Abstract
Streptomyces not only exhibits complex morphological differentiation but also produces a plethora of secondary metabolites, particularly antibiotics. To improve our general understanding of the complex network of undecylprodigiosin (Red) biosynthesis regulation, we used an in vivo transposition system to identify novel regulators that influence Red production in Streptomyces coelicolor M145. Using this screening system, we obtained 25 Red-deficient mutants. Twenty-four of these mutants had a transposon inserted in the previously described Red biosynthetic gene cluster and produced different amounts of another secondary metabolite, actinorhodin (Act). One mutant was shown to have an insertion in a different region of the chromosome upstream of the previously uncharacterized gene rrdA (regulator of redD, sco1104), which encodes a putative TetR family transcription factor. Compared with wild-type strain M145, the rrdA null mutant exhibited increased Red production and decreased Act production. A high level of rrdA expression resulted in a severe reduction in Red production and Act overproduction. Reverse transcription-PCR analysis showed that RrdA negatively regulated Red production by controlling redD mRNA abundance, while no change was observed at the transcript level of the Act-specific activator gene, actII-orf4. The effects on Act biosynthesis might arise from competition for precursors that are common to both pathways.
In addition to its complex morphological differentiation, the gram-positive genus Streptomyces is notable for its ability to produce a wide range of secondary metabolites during its life cycle. These metabolites include the majority of known pharmaceutically important secondary metabolites that exhibit antibacterial, anticancer, and immunosuppressive activities (6, 7). The genes responsible for the biosynthesis of secondary metabolites are often physically clustered in the genome and coordinately regulated by pathway-specific transcriptional activators (1, 4, 10, 12, 29, 33). These specific regulators are controlled by various higher-level pleiotropic regulators, and their expression is typically affected by a variety of environmental and physiological cues, including the nature and levels of carbon and nitrogen sources and the availability of phosphate and small signaling molecules, such as ppGpp and γ-butyrolactone (5).
Streptomyces coelicolor A3 (2) has been used for many years as a model organism in morphological and physiological differentiation studies, particularly in studies of the regulation of antibiotic biosynthesis (7). S. coelicolor produces four antibiotics: actinorhodin (Act), undecylprodigiosin (Red), methylenomycin, and calcium-dependent antibiotic. It has been shown that certain regulators are involved in the pleiotropic control of antibiotic production in S. coelicolor, including AbsA1/A2, AfsR/K, AtrA, and PhoR/P (18, 24, 31, 32). Recently, mutational analysis and adventitious overexpression of key regulators in S. coelicolor revealed cross-regulation at the transcriptional level among disparate antibiotic biosynthetic pathways (15). The diversity of these regulatory elements suggests that the regulation of antibiotic production is a complicated process, and many genes remain to be identified.
Red is one of the prodiginine secondary metabolites, and this group of compounds is attracting increasing interest due to its immunosuppressive and anticancer activities (34). In this study, we used an insertion mutagenesis system to identify new genes that regulate Red biosynthesis in S. coelicolor M145. We discovered a novel TetR-like transcription factor gene, rrdA (regulator of redD, sco1104), that was involved in the regulation of secondary metabolism. In this study, we also report a link between Red production and Act production that probably occurs at the level of the metabolites utilized that are common to both pathways in S. coelicolor.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α (22) was used for plasmid propagation. E. coli Rosetta-gami (Novagen) was used to express recombinant proteins. Mannitol soy flour (MS) (16) agar was used to generate spores and select Streptomyces exconjugants. YBP agar (2 g yeast extract, 2 g beef extract, 4 g Bacto peptone, 1 g MgSO4, 15 g NaCl, 15 g agar, and 10 g glucose in 1 liter [final volume] H2O) was used for phenotype screening and RNA preparation. YBP liquid medium was used for quantitative antibiotic assays. YEME (16) was used to cultivate mycelia for genomic DNA preparation. Conjugation of E. coli ET12567/pUZ8002 with Streptomyces was performed as described previously (16). Depending on the requirements, antibiotics were added at the following final concentrations: ampicillin, 50 μg ml−1; chloramphenicol, 33 μg ml−1; kanamycin, 30 μg ml−1; tetracycline, 12.5 μg ml−1; thiostrepton, 25 μg ml−1; and apramycin, 20 μg ml−1.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description | Reference or source |
|---|---|---|
| S. coelicolor strains | ||
| M145 | Prototroph SCP1− SCP2− | 16 |
| ΔrrdA | M145 with rrdA gene disrupted | This study |
| ΔrrdA/pFDZ16 | ΔrrdA carrying integrative plasmid pFDZ16 | This study |
| ΔrrdA/pFDZ16*-rrdA | ΔrrdA carrying integrative plasmid pFDZ16*-rrdA | This study |
| WT/pFDZ16 | M145 carrying integrative plasmid pFDZ16 | This study |
| WT/pFDZ16-rrdA | M145 carrying integrative plasmid pFDZ16-rrdA | This study |
| E. coli strains | ||
| DH5α | F−recAlacZΔM15 | 22 |
| ET12567 | dam dcm hsdS | 16 |
| Rosetta-gami | Δara-leu7697 ΔlacX74 ΔphoA(PvuII) phoRaraD139 ahpCgalEgalKrpsL F− [lac+ (lacIq) pro] gor522::Tn10(Tcr) trxB::Kan (DE3)/pRARE (Cmr) | Novagen |
| Plasmids | ||
| pFDZ315 (Tn315) | Conjugative plasmid that does not replicate in Streptomyces strains harboring transposon Tn315, which uses the transposase of insertion element IS204 as a trans factor and contains a kanamycin resistance gene, origin of pUC plasmids, and ermE promoter in the terminal inverted repeats; Kanr Ampra | X. Zhang et al., unpublished |
| pFDZ15 | E. coli-Streptomyces integrative shuttle vector; vector pRT802 was cut with SmaI and EcoRV, resulting in a 3.8-kb segment that was subsequently inserted into the NaeI site of pBluescript II KS+ (Stratagene), yielding pFDZ15; Kanr Amprb | This study |
| pFDZ16 | E. coli-Streptomyces integrative shuttle vector containing tipA promoter; the 5.4-kb ApaI/BclI fragment of pFDZ15 was ligated with the 5.0-kb BamHI/ApaI segment of pIJ6021 to obtain pFDZ16; Kanr Thior Amprc | This study |
| pFDZ16*-rrdA | Derivative obtained from pFDZ16 depletion of PtipA, containing the rrdA gene and its promoter; Kanr Ampr Thior | This study |
| pFDZ16-rrdA | Derivative obtained from pFDZ16, containing the rrdA gene located downstream of the tipA promoter; Kanr Ampr Thior | This study |
| pUCm-T | 2.7-kb cloning vector; Ampr | Shenggong Company |
| pBC-AM | Donor of aac(3)-IV; the 1.5-kb HindIII/EcoRI fragment of pULVK2A was inserted between the corresponding sites in vector pBC-SK(−) (Stratagene) to produce pBC-AM; Aprar Cmrd | This study |
| pUC-LR | DNA fragment containing the rrdA gene, cloned in pUCm-T; Ampr | This study |
| pUC-LAR | DNA fragment aac(3)-IV containing apramycin resistance gene, cloned in pUC-LR; Ampr | This study |
| pHZ1358 | E. coli-Streptomyces shuttle vector; Thior Aprar | 26 |
| pHZ1358-LAR | Derivative obtained from pHZ1358, containing the rrdA gene replacement cassette; Ampr Thior Aprar | This study |
| pET28a | Expression vector; Kanr | Novagen |
| pET28-RrdA | RrdA overexpression vector resulting in an N-terminal His6 oligopeptide fusion; Kanr | This study |
| pET22a | Expression vector; Ampr | Novagen |
| pET22-RrdA | RrdA overexpression vector resulting in an N-terminal His6 oligopeptide fusion; Ampr Cmr Tcr Kanr | This study |
| Primers | ||
| Oxj130 | 5′ ATCCATGGCGCATATGTCCCCGCGCAGCGCC 3′ | |
| Oxj131 | 5′ ATAAGCTTACCCGGCCAAGCGGGTCC 3′ | |
| Oxj144 | 5′ TCTAGACCGCCGTTCTGTTCGACTT 3′ | |
| Oxj145 | 5′ GGATCCACATCACCGTCCTGCCCTC 3′ | |
| Oxj161 | 5′ TCTGCCCTCTGACCGCGG 3′ | |
| Oxj162 | 5′ GCCGTCCTCGCGCGTTCT 3′ | |
| Oxj163 | 5′ CTGGCTCCTGGGCGGTCT 3′ | |
| Oxj164 | 5′ ACCAACCCTGGCGTCTCC 3′ | |
| Oxj185 | 5′ CGCCGAAGGAGGAACCGA 3′ | |
| Oxj186 | 5′ CCCGTCATCCACCGAACG 3′ | |
| Oxj187 | 5′ CCCAGGCGCTGGTAGACCT 3′ | |
| Oxj188 | 5′ CCGTCGAGCCGAAAGAGGA 3' | |
| Oxj201 | 5′ CCGGAGCCAGCCAAAGATC 3′ | |
| Oxj202 | 5′ GGAGGGCGTTGAGGACGTT 3′ | |
| Oxj203 | 5′ TGCTGACCAAGCCCGAGAA 3′ | |
| Oxj204 | 5′ CGGTGTACGTGGGACCTGAC 3′ | |
| Oxj205 | 5′ TGGTGCTGCTGCTCCTCAG 3′ | |
| Oxj206 | 5′ ATCCAGTCCCGCGTCCAA 3′ | |
| Oxj211 | 5′ GCGCCTCGGTCAATGAAGA 3′ | |
| Oxj212x | 5′ CGCTTTCCGGGGAAGTAGTAC 3′ | |
| Oxj237 | 5′ CTCTGTCATGGCGCTCATTGA 3′ | |
| Oxj238 | 5′ TTCGCTGCGACGCTCTTT 3′ |
Mutagenesis of S. coelicolor M145.
Insertional mutagenesis of M145 was conducted by in vivo transposition using Tn315 (Table 1) (X. Zhang, Y. Bao, X. Ou, P. Zhou, G. Zhao, and X. Ding, unpublished data) harbored by pFDZ315, which is a conjugative plasmid that does not replicate in Streptomyces. Tn315, which was derived from insertion element IS204 of Nocardia asteroides YP21 (35), contains a kanamycin resistance gene, the replication origin of pUC plasmids, and an ermE promoter in the terminal inverted repeats. After plasmid pFDZ315 was introdued into S. coelicolor M145 by conjugation, a single copy of transposon Tn315 was inserted into the chromosome, and exconjugants were selected by growth on MS media flooded with kanamycin (30 μg ml−1). The chromosomal locations of the Tn315 insertions were determined by sequencing the transposon-flanking DNA by plasmid rescue. Briefly, genomic DNA from the mutants was isolated and digested with ApaI, a site for which is not present in Tn315. The DNA was then self-ligated and transformed in E. coli DH5α using a standard protocol (16). Colonies were selected for kanamycin resistance, and the rescued plasmids were sequenced.
Gene disruption, complementation, and overexpression.
Targeted gene replacement mediated by homologous recombination was used to generate an rrdA null mutant. A 2.2-kb DNA fragment containing the rrdA gene was amplified by PCR using primers Oxj144 and Oxj145 (Table 1) and cloned into pUCm-T (Shenggong Company, Shanghai, China) by T/A cloning to form pUC-LR. The 1.5-kb SmaI fragment containing the aac(3)-IV gene from plasmid pBC-AM (Table 1) was subsequently inserted into EcoNI-linearized pUC-LR, and the rrdA gene was separated into 1.1-kb left and 1.1-kb right arms for homologous recombination to produce pUC-LAR. pUC-LAR was further cut with XbaI/ScaI to generate the 3.7-kb gene replacement cassette LAR. LAR was then inserted between the corresponding sites in the Streptomyces-E. coli shuttle vector pHZ1358 (26), which is a very unstable vector in streptomycetes, yielding the inactivation construct pHZ1358-LAR. This construct was introduced by conjugation from the donor E. coli ET12567/pUZ8002 into the recipient S. coelicolor M145 (16). The Aprar Thios double-crossover colonies were screened as possible mutant candidates in which the rrdA gene was disrupted by aac(3)-IV through homologous recombination. The mutants were confirmed by PCR using primers Oxj163 and Oxj164 (Table 1), which are located outside the homologous recombination regions, and this was followed by DNA sequencing.
The DNA fragment encompassing the complete rrdA gene and its possible promoter was amplified by PCR using primers Oxj144 and Oxj145. The product was cut with SacI/XhoI to produce a 1.4-kb fragment that was then inserted between the corresponding sites in vector pFDZ16 (Table 1), which is a Streptomyces-E. coli single-copy integration shuttle vector. This resulted in pFDZ16*-rrdA. This plasmid was subsequently conjugated with the rrdA null mutant for genetic complementation from the donor E. coli ET12567/pUZ8002. Exconjugants were selected by growth on MS media flooded with thiostrepton (25 μg ml−1).
The rrdA gene was amplified by PCR using primers Oxj130 and Oxj131 (Table 1), cut with NdeI/HindIII, and inserted between the corresponding sites in vector pFDZ16, which is a Streptomyces-E. coli single-copy integration shuttle vector carrying the tipA promoter. This resulted in pFDZ16-rrdA. This plasmid was then conjugated from the donor E. coli ET12567/pUZ8002 into wild-type strain M145 to overexpress rrdA. The exconjugants were selected by growth on MS media flooded with thiostrepton (25 μg ml−1).
Quantification of antibiotics.
Act and Red were assayed as previously described (16). Briefly, a culture grown in 40 ml YBP liquid medium was filtered, and the supernatant and pellet were separated. For Act, KOH was added to the supernatant to a final concentration of 1 M, and the optical density at 640 nm was determined. For Red, the mycelial pellet was dried under a vacuum and extracted with 10 ml methanol (adjusted to pH 2) overnight at room temperature. The optical density at 530 nm was then determined. Measurements were always obtained for three independent cultures.
RT-PCR analysis.
Mycelia grown on cellophane disks in YBP medium at 30°C for different time periods were scraped, and the RNA was isolated using a modified Kirby mixture. The isolated RNA was subjected to phenol-chloroform extraction and DNase I treatment, as described previously (16). Reverse transcription (RT) was performed with a high-fidelity RNA PCR kit (Takara, Japan) used according to the manufacturer's instructions. The primers used for RT-PCR are shown in Table 1. The following primers were used: for redZ, Oxj203 and Oxj204; for redD, Oxj201 and Oxj202; for actII-orf4, Oxj205 and Oxj206; for hrdB, Oxj237 and Oxj238; and for rrdA, Oxj211 and Oxj212x. The PCR conditions were as follows: 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The numbers of PCR cycles used were 27 for redZ, 29 for redD, and 26 for actII-orf4, rrdA, and hrdB. Two independent cultures were used for each condition, and the results were found to be consistent. RNA that was not reverse transcribed was used as a control, and the results obtained with this RNA were negative.
Production and purification of recombinant RrdA and electrophoretic mobility shift assays.
A DNA fragment encoding the predicted 233-amino-acid sequence of RrdA was generated using primers Oxj130 and Oxj131. The PCR fragment was cut with NdeI and HindIII and inserted between the corresponding sites in the expression vector pET28a (Novagen) to generate pET28-RrdA. pET28-RrdA was cut with XbaI/HindIII to produce an 855-bp segment, and this segment was inserted between the corresponding sites in pET22a (Novagen) to obtain the final RrdA expression vector pET22-RrdA. The recombinant RrdA protein was tagged at the N terminus with a His6 oligopeptide and was purified on an Ni-nitrilotriacetic acid spin column used according to the vendor's instructions (Qiagen). Electrophoretic mobility shift assays were performed using the method of Uguru et al. (31). DNA and protein were mixed to obtain a final volume of 20 μl in TGEK buffer (10 mM Tris-Cl [pH 7.9], 10% [vol/vol] glycerol, 0.1 mM EDTA, 50 mM KCl) at 30°C for 20 min. Samples were run on native 5% acrylamide-bisacrylamide (80:1) gels at 120 V for 1 h and subsequently visualized. The primer pairs used to generate the various redD DNA probes were Oxj161/Oxj162 (402 bp), Oxj185/Oxj186 (408 bp), and Oxj187/Oxj188 (410 bp).
RESULTS AND DISCUSSION
Mutagenesis of S. coelicolor to identify genes that affect Red production.
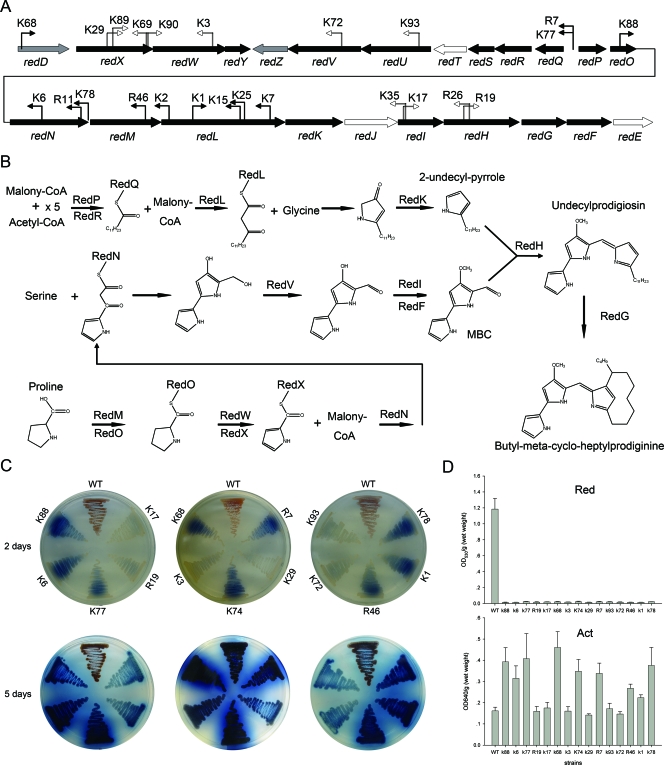
The genes involved in Red production reside in a single cluster (sco5877 [redD] to sco5899 [redE]) that is organized into four transcriptional units (redDXWY, redZ, redQRSTUV, and redPONMLKJIHGFE) (Fig. 1A) (34). Production begins with the synthesis of 4-methoxy-2,2′-bipyrrole-5-carbaldehyde (MBC) and 2-undecyl-pyrrole, and condensation of the monopyrrole with MBC leads to the formation of the tripyrrole pigments of Red (Fig. 1B).
FIG. 1.
Phenotypes of Red-deficient mutants and locations of the insertions. (A) Locations of the mutations in the Red biosynthetic gene cluster. The black arrows indicate genes that are required for Red biosynthesis, while the open arrows indicate genes that are not essential. The gray arrows indicate the redD and redZ regulatory genes. The bent open arrows indicate mutations that influence only Red production, while the filled bent arrows indicate mutations that affect both Red production and Act production. The directions of the bent arrows indicate the transcript orientation of PermE in Tn315 in the mutants. (B) Red biosynthetic pathway. RedU is required for activation of the acyl carrier protein domains of RedO. The function of RedY is unknown (34). (C) Mutants and wild-type strain M145 were grown on YBP agar at 30°C for 2 days and 5 days. The bottoms of the plates are shown. WT, wild type. (D) Antibiotic production assay. Act production and Red production by the mutants and wild-type strain M145 were assayed in cultures grown in YBP liquid medium at 30°C for 88 h. The bars indicate the averages of three independent determinations, and the error bars indicate the standard errors. OD530, optical density at 530 nm; OD640, optical density at 640 nm.
We used an in vivo transposition system to generate a new collection of mutants that were defective in Red production (see Materials and Methods for details). We succeeded in obtaining 25 insertion mutants that did not produce red-pigmented antibiotics. In each of these mutants, we sequenced the DNA flanking the transposon Tn315 and compared the sequences with the complete S. coelicolor genome (http://strepdb.streptomyces.org.uk) by BLAST analysis. This led to identification of genes that were disrupted or influenced. Of the 25 mutations, 24 were localized in 11 genes and two gene spacers within the Red biosynthetic gene cluster (Fig. 1A), while only 1 insertion mutation in K74 was outside this cluster. The phenotypes of 15 representative insertion mutants are shown in Fig. 1C and 1D. Compared with wild-type strain M145, insertion mutants K29, K69, K89, K90, K3, K72, K93, K17, K35, R19, and R26 (with mutations in the redX, redW, redV, redU, redI, and redH genes) showed no apparent change in blue-pigmented antibiotics, while mutants K68, K77, R7, K88, R11, R6, K78, R46, K1, K2, K7, K15, and K25 (with mutations in redD, the redQ-redP gene spacer, redO, redN, the redN-redM gene spacer, and redL) produced large amounts of blue-pigmented Act but no Red (Fig. 1C and 1D). The genes that had little effect on Act production (redX, redW, redV, redU, redI, and redH) encoded proteins that synthesize MBC or condense MBC and 2-undecyl-pyrrole into Red; however, these genes do not participate in the biosynthesis of 2-undecyl-pyrrole. Of the genes that significantly influence Act production, redD encodes the direct regulator that activates the transcription of Red biosynthetic genes, while redP, redQ, and redL encode proteins that participate in the biosynthesis of 2-undecyl-pyrrole. The redO, redN, and redM genes are located upstream of redL but are in the same transcriptional unit (Fig. 1A).
Analysis of mutant K74 led to identification of rrdA (sco1104), a tetR family gene.
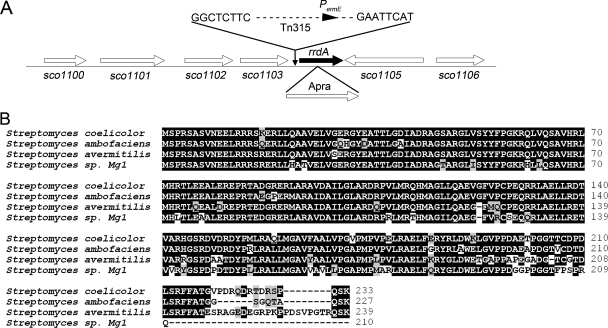
The insertion in mutant K74 was located 40 bp upstream of the translation initiation codon of rrdA (sco1104) (Fig. 2A). Since the IS204-based transposon Tn315 contains an internal ermE promoter, it is likely that the loss of Red production is due to a high level of rrdA expression rather than a polar effect caused by Tn315 integration in the K74 mutant. The predicted 233-amino-acid protein encoded by the rrdA gene, which is not clustered with either the Red biosynthesis or Act biosynthesis genes (it is separated from the Red biosynthesis and Act biosynthesis genes by approximately 5,270 kb and 4,360 kb, respectively) exhibited sequence similarity to the TetR family of transcriptional regulators, and its function has not been determined.
FIG. 2.
RrdA is highly conserved in Streptomyces. (A) rrdA (sco1104) locus in the S. coelicolor genome. The insertion site of transposon Tn315 in mutant K74 is indicated. The aac(3)-IV fragment (1.5 kb, conferring apramycin resistance) used to disrupt the rrdA gene is indicated below the map. (B) Multiple-amino-acid-sequence alignment of RrdA from S. coelicolor and the homologous proteins from S. avermitilis MA-4680 (SAV1503), S. ambofaciens ATCC 23877 (accession number CAJ90078.1), and Streptomyces sp. strain Mg1 (accession number EDX26764.1) obtained using BioEdit 7.0.0 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html/).
The TetR family is a common class of transcriptional factors that regulate a wide range of cellular activities in different bacteria. These activities include antibiotic production, multidrug resistance, amino acid metabolism, pathogenicity, and development (3, 13). This family of proteins typically possesses an N-terminal helix-turn-helix DNA-binding domain and a C-terminal ligand-binding domain. These proteins activate or repress their target genes by binding to their promoters. Genome analysis revealed that S. coelicolor contains 151 genes encoding proteins that belong to the TetR family of transcriptional regulators (2, 21), but only a few of these genes have been characterized in detail. ActR regulates Act export by repressing the transcription of the efflux pump ActA (27). Pip and PqrA control pristinamycin and paraquat resistance genes by repressing the transcription of drug efflux pumps (8, 9). CprA and CprB, together with the products of unknown target genes, control secondary metabolite formation and sporulation (20). ScbR controls γ-butyrolactone synthesis by activating the transcription of ScbA (28), while AtrA controls Act biosynthesis by activating ActII-ORF4 transcription (31).
Functional domain analysis using the simple modular architecture research tool (SMART; http://smart.embl-heidelberg.de/) revealed that the region corresponding to 20 to 66 amino acid residues of RrdA is a helix-turn-helix DNA-binding domain, while the C terminus of RrdA does not appear to be similar to the C-terminal ligand-binding domain of most TetR family proteins. As shown in Fig. 2B, RrdA and proteins homologous to it are highly conserved, and so far, these proteins have not been identified in any organism other than Streptomyces. SAV1503 from Streptomyces avermitilis MA-4680 and hypothetical proteins from Streptomyces ambofaciens ATCC 23877 (accession number CAJ90078.1) and Streptomyces sp. strain Mg1 (accession number EDX26764.1) have 83%, 90%, and 81% identity to RrdA, respectively. However, the genes surrounding rrdA are conserved in many divergent bacteria, including Streptomyces, Saccharopolyspora, Salinispora, Solibacter, Erythrobacter, and Nocardia. The products of the sco1100, sco1101, and sco1102 genes showed amino acid sequence similarity to integral membrane proteins. The sco1103, sco1105, and sco1106 genes encode a hydrolase, a secreted protein, and a lipoprotein, respectively. The specific functions of these genes have not been determined yet, and no obvious relationships among these predicted proteins have been identified.
RrdA influences Red and Act production in different ways.
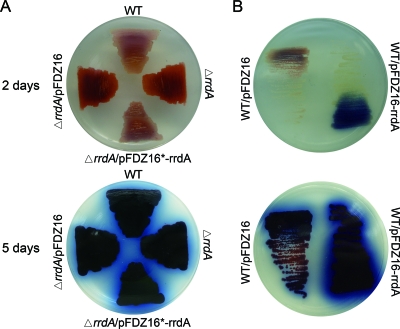
To assess the role of rrdA in S. coelicolor development, we attempted to knock out rrdA in S. coelicolor M145. We succeeded in constructing an rrdA mutant by partially replacing rrdA with the apramycin resistance cassette aac(3)-IV using a homologous recombination strategy. Four independent double recombinants were selected and confirmed by PCR and sequencing (data not shown). When the organisms were grown on solid YBP medium, Red production was significantly increased, while Act production was apparently unchanged in the rrdA mutant compared with wild-type strain M145 (Fig. 3A). The phenotype was complemented by pFDZ16*-rrdA, an integrating plasmid containing only rrdA and its probable 0.7-kb upstream promoter DNA fragment. We also tried to overexpress rrdA in wild-type strain M145 by introducing pFDZ16-rrdA, a plasmid containing the PtipA promoter and rrdA coding sequence. Previous studies have reported that genes cloned with the tipA promoter appear to be expressed at basal levels even in the absence of thiostrepton induction (23, 30). Transformants of S. coelicolor containing pFDZ16-rrdA produced no red-pigmented antibiotics either with or without thiostrepton induction, so observation of the phenotype and RT-PCR analysis (see below) of the rrdA high-expression strain were performed in the absence of thiostrepton induction. As shown in Fig. 3B, a high level of rrdA expression resulted in loss of Red production and production of amounts of Act larger than those in M145; thus, the behavior of this mutant was the same as that of the K74 mutant.
FIG. 3.
Phenotypes of the rrdA null mutant and high-expression strain. The bottoms of the plates are shown. (A) Colonies of the parent strain S. coelicolor M145 (WT), the rrdA null mutant (ΔrrdA), the rrdA mutant harboring the empty vector pFDZ16 (ΔrrdA/pFDZ16), and the complemented strain (ΔrrdA/pFDZ16*-rrdA) were grown on YBP medium at 30°C for 2 days and 5 days. (B) Colonies of the parent strain M145 harboring the empty vector pFDZ16 (WT/pFDZ16) and the rrdA high-expression strain (WT/pFDZ16-rrdA) were grown on YBP medium at 30°C for 2 days and 5 days.
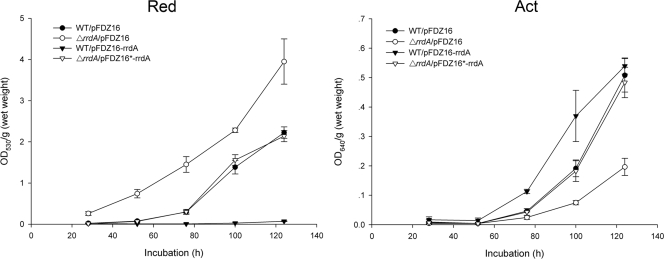
Production of Act and Red by the rrdA high-expression strain, a null mutant, a complemented strain, and M145 was measured in YBP liquid media at fixed intervals. As shown in Fig. 4, the rrdA null mutant produced Red earlier and at much higher level than the parent strain did, but the level of Act production was lower than that in the parent strain. In the rrdA high-expression strain, Red production dramatically decreased to levels that could barely be detected, and the level of Act production was higher than that in M145 on days 3 and 4. On day 5, the difference in Act production between M145 and the rrdA high-expression strain did not appear to be as significant as the difference observed when the organisms were cultured on solid media (Fig. 1A). The slight but notable difference in Act production between liquid media and solid media might be explained by differences in the developmental stages of the rrdA high-expression strain under these two different culture conditions. These results underline the fact that RrdA is a negative regulator of Red production and has a positive effect on Act production.
FIG. 4.
Antibiotic production by the M145 strain harboring the empty vector pFDZ16, the rrdA null mutant harboring the empty vector pFDZ16, the complemented strain, and the rrdA high-expression strain. Incubation was carried out in the YBP liquid medium at 30°C. The symbols indicate the averages of three independent determinations, and the error bars indicate the standard errors. OD530, optical density at 530 nm; OD640, optical density at 640 nm.
RrdA negatively regulates Red production by controlling the abundance of RedD mRNA.
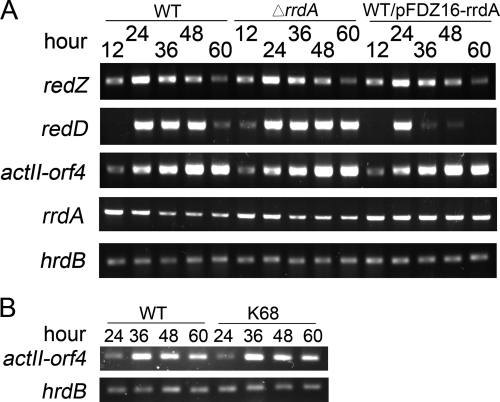
The expression of antibiotic biosynthetic clusters is normally regulated by pathway-specific activators (4, 10, 29). In S. coelicolor, Act biosynthesis and Red biosynthesis have been shown to depend on the transcriptional activation of the Act and Red biosynthetic clusters by ActII-ORF4 and the RedD and RedZ proteins, respectively. RedD, which is the direct transcriptional activator for the Red biosynthetic cluster, is RedZ dependent (12, 33). The transcription of rrdA, actII-orf4, redD, and redZ in M145, an rrdA high-expression strain, and a null mutant was therefore analyzed by RT-PCR. Total RNA was isolated both from M145 cultures at different stages of development and from mutants grown on cellophane placed on YBP solid medium. As shown in Fig. 5A, the results correlated well with the results of Red production in liquid media or on solid YBP medium. The level of transcription of redD was slightly higher in the rrdA null mutant than in M145 and significantly lower at 36 to 60 h in the rrdA high-expression strain than in M145. However, at 24 h, redD seemed to be transcribed at the same level in the rrdA high-expression strain and the wild type. The levels of transcription of actII-orf4 and redZ appeared to be the same as those in M145. The level of the rrdA transcript peaked at 12 h and then decreased to a stable level after 36 h. At this point, M145 became pink, which indicates the onset of Red production. Since the primer sequences of rrdA are located in the upstream region of the insertion site of the aac(3)-IV fragment used for gene disruption, the rrdA transcription profile of the rrdA null mutant could also be detected, and it was very similar to that of M145, while in the rrdA high-expression strain the rrdA transcript level was higher in all distinct phases during development. These results indicated that RrdA negatively regulated Red production by controlling the abundance of RedD mRNA.
FIG. 5.
RT-PCR results for wild-type strain M145, the rrdA null mutant, the rrdA high-expression strain, and redD null mutant K68. (A) Transcriptional levels of the pathway-specific activators, including redD (primers Oxj201 and Oxj202, 368 bp), redZ (primers Oxj203 and Oxj204, 322 bp), actII-orf4 (primers Oxj205 and Oxj206, 320 bp), rrdA [primers Oxj211 and Oxj212x, 166 bp; the primer sequences are located in the upstream region of the insertion site of the aac(3)-IV fragment used for gene disruption], and hrdB (control) (primers Oxj237 and Oxj238, 207 bp). (B) RT-PCR results for the redD null mutant K68 and wild-type strain M145 for transcriptional detection of actII-orf4. WT, wild type.
Since rrdA encodes a putative TetR family transcription factor, we performed electrophoretic mobility shift assays to examine the binding of the purified recombinant RrdA protein to three different DNA probes (nucleotides −296 to 115, −94 to 315, and 61 to 462 relative to the transcriptional start site of redD) (19) in TGEK buffer. The effects of Na+, Mg2+, and Mn2+ on RrdA binding were also tested. Unfortunately, no specific binding of RrdA to the DNA probes was observed under any of the test conditions used (data not shown). It is possible that the activated binding of RrdA to the redD promoter requires additional factors or that RrdA indirectly regulates redD transcription and one or more as-yet-unidentified proteins may participate in this regulation.
Cross-regulation of Red and Act production might occur through a mechanism involving competition for common precursors.
Huang et al. reported that deletion of the Red-specific regulator gene redZ caused delayed transcription of the actII-orf4 and Act biosynthesis genes in S. coelicolor (15). RrdA, which upregulates Act production and downregulates Red production, affects transcription of only the Red-specific regulator gene redD. We hypothesized that another mechanism may be involved in the cross-regulation of Red biosynthesis and Act biosynthesis. Therefore, we determined the transcriptional level of actII-orf4 in the redD null mutant K68. However, no difference was observed between K68 and wild-type strain M145 (Fig. 5B). The results indicated that the positive effect of the redD mutation on Act production was not due to any alteration at the transcriptional level of actII-orf4. Together with the observation that mutations in Red biosynthesis genes affect Act production, this suggests that Red production and Act production are interlinked at a nontranscriptional level.
It is known that Act production begins with the synthesis of a 16-carbon polyketide backbone by a type II polyketide synthase complex that uses malonyl coenzyme A (malonyl-CoA) and acetyl-CoA as the precursors (27). These precursors are also necessary for the production of 2-undecyl-pyrrole, which is an intermediate metabolite in the Red biosynthesis pathway (34). The redD mutation eliminated transcription of most biosynthetic gene clusters (14, 19), and mutations in the redQ-redP gene spacer, redO, redN, the redN-redM gene spacer, and redL disrupted or influenced, probably due to a polar effect, the expression of key enzymes (RedP, RedQ, and RedL) involved in the synthesis of 2-undecyl-pyrrole. This in turn diverted malonyl-CoA and acetyl-CoA from Red biosynthesis to Act biosynthesis. This hypothesis can be used to provide a simple explanation for the positive effect of RrdA on Act production. Thus, a decrease in Red production could result in an increase in Act production, and an increase in Red production could lead to a decrease in Act production due to the levels of the metabolic precursors utilized by both biosynthetic pathways (Fig. 4). A similar effect was also observed for nanchangmycin production by Streptomyces nanchangensis. A DNA fragment deletion in cluster C of S. nanchangensis resulted in at least a threefold increase in nanchangmycin production because biosynthesis of the compounds involved similar precursors (26). redX, redW, redV, redU, redI, and redH encode proteins that participate in the biosynthesis of MBC or in the final step of Red biosynthesis. Liquid chromatography-mass spectrometry analysis of the redU mutant showed that Red was eliminated and 2-undecyl-pyrrole accumulated (25). Therefore, disruption of these genes had little effect on the accumulation of malonyl-CoA and acetyl-CoA and had no obvious influence on production of blue-pigmented Act.
In conclusion, our study revealed that the nontranscriptional cross-regulation of Act biosynthesis and Red biosynthesis might involve competition for common precursors. We also partially characterized the TetR-like protein gene rrdA and found that it encodes a novel negative regulator of Red biosynthesis in S. coelicolor. These observations should be useful in fermentation engineering of undecylprodigiosin, which is a candidate drug for cancer therapy (34).
Acknowledgments
We thank Zixin Deng, Maggie Smith, and Juan F. Martin for providing strains and plasmids.
This work was supported by grants from the National Natural Science Foundation of China (grants 30600009 and 30830002) and the China National Basic Research Program (973 program grant 2009CB522605).
Footnotes
Published ahead of print on 5 February 2009.
REFERENCES
- 1.Aceti, D. J., and W. C. Champness. 1998. Transcriptional regulation of Streptomyces coelicolor pathway-specific antibiotic regulators by the absA and absB loci. J. Bacteriol. 180:3100-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bertrand, K. P., K. Postle, J. Lewis, V. Wray, and W. S. Reznikoff. 1983. Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene 23:149-156. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, M. 1996. 1995 Colworth Prize Lecture. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142:1335-1344. [DOI] [PubMed] [Google Scholar]
- 5.Bibb, M. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208-215. [DOI] [PubMed] [Google Scholar]
- 6.Challis, G. L., and D. A. Hopwood. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14555-14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. [DOI] [PubMed] [Google Scholar]
- 8.Cho, Y.-H., E.-J. Kim, H.-J. Chung, J.-H. Choi, K. F. Chater, B.-E. Ahn, J.-H. Shin, and J.-H. Roe. 2003. The pqrAB operon is responsible for paraquat resistance in Streptomyces coelicolor. J. Bacteriol. 185:6756-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folcher, M., R. P. Morris, G. Dale, K. Salah-Bey-Hocini, P. H. Viollier, and C. J. Thompson. 2001. A transcriptional regulator of a pristinamycin resistance gene in Streptomyces coelicolor. J. Biol. Chem. 276:1479-1485. [DOI] [PubMed] [Google Scholar]
- 10.Gramajo, H. C., E. Takano, and M. J. Bibb. 1993. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7:837-845. [DOI] [PubMed] [Google Scholar]
- 11.Gregory, M. A., R. Till, and M. C. M. Smith. 2003. Integration site for Streptomyces phage φBT1 and development of site-specific integrating vectors. J. Bacteriol. 185:5320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guthrie, E., C. Flaxman, J. White, D. Hodgson, M. Bibb, and K. F. Chater. 1998. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology 144:727-738. [DOI] [PubMed] [Google Scholar]
- 13.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 14.Huang, J., C.-J. Lih, K.-H. Pan, and S. N. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, J., J. Shi, V. Molle, B. Sohlberg, D. Weaver, M. J. Bibb, N. Karoonuthaisiri, C. J. Lih, C. M. Kao, M. J. Buttner, and S. N. Cohen. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58:1276-1287. [DOI] [PubMed] [Google Scholar]
- 16.Kieser, T., M. J. Bibb, M. J. Butter, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 17.Kumar, C. V., J. J. Coque, and J. F. Martin. 1994. Efficient transformation of the cephamycin C producer Nocardia lactamdurans and development of shuttle and promoter-probe cloning vectors. Appl. Environ. Microbiol. 60:4086-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenzie, N., and J. Nodwell. 2007. Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J. Bacteriol. 189:5284-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narva, K. E., and J. S. Feitelson. 1990. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2). J. Bacteriol. 172:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onaka, H., T. Nakagawa, and S. Horinouchi. 1998. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol. Microbiol. 28:743-753. [DOI] [PubMed] [Google Scholar]
- 21.Ramos, J. L., M. Martinez-Bueno, A. J. Molina-Henares, W. Teran, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Schmitt-John, T., and J. W. Engels. 1992. Promoter constructions for efficient secretion expression in Streptomyces lividans. Appl. Microbiol. Biotechnol. 36:493-498. [DOI] [PubMed] [Google Scholar]
- 24.Sola-Landa, A., R. S. Moura, and J. F. Martin. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. USA 100:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanley, A. E., L. J. Walton, M. K. Zerikly, C. Corre, and G. L. Challis. 2006. Elucidation of the Streptomyces coelicolor pathway to 4-methoxy-2,29-bipyrrole-5-carboxaldehyde, an intermediate in prodiginine biosynthesis. Chem. Commun. 38:3981-3983. [DOI] [PubMed] [Google Scholar]
- 26.Sun, Y., X. Zhou, J. Liu, K. Bao, G. Zhang, G. Tu, T. Kieser, and Z. Deng. 2002. ‘Sreptomyces nanchangensis,’ a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 148:361-371. [DOI] [PubMed] [Google Scholar]
- 27.Tahlan, K., S. K. Ahn, A. Sing, T. D. Bodnaruk, A. R. Willems, A. R. Davidson, and J. R. Nodwell. 2007. Initiation of actinorhodin export in Streptomyces coelicolor. Mol. Microbiol. 63:951-961. [DOI] [PubMed] [Google Scholar]
- 28.Takano, E., R. Chakraburtty, T. Nihira, Y. Yamada, and M. J. Bibb. 2001. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015-1028. [DOI] [PubMed] [Google Scholar]
- 29.Takano, E., H. C. Gramajo, E. Strauch, N. Andres, J. White, and M. J. Bibb. 1992. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 6:2797-2804. [DOI] [PubMed] [Google Scholar]
- 30.Takano, E., J. White, C. J. Thompson, and M. J. Bibb. 1995. Construction of thiostrepton-inducible, high-copy-number expression vectors for use in Streptomyces spp. Gene 166:133-137. [DOI] [PubMed] [Google Scholar]
- 31.Uguru, G. C., K. E. Stephens, J. A. Stead, J. E. Towle, S. Baumberg, and K. J. McDowall. 2005. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol. Microbiol. 58:131-150. [DOI] [PubMed] [Google Scholar]
- 32.Umeyama, T., and S. Horinouchi. 2001. Autophosphorylation of a bacterial serine/threonine kinase, AfsK, is inhibited by KbpA, an AfsK-binding protein. J. Bacteriol. 183:5506-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White, J., and M. Bibb. 1997. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J. Bacteriol. 179:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson, N. R., P. C. Fineran, F. J. Leeper, and G. P. C. Salmond. 2006. The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4:887-899. [DOI] [PubMed] [Google Scholar]
- 35.Yao, W. S., Y. L. Yang, and J. S. Chiao. 1994. IS204: an insertion sequence from Nocardia asteroides (mexicana) Yp21. Plasmid 32:262-269. [DOI] [PubMed] [Google Scholar]