Abstract
Oenococcus oeni strains are well-known for their considerable phenotypic variations in terms of tolerance to harsh wine conditions and malolactic activity. Genomic subtractive hybridization (SH) between two isolates with differing enological potentials was used to elucidate the genetic bases of this intraspecies diversity and identify novel genes involved in adaptation to wine. SH revealed 182 tester-specific fragments corresponding to 126 open reading frames (ORFs). A large proportion of the chromosome-related ORFs resembled genes involved in carbohydrate transport and metabolism, cell wall/membrane/envelope biogenesis, and replication, recombination, and repair. Six regions of genomic plasticity were identified, and their analysis suggested that both limited recombination and insertion/deletion events contributed to the vast genomic diversity observed in O. oeni. The association of selected sequences with adaptation to wine was further assessed by screening a large collection of strains using PCR. No sequences were found to be specific to highly performing (HP) strains alone. However, there was a statistically significant positive association between HP strains and the presence of eight gene sequences located on regions 2, 4, and 5. Gene expression patterns were significantly modified in HP strains, following exposure to one or more of the common stresses in wines. Regions 2 and 5 showed no traces of mobile elements and had normal GC content. In contrast, region 4 had the typical hallmarks of horizontal transfer, suggesting that the strategy of acquiring genes from other bacteria enhances the fitness of O. oeni strains.
Oenococcus oeni is the most common species of lactic acid bacteria (LAB) associated with malolactic fermentation (MLF) in wine. After alcoholic fermentation (AF), most red and some white wines undergo MLF to improve their organoleptic properties and microbiological stability. From an evolutionary point of view, O. oeni may be considered a specialized microorganism, as it has a small genome of approximately 1.8 Mb (27) and occupies a very narrow ecological niche. It is one of the naturally occurring LAB in grape must. As a result of natural selection, thanks to its remarkable tolerance for the fluctuating environmental conditions during AF, O. oeni becomes the dominant species among those triggering MLF (for a review, see reference 42).
Oenococcus oeni has been described as a relatively homogeneous species, and a variety of molecular approaches failed to clearly differentiate strains on a molecular level (23, 39, 45, 48, 49). Discrimination was accomplished only recently, using fine techniques, such as differential display PCR, restriction endonuclease analysis-pulsed-field gel electrophoresis (18, 22, 50), and multilocus sequence typing (7, 8). Multilocus sequence typing clearly demonstrated an unexpectedly high level of allelic diversity in O. oeni. This was recently partially elucidated by Marcobal et al. (26), who demonstrated the hypermutable status in the genus, due to the absence of the mismatch repair (MMR) genes mutS and mutL. As reported for other bacteria, the lack of mut genes results in the accumulation of spontaneous errors in DNA replication, or in reduced stringency in recombination, thus generating high levels of polymorphism (35). More interestingly, the lack of MMR also facilitates the generation of isolates with beneficial mutations, resulting in increased fitness for the environment. This point is of tremendous importance in selecting commercial O. oeni starters. The considerable phenotypic variation in resistance to wine conditions among indigenous O. oeni isolates coexisting in tanks in the early stages of fermentation is well established and was simply thought to account for the unpredictability of MLF. Commercial starter strains are now widely used in winemaking for greater control over this conversion. However, although progress has been made in selecting and preparing O. oeni strains, commercial starters for direct inoculation in wines are not always effective (6).
A comprehensive catalogue of the genomic differences contributing to plasticity in O. oeni populations is now essential to provide relevant data on fundamental aspects of its evolution. A further aim is to identify genetic markers predictive of enological performance to guide industrial strain selection strategies. A first step in this direction identified correlations between enhanced enological properties and genomic variations in nine target genes (7). Another interesting avenue to explore is the possible contribution of high-level polymorphism to fitness. Genetic recombination creates rearrangements (deletions, duplications, and inversions) and mediates the integration of horizontally acquired mobile elements, such as plasmids, bacteriophages, genomic islands, and transposable elements. The introduction of genes with novel functions into genomes or their replacement with functionally similar orthologs or paralogs is recognized as an important mechanism for adaptation to specific niches in prokaryotes and LAB in particular (24, 25, 32).
The major aim of this study was to explore the genome diversity in O. oeni populations and extend our knowledge of the global gene repertoire of this species. Comparative genome analyses using subtractive hybridization (SH) were used to identify genetic differences between two O. oeni isolates with different tolerances for the wine environment. These data were combined with the publicly available genome sequence of the O. oeni PSU-1 strain, as well as that of the ATCC BAA-1163 strain, completely sequenced in our laboratory (A. Lonvaud, J. Guzzo, et al., unpublished data). We present the first comparative genomic analysis of O. oeni, showing great genetic diversity among the various strains and highlighting the role of insertion/deletion (indel) events in creating diversity. Furthermore, we propose two hypotheses: variable gene distribution may be related to enological potential and some of these sequences have been acquired on small islets through horizontal gene transfer (HGT).
MATERIALS AND METHODS
Bacterial strains, selection, and culture conditions.
Strains of O. oeni expected to be as diverse as possible were selected in 2000 on the basis of their origin—different vineyards in France, the United States, and Portugal—and type of product—red wine, white wine, champagne, and liquor wines. Species identification was carried out by PCR using primers OO1 and OO2, targeting the gene encoding the malolactic enzyme specific to O. oeni (12). Our study also included five commercial malolactic starters and the laboratory strains IOEB 8413 and ATCC BAA-1163. To avoid redundancy in our collection, the isolates were analyzed by pulsed-field gel electrophoresis using NotI, as previously described, and strains with distinct pulsotypes were retained (2). In addition, strains were analyzed using an insertion element (IS)-based PCR strategy, currently developed in our laboratory to type O. oeni isolates. This PCR targets the junctions between the different IS30 elements identified in the genome of O. oeni and the adjacent sequences (C. Le Marrec, et al., unpublished data). The selected 79 strains are presented in Table 1. All O. oeni isolates were routinely grown at 25°C in modified MRS (Difco) containing 1% dl-malate and adjusted to pH 5.0 (M-MRS).
TABLE 1.
O. oeni strains used in this work and their originsa
| Enological potential and strain(s) | Origin |
|---|---|
| HP | |
| B1 (Microenos MBR B); IOEB Sarco 450 | Laffort, commercial strains |
| PSU-1 | California, commercial strain |
| VO (Viniflora oenos) | CHR Hansen, commercial strain |
| VF (Vitilactic F) | Martin Vialatte, commercial strain |
| IOEB Sarco 347, 438a | Red wine-MLF, Gironde (La Roquille, Arveyres), France |
| IOEB Sarco 433a | Red wine-MLF, Lot (Cahors), France |
| IOEB Sarco 384 | White wine-MLF, Savoie, France |
| IOEB Sarco 396, 422 | White wine-MLF, Jura, France |
| IOEB Sarco 451, 453, 1491 | Red wine, Gironde, France |
| IP | |
| IOEB Sarco 7, 10 | Red wine, Landes (Tursan), France |
| IOEB Sarco 114, 394, 417, 419, 452, 456, 457, 459, 461 | Red wine-MLF, Gironde (Bordeaux), France |
| IOEB Sarco 237, 238, 239, 240, 241 | Champagne, Champagne, France |
| IOEB Sarco 386, 387 | Red wine, Gironde (Margaux), France |
| IOEB Sarco 388 | Red wine, Pyrénées Atlantiques (Crouseilles), France |
| IOEB Sarco 390, 392, 393, 448, 449 | White wine, Chablis, France |
| IOEB Sarco 395, 397b | White wine, MLF, Savoie, France |
| IOEB Sarco 401 | White wine, Lot et Garonne (Bazens), France |
| IOEB Sarco 410, 411, 412, 413 | White wine, Gironde, France |
| IOEB Sarco 414 | White wine, Bourgogne, France |
| IOEB Sarco 416 | Red wine, southern France |
| IOEB Sarco 420, 421, 423 | White wine, MLF, Jura, France |
| IOEB Sarco 415, 426, 427, 429, 431, 432 | Red wine, Gironde (Quinsac, St. Germain du Puch, Parempuyre, Libourne, Blanquefort, Rauzan), France |
| IOEB Sarco 442 | Floc, Gascogne, France |
| IOEB Sarco 443, 444 | Pineau, Charentes, France |
| LP | |
| IOEB 8413, ATCC BAA-1163 | Red wine-MLF, Aquitaine, France |
| IOEB Sarco 37, 39a, 171, 428, 440, 447 | Red wine-MLF, Gironde (St. Seurin sur l'isle, Petit Palais, Sadirac, Libourne, Saint Emilion, Bordeaux), France |
| IOEB Sarco 436b, 446 | Red wine-MLF, Gironde (Arveyres), France |
| IOEB Sarco 399 | Red wine-MLF, Gers (Vic Fezensac), France |
| IOEB Sarco 409 | White wine-MLF, Charentes, France |
| IOEB Sarco 4 | White wine-MLF, Jura, France |
| IOEB Sarco 433b | Red wine-MLF, Lot (Cahors), France |
| IOEB Sarco 434 | Red wine, Portugal |
| IOEB Sarco 441 | Floc, Gascogne, France |
| IOEB Sarco 445 | Banyuls, Côte Vermeille, France |
| IOEB Sarco 454, 455, 462 | White wine, Gironde (Bordeaux), France |
The type of product, the geographical region, and the city (in parentheses) are indicated. Strains were classified into three groups referring to their enological potential (high, intermediate, or low) to induce MLF in three different wines. IOEB, Institut d'Oenologie de Bordeaux (France).
An assessment of the resistance to wine and the malolactic efficiency of the 79 strains was performed as previously described (7). Briefly, precultures grown in modified acidic grape medium (11) were used to inoculate three different wines. Microvinifications were done in duplicate using independent precultures. Assays were conducted at 20°C and monitored for a 3-week period for their residual malic acid content. The consumption of malic acid was checked weekly using a thin-layer chromatography method (Roche, France). Briefly, a 5-μl sample of wine was spotted and allowed to separate using a solvent containing 67% (vol/vol) methanol, 17% (vol/vol) acetic acid, and 0.1% (wt/vol) bromophenol blue. The completion of the MLF was further confirmed by determining the malic acid concentration (l-malic acid UV test; R-Biopharm). Strains which completed MLF in the three tested wines (six assays for each strain) were further considered isolates of high potential (HP) (Table 1). Isolates which did not perform MLF in any assay formed the low potential (LP) group. The remaining strains completed MLF in one or two out of the three tested wines, with low reproducibility, and were further considered isolates of intermediate potential (IP) (Table 1).
Genomic subtraction and cloning of fragments.
The isolation of genomic DNA and genomic subtraction was performed through the Genome Express proprietary protocol (Cogenics-Genome Express S.A., Meylan, France), adapted from the initial Straus and Ausubel SH protocol (43) in which strains IOEB Sarco 1491 and IOEB 8413 were used as the tester and driver, respectively (named hereafter SH-1491). The restriction enzymes Sau3A and RsaI were used for the digestion of genomic DNA. Similarly, the opposite SH experiment was conducted, in which strains IOEB 8413 and IOEB Sarco 1491 were used as the tester and driver, respectively (SH-8413). Subtractive PCR probes originating from SH-1491 and SH-8413 were amplified, respectively, from 1,508 clones and 1,403 clones (see Table S1 in the supplemental material) and sequenced in a single pass using the M13 primer according to standard protocols. All sequences were performed on Amersham Pharmacia Biotech Mega BACE4000 DNA sequencers.
Bioinformatics analysis.
(i) A set of 786 filtered sequences (466 and 320 for the Sau3AI and RsaI tester digestions, respectively) with a mean size of 616 ± 172 bp was provided by SH-1491 and assembled using the phred/phrap programs (14) with their default parameters. A total of 127 contigs and 55 singlets with mean sizes of 641 bp and 415 bp, respectively, were obtained from both digestions (see Table S1 in the supplemental material), totaling about 104 kb (i.e., 5.8% of the genome size) and with a GC content of 38.7%. The sequences of the 182 SH-1491 genomic fragments were then tested for their strain specificities, annotated, and ordered to reconstitute the original genome through an in-house bioinformatics strategy.
(ii) The 132 sequences obtained from SH-8413 were first trimmed of vector sequences and then tested by BLASTN and BLASTX cross-searches against the genome sequence of O. oeni ATCC BAA-1163 (not yet public at the time of this writing; a draft version has been released by A. Lonvaud, J. Guzzo, and coworkers under the GenBank accession number AAUV00000000.1). Since the 132 SH-8413 sequences were 99.8% identical to their ATCC BAA-1163 counterparts, both strains were then considered closely related isolates and made the complete genome sequence of strain ATCC BAA-1163 a useful reference for comparative analyses and the rapid and efficient detection of false positives. SH-1491 sequences exhibiting more than 93% identity to their ATCC BAA-163 counterparts were considered false positives and removed from the SH-1491-specific set. Because some of these discarded sequences may also reflect additional gene copies, i.e., paralogous genes in the IOEB Sarco 1491 strain with respect to the control strain, this collection of sequences was kept for further bioinformatic investigations.
(iii) The rest of the specific SH-1491 sequences were submitted to syntactic and functional annotation. BLASTX homology searches were performed against the nonredundant GenBank protein sequence database available at NCBI, and best BLAST hits (BBH) were recorded. A total of 14 SH-1491 orphan sequences, i.e., without any detectable homologs, were set apart and not submitted to the rest of the procedure. Mobile elements were identified using the ISfinder database (41) and manual expertise.
(iv) Because two restriction enzymes were used during SH, some genetic elements needed to be locally microassembled to reconstitute their original architecture as well as to reconstitute a given region. The analysis was completed with the aid of the genome annotation data from O. oeni PSU-1 strain (CP000411), Lactobacillus plantarum WCFS1 (AL935263), and Streptococcus agalactiae A909 (NC_007432). Sequences were orientated and ordered along the corresponding homolog sequences using either T-COFFEE (31) or CAP2 (19). The presence of at least three consecutive genetic elements was taken as a criterion to define the presence of a plasticity region in the IOEB Sarco 1491 O. oeni strain. All details are given in Tables S1 in the supplemental material.
Genomic PCR.
The validation of each subtracted sequence as tester specific was performed by PCR, using the DNA of the tester and driver strains as well as the DNA of the ATCC BAA-1163 strain as controls. PCR was also used to (i) assess SH-1491 sequence distribution in our collection of O. oeni isolates, using internal primers, and (ii) characterize the presence and gene arrangement in the plasticity regions when necessary. The primer design was achieved using the eprimer3 and Oligo Analyser 1.0.3 software. Oligonucleotides were purchased from Sigma-Aldrich. They are described in Table 2.
TABLE 2.
Primers used in this study
| Name | Location/purpose | PCR primer sequence (5′-3′) |
|---|---|---|
| Reg.1 F | OEOE_0684, upstream of region 1 | TACGAAGATGGTATTAAAGCGGTTA |
| Reg 1 R | OEOE_0691, downstream of region 1 | TATCAAAATCAAGATCTTTTTCCAA |
| 2.1 F | Region 2, checking of 3-oxo-acyl carrier protein reductase | CATCTTTTGAAAGGAGACAA |
| 2.1 R | Region 2, checking of 3-oxo-acyl carrier protein reductase | TTAAGGCACCAAGATTTTTA |
| 2.2 F | Region 2, checking of HP YP_809753 | GGCTGCTGAGAAATCTATTA |
| 2.2 R | Region 2, checking of HP YP_809753 | TTTTAGAAAACGGTAAACCA |
| Reg 4 F | Region 4, left end, IS30 | ACACCATTTTAAAGGAAGC |
| Reg 4 R | Region 4, right end, intergenic fnr/IS30 | AGGAGCTGTTAGTGCCTGTA |
| 4.1. F | Region 4, checking of HP NP_786185 | GTCCTATGCCGACTTCCA |
| 4.1. R | Region 4, checking of HP NP_786185 | ACTGAAGACTGCGTTAGCTC |
| 4.2. F | Region 4, checking of cadA ATPase | TCAAGATACCAAGTCCAAA |
| 4.2. R | Region 4, checking of cadA ATPase | TAAGACTGCTAAAACGAGGA |
| 4.3. F | Region 4, checking of dpsB | ACGGACATTCTGATCTAACA |
| 4.3. R | Region 4, checking of dpsB | TTACGTTAAGTTGACCCAAG |
| 4.4. F | Region 4, checking of copper chaperone | TTAACCTTGACGTTTTCAAC |
| 4.4. R | Region 4, checking of copper chaperone | AGCAAGGTAAAGGCTAACTT |
| 4.5. F | Region 4, checking of fnr | CTTAGTACCGACGCATGTTT |
| 4.5. R | Region 4, checking of fnr | GTTTAAGACCTTGATGCTGA |
| Reg 5 F | Region 5, left end | AAACGCATTTTGTTTAATAGCATCT |
| Reg 5 R | Region 5, right end | CTACGCATGATTCCTCATTATCTCT |
| Reg 6 R | Region 6, right end (PTS IIA) | AGGGAGATGTTTCCAATGAAGA |
| OmrA F | RT-qPCRa | GTACCGGTTACAGCTCTA |
| OmrA R | RT-qPCR | GCCATAGCATCCTGAGT |
| Dldh F | RT-qPCR | TGAACAATGGGGAAAAGAGC |
| Dldh R | RT-qPCR | AATTGCATCATAGCCCTTGG |
RT-qPCR, reverse transcription-quantitative PCR.
RNA isolation.
Exponential phase cultures grown in M-MRS were exposed to the following stresses: 10 min at 52°C, 1 h in the presence of 12% alcohol, or 1 h at an acidic pH (pH 3.8). The cells were harvested by centrifugation, resuspended in 1 ml of Tri-reagent (Sigma) and mechanically broken up with glass beads as described before (13). After centrifugation, RNA was extracted from supernatants as recommended by the manufacturer. Purified RNA was suspended in appropriate volume of 0.1% diethyl pyrocarbonate-treated water. Contaminating chromosomal DNA was removed by digestion with RNase-free DNase (1 U/μl; Ambion). The determination of the sample concentration and quality was performed by optical density readings at 260 and 280 nm (SmartSpec Plus spectrophotometer; Bio-Rad, France) and by agarose gel electrophoresis. RNA preparations were stored at −80°C until used. Extractions were performed from two independent cultures.
Reverse transcription- and real-time PCR.
cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad) as recommended. The 25-μl reaction mixture (4 μl of 5× iScript reaction mix, 1 μl of iScript reverse transcriptase, and 1 μg of RNA) was incubated for 5 min at 25°C, 30 min at 42°C, and 5 min at 85°C. Real-time PCR was carried out on a Bio-Rad iCycler with the iQ SYBR green supermix (Bio-Rad) in 96-well plates. A volume of 5 μl containing 10 pg of cDNA was added to 20 μl of PCR mixture (12.5 μl of iQ SYBR green supermix, 0.05 μl of each primer at 100 μM, and 6.4 μl of RNase-free water). Specific cDNA was amplified by real-time PCR with specific primers that were designed based on internal regions of the stress-responsive omrA gene (3) and of the eight SH fragments of interest identified in our study (see Table S1 in the supplemental material). In each run, negative and positive controls were included. Thermal cycling conditions were designated as follows: the reaction mixture was preheated for an initial denaturation at 95°C for 5 min, and 30 cycles were then carried out, each consisting of a denaturation step (30 s, 95°C), an annealing step (20 s, 52°C), and an extension step (20 s, 72°C). For each measurement, the threshold cycle value was determined with a baseline set manually at 100 relative fluorescence units (RFU). The differences in gene expression between the M-MRS standard condition and wine mimetic stresses (1-h shock in ethanol, acid, or heat) were analyzed using the comparative critical threshold method, in which the amount of target RNA is adjusted to a reference relative to an internal calibrated target RNA as previously described (13). The results were normalized by using the ldhD gene coding for lactate dehydrogenase, as its transcript level is weakly affected by the three tested stress conditions (10). Manipulations were done in duplicate. We considered that genes were significantly down- or upregulated if their relative expression levels were found to be at least twofold lower or higher than that of the control reference (10, 13).
Statistical analysis.
A chi-square test was computed considering the number of positive PCR amplifications obtained in the strain collection for each couple of primers and the total numbers of strains in each group of enological potential. A sequence was considered statistically overrepresented in the HP group if the chi-square test was significant at the 5% level.
Nucleotide sequence accession numbers.
The nucleotide sequence data corresponding to regions 4, 5, and 6 from the IOEB Sarco 1491 strain have been deposited under the accession numbers FJ625794, FJ613272, and FJ613273, respectively.
RESULTS
Analysis of the subtracted library.
SH-1491 revealed 182 tester-specific fragments, containing a total of 140 genetic elements. A total of 14 sequences had no significant BLASTX match in the database (Table 3). The remaining fragments corresponded to 126 putative open reading frames (ORFs), of which 81 had a significant BLASTX match with sequences originating from the O. oeni gene repertoire. In contrast, the IOEB Sarco 1491 strain had retained 45 sequences, displaying no BLASTX matches with known O. oeni sequences but with homologs in other bacteria (Table 3). Details of these sequences are given in Tables S1, S2, and S3 in the supplemental material.
TABLE 3.
Identification of sequences obtained by SH experiments
| BlastX query result | Total no. of sequences | No. of sequences with indicated characteristics
|
|||
|---|---|---|---|---|---|
| Mobile elements
|
Other genetic elements | ||||
| Phages | Plasmids | ISs | |||
| No significant hit | 14 | ||||
| Hit in O. oeni | 81 | 22 | 0 | 2 | 57 |
| Hit outside O. oeni | 45 | 14 | 0 | 2 | 29 |
| Total | 140 | ||||
Among the first group of 81 sequences, 71% showed homology with O. oeni chromosomal DNA (see Table S2 in the supplemental material). A total of 34 sequences had no matches in ATCC BAA-1163, but orthologs were present in other O. oeni isolates with high identity levels (80 to 100%). The remaining nucleotide sequences matched protein sequences of both the PSU-1 and ATCC BAA-1163 strains, with similar identity levels, except for a peptide methionine sulfoxide reductase (91.8 and 76.3% identity in the two isolates, respectively). It is noteworthy that 11 of those sequences shared less than 80% identity with their O. oeni counterparts (see Table S2 in the supplemental material). In this first group, a total of 13 sequences were related to hypothetical proteins. Other predicted protein sequences derived from the SH fragments matched proteins related to carbohydrate transport and metabolism (n = 6), cell wall/membrane/envelope biogenesis (n = 5), and replication recombination (n = 4). Interestingly, six sequences matched components of the arc operon involved in the arginine degradation pathway in O. oeni ATCC 23279 (12, 29, 47): an ornithine carbamoyltransferase (ArcB), a carbamate kinase (ArcC), two arginine/ornithine ArcD1 and ArcD2 antiporters, an arginine deiminase (ArcA), and an arginyl-tRNA synthase (ArgS2). The first four genes were absent from the PSU-1 and ATCC BAA-1163 genomes, while orthologous arcA and argS2 sequences were detected in both strains. The argS2-related sequence is not part of the ADI operon but corresponds to the argS1 divergent ectopic copy of the gene found in both arc+ and arc-lacking strains (29). As the IOEB Sarco 1491 strain degrades arginine, unlike IOEB 8413 (47), this result illustrates the effectiveness of SH, as used in our laboratory, at retrieving IOEB Sarco 1491 strain-specific genes. This first group of 81 Oenococcus-related sequences also contained mobile/extrachromosomal sequences, consisting of two IS-related elements, as well as 22 phage-related sequences (see Table S2 in the supplemental material).
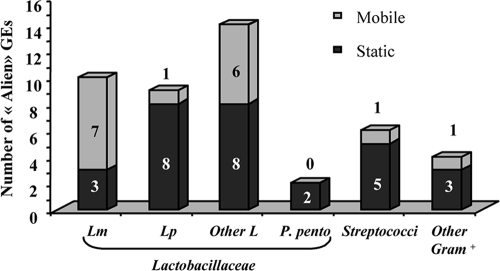
SH-1491 also revealed a second group of 45 sequences (29%) with best BLASTX matches outside the species, rather than with previously sequenced O. oeni genes/proteins (Table 3; see also Table S3 in the supplemental material). These novel gene sequences represented approximately 2% of the total number of ORFs in the genomes of O. oeni PSU-1 and ATCC BAA-1163 and were, therefore, added to the global gene repertoire of O. oeni. This group contained 29 sequences corresponding to chromosomally encoded proteins, mainly dedicated to sugar transport and metabolism, particularly incomplete transport systems for cellobiose (phosphotransferase system [PTS] subunit A) and fructose (PTS IIABC). The remaining novel sequences consisted of 14 phage- and two transposon-associated genes. The BBH of the 45 alien sequences were associated with other LAB usually present in wine environments, such as lactobacilli (55%), Pediococcus pentosaceus, and Leuconostoc mesenteroides (Fig. 1).
FIG. 1.
Origins of the BBH of the 45 alien sequences identified in O. oeni IOEB Sarco 1491. Lm, L. mesenteroides; Lp, L. plantarum; L, lactobacilli; P. pento, P. pentosaceus; GEs, genetic elements.
Contribution of phages to plasticity.
One striking finding was the subtraction of 36 phage-related sequences (26%) (see Tables S2 and S3 in the supplemental material). Analysis revealed that the IOEB Sarco 1491 strain contained at least part of the temperate oenophages fOg44, fOg30, and fOgPSU1 (12, 2, and 3 matching sequences, respectively), as well as smaller tracts of ΦL10 (n = 3) and Φ10MC (n = 1) (16, 17, 38, 44). Another 14 phage-related sequences had best BLASTX matches (>30% identity) with structural proteins from phages infecting other LAB: a ΦC31-like phage (7 matching sequences), ΦAT3 (n = 4), ΦSfi19 (n = 1), Φbil309 (n = 1), and ΦLp2 (n = 1), infecting L. mesenteroides, Lactobacillus casei, Streptococcus thermophilus, Lactococcus lactis, and L. plantarum, respectively. There was only one exception: a sequence matching a replisome protein from a more distantly related phage, infecting Clostridium beijerinckii (Fig. 1).
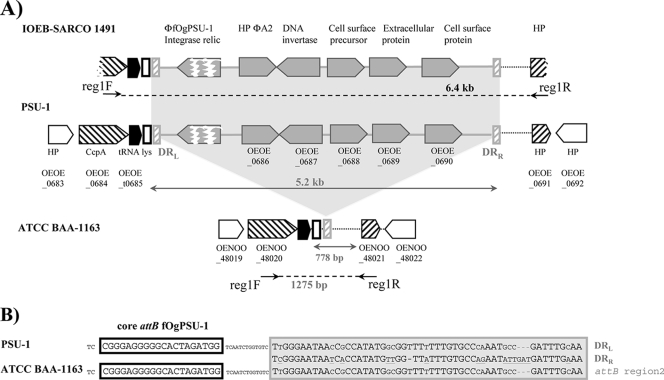
Among the phage-related sequences identified above, we focused on a DNA fragment related to a hypothetical protein from ΦA2, infecting L. casei, and found an orthologous sequence in the genome of strain PSU-1 (OEOE_0686). In this strain, the phage-related sequence was located upstream from four ORFs corresponding to a putative DNA invertase gene, recently identified on plasmids isolated from S. thermophilus (15) and Lactobacillus paracasei (9), and three putative cell surface proteins, whose sequences have few BLAST hits in the LAB family (Fig. 2). Remarkably, sequences corresponding to the latter three sequences were also subtracted from the genome of the IOEB Sarco 1491 strain (Table 4). The sizes and organizations of the locus referred to as region 1 were the same in the genomes of both PSU-1 and IOEB Sarco 1491 isolates, as revealed by an analysis of the 6.4-kb amplicon obtained from a PCR with flanking primers reg1F and reg1R, based in ORFs OEOE_0684 and OEOE_0691, respectively. Only a 778-bp amplicon was obtained from the genomic DNA of both IOEB 8413 and ATCC BAA-1163, indicating that region 1 was absent from these isolates. To better characterize this indel event, we compared the corresponding 778-bp intergenic region from the ATCC BAA-1163 strain with the boundaries of region 1 in the PSU-1 strain. Region 1 was, therefore, located between genes coding for a tRNA lysine and a hypothetical protein (Fig. 2). This analysis also led to the identification of (i) two imperfect ∼50-bp direct repeats (DR) flanking region 1 and (ii) a third copy in the 778-bp intergenic region in ATCC BAA-1163 (Fig. 2). Interestingly, the leftward DR was just downstream from the core attB site proposed for fOgPSU-1 prophage establishment in the PSU-1 strain (38). Despite the presence of five fOgPSU-1-related sequences among the O. oeni IOEB Sarco 1491-specific genes, amplification of a 6.4-kb amplicon with reg1F and reg1R primers showed that no lysogenization by any fOg-related phage occurred at this particular attB site in this isolate. A fOgPSU-1-related element is, therefore, likely to be integrated at another site in the IOEB Sarco 1491 strain. The existence of multiple loci as targets for prophage integration has been demonstrated in O. oeni (38). One potential target is attBfog44, located nearby, downstream from OEOE_0696. Considering all these data, the chromosomal region under investigation is, therefore, likely to be a spot of high prophage integration in O. oeni.
FIG. 2.
Organization of region 1, a possible tract of phage origin. (A) Schematic representation of region 1 in the IOEB Sarco 1491, PSU-1, and ATCC BAA-1163 strains. The core attB site used by fOgPSU-1 and overlapping the tRNA gene (OEOE_t0685) is represented by a black box. The gray hatched boxes represent the putative junctions (PSU-1) and core attachment sites (ATCC BAA-1163), considered to characterize the site-specific integration of region 1 in the O. oeni genome. (B) Alignments of the putative left, right and bacterial attachment sites found in the PSU-1 and ATCC BAA-1163 strains, respectively. DRL, leftward DR; DRR, rightward DR.
TABLE 4.
Characteristics of the six plasticity regions identified in this study and prevalence of selected specific genomic sequences in a collection of strains with LP, IP, or HP to induce MLF
| Region | Accession no.a | Predicted functionb | Organism | Size (aa)c | Coordinates (aa)d | E value | % of identity | % of coverage | Total % | Prevalence in HP groupe | Mobilityf |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | YP_810287 | FA2-related hypothetical protein | O. oeni PSU-1 | 339 | 1-91, 125-210 | 1E-37, 6E-22 | 73.4 | 52.2 | Phage-related sequences | ||
| YP_810289* | Hypothetical protein | O. oeni PSU-1 | 191 | 39-191 | 2E-80 | 100 | 80.1 | 88.5 | ns | Flanking DR | |
| YP_810290* | Hypothetical protein | O. oeni PSU-1 | 166 | 26-166 | 4E-65 | 90.7 | 84.9 | 87.2 | ns | 50 bp, imperfect | |
| YP_810291 | Cell surface protein | O. oeni PSU-1 | 164 | 1-25 | 2E-05 | 96.0 | 15.2 | ||||
| 2 | YP_809730 | Hypothetical protein | O. oeni PSU-1 | 123 | 1-123 | 7E-54 | 86.1 | 100 | ni | ||
| YP_809731 | Transcriptional regulator | O. oeni PSU-1 | 453 | 1-48, 298-453 | 2E-26, 1E-82 | 100 | 45.0 | ||||
| YP_809743* | Arabinose efflux permease | O. oeni PSU-1 | 387 | 1-40, 249-387 | 4E-13, 1E-62 | 93.8 | 46.3 | 77.7 | ns | ||
| YP_809745* | 3-Oxo-acyl carrier protein reductase | O. oeni PSU-1 | 252 | 1-57 | 7E-25 | 100 | 22.6 | 79.5 | 4.40 | ||
| YP_809747* | Galactoside-O-acetyltransferase | O. oeni PSU-1 | 177 | 1-150 | 2E-80 | 98.6 | 84.7 | 78.2 | |||
| YP_809748* | NADH quinone reductase | O. oeni PSU-1 | 300 | 88-143, 174-300 | 1E-24, 1E-68 | 100 | 61.0 | 85.9 | ns | ||
| YP_809752* | Hypothetical protein | O. oeni PSU-1 | 129 | 101-129 | 8E-08 | 100 | 21.7 | 73.0 | ns | ||
| YP_809753* | Hypothetical protein | O. oeni PSU-1 | 78 | 1-78 | 7E-27 | 80.7 | 100 | 78.0 | |||
| YP_809754* | Phosphoglycerate mutase | O. oeni PSU-1 | 246 | 1-246 | 1E-134 | 98.8 | 100 | 85.9 | ns | ||
| YP_809755 | Hypothetical protein | O. oeni PSU-1 | 66 | 1-66 | 5E-31 | 100 | 100 | ||||
| 3 | YP_811244* | Polysaccharide transporter, PST family | O. oeni PSU-1 | 479 | 1-74, 160-478 | 3E-23, 1E-136 | 75.3 | 81.6 | 58.9 | ns | ni |
| ATCC BAA-1163 | 479 | 1-74, 160-478 | 3E-23, 1E-136 | 75.0 | |||||||
| YP_811245* | Predicted glycosyltransferase | O. oeni PSU-1 | 269 | 226-268 | 2E-11 | 65.0 | 16.0 | 65.4 | ns | ||
| ATCC BAA-1163 | 269 | 226-268 | 2E-11 | 65.0 | |||||||
| YP_811247* | Glycosyltransferase-related enzyme | O. oeni PSU-1 | 312 | 1-59 | 1E-18 | 71.0 | 18.9 | 57.7 | ns | ||
| ATCC BAA-1163 | 312 | 1-59 | 1E-18 | 71.0 | |||||||
| YP_811251* | Predicted glycosyltransferase | O. oeni PSU-1 | 306 | 6-302 | 1E-80 | 54.3 | 97.1 | 65.4 | ns | ||
| ATCC BAA-1163 | 306 | 6-302 | 1E-80 | 54.3 | |||||||
| YP_811248 | Hypothetical protein | O. oeni PSU-1 | 455 | 1-29, 315-448 | 1E-04, 8E-51 | 82.7 | 36.0 | ns | ns | ||
| ATCC BAA-1163 | 455 | 1-29, 315-448 | 1E-04, 8E-51 | 82.6 | |||||||
| YP_811249* | Capsular polysaccharide biosynthesis protein | O. oeni PSU-1 | 181 | 72-179 | 6E-36 | 66.6 | 59.7 | 60.2 | ns | ||
| ATCC BAA-1163 | 181 | 72-179 | 6E-36 | 66.6 | |||||||
| 4 | NP_786654* | Hypothetical protein | L. plantarum | 323 | 1-323 | 1E-165 | 94.0 | 100 | 16.7 | ns | Flanking IS |
| NP_786185* | Hypothetical protein | L. plantarum | 117 | 58-117 | 3E-20 | 83.3 | 51.3 | 14.0 | 3.6 | ||
| YP_809989 | Hypothetical protein | O. oeni PSU-1 | 67 | 1-67 | 6E-17 | 75.7 | 100 | ||||
| YP_809985* | Cadmium transport ATPase | O. oeni PSU-1 | 634 | 1-551 | 0.0 | 84.2 | 86.9 | 21.8 | 4.6 | ||
| YP_809986* | GlpF aquaglyceroporin | O. oeni PSU-1 | 250 | 1-112 | 6E-57 | 92.0 | 44.8 | 20.5 | ns | ||
| NP_786657* | Thioredoxin | L. plantarum | 106 | 9-106 | 3E-50 | 97.9 | 92.5 | 35.9 | ns | ||
| YP_809988* | Hypothetical protein | L. plantarum | 89 | 1-82 | 1E-33 | 90.2 | 92.1 | 29.5 | ns | ||
| YP_809990* | Dps | O. oeni PSU-1 | 184 | 1-184 | 6E-92 | 98.2 | 100 | 16.7 | 4.3 | ||
| YP_809991* | Copper chaperone | O. oeni PSU-1 | 74 | 1-57 | 9E-24 | 96.4 | 100 | 23.0 | 5.0 | ||
| YP_809992* | Fnr-like protein | O. oeni PSU-1 | 221 | 1-221 | 1E-116 | 98.6 | 100 | 33.3 | 5.7 | Flanking IS | |
| 5 | NP_784135 | Acetyltransferase | L. plantarum | 161 | 1-113 | 3E-23 | 47.7 | 70.2 | ni | ||
| NP_786731* | 6-Phospho-beta-glucosidase | L. plantarum | 480 | 1-476 | 0 | 72.6 | 99.2 | 48.7 | ns | ||
| YP_279925* | PTS, beta-glucoside-specific IIABC | S. pyogenes | 620 | 201-602 | 1E-116 | 51.3 | 64.8 | 42.3 | ns | ||
| NP_786735* | Maltose phosphorylase | L. plantarum | 734 | 590-734 | 2E-26 | 50.0 | 19.7 | 36.9 | 5.0 | ||
| 6 | YP_330359* | PTS, fructose-specific, IIC | S. agalactiae | 367 | 1-338 | 1E-119 | 63.0 | 92.1 | 5.1 | ns | ni |
| YP_330360* | PTS, fructose-specific, IIA | S. agalactiae | 149 | 8-149 | 9E-32 | 53.2 | 95.3 | 5.1 | ns | ||
| YP_330361 | PTS, fructose-specific, IIB | S. agalactiae | 103 | 1-87 | 8E-10 | 44.8 | 84.5 |
Order as found in O. oeni IOEB Sarco 1491. Asterisks indicate sequences used in the distribution analysis.
A statistically significant positive association was found between HP strains and the presence of the sequences with the functions highlighted in bold.
aa, amino acids.
Amino acid position in BBH.
Chi-square value (see Materials and Methods) expressing the preferred distribution of each of the eight tester-specific sequences in the HP group and IP plus LP groups. ns, nonsignificant.
Mobility-associated features (phage-related sequences, DRs, or ISs). ni, not identified.
The hypothesis of a possible phage origin for region 1 was further supported by the observation that the noncoding region immediately downstream from the leftward DR exhibited a significant homology to a remnant fOgPSU-1-related integrase gene. Phage integrase gene and attachment sites are known to be in close proximity on viral genomes. Region 1 may, therefore, represent a remnant prophage and the DR the attachment sites used for its site-specific integration in the O. oeni genome. The distribution of region 1 was assessed in our collection, using two sets of primers internal to the OEOE_0688 and OEOE_0689 genes. The ratios of positive isolates for the traits were 88.5 and 87.2%, respectively (Table 4). This may indicate an ancient acquisition of a prophage in O. oeni, which may have undergone many evolutionary stages, resulting in the current structure of region 1. The presence of a defective integrase and imperfect direct repeats flanking the locus may be part of the mechanism promoting stabilization in the species. It remains unclear whether the absence of region 1 in strains ATCC BAA-1163 and IOEB 8413 is due to a lack of lysogenization or a deletion event, based on an illegitimate recombination process, using the short stretches of homologous DNA bordering the region.
Other plasticity regions.
A total of 33 additional SH-1491 sequences (absent or highly divergent in the driver) mapped to five distinct plasticity regions, in which synteny was conserved in O. oeni (PSU-1 and/or ATCC BAA-1163) or other LAB (Table 4).
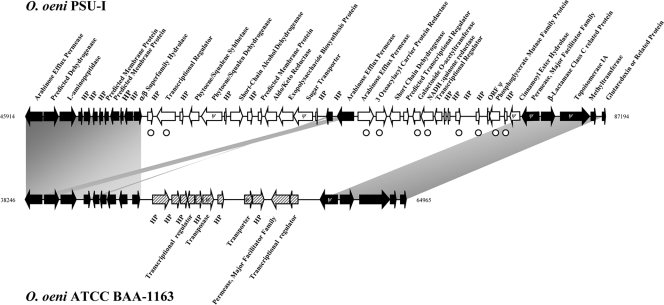
A set of 10 SH-1491 sequences mapped to region 2 (30 ORFs; ∼30 kb) in the genome of the PSU-1 strain (Table 4). This region, located between the OEOE_0060 and OEOE_0091 genes, is presented in Fig. 3. It consists of a variety of genes, including those coding for proteins involved in carotenoid biosynthesis. Various genes dedicated to sugar transport and metabolism are also interspersed in the region, including those encoding a galactoside acetyltransferase and a putative arabinose permease. The region also contains genes encoding hypothetical proteins whose functions have yet to be elucidated. Region 2 as a whole was absent from the genome of the ATCC BAA-1163 strain, replaced by a different set of 11 genes, including seven encoding hypothetical proteins (Fig. 3). PCR analysis revealed the distribution of seven subtracted sequences, indicating the prevalence of region 2 among our strain panel of 79 isolates. Positive strains were in the 73 to 85.9% range (Table 4). The GC content percentage of the region was characteristic of the O. oeni genome as a whole. Last, no common hallmarks of laterally transferred genes (unusual GC bias, anomalous codon usage, presence of tRNA genes at the 3′ end, tendency to be flanked by DR, and inclusion of mobile genetic elements) were found in its vicinity in strain PSU-1. Region 2 may, therefore, result from an orthologous replacement event, occurring during the evolutionary history of the species (32).
FIG. 3.
Organization of region 2 in O. oeni PSU-1 and ATCC BAA-1163. Black arrows indicate genes common to O. oeni PSU-1 and ATCC BAA-1163, white arrows indicate genes specific to PSU-1, and hatched arrows indicate genes specific to ATCC BAA-1163. Ψ, pseudogene; ○, SH-1491-specific sequences.
Region 3 corresponded to six genes encoding proteins required for the biosynthesis of capsular polysaccharides, including three putative glycosyltransferases. Unlike the other plasticity regions, variability did not consist of different genes, but rather homologs with low identity. Hence, although four of the SH-1491-specific sequences had a best, identical BLASTX match with proteins from both O. oeni PSU-1 and ATCC BAA-1163, they shared less than 80% identity with the matching proteins. In addition, the gene ordering differed from that of strain PSU-1 (Table 4). The reason for this divergence, observed only in the IOEB Sarco 1491 strain, is not known. Among the various hypotheses is that the region subtracted from the IOEB Sarco 1491 strain may result from a duplication event and/or a substantial intragenic recombination, as recently proposed by de Las Rivas et al. (8).
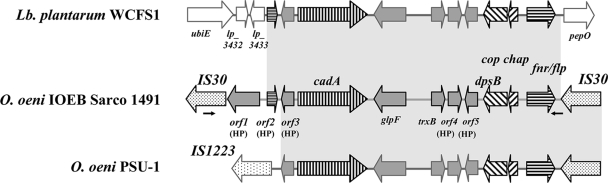
Three additional plasticity regions (4, 5, and 6) were detected. They differed from the above loci as they harbored gene sequences with deduced protein sequences displaying BBH outside O. oeni. The most interesting of these was region 4, which comprised 11 ORFs in the IOEB Sarco 1491 strain, including four with a BBH in L. plantarum (Fig. 4). The locus, presenting a GC bias, was sandwiched between two copies of IS30, whose sequences were also partially subtracted. In addition, part of the intervening sequence exceeded 90% identity with L. plantarum DNA, while synteny and gene order were conserved between the corresponding orthologous regions (Fig. 4). All these data indicate a recent HGT event (Fig. 4). Identifiable functions corresponded to aquaglyceroporin (GlpF) and P1-type ATPase (CadA) dedicated, respectively, to the efflux of water and cadmium (Cd2+) in bacteria. The next gene corresponded to trxB, the second thioredoxin gene identified in O. oeni (20). Thioredoxins are key players in maintaining intracellular protein disulfides in reduced form. Region 4 also harbored a gene coding for a Dps-like protein in the mini-ferritin family. These proteins are particularly important during oxidative stress or the depletion of environmental nutrients (46). The next genes were a putative copper chaperone and a transcriptional regulator in the fumarate-nitrate-reductase (FNR) transcription regulator family. FNR-like regulators respond to a broad spectrum of intracellular and exogenous signals, including cyclic AMP, anoxia, redox state, oxidative stress, and temperature (21). Region 4 was present in the commercial O. oeni PSU-1 genome but displayed slight modifications at its leftward extremity. ORF1 and ORF2 were missing, while the left IS30 was replaced by an IS1223-related element (Fig. 4). IS1223 elements belong to the IS3 family and were recently identified in the genome of Lactobacillus salivarius UCC118 (41). Using Reg 4 F/R primers revealed that the strains in our collection that harbored region 4 all exhibited the structure described for the IOEB Sarco 1491 strain. The results of the distribution analysis of selected sequences internal to nine ORFs, showing that positive isolates varied from 14 to 36% (Table 2), also indicated polymorphism within the region.
FIG. 4.
Architecture of region 4 in O. oeni (IOEB Sarco 1491 and PSU-1) and L. plantarum. In the IOEB Sarco 1491 strain, the 7-kb region contains 11 genes flanked by two IS30 sequences. A statistically significant positive association was found between HP strains and the presence of sequences indicated by hatched arrows. Primers Reg 4 F and Reg 4 R are represented by black arrows.
Regions 5 and 6 contained ∼5 kb and 4 kb, respectively. These loci included an acetyltransferase, a glucosidase, and a maltose phosphorylase, as well as genes involved in sugar transport (Table 4). Matching proteins were not found in O. oeni but were found in L. plantarum/Streptococcus pyogenes (region 5) and S. agalactiae (region 6), with identity levels from 47 to 76%. BLASTN searches revealed no hits in the databanks. An examination of the local and relative GC content of the O. oeni genome did not reveal any significant variations or hallmarks of horizontal transfer (using Island Finder). Significantly, distribution analysis of five sequences representing both regions in our collection revealed low scores, also indicating recent horizontal transfer (Table 4). This was striking for region 6, as only 5% of the 79 isolates screened shared the sequences corresponding to the putative PTS-fructose-specific IIC and IIA.
Plasticity regions and association with bacterial fitness.
To investigate the involvement of the plasticity regions identified in the increased fitness of O. oeni, the distribution of selected sequences among groups previously characterized for their enological potential was analyzed, as well as gene expression under enological stresses. The distribution of 28 selected sequences representing the six plasticity regions described above, as well as randomly selected singletons, was assessed using PCR (see Table 4). Their prevalence was determined in a strain collection including 19 LP, 44 IP, and 14 HP isolates (the 10 wild strains described above and the four commercial malolactic starters B1, PSU-1, VO, and VF). None of the SH-1491 sequences were unequivocally present in HP isolates. However, the prevalence of eight individual SH-1491 sequences was significantly higher in the HP group than in the rest of the O. oeni collection. The greatest statistically significant differences between the groups concerned the putative dps and copper chaperone sequences, both from region 4. These were present in 43% of HP and 11% of IP and LP isolates and 50% of HP and 17% of IP and LP isolates, respectively. Three additional SH-1491 fragments belonging to the same 7-kb region, related to the hypothetical protein NP_7876185, the P-type ATPase, and Fnr, were also prevalent. The three remaining prevalent sequences were related to the 3-oxo-acyl carrier protein reductase and hypothetical protein YP_809753, both from region 2, as well as a maltose phosphorylase from region 5.
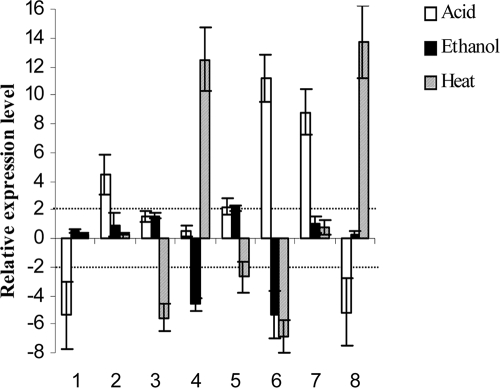
Assuming that the eight genes were novel factors involved in adaptation to wine, we investigated whether any of the genes were actively expressed under normal (laboratory) culture conditions as well as during ethanol and acid shocks, two major stress conditions in winemaking identified during our selection of HP strains (see Materials and Methods). Heat shock was also tested, as it is also a frequent stress factor during winemaking. RNA was extracted from stressed and control cells during the exponential growth phase in MRS broth and the relative expression of the eight genes was determined by reverse transcription followed by quantitative PCR (Fig. 5). Six HP strains were tested, including five that were positive for the eight sequences of interest (IOEB Sarco 1491, 438, 433a, 384, and B1), while the commercial PSU-1 strain was positive for only six potential-associated fragments. Transcripts were obtained for all genes tested, in both stressed and control cells. Remarkably, each performance-associated fragment corresponded to a stress-responsive gene and was induced or repressed by a factor greater than two following at least one of the wine stresses tested. Variations in expression were observed following one (dpsB, copper chaperone, HP NP_786185, and oxo-acyl carrier protein reductase), two (HP YP_809753 and fnr), or all three stresses (maltose phosphorylase and P-ATPase) applied to the O. oeni strains.
FIG. 5.
Expression of the eight enological potential-associated fragments during acid (unfilled bars), ethanol (filled bars), and heat (hatched bars) shocks. 1, Dps; 2, copper chaperone; 3, HP NP_786185; 4, HP YP_809753; 5, maltose phosphorylase; 6, CadA; 7, 3-oxo-acyl carrier protein reductase; 8, Fnr.
DISCUSSION
The major aim of this study was to explore genome diversity among O. oeni strains and extend the global gene repertoire for this species. Various divergent sequences as well as novel sequences representing about 2% of the total number of ORFs present in the complete sequenced genomes of O. oeni PSU-1 and ATCC BAA-1163 were identified by SH. Our data are therefore consistent with recent developments in the study of bacterial genomes, which have made it abundantly clear that the sequence of a few genomes is unlikely to provide full information about the species.
From the analysis of the subtracted sequences, some hypotheses concerning the mechanisms generating plasticity in O. oeni could be proposed. The findings of numerous variable sequences support the role of recombination in creating diversity in O. oeni (8). SH identified major variations in singlets, and some also mapped to discrete areas. This is true for region 3, a small polymorphic locus related to polysaccharide biosynthesis. We believe that this locus is part of a larger region of plasticity, since it is immediately upstream from the recently described dpsA variable region (2).
Another important point highlighted by our study was the occurrence of indel events in O. oeni. Region 2 (30 ORFs) was present in strains IOEB Sarco 1491 and PSU-1 but absent in strain ATCC BAA-1163. No hallmarks of transfer were found, suggesting a possible loss of the region.
High abundance and diversity of sequences of foreign origin were found. A substantial proportion could be attributed to phage-related sequences (26% of the subtracted sequences). Some of the matching proteins were related to phages known to be active with O. oeni (Φfog44, Φfog30, ΦfogPSU-1, ΦL10, and Φ10MC), suggesting the lysogenization of the IOEB Sarco 1491 strain. However, matches with phages infecting other LAB, as well as more distantly related gram-positive bacteria, were also identified in this study. Similar findings were recently reported in O. oeni BIFI-83 (25). Phages evolve mainly by exchanging modules, and it is now widely accepted that these exchanges are not restricted to phages infecting the same bacterial species (5). The significant contribution of phage-related sequences to the dispensable gene reservoir of O. oeni is consistent with numerous reports of a high incidence of lysogeny in the species (1, 34). However, these findings contrast with the situation recently described in the genomes of the ATCC BAA-1163 and PSU-1 strains, which do not contain any intact temperate bacteriophages or large tracts (>10 kb) of obvious bacteriophage origin (27, 37, 38). The apparent discrepancy with the genome data may be related to the fact that the IOEB Sarco 1491 strain was only recently isolated from wine (2000) and moderately propagated in the laboratory. In contrast, two long-established laboratory strains have been chosen for sequencing projects, and this is also true of the IOEB 8413 driver strain. Many genetic changes occur during multiple laboratory subcultures, including the loss of mobile elements. These events have probably resulted in an underestimation of phage sequences in the previously available complete genome data of O. oeni. An assessment of the stability of phage-related sequences during the prolonged cultivation of bacterial strains on synthetic media, in the absence of any phage pressure, is now required.
Our results confirm previous findings, indicating the presence of strong, widespread pressure due to bacteriophage predation in the environment of O. oeni. This phenomenon has major repercussions on virus evolution. Thus, virulent bacteriophages have the potential to activate dormant prophages, leading to rapid evolution. The evolution of temperate bacteriophages via host recombination results in chimeric phages with novel phenotypes. Phages also exert considerable influence on the genomes of their bacterial hosts through various mechanisms, e.g., prophages also play a prominent role in shaping bacterial genome architecture. An interesting mechanism, by which Campylobacter jejuni bacteria were infected by virulent bacteriophage CP34, led to major genomic rearrangements in the surviving bacteria (40), leading to the hypothesis that these bacteria exist in vivo as families of related metagenomes generated to survive local environmental pressures.
Among the putative sequences of foreign origin, we identified 21 genes that were physically clustered in four loci (regions 1, 4, 5, and 6). Region 1 corresponded to a remnant prophage. Regions 2, 4, and 5 were particular as these sequences had best BLASTX matches outside the species, while gene order was conserved between the orthologous regions. In addition, sequences displayed a restricted distribution among our strain panel. These data may denote their acquisition through lateral gene transfer. Accordingly, region 4 had additional hallmarks of transfer (atypical patterns of codon usage and presence of flanking IS elements). An increase of HGT is one of the consequences of a defect in the MMR system in bacteria.
We identified six different regions of plasticity in this study, resulting from recombination or indel events. We next investigated their involvement in the increased fitness of O. oeni in wine. The bacterial species is not amenable to genetic transformations enabling gene inactivation. Therefore, we analyzed the distribution of selected sequences from each region among groups previously characterized for their enological potential (high, intermediate, or low). We also analyzed gene expression under enological stresses. There was a statistically significant positive association between HP strains and the presence of eight genes. The eight genes were also stress-responsive in wine in HP strains. Although these data do not represent formal proof, they suggest that the presence of these particular sequences contribute to the fitness of O. oeni in wine. All eight sequences belonged to regions 2, 4, and 5, expected to result from indel events. A sequence related to an oxo-acyl carrier protein reductase was part of region 2, a locus containing 30 ORFs that is proposed to be deleted in some O. oeni strains. Further work is now needed to understand how this sequence could benefit O. oeni in wine. Interestingly, this locus also contained a sequence related to an arabinose permease. Arabinose is one of the sugars detected in wines at the end of AF, when glucose and fructose have been degraded by yeasts. It is well-known that some O. oeni isolates do not utilize arabinose (A. Lonvaud-Funel, unpublished data). The correlation of this phenotype with the absence of region 2 is currently being investigated in the laboratory, even though no prevalence of this sequence among HP strains was observed in the current study. Other potential-associated sequences were found in regions 4 and 5, proposed to be acquired by HGT. Hence, five out the nine tested genes from region 4 were associated to fitness and were stress responsive in wine. Although their functionality and role remain to be assessed, it is noteworthy that one of these potential-associated genes was a Dps gene homolog. A first member of this family was recently identified on a genomic island in O. oeni and shown to protect E. coli from the deleterious effects of wine, copper, and ferric ions (2). Another remarkable potential-associated sequence is related to cadA, which encodes a putative cadmium efflux system. Its significant induction under acid stress was consistent with data from Penaud et al. (33), who demonstrated this trait for various P-type ATPases detected in the genome of Lactobacillus bulgaricus. Cadmium is a major environmental hazard and contributes indirectly to oxidative stress by affecting the cellular thiol redox balance of bacteria. Cadmium is not commonly found on grapes or in wine and originates mostly from environmental and technological contamination. However, the prevalence of this stressor in the environment of O. oeni is all the more questionable, as cadA was found close to a thioredoxin gene, trxB. Trx has been observed to play a protective role against Cd2+ in yeasts, while Trx from E. coli has recently been demonstrated to be a potent Cd2+ chelator upon acute Cd2+ exposure (36).
A prophage was recently shown to modify S. thermophilus fitness through lysogenic conversion (5). This was not the case for region 1, as the genes encoding the cell surface proteins present in this putative remnant prophage were not prevalent among HP strains. Similarly, we observed no potential-associated sequences in region 6, related to a putative PTS-fructose system in O. oeni. This region needs further study. Hence, fructose in combination with arginine was previously shown to trigger the expression of a subset of stress-responsive genes, such as arcR, omrR, and ftsH, while the cultivation of O. oeni in a fructose- and arginine-supplemented medium prior to wine exposure protected the bacteria from subsequent wine shock (4).
The acquisition of novel genes is proposed to be a strategy used by O. oeni to adapt to its environment. Our study raises additional questions about the mechanisms of transfer and the nature of the donors. An examination of region 4 provides some interesting clues. The locus is flanked by IS30 elements, suggesting a transfer through transposition. Members of this IS family have been widely transferred among LAB, where they are linked to genes involved in cold-shock adaptation, bacteriocin production, sugar utilization, and antibiotic resistance. It has, therefore, been suggested that IS30-related elements play a role in LAB genome plasticity and environmental adaptation (30, 32). While it is likely that this 7-kb locus is acquired, it is difficult to assess the relevancy of proposed donors, as the genes detected for potential horizontal transfer have generally undergone amelioration. Accordingly, the different structures observed in the PSU-1 and IOEB Sarco 1491 strains may be indicators of this type of rapid amelioration in the O. oeni species. As BBH were found for some sequences in Lactobacillus plantarum, frequently isolated from wine environments, this LAB is a candidate donor. Both species share physical as well as phylogenetic proximity, considered to influence lateral transfers. The locus is not flanked by IS elements in L. plantarum WFSC1. On the other hand, Molenaar and coworkers (28) did not identify this locus as a region of flexibility during exploration of L. plantarum genome diversity. However, the microarrays used only tested 81% of the total number of bases in the WCFS1 isolate. One alternative hypothesis is that O. oeni and L. plantarum are both recent recipients from an unknown donor. We observed that the BBH of 19 other SH sequences (singlets) were also associated with LAB present in the environment of O. oeni (lactobacilli and L. mesenteroides). The availability of complete genomic sequences will soon provide the opportunity to measure and compare the amount of laterally transferred sequences between O. oeni and the other bacteria found in the complex ecosystems present in must and wines.
This work provided some evidence for an association between the presence of particular sequences and the enological potential. However, it should be noted that O. oeni is present on the surface of grape berries. We cannot rule out that some of the subtracted sequences in this study may be relevant for the survival of O. oeni in a plant-associated environment. For example, divergence in the sequences harbored in region 3 may modulate the synthesis and/or structure of extracellular compounds, resulting in variations in surface interactions and, possibly, the ability to persist in the niche.
Supplementary Material
Acknowledgments
Our colleagues Cécile Miot-Sertier, Olivier Claisse, Patrick Lucas, Marguerite Dols, and Pascal Durrens are gratefully acknowledged for critical reading of the manuscript.
A.D. received a grant from the French Ministry of Education.
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arendt, E. K., A. Lonvaud, and W. P. Hammes. 1991. Lysogeny in Leuconostoc oenos. J. Gen. Microbiol. 137:2135-2139. [DOI] [PubMed] [Google Scholar]
- 2.Athané, A., E. Bilhère, E. Bon, P. Lucas, A. Lonvaud, and C. Le Marrec. 2008. Characterization of an acquired dps-containing gene island in the lactic acid bacterium Oenococcus oeni. J. Appl. Microbiol. 105:1866-1875. [DOI] [PubMed] [Google Scholar]
- 3.Bourdineaud, J. P., B. Nehme, S. Tesse, and A. Lonvaud-Funel. 2004. A bacterial gene homologous to ABC transporters protects Oenococcus oeni from ethanol and other stress factors in wine. Int. J. Food Microbiol. 92:1-14. [DOI] [PubMed] [Google Scholar]
- 4.Bourdineaud, J. P. 2006. Both arginine and fructose stimulate pH-independent resistance in the wine bacteria Oenococcus oeni. Int. J. Food Microbiol. 107:274-280. [DOI] [PubMed] [Google Scholar]
- 5.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversions. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coucheney, F., N. Desroches, M. Bou, R. Tourdot-Maréchal, L. Dulau, and J. Guzzo. 2005. A new approach for selection of Oenococcus oeni strains in order to produce malolactic starters. Int. J. Food Microbiol. 105:463-470. [DOI] [PubMed] [Google Scholar]
- 7.Delaherche, A., E. Bon, A. Dupe, M. Lucas, B. Arveiler, A. De Daruvar, and A. Lonvaud-Funel. 2006. Intraspecific diversity of Oenococcus oeni strains determined by sequence analysis of target genes. Appl. Microbiol. Biotechnol. 73:394-403. [DOI] [PubMed] [Google Scholar]
- 8.de Las Rivas, B., A. Marcobal, and R. Munoz. 2004. Allelic diversity and population structure in Oenococcus oeni as determined from sequence analysis of housekeeping genes. Appl. Environ. Microbiol. 70:7210-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desmond, C., R. P. Ross, G. Fitzgerald, and C. Stanton. 2005. Sequence analysis of the plasmid genome of the probiotic strain Lactobacillus paracasei NFBC338 which includes the plasmids pCD01 and pCD02. Plasmid 54:160-175. [DOI] [PubMed] [Google Scholar]
- 10.Desroche, N., C. Beltramo, and J. Guzzo. 2005. Determination of an internal control to apply reverse transcription quantitative PCR to study stress response in the lactic acid bacterium Oenococcus oeni. J. Microbiol. Methods 60:325-333. [DOI] [PubMed] [Google Scholar]
- 11.Dicks, L. M. T., H. J. J. Van Vuuren, and F. Dellaglio. 1990. Taxonomy of Leuconostoc species, particularly Leuconostoc oenos, as revealed by numerical analysis of total soluble cell protein patterns, DNA base composition and DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 40:83-91. [Google Scholar]
- 12.Divol, B., T. Tonon, S. Morichon, E. Gindreau, and A. Lonvaud-Funel. 2003. Molecular characterization of Oenococcus oeni genes encoding proteins involved in arginine transport. J. Appl. Microbiol. 94:738-746. [DOI] [PubMed] [Google Scholar]
- 13.Dols-Lafargue, M., H. Y. Lee, C. Le Marrec, A. Heyraud, G. Chambat, and A. Lonvaud-Funel. 2008. Studies on gtf, a glucosyltransferase gene in the genome of Pediococcus parvulus and Oenococcus oeni, two bacterial species commonly found in wine. Appl. Environ. Microbiol. 74:4079-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 15.Geis, A., H. A. El Demerdash, and K. J. Heller. 2003. Sequence analysis and characterization of plasmids from Streptococcus thermophilus. Plasmid 50:53-69. [DOI] [PubMed] [Google Scholar]
- 16.Gindreau, E., S. Torlois, and A. Lonvaud-Funel. 1997. Identification and sequence analysis of the region encoding the site-specific integration system from Leuconostoc oenos (Oenococcus oeni) temperate bacteriophage phi 10MC. FEMS Microbiol. Lett. 147:279-285. [DOI] [PubMed] [Google Scholar]
- 17.Gindreau, E., and A. Lonvaud-Funel. 1999. Molecular analysis of the region encoding the lytic system from Oenococcus oeni temperate phi 10MC. FEMS Microbiol. Lett. 171:231-238. [DOI] [PubMed] [Google Scholar]
- 18.Guerrini, S., A. Bastianini, G. Blaiotta, L. Granchi, G. Moschetti, S. Coppola, S. Romano, and M. Vincenzini. 2003. Phenotypic and genotypic characterization of Oenococcus oeni strains isolated from Italian wines. Int. J. Food Microbiol. 83:1-14. [DOI] [PubMed] [Google Scholar]
- 19.Huang, X. 1996. An improved sequence assembly program. Genomics 133:21-31. [DOI] [PubMed] [Google Scholar]
- 20.Jobin, M. P., D. Garmyn, C. Diviès, and J. Guzzo. 1999. Expression of the Oenococcus oeni trxA gene is induced by hydrogen peroxide and heat shock. Microbiology 145:1245-1251. [DOI] [PubMed] [Google Scholar]
- 21.Körner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:589-592. [DOI] [PubMed] [Google Scholar]
- 22.Lechiancole, T., G. Blaiotta, D. Messina, V. Fusco, F. Vilani, and G. Salzano. 2006. Evaluation of intra-specific diversities in Oenococcus oeni through analysis of genomic and expressed DNA. Syst. Appl. Microbiol. 29:375-381. [DOI] [PubMed] [Google Scholar]
- 23.Le Jeune, C., and A. Lonvaud-Funel. 1997. Sequence of DNA 16S/23S spacer region of Leuconostoc oenos (Oenococcus oeni): application to strain differentiation. Res. Microbiol. 148:79-86. [DOI] [PubMed] [Google Scholar]
- 24.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, et al. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcobal, A., B. de Las Rivas, M. W. Moreno-Arribas, and R. Munoz. 2006. Evidence for horizontal gene transfer as origin of putrescine production in Oenococcus oeni RM83. Appl. Environ. Microbiol. 72:7954-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcobal, A. M., D. A. Sela, Y. I. Wolf, K. S. Makarova, and D. A. Mills. 2008. Role of hypermutability in the evolution of the genus Oenococcus. J. Bacteriol. 190:564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills, D. A., H. Rawsthorne, C. Parker, D. Tamir, and K. Makarova. 2005. Genomic analysis of Oenococcus oeni PSU-1 and its relevance to winemaking. FEMS Microbiol. Rev. 29:465-475. [DOI] [PubMed] [Google Scholar]
- 28.Molenaar, D., F. Bringel, F. H. Schuren, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 187:6119-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nehmé, B., M. A. Ganga, and A. Lonvaud-Funel. 2006. The arginine deiminase locus of Oenococcus oeni includes a putative arginyl-tRNA synthetase ArgS2 at its 3′-end. Appl. Microbiol. Biotechnol. 70:590-597. [DOI] [PubMed] [Google Scholar]
- 30.Nicoloff, H., and F. Bringel. 2003. ISLp11 is a functional IS30-related insertion element in Lactobacillus plantarum that is also found in other lactic acid bacteria. Appl. Environ. Microbiol. 69:6032-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 32.Novichkov, P. S., M. V. Omelchenko, M. S. Gelfan, A. A. Mironov, Y. I. Wolf, and E. V. Koonin. 2004. Genome-wide molecular clock and horizontal gene transfer in bacterial evolution. J. Bacteriol. 186:6575-6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penaud, S., A. Fernandez, S. Boudebouzze, S. D. Erhlich, E. Maguin, and M. van de Guchte. 2006. Induction of heavy-metal-transporting CPX-type ATPases during acid adaptation in Lactobacillus bulgaricus. Appl. Environ. Microbiol. 72:7445-7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poblet-Icart, M., A. Bordons, and A. Lonvaud-Funel. 1998. Lysogeny of Oenococcus oeni and study of their induced bacteriophages. Curr. Microbiol. 36:365-369. [DOI] [PubMed] [Google Scholar]
- 35.Prunier, A. L., and R. Leclercq. 2005. Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. J. Bacteriol. 187:3455-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rollin-Genetet, F., C. Berthomieu, A. H. Davin, and E. Quéméneur. 2004. Escherichia coli thioredoxin inhibition by cadmium: two mutually exclusive binding sites involving Cys32 and Asp26. Eur. J. Biochem. 271:1299-1309. [DOI] [PubMed] [Google Scholar]
- 37.Santos, R., C. Sao-José, G. Vieira, H. Paveia, and M. A. Santos. 1998. Genome diversity in temperate bacteriophages of Oenococcus oeni. Arch. Virol. 143:523-536. [DOI] [PubMed] [Google Scholar]
- 38.Sao-José, C., S. Santos, J. Nascimento, A. G. Brito-Madurro, and M. A. Santos. 2004. Diversity in the lysis-integration region of oenophage genomes and evidence for multiple tRNA loci, as targets for prophage integration in Oenococcus oeni. Virology 325:82-95. [DOI] [PubMed] [Google Scholar]
- 39.Sato, H., F. Yanagida, T. Shinohara, M. Suzuki, K. Suzuki, and K. Yokotsuka. 2001. Intraspecific diversity of Oenococcus oeni isolated during red wine-making in Japan. FEMS Microbiol. Lett. 202:109-114. [DOI] [PubMed] [Google Scholar]
- 40.Scott, A. E., A. R. Timms, P. L. Connerton, C. Loc Carrillo, K. A. Radzum, and I. F. Connerton. 2007. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 3:1142-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siguier, P., J. Perochon, L. Lestrade, J. Mahillon, and M. Chandler. 2006. Isfinder: the reference center for bacterial insertion sequences. Nucleic Acids Res. 34:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spano, G., and S. Massa. 2006. Environmental stress response in wine lactic acid bacteria: beyond Bacillus subtilis. Crit. Rev. Microbiol. 32:77-86. [DOI] [PubMed] [Google Scholar]
- 43.Straus, D. F., and M. Ausubel. 1990. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc. Natl. Acad. Sci. USA 87:1889-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutherland, M., H. J. van Vuuren, and M. M. Howe. 1994. Cloning, sequence and in vitro transcription/translation analysis of a 3.2-kb EcoRI-HindIII fragment of Leuconostoc oenos bacteriophage L10. Gene 148:125-129. [DOI] [PubMed] [Google Scholar]
- 45.Tenreiro, R., M. A. Santos, H. Paveia, and G. Vieira. 1994. Inter-strain relationships among wine leuconostocs and their divergence from other Leuconostoc species, as revealed by low frequency restriction fragment analysis of genomic DNA. J. Appl. Bacteriol. 77:271-280. [DOI] [PubMed] [Google Scholar]
- 46.Theil, E. C. 2007. Coordinating responses to iron and oxygen stress with DNA and mRNA promoters: the ferritin story. Biometals 20:513-521. [DOI] [PubMed] [Google Scholar]
- 47.Tonon, T., J. P. Bourdineaud, and A. Lonvaud-Funel. 2001. The arcABC gene cluster encoding the arginine deiminase pathway of Oenococcus oeni, and arginine induction of a CRP-like gene. Res. Microbiol. 152:653-661. [DOI] [PubMed] [Google Scholar]
- 48.Wydau, S., R. Dervyn, J. Anba, S. Dusko Ehrlich, and E. Maguin. 2006. Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiol. Lett. 257:32-42. [DOI] [PubMed] [Google Scholar]
- 49.Zapparoli, G., C. Reguant, A. Bordons, S. Torriani, and F. Dellaglio. 2000. Genomic DNA fingerprinting of Oenococcus oeni strains by pulsed-field gel electrophoresis and randomly amplified polymorphic DNA-PCR. Curr. Microbiol. 40:351-355. [DOI] [PubMed] [Google Scholar]
- 50.Ze-Ze, L., R. Tenreiro, and H. Paveia. 2000. The Oenococcus oeni genome: physical and genetic mapping of strain GM and comparison with the genome of a “divergent” strain, PSU-1. Microbiology 146:3195-3204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.