responsible for cork taint in wine. Aust. N. Z. Wine Ind. J. 4:62-69. [Google Scholar]
- 5.Anderson, I. C., C. D. Campbell, and J. I. Prosser. 2003. Diversity of fungi in organic soils under a moorland-Scots pine (Pinus sylvestris L.) gradient. Environ. Microbiol. 5:1121-1132. [DOI] [PubMed] [Google Scholar]
- 6.Besson, I., C. Creuly, J. B. Gros, and C. Larroche. 1997. Pyrazine production by Bacillus subtilis in solid-state fermentation on soybeans. Appl. Microbiol. Biotechnol. 47:489-495. [Google Scholar]
- 7.Boutou, S., and P. Chatonnet. 2007. Rapid headspace solid-phase microextraction/gas chromatographic/mass spectrometric assay for the quantitative determination of some of the main odorants causing off-flavours in wine. J. Chromatogr. A 1141:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Brodie, E., S. Edwards, and N. Clipson. 2003. Soil fungal community structure in a temperate upland grassland soil. FEMS Microbiol. Ecol. 45:105-114. [DOI] [PubMed] [Google Scholar]
- 9.Chatonnet, P., S. Bonnet, S. Boutou, and M. D. Labadie. 2004. Identification and responsibility of 2,4,6-tribromoanisole in musty, corked odors in wine. J. Agric. Food Chem. 52:1255-1262. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, T., and G. A. Reineccius. 1991. A study of factors influencing 2-methoxy-3-isopropylpyrazine production by Pseudomonas perolens using acid trap and UV spectroscopy. Appl. Microbiol. Biotechnol. 36:304-308. [Google Scholar]
- 11.Chu, C. 1998. Pyrazine formation from amino acids and reducing sugars, a pathway other than Strecker degradation. J. Agric. Food Chem. 46:1515-1517. [Google Scholar]
- 12.Cocolin, L., L. F. Bisson, and D. A. Mills. 2000. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 189:81-87. [DOI] [PubMed] [Google Scholar]
- 13.Coque, J. J. R., M. L. Álvarez-Rodríguez, and G. Larriba. 2003. Characterization of an inducible chlorophenol O-methyltransferase from Trichoderma longibrachiatum involved in the formation of chloroanisoles and determination of its role in cork taint of wines. Appl. Environ. Microbiol. 69:5089-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cserjesi, A. J., and E. L. Johnson. 1972. Methylation of pentachlorophenol by Trichoderma virgatum. Can. J. Microbiol. 18:45-47. [DOI] [PubMed] [Google Scholar]
- 15.Danesh, P., F. M. V. Caldas, J. J. F. Marques, and M. V. San-Romão. 1997. Mycobiota in Portuguese ‘normal’ and ‘green’ cork throughout the manufacturing process of stoppers. J. Appl. Microbiol. 82:689-694. [DOI] [PubMed] [Google Scholar]
- 16.Dickschat, J. S., H. Reichenbach, I. Wagner-Döbler, and S. Schulz. 2005. Novel pyrazines from the myxobacterium Chondromyces crocatus and marine bacteria. Eur. J. Org. Chem. 19:4141-4153. [Google Scholar]
- 17.Doare-Lebrun, E., A. El Arbi, M. Charlet, L. Guerin, J. J. Pernelle, J. C. Ogier, and M. Bouix. 2006. Analysis of fungal diversity of grapes by application of temporal temperature gradient gel electrophoresis: potentialities and limits of the method. J. Appl. Microbiol. 101:1340-1350. [DOI] [PubMed] [Google Scholar]
- 18.Gallois, A., and P. A. Grimont. 1985. Pyrazines responsible for the potatolike odor produced by some Serratia and Cedecea strains. Appl. Environ. Microbiol. 50:1048-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardes, M., and T. D. Bruns. 1993. Its primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 20.Gerber, N. N. 1977. Three highly odorous metabolites from an actinomycete: 2-isopropyl-3-methoxy-pyrazine, methylisoborneol, and geosmin. J. Chem. Ecol. 3:475-482. [Google Scholar]
- 21.Goswami, M., E. Recio, S. Campoy, J. F. Martin, and J. J. R. Coque. 2007. Environmental significance of O-demethylation of chloroanisoles by soil bacterial isolates as a mechanism that improves the overall biodegradation of chlorophenols. Environ. Microbiol. 9:2512-2521. [DOI] [PubMed] [Google Scholar]
- 22.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 23.Haruta, S., S. Ueno, I. Egawa, K. Hashiguchi, A. Fujii, M. Nagano, M. Ishii, and Y. Igarashi. 2006. Succession of bacterial and fungal communities during a traditional pot fermentation of rice vinegar assessed by PCR-mediated denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 109:79-87. [DOI] [PubMed] [Google Scholar]
- 24.Insa, S., E. Besalú, C. Iglesias, V. Salvadó, and E. Anticó. 2006. Ethanol-water extraction combined with solid-phase extraction and solid-phase microextraction concentration for the determination of chlorophenols in cork stoppers. J. Agric. Food Chem. 54:627-632. [DOI] [PubMed] [Google Scholar]
- 25.Jäger, J., J. Diekman, D. Lorenz, and L. Jakob. 1996. Cork-borne bacteria and yeasts as potential producers of off-flavours in wine. Aust. J. Grape Wine Res. 2:35-41. [Google Scholar]
- 26.Juanola, R., L. Guerrero, D. Subirà, V. Salvadó, S. Insa, J. A. G. Regueiro, and E. Anticó. 2004. Relationship between sensory and instrumental analysis of 2,4,6-trichloroanisole in wine and cork stoppers. Anal. Chim. Acta 513:291-297. [Google Scholar]
- 27.Kiener, A. 1992. Enzymatic oxidation of methyl groups on aromatic heterocycles: a versatile method for the preparation of heteroaromatic carboxylic acids. Angew. Chem. Int. Ed. Engl. 34:774-775. [Google Scholar]
- 28.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 29.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley, Chichester, England.
- 30.Lee, T. H., and R. F. Simpson. 1993. Microbiology and chemistry of cork taints in wine, p. 353-372. In G. H. Fleet (ed.), Wine microbiology and biotechnology, vol. 1. G. Harwood Academic Publishers, Chur, Switzerland. [Google Scholar]
- 31.Maggi, L., V. Mazzoleni, M. D. Fumi, and M. R. Salinas. 2008. Transformation ability of fungi isolated from cork and grape to produce 2,4,6-trichloroanisole from 2,4,6-trichlorophenol. Food Addit. Contam. A 25:265-269. [DOI] [PubMed] [Google Scholar]
- 32.Martínez-Uruñuela, A., J. M. González-Sáiz, and C. Pizarro. 2005. Multiple solid-phase microextraction in a non-equilibrium situation: application in quantitative analysis of chlorophenols and chloroanisoles related to cork taint in wine. J. Chromatogr. A 1089:31-38. [DOI] [PubMed] [Google Scholar]
- 33.Mills, D. A., E. A. Johannsen, and L. Cocolin. 2002. Yeast diversity and persistence in botrytis-affected wine fermentations. Appl. Environ. Microbiol. 68:4884-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mottram, D. S., and L. S. Patterson. 1984. 2,6-Dimethyl-3-methoxypyrazine: a microbiologically-produced compound with an obnoxious musty odour. Chem. Ind. 1984:448-449.
- 35.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek 73:127-150. [DOI] [PubMed] [Google Scholar]
- 36.Neilson, A. H., C. Lindgren, P. A. Hynning, and M. Remberger. 1988. Methylation of halogenated phenols and thiophenols by cell-extracts of gram-positive and gram-negative bacteria. Appl. Environ. Microbiol. 54:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira, A. C., C. M. Peres, J. M. Correia Pires, C. Silva Pereira, S. Vitorino, J. J. Figueiredo Marques, M. T. Barreto Crespo, and M. V. San Romão. 2003. Cork stoppers industry: defining appropriate mould colonization. Microbiol. Res. 158:117-124. [DOI] [PubMed] [Google Scholar]
- 38.Pla, M., A. Jofré, M. Martell, M. Molinas, and J. Gómez. 2000. Large accumulation of mRNA and DNA point modifications in a plant senescent tissue. FEBS Lett. 472:14-16. [DOI] [PubMed] [Google Scholar]
- 39.Prak, S., Z. Gunata, J. P. Guiraud, and S. Schorr-Galindo. 2007. Fungal strains isolated from cork stoppers and the formation of 2,4,6-trichloroanisole involved in the cork taint of wine. Food Microbiol. 24:271-280. [DOI] [PubMed] [Google Scholar]
- 40.Prat, C., L. Bañeras, and E. Anticó. 2008. Screening of musty-earthy compounds from tainted cork using water-based soaks followed by headspace solid-phase microextraction and gas chromatography-mass spectrometry. Eur. Food Res. Technol. 227:1085-1090. [Google Scholar]
- 41.Rappert, S., and R. Muller. 2005. Microbial degradation of selected odorous substances. Waste Manag. 25:940-954. [DOI] [PubMed] [Google Scholar]
- 42.Schulz, S., and J. S. Dickschat. 2007. Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24:814-842. [DOI] [PubMed] [Google Scholar]
- 43.Sefton, M. A., and R. F. Simpson. 2005. Compounds causing cork taint and the factors affecting their transfer from natural cork closures to wine: a review. Aust. J. Grape Wine Res. 11:226-240. [Google Scholar]
- 44.Serra, R., S. Peterson, and A. Venancio. 2008. Multilocus sequence identification of Penicillium species in cork bark during plank preparation for the manufacture of stoppers. Res. Microbiol. 159:178-186. [DOI] [PubMed] [Google Scholar]
- 45.Silva, A., M. Lambri, and M. D. De Faveri. 2003. Evaluation of the performances of synthetic and cork stoppers up to 24 months post-bottling. Eur. Food Res. Technol. 216:529-534. [Google Scholar]
- 46.Silva Pereira, C., J. J. Figuereido Marques, and M. V. San Romão. 2000. Cork taint in wine: scientific knowledge and public perception: a critical review. Crit. Rev. Microbiol. 26:147-162. [DOI] [PubMed] [Google Scholar]
- 47.Simpson, R. F., D. L. Capone, and M. A. Sefton. 2004. Isolation and identification of 2-methoxy-3,5-dimethylpyrazine, a potent musty compound from wine corks. J. Agric. Food Chem. 52:5425-5430. [DOI] [PubMed] [Google Scholar]
- 48.Simpson, R. F., D. L. Capone, B. C. Duncan, and M. A. Sefton. 2005. Incidence and nature of “fungal must” taint in wine corks. Aust. N. Z. Wine Ind. J. 20:26-31. [Google Scholar]
- 49.Soleas, G. J., J. Yan, T. Seaver, and D. M. Goldberg. 2002. Method for the gas chromatographic assay with mass selective detection of trichloro compounds in corks and wines applied to elucidate the potential cause of cork taint. J. Agric. Food Chem. 50:1032-1039. [DOI] [PubMed] [Google Scholar]
- 50.Spanish Association for Standardisation and Certification. 2004. Cork stoppers: sensory analysis, p. 1-9. In UNE 56928. Spanish Association for Standardisation and Certification, Madrid, Spain.
- 51.Thompson, M. J., and R. L. Sinsabaugh. 2000. Matric and particulate phosphatase and aminopeptidase activity in limnetic biofilms. Aquat. Microb. Ecol. 21:151-159. [Google Scholar]
- 52.Vainio, E. J., and J. Hantula. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104:927-936. [Google Scholar]
- 53.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal rRNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA.