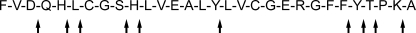Abstract
Nearly all high-molecular-weight (HMW) dissolved organic nitrogen and part of the particulate organic nitrogen in the deep sea are present in hydrolysis-resistant amides, and so far the mechanisms of biodegradation of these types of nitrogen have not been resolved. The M12 family is the second largest family in subclan MA(M) of Zn-containing metalloproteases and includes most enzymes from animals and only one enzyme (flavastacin) from a human-pathogenic bacterium (Flavobacterium meningosepticum). Here, we characterized the novel M12 protease myroilysin with elastinolytic activity and collagen-swelling ability from the newly described deep-sea bacterium Myroides profundi D25. Myroilysin is a monomer enzyme with 205 amino acid residues and a molecular mass of 22,936 Da. It has the same conserved residues at the four zinc ligands as astacin and very low levels of identity (≤40%) to other metalloproteases, indicating that it is a novel metalloprotease belonging to subfamily M12A. Myroilysin had broad specificity and much higher elastinolytic activity than the bacterial elastinase pseudolysin. To our knowledge, it is the first reported elastase in the M12 family. Although it displayed very low activity with collagen, myroilysin had strong collagen-swelling ability and played a synergistic role with collagenase in collagen hydrolysis. It can be speculated that myroilysin synergistically interacts with other enzymes in its in situ biotic assemblage and that it may play an important role in the degradation of deep-sea HMW organic nitrogen.
Although the chemical dynamics of marine dissolved organic nitrogen (DON) have yet to be resolved, it has been demonstrated that nearly all deep-sea high-molecular-weight (HMW) DON is present in amides that resist both chemical hydrolysis and biological degradation (1). It was estimated that the total input of particular organic nitrogen (PON) to the deep-sea sediment is 36 ± 21 μmol m−2 day−1 on average (5). Although it is not clear what kinds of proteins deep-sea HMW organic nitrogen contains, hydrolysis-resistant amides have been observed in sedimentary PON (13). Insoluble collagen and elastin, which are abundant in marine animals, are recognized to be hydrolysis resistant and biologically refractory, suggesting that they are probably components of deep-sea PON and HMW DON. How deep-sea PON and HMW DON are degraded is still poorly understood because little is known about the bacterial species and the kinds of proteases participating in the hydrolysis process (11). Since there are lots of hydrolysis-resistant proteins in deep-sea PON and HMW DON, it is reasonable to speculate that some bacterial proteases participating in the degradation of these PON and HMW DON have special functions or unknown mechanisms of action.
Metalloproteases are grouped into 15 clans in the latest release (release 8.2) of the MEROPS database (the peptidase database at http://merops.sanger.ac.uk/) (25). Clan MA, the largest clan of metalloproteases, contains zinc-dependent metalloproteases in which the first and second zinc ligands are the two histidines in the motif HEXXH (24). Based on the nature of the third zinc ligand, clan MA is divided into two subclans, MA(E) (gluzincins) with a glutamate residue as the third zinc ligand and MA(M) (metzincins) with a histidine or aspartate residue as the third zinc ligand in the extended motif HEXXHXXGXXH/D. In the MA(M) subclan, family M12 is the second largest family and is divided into two subfamilies, subfamilies M12A and M12B (24). The members of M12A are astacin-like metalloproteases with the conserved M-turn sequence SIMHY at the fourth zinc ligand (16, 24). Interestingly, most members of family M12 are from animals, function in the extracellular space, and occur as either membrane-bound or secreted enzymes (17, 24). Flavastacin is the only bacterial protease in the M12 family that has been purified and characterized to date, and this enzyme is a glycosylated protease secreted by the pathogenic organism Flavobacterium meningosepticum that was reported in 1995 (23, 26, 29, 30). Because no bacterial protease belonging to M12 other than flavastacin has been reported since 1995, it has been debated whether the occurrence of flavastacin demonstrates that the zinc-binding motif of the astacin subfamily appeared long ago before the divergence of prokaryotes and eukaryotes or is the result of a horizontal transfer (30).
Myroides profundi D25 is a newly described protease-producing barotolerant bacterium which was isolated from the deep-sea sediment at a water depth of 1,245 m at site MD05-2907 (24°47.19′ N, 122°29.30′ E) near southern Okinawa (33). In this paper, the protease produced by M. profundi D25, designated myroilysin, is described. Gene cloning, sequence analysis, purification, characterization of the specificity, the caseinolytic and elastinolytic activities, and the collagen-swelling ability of myroilysin are described. The results showed that this enzyme is a novel astacin-like metalloprotease belonging to subfamily M12A that has elastinolytic activity and plays a synergistic role with collagenase in collagen hydrolysis since it swells collagen.
MATERIALS AND METHODS
Bacterial strains and materials.
M. profundi D25 isolated from deep-sea sediment and preserved in 15% glycerol at −80°C was used for protease production. Insoluble type I collagen fiber (bovine Achilles tendon), fibrinogen (bovine plasma), and the collagenase from Clostridium histolyticum were purchased from Worthington Biochemical Co., and gelatin was purchased from Boston Biomedical Inc. Fibrous elastin and elastinorcein were purchased from Sigma. Various synthetic peptides were purchased from Bachem Co. DEAE-Sepharose Fast Flow gel, the materials used for polyacrylamide gel electrophoresis (PAGE), and the protein marker used in sodium dodecyl sulfate (SDS)-PAGE were all purchased from Amersham (Sweden).
Protease purification.
Strain D25 was grown in a fermentation medium composed of 2.0% corn powder, 2.0% bean powder, 1.0% wheat bran, 0.4% Na2PO4, 0.03% KH2PO4, 0.1% CaCl2, and artificial seawater (pH 8.0) with shaking for 72 h at 15°C as previously described (6). The culture was centrifuged at 10,000 × g for 15 min at 4°C. The proteins in the supernatant were precipitated by slowly adding ammonium sulfate powder to a concentration of 55%. The precipitate was collected by centrifugation (8,500 × g, 10 min) and dissolved in 50 mM Tris-HCl buffer (pH 8.9). The sample was put on a DEAE-Sepharose Fast Flow chromatography column (1.6 cm by 20 cm) preequilibrated with the same buffer. Then the column was eluted with a linear gradient of 0 to 0.8 M NaCl. The fraction with protease activity was collected. The enzyme purity was analyzed by 12.5% SDS-PAGE. The molecular mass was analyzed using matrix-assisted laser desorption ionization—time of flight mass spectrometry (Applied Biosystems Voyager DE Pro; ABI, United States).
For overexpression and purification of pseudolysin, the gene encoding pseudolysin was cloned by PCR from the genome DNA of Pseudomonas aeruginosa PAO1, which was generously provided by Li-yan Yu (National Laboratory for Screening New Microbial Drugs, Institute of Medicinal Biotechnology). Then it was overexpressed in E. coli BL21(DE3) with pET-22b as the expression vector and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) as an inducer. After cultivation at 37°C overnight, cells were collected and disrupted by sonication in an ice bath. Then the cell extract was incubated at 30°C for 2 h to promote the maturation of pseudolysin. After incubation, the enzyme was purified by precipitation with 60% solid ammonium sulfate, followed by elution of a DEAE Sepharose Fast Flow column with 0 to ∼0.5 M NaCl in 50 mM Tris-HCl buffer (pH 9.5). Fractions with high protease activities were collected, dialyzed against 50 mM Tris-HCl buffer (pH 7.9), and analyzed by 12.5% SDS-PAGE.
N-terminal amino acid sequence analysis of myroilysin.
Purified myroilysin run in an SDS-PAGE gel was transferred to a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad). Its N-terminal amino acid sequence was analyzed by Edman degradation with PROCISE491 (Applied Biosystems), and the result was the sequence GAVVRSTKWPNGSVITVGLYGGTPYVRSKVKQYAQEWSNY. A BLAST analysis in NCBI showed that this sequence had the highest levels of identity to the sequences of two Zn-containing metalloproteases (accession numbers ABK97392.1 and AAZ45577.1).
Cloning of the gene encoding myroilysin.
Genomic DNA of M. profundi D25 was extracted by the NaCl-cetyltrimethylammonium bromide method (21). Based on the N-terminal amino acid sequence of myroilysin and the conserved sequence of the catalytic center of Zn-containing metalloproteases, two primers were designed and synthesized. With the genomic DNA as template, PCR amplification was performed by using Taq DNA polymerase for 30 cycles consisting of 94°C for 30 s, 50°C for 1 min, and 72°C for 2 min. A 350-bp product was amplified and sequenced. Then six specific primers were designed based on the 5′ terminal sequence and 3′ terminal sequence of this product, and two general primers were designed, one containing a protein initiation codon and one containing a stop codon. Chromosome walking was used to amplify the neighboring sequence of the 350-bp product by thermal asymmetric interlaced PCR (15). A 1,050-bp upstream sequence and a 547-bp downstream sequence of the 350-bp product were amplified from the genomic DNA and sequenced. Through assembly, a 1,947-bp sequence containing an 822-bp open reading frame that encodes myroilysin was obtained. After it was verified by PCR, the sequence of this gene was submitted to GenBank.
Protein determination and enzyme assay.
Proteins were assessed by Bradford's method with bovine serum albumin as the standard (4). The caseinolytic activity was determined as previously described (6). One unit of enzyme activity was defined as the amount of enzyme that catalyzed the formation of 1 μg tyrosine per min. The elastinolytic activity was assayed by the method of Sachar et al. (28), with a slight modification. The enzyme was incubated with 5 mg elastinorcein in 0.25 ml 50 mM Tris-HCl with continuous stirring for 1 h, and then the residual elastinorcein was removed by centrifugation. The absorption at 590 nm of the supernatant was measured. One unit of enzyme activity was defined as the amount of enzyme that caused an increase of 0.01 U of absorbance at 590 nm per min. The collagenolytic activities with type I collagen and gelatin (1%) were determined by the method provided by Worthington Biochemical Co. (10). The reaction times were 5 h for type I collagen fiber and 0.5 h for gelatin. For type I collagen fiber, 1 U is defined as the release of 1 μmol of l-leucine equivalents from collagen in 5 h. For gelatin, 1 U is defined as the release of 1 μmol of l-leucine equivalents from gelatin in 1 min. The amidolytic activities with N-succinyl-Phe-Ala-Ala-Phe-p-nitroanilide (Su-FAAF-pNa), N-succinyl-Ala-Ala-Ala-p-nitroanilide (Su-AAA-pNa), and N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (Su-AAPF-pNa) were determined by the method of Peek et al. (22). One unit of enzyme activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of p-nitroaniline per min. Fibrin and fibrinogen degradation was determined electrophoretically by the method of Datta et al. (7). The proteolytic activity with furylacryloyl-Gly-Leu-NH2 and furylacryloyl-Gly-Phe-NH2 was measured by using Feder's method (9).
Analysis of the myroilysin cleavage sites on the oxidized insulin B chain.
Myroilysin was incubated with oxidized insulin B chain (1:40, by mass) in 50 mM Tris-HCl (pH 9.0) at 37°C for 15, 30, and 50 min, and then the reaction was stopped by adding 1% trifluoroacetic acid. The hydrolytic products were separated on a C18 column (Venusil MP C18), using a high-performance liquid chromatography (HPLC) system (LC-10ADVP), by the method described by Authier et al. (2). The molecular masses of the hydrolytic products were analyzed by mass spectrometry using Thermo LTQ mass spectrometry. The sequence of every hydrolytic product was analyzed by using proteomics and sequence analysis tools at the expasy website (http://www.expasy.org) based on its molecular mass.
Observation of hydrolysis of fibrous elastin by myroilysin.
One milliliter of 50 mM Tris-HCl (pH 9.0) containing 100 μg/ml myroilysin was incubated with 10 mg fibrous elastin at 37°C with continuous stirring. Fibrous elastin in 50 mM Tris-HCl (pH 9.0) without myroilysin was used as a control. After treatment for 1, 2, and 3 h, the samples were observed with an inverted microscope (Olympus IX71) at room temperature.
Analysis of the collagen-swelling ability of myroilysin.
Ten milligrams of type I collagen fiber was added to 2 ml 50 mM Tris-HCl (pH 9.0) containing different concentrations (0, 30, 50, 80, and 100 μg/ml) of myroilysin or 6 M urea in tubes. The samples were incubated at 37°C for 1 h with continuous stirring and then were photographed. To analyze the collagen-swelling ability of myroilysin in seawater at 4°C, a tube containing 10 mg collagen in artificial seawater and 100 μg/ml myroilysin was incubated for 5 h at 4°C with continuous stirring. Samples treated with 0 and 100 μg/ml myroilysin were observed by scanning electron microscopy (SEM) (Hitachi S-570) by using the method of Usha and Ramasami (32).
Analysis of the synergistic role of myroilysin with collagenase in collagen hydrolysis.
Five milligrams of type I collagen fiber in 50 mM Tris-HCl (pH 9.0) in the presence and absence of 100 μg/ml myroilysin was incubated at 37°C for 1 h with continuous stirring. Then 20 μg/ml collagenase from C. histolyticum was added, and the samples were incubated at 37°C for 5 h with continuous stirring. After incubation, the samples were centrifuged, and the collagenolytic activity in the supernatant was measured. The sample without either myroilysin or collagenase and the sample with myroilysin but no collagenase were used as controls.
Nucleotide sequence accession number.
The sequence of the gene that encodes myroilysin has been deposited in the GenBank database under accession no. EU883966.
RESULTS
Analysis of the role of the protease secreted by M. profundi D25.
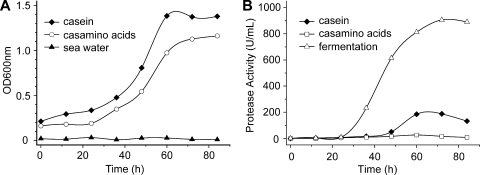
To analyze the role of the protease secreted by strain D25, strain D25 was inoculated into three media, artificial seawater, artificial seawater with 0.4% Casamino Acids (Casamino Acids medium), and artificial seawater with 0.4% casein (casein medium), and cultured at 15°C. The strain did not grow visibly in artificial seawater, grew well in Casamino Acids medium with little protease secretion, and grew well in casein medium with a high level of protease secretion (Fig. 1A and 1B). Therefore, a protein-enriched and amino acid-free medium could induce the production and secretion of the extracellular protease of strain D25, which indicated that the biophysiological role of the protease secreted by strain D25 is to degrade surrounding proteins to obtain nutrients.
FIG. 1.
Growth of (A) and protease production by (B) M. profundi D25 in different media at 15°C. Seawater medium was artificial seawater (pH 8.0); casein medium was artificial seawater containing 0.4% casein (pH 8.0); Casamino Acids medium was artificial seawater containing 0.4% Casamino Acids; and fermentation medium was prepared as described in Materials and Methods. OD600nm, optical density at 600 nm.
Purification and physicochemical characterization of the protease secreted by strain D25.
The production of the protease secreted by strain D25 in casein medium and the production in fermentation medium were compared (Fig. 1B). The protease activity in fermentation medium was about four times higher than that in casein medium. Consequently, the fermentation medium was used for production and purification of the protease. The protease secreted by strain D25 was purified using ammonium sulfate precipitation and ion-exchange chromatography as described in Materials and Methods, which resulted in 25-fold purification and a final yield of 24.6% (see Fig. S1 and Table S1 in the supplemental material). SDS-PAGE suggested that the enzyme was >98% pure and had a molecular mass of more than 25,000 Da (see Fig. S2 in the supplemental material), while mass spectrometry indicated that its molecular mass was 22,936 Da (see Fig. S3 in the supplemental material). This purified protease was designated myroilysin in this study. It was deduced from Fig. S2 in the supplemental material that myroilysin was the main protease secreted by strain D25. Analysis of the form of myroilysin by gel filtration chromatography showed that this enzyme is a monomer enzyme (Table 1; see Fig. S4 in the supplemental material). The matrix-assisted laser desorption ionization—time of flight mass spectrometry spectrum also showed that it is a monomer enzyme because no peak with a molecular mass higher than 22,936 Da appeared in the spectrum (see Fig. S3 in the supplemental material). The apparent melting temperature of myroilysin determined by circular dichroism spectroscopy was 42.6°C (Table 1; see Fig. S5 in the supplemental material). Its N-terminal 40 amino acid sequence analyzed by Edman degradation is GAVVRSTKWPNGSVITVGLYGGTPYVRSKVKQYAQEWSNY. BLAST analysis of this N-terminal sequence with the NCBI database showed that it had 36% identity to the Zn-containing metalloprotease from Dechloromonas aromatica RCB (accession number AAZ45577.1) and 34% identity to the Zn-containing metalloprotease from an uncultured bacterium (accession number ABK97392.1), suggesting that myroilysin is a Zn-containing metalloprotease.
TABLE 1.
Physicochemical characteristics of myroilysin
| Characteristic | Result(s) |
|---|---|
| Length of DNA sequence (bp) | 822 |
| No. of amino acid residues in mature protein | |
| sequence | 205 |
| No. of amino acid residues in signal sequence | |
| and prosequence | 68 |
| Molecular mass (sequence) (Da) | 22,963.2 |
| Molecular mass (mass spectrometry) (Da) | 22,936.6 |
| Isoelectric point (sequence) | 6.3 |
| Form in Tris-HCl buffer | Monomer |
| Apparent melting temp (°C) (scan rate, 1°C/min)a | 42.6 |
| Half-time at 40°C (min)b | 28 |
| Half-time at 45°C (min)b | 5 |
| Optimum pH with casein (50°C)c | 9.5 |
| Optimum pH with elastinorcein (40°C)c | 9.0 |
| Optimum temp with casein (°C)d | 50 |
| Optimum temp with elastinorcein (°C)d | 40-∼45 |
The apparent melting temperature was calculated based on the thermal unfolding curve for myroilysin detected with a circular dichroism spectropolarimeter (Jasco J810) at 222 nm with an enzyme concentration of 0.7 mg/ml (see Fig. S5 in the supplemental material).
Myroilysin (0.1 mg/ml) was incubated at 35°C, 40°C, and 45°C. Samples were taken at intervals for the activity assay. The half-time was the time that it took to eliminate 50% of the activity of myroilysin with casein at a given temperature.
The optimum pHs were determined by measuring the activities of myroilysin with casein (50°C) and elastinorcein (40°C) in 50 mM Tris buffer at pHs ranging from 6 to ∼11.
The optimum temperatures were determined by measuring the activities of myroilysin with casein and elastinorcein in 50 mM Tris buffer (pH 9.0) at temperatures ranging from 0 to ∼60°C.
Gene cloning and sequence analysis of myroilysin.
Based on the N-terminal sequence of myroilysin and the conserved sequence of the catalytic center of Zn-containing metalloproteases, 350 bp of the gene sequence of myroilysin, encoding the N terminus to the catalytic center, was cloned. Then the whole gene, together with the upstream 846 bp, was cloned by thermal asymmetric interlaced PCR (see Fig. S6 in the supplemental material). The open reading frame of this gene has 822 bp, from an ATG start codon to a TAA stop codon. It encodes a protein containing 273 amino acid residues with a calculated molecular mass of 30,243.6 Da, which is the precursor of myroilysin. This precursor contains a signal peptide sequence consisting of 23 amino acid residues (M-68 to N-46) or 25 amino acid residues (M-68 to N-44), as predicted by signalP (3). Based on the determined N-terminal sequence of mature myroilysin, a propeptide consisting of 45 (D-45 to R-1) or 43 (D-43 to R-1) amino acid residues between the signal peptide and the mature enzyme could be determined. Both the signal peptide and the propeptide were cleaved off during enzyme maturation. Mature myroilysin contains 205 amino acid residues (G1 to N205) and has a calculated molecular mass of 22,963 Da, in agreement with the molecular mass determined by mass spectrometry, which shows that it is a protein without glycosylation. The BLAST search of the NCBI Conserved Domain Database (CDD v2.14; http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) revealed that myroilysin is a Zn-dependent metalloprotease belonging to MA(M) subclan. The conserved amino acid residues for the four zinc ligands of myroilysin are HEXXHXXGXXHE…SIMHY, which is consistent with those of astacin and astacin-like proteases, indicating that myroilysin should be a member of the astacin subfamily (subfamily M12A). The signal peptide and propeptide of the precursor have little identity to those of other metalloproteases. As observed for mature myroilysin, other than the residues at or near the four zinc ligands, the amino acid residues have little identity to those of other Zn-dependent metalloproteases. Myroilysin shows the highest identity (40%) to the metalloprotease from an uncultured bacterium (accession number ABK97392.1) (14), 33% identity to the deduced metalloprotease from D. aromatica RCB (accession number AAZ45577.1), 18% identity to astacin, and 15% identity to flavastacin. These results show that myroilysin is a novel M12A metalloprotease.
Analysis of the substrate specificity of myroilysin.
The substrate specificity of myroilysin with various proteins and synthetic peptides was assayed and compared with that of pseudolysin, a well-characterized bacterial elastase in family M4 (12, 18-20), because no elastase in family M12 has been reported. Myroilysin could hydrolyze various proteins, including casein, fibrous elastin, fibrinogen, fibrin, and gelatin (Table 2). It had 6.78 U/mg elastinolytic activity in artificial seawater at 4°C, suggesting that it probably can degrade elastin in the deep sea. When it hydrolyzed fibrinogen and fibrin, it produced peptides different from those produced by pseudolysin (see Fig. S7 in the supplemental material), showing that its cleavage sites on fibrinogen and fibrin were different from those of pseudolysin. Myroilysin had little activity with type I collagen fibers. For synthetic peptides, it had slight amidolytic activity with Su-FAAF-pNa, Su-AAA-pNa, and Su-AAPF-pNa and no proteolytic activity with furylacryloyl-Gly-Leu-NH2 and furylacryloyl-Gly-Phe-NH2. In contrast, pseudolysin had no amidolytic activity with Su-FAAF-pNa, Su-AAA-pNa, and Su-AAPF-pNa and high proteolytic activity with furylacryloyl-Gly-Leu-NH2 and furylacryloyl-Gly-Phe-NH2 (Table 2).
TABLE 2.
Substrate specificity of myroilysin with various proteins and synthetic peptides
| Substrate | Function or activity (U/mg)a
|
|
|---|---|---|
| Myroilysin | Pseudolysin | |
| Fibrinb | Hydrolyzes α, β, and γ | Hydrolyzes α, β, and γ |
| Fibrinogenb | Hydrolyzes Aα and Bβ | Hydrolyzes Aα and Bβ |
| Casein | 8,541 | 7,061 |
| Elastinorcein | 243.3 | 91.1 |
| Insoluble type I collagen fiber | 3.82 | 0 |
| Gelatin | 4.46 | 4.97 |
| Su-AAA-pNa | 2.78 | 0 |
| Su-FAAF-pNa | 7.98 | 0 |
| Su-AAPF-pNa | 3.55 | 0 |
| Furylacryloyl-Gly-Leu-NH2 | 0 | 642c |
| Furylacryloyl-Gly-Phe-NH2 | 0 | 3,364c |
Unless otherwise indicated, the values are specific activities (in U/mg). The activities of myroilysin and pseudolysin with casein, gelatin, and synthetic substrates were measured at 50°C and the activities with elastinorcein and insoluble type I collagen were measured at 37°C as described in Materials and Methods. The data are the means of three experiments, and the standard deviations were ≤5%.
Hydrolysis of fibrin and fibrinogen by myroilysin and pseudolysin was performed at 50°C, and the results were analyzed by SDS-PAGE, as shown in Fig. S7 in the supplemental material.
The proteolytic activities of myroilysin and pseudolysin with furylacryloyl-Gly-Leu-NH2 and furylacryloyl-Gly-Phe-NH2 were measured with Feder's method (9, 22); the data are the values of kcat/Km (in M−1 s−1).
The pattern of cleavage of the oxidized insulin B chain by myroilysin was analyzed by HPLC and mass spectrometry (Fig. 2; see Fig. S8 to S10 and Table S2 in the supplemental material). Based on the HPLC spectra of the cleavage patterns of the oxidized insulin B chain hydrolyzed by myroilysin in 15, 30, and 50 min (see Fig. S8 in the supplemental material), the T27-P28 peptide bond was the most susceptible cleavage site, and the highest hydrolytic rate was observed for this bond. The hydrolytic rates for peptide bonds S9-H10, H10-L11, F25-Y26, and Y26-T27 were also comparatively high, and the hydrolytic rates for peptide bonds N3-Q4, H5-L6, L6-C7, Y16-L17, and K29-A30 were comparatively low.
FIG. 2.
Cleavage of oxidized insulin B chain by myroilysin. The cleavage pattern of myroilysin with the oxidized insulin B chain was determined as described in Materials and Methods.
Characterization of the caseinolytic and elastinolytic activities of myroilysin.
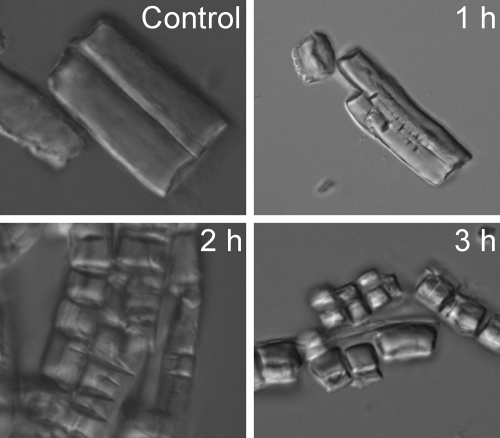
Figure 3 clearly shows that myroilysin could cut fibrous elastin into fragments, suggesting that it is an endopeptidase with elastin. Myroilysin had the highest activity at 50°C and pH 9.5 with casein and the highest activity at 40 to ∼45°C and pH 9 with elastinorcein. Its thermostability was rather low. A 100-μg/ml myroilysin solution in 50 mM Tris-HCl (pH 9.0) lost 50% of its activity at 45°C in 5 min and at 40°C in 28 min (Table 1; see Fig. S11 in the supplemental material). Therefore, the longer reaction time for elastinorcein hydrolysis (1 h) than for casein hydrolysis (10 min) might result in the lower optimum temperature with elastinorcein than with casein. At 50°C with casein as the substrate, the Km and Vmax of myroilysin were 0.38% and 57.8 μM/min, respectively (see Fig. S12 in the supplemental material). The metal ions Fe2+, Co2+, Ni2+, and Zn2+ were found inhibit the caseinolytic and elastinolytic activities of myroilysin, whereas Li+, K+, Mg2+, Ca2+, Sr2+, Ba2+, and Na+ had a little or no effect. The metal-chelating agent 1,10-phenanthroline could completely inhibit the caseinolytic and elastinolytic activities, while EGTA and EDTA had moderate inhibitory effects and leuhistin had little effect (Table 3).
FIG. 3.
Hydrolysis of fibrous elastin by myroilysin observed using an inverted microscope. Ten milligrams of fibrous elastin in 1 ml 50 mM Tris-HCl (pH 9.0) containing 100 μg/ml myroilysin was incubated at 37°C with continuous stirring. Fibrous elastin in 50 mM Tris-HCl (pH 9.0) without myroilysin was used as a control. After treatment for 1, 2, and 3 h, the samples were photographed with an inverted microscope (Olympus IX71) at room temperature. Magnification, ×960.
TABLE 3.
Effects of metal ions and protease inhibitors on the caseinolytic and elastinolytic activities of myroilysin
| Metal ion or inhibitor | Relative activity (%)a
|
Residual activity (%)b
|
||
|---|---|---|---|---|
| Casein | Elastinorcein | Casein | Elastin | |
| Control | 100 | 100 | ||
| Metal ions | ||||
| Mg2+ | 105.1 | 90.2 | ||
| K+ | 100 | 100 | ||
| Na+ | 100.2 | 99.4 | ||
| Li+ | 100 | 94.6 | ||
| Ba2+ | 90.2 | 87.9 | ||
| Ca2+ | 89.4 | 87.1 | ||
| Cu2+ | 88.5 | 100.1 | ||
| Sr2+ | 87.9 | 83.1 | ||
| Co2+ | 68.5 | 20.1 | ||
| Ni2+ | 33.4 | 8.3 | ||
| Fe2+ | 21.1 | 15.4 | ||
| Zn2+ | 22.9 | 1.4 | ||
| Inhibitors | ||||
| Control | 100 | 100 | ||
| 1,10-Phenanthroline | 0 | 0.1 | ||
| EGTA | 51.2 | 42.6 | ||
| EDTA | 67.6 | 64.4 | ||
| Leuhistin | 98.1 | 95.1 | ||
Enzyme activity was measured at 50°C for casein and at 37°C for elastinorcein. The activity of myroilysin without any metal ion was used as a control (100%). Activity was expressed as the activity of myroilysin with a metal ion relative to the activity with the control. The data are the means of three experiments, and the standard deviations were ≤5%.
Myroilysin was incubated with each inhibitor at 25°C for 20 min, and then the enzyme activity was measured at 50°C for casein and at 37°C for elastinorcein. The activity of myroilysin without any inhibitor was used as the control (100%). Activity was expressed as the activity of myroilysin treated with an inhibitor relative to the control activity. The data are the means of three experiments, and the standard deviations were ≤5%.
Analysis of the collagen-swelling ability of myroilysin.
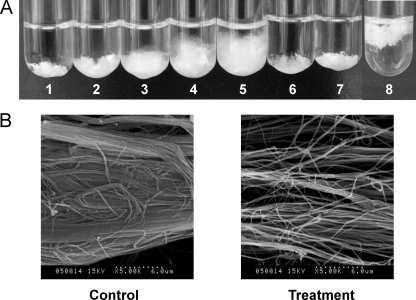
Myroilysin had strong collagen-swelling ability, which was concentration dependent. At 37°C, 100 μg/ml myroilysin could make 10 mg collagen fiber swell noticeably in 1 h. Even in artificial seawater at 4°C, 100 μg/ml myroilysin could make 10 mg collagen fiber swell noticeably in 5 h, suggesting that myroilysin probably has collagen-swelling ability in the deep sea (Fig. 4). Moreover, the collagen-swelling ability of myroilysin was much greater than that of 6 M urea, which is a collagen-swelling agent (32). A 4 mM 1,10-phenanthroline solution noticeably inhibited the collagen-swelling ability of myroilysin, suggesting that this ability may be associated with catalysis. SEM showed that the collagen fibers were very loose after treatment with myroilysin (Fig. 4), suggesting that some interfiber linkages were destroyed.
FIG. 4.
(A) Collagen swelling with myroilysin and urea. Ten milligrams of collagen in 2 ml 50 mM Tris-HCl buffer (pH 9.0) was incubated for 1 h at 37°C with continuous stirring. Tubes 1 to 5 contained 0, 25, 50, 75, and 100 μg/ml myroilysin, respectively. Tube 6 contained 6 M urea. Tube 7 contained 100 μg/ml myroilysin and 4 mM 1,10-phenanthroline. Tube 8 containing 10 mg collagen in artificial seawater and 100 μg/ml myroilysin was incubated for 5 h at 4°C with continuous stirring. (B) Collagen swollen by myroilysin as observed by SEM (Hitachi S-570). The control was 10 mg of collagen in 2 ml 50 mM Tris-HCl buffer that was incubated at 37°C for 1 h. For treatment, 10 mg of collagen in 2 ml 50 mM Tris-HCl buffer containing 100 μg/ml myroilysin was incubated at 37°C for 1 h. The samples were observed by SEM (Hitachi S-570) by using the method of Usha and Ramasami (22).
Analysis of the synergistic role of myroilysin with collagenase in collagen hydrolysis.
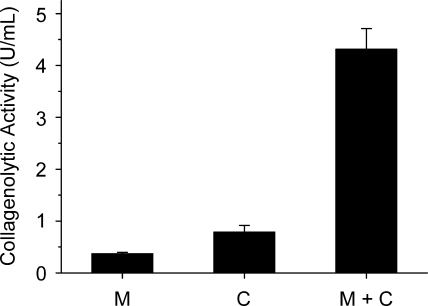
The effect of the collagen-swelling ability of myroilysin on the collagen-catalyzing efficiency of collagenase was investigated. Type I collagen fiber was treated with myroilysin for 1 h at 37°C. Then the collagenase from C. histolyticum was added to treated and untreated collagen fiber. The catalytic efficiency with the myroilysin-treated collagen was four times higher than that with the untreated collagen (Fig. 5), showing that myroilysin played a synergistic role with collagenase in collagen hydrolysis.
FIG. 5.
Synergistic action of collagenase and myroilysin for collagen hydrolysis. Five milligrams of type I collagen fiber in 1 ml 50 mM Tris-HCl buffer was incubated at 37°C with continuous stirring. Bar M, sample containing 100 μg/ml myroilysin that was incubated for 6 h; bar C, sample that was incubated for 1 h, after which 20 μg/ml collagenase was added and the preparation was incubated for 5 h; bar M+C, sample containing 100 μg/ml myroilysin that was incubated for 1 h, after which 20 μg/ml collagenase was added and the preparation was incubated for 5 h.
DISCUSSION
Although family M12 is the second largest family in subclan MA(M), only one bacterial protease in this family, flavastacin, has been reported previously (23, 26, 29, 30). In this paper, another M12 protease from the deep-sea bacterium M. profundi D25, myroilysin, is described. Myroilysin has the same conserved sequence (HEXXHXXGXXHE…SIMHY) at the four zinc ligands as astacin, suggesting that myroilysin should be a member of subfamily M12A. Moreover, purified myroilysin only has one catalytic domain with a molecular mass of 22,936 Da, just like astacin. However, it only has 18% identity to astacin, and the highest level of identity (40%) is with the metalloprotease from an uncultured bacterium (accession number ABK97392.1), which consists of 359 amino acids with a molecular mass of 39 kDa (14). Myroilysin also has a very low level of identity (15%) to flavastacin. Flavastacin is an O-glycosylated protein with 352 amino acid residues (23, 26, 29), whereas myroilysin has only 205 amino acid residues and is not glycosylated. Therefore, myroilysin is a novel bacterial protease in the astacin subfamily (M12A). Besides the characterized enzymes flavastacin and myroilysin, 17 proteases deduced from bacteria and eight proteases from fungi in M12 could be searched in the MEROPS database (25). The presence of these microbial proteases suggests that the zinc-binding motif of the astacin family probably appeared long ago, before the divergence of prokaryotes and eukaryotes.
Myroilysin had broad specificity with various proteins, such as casein, fibrous elastin, fibrinogen, fibrin, and gelatin. It had much higher specific activity with elastin than pseudolysin, a well-characterized bacterial elastase belonging to family M4, had (12, 18-20). Microscope observation also showed it could cleave fibrous elastin. These findings show that myroilysin is an elatase. To our knowledge, this enzyme is the first reported elastase in family M12. The cleavage pattern of myroilysin with the oxidized insulin B chain also showed that it had broad specificity and that it shared some, but not all, cleavage sites with pseudolysin and porcine pancreatic elastase (8). These results show the difference between myroilysin and pseudolysin in terms of substrate specificity. The role of myroilysin in strain D25 is to degrade surrounding proteins for nutrients. The broad specificity of myroilysin enables it to be versatile for protein degradation. Although myroilysin had high optimum temperatures with casein (50°C) and elastin (40 to ∼45°C), its thermostability is rather low, which may be the result of its long-term adaptation to the permanently cold environment in the deep sea.
It has been reported that animal elastase has a synergistic effect with collagenase in collagen degradation (27, 34). Although some bacterial elastases have been studied (8, 18, 31), there has been no report that a bacterial elastase has a synergistic effect with collagenase in collagen degradation. Myroilysin did not readily hydrolyze type I collagen fiber, yet its collagen-swelling activity exceeded that of 6 M urea. It can be speculated that myroilysin synergistically interacts with other enzymes in the in situ biotic assemblage and that it may play an important role in the degradation of deep-sea HMW organic nitrogen.
Supplementary Material
Acknowledgments
This work was supported by the Hi-Tech Research and Development Program of China (grants 2006AA09Z414, 2007AA091903, and 2007AA021306), the National Natural Science Foundation of China (grant 30770040), the Program for New Century Excellent Talents in University (grant NCET-06-0578), the Foundation for Young Excellent Scientists in Shandong Province (grant 2006BS02002), and the COMRA Program (grant DYXM-115-02-2-6).
Footnotes
Published ahead of print on 5 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aluwihare, L. I., D. J. Repeta, S. Pantoja, and C. G. Johnson. 2005. Two chemically distinct pools of organic nitrogen accumulate in the ocean. Science 308:1007-1010. [DOI] [PubMed] [Google Scholar]
- 2.Authier, F., M. Metioui, S. Fabrega, M. Kouach, and G. Briand. 2002. Endosomal proteolysis of internalized insulin at the C-terminal region of the B chain by cathepsin D. J. Biol. Chem. 277:9437-9446. [DOI] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. Von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Brunnegard, J., S. Grandel, H. Stahl, A. Tengberg, and P. O. J. Hall. 2004. Nitrogen cycling in deep-sea sediments of the Porcupine Abyssal Plain, NE Atlantic. Prog. Oceanogr. 63:159-181. [Google Scholar]
- 6.Chen, X.-L., Y.-Z. Zhang, P.-J. Gao, and X.-W. Luan. 2003. Two different proteases produced by a deep-sea psychrotrophic strain Pseudoaltermonas sp. SM9913. Mar. Biol. 143:989-993. [Google Scholar]
- 7.Datta, G., A. Dong, J. Witt, and A. T. Tu. 1995. Biochemical characterization of basilase: a fibrinolytic enzyme from Crotalus basiliscus. Arch. Biochem. Biophys. 317:365-373. [DOI] [PubMed] [Google Scholar]
- 8.Dumont, L., B. Verneuil, J. Wallach, and R. Julien. 1994. Purification and characterization of an alkaline elastase from Myxococcus xanthus. Eur. J. Biochem. 223:775-782. [DOI] [PubMed] [Google Scholar]
- 9.Feder, J. 1968. A spectrophotometric assay for neutral protease. Biochem. Biophy. Res. Commun. 32:326-332. [DOI] [PubMed] [Google Scholar]
- 10.Freehold, N. J. 1972. Worthington enzyme manual, p. 43-45. Worthington Biochemical Corporation, Lakewood, NJ.
- 11.Jorgensen, B. B., and A. Boetius. 2007. Feast and famine—microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5:770-781. [DOI] [PubMed] [Google Scholar]
- 12.Kessler, E., and D. Ohman. 2004. Pseudolysin, p. 401-409. In A. J. Barrett, N. D. Rawlings, and J. F. Woessner (ed.), Handbook of proteolytic enzymes, 2nd ed. Elsevier Press, London, United Kingdom.
- 13.Knicker, H., and P. G. Hatcher. 1997. Survival of protein in an organic-rich sediment: possible protection by encapsulation in organic matter. Naturwissenschaften 84:231-234. [Google Scholar]
- 14.Lee, D.-G., J. H. Jeon, M. K. Jang, N. Y. Kim, J. H. Lee, J.-H. Lee, S.-J. Kim, G.-D. Kim, and S.-H. Lee. 2007. Screening and characterization of a novel fibrinolytic metalloprotease from a metagenomic library. Biotechnol. Lett. 29:465-472. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y. G., and R. F. Whittier. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674-681. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 17.Mohrlen, F., M. Maniura, G. Plickert, M. Frohme, and U. Frank. 2006. Evolution of astacin-like metalloproteases in animals and their function in development. Evol. Dev. 8:223-231. [DOI] [PubMed] [Google Scholar]
- 18.Morihara, K., H. Tsuzuki, T. Oka, H. Inoue, and M. Ebata. 1965. Pseudomonas aeruginosa elastase. Isolation, crystallization, and preliminary characterization. J. Biol. Chem. 240:3295-3304. [PubMed] [Google Scholar]
- 19.Morihara, K., and H. Tsuzuki. 1971. Comparative study of various neutral proteinases from microorganisms: specificity with oligopeptides. Arch. Biochem. Biophys. 146:291-296. [DOI] [PubMed] [Google Scholar]
- 20.Morihara, K. 1995. Pseudolysin and other pathogen endopeptides of thermolysin family. Methods Enzymol. 248:242-253. [DOI] [PubMed] [Google Scholar]
- 21.Murray, A. W., and W. F. Thompson. 1980. Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 8:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peek, K., D. P. Veitch, M. Prescott, R. M. Daniel, B. MacIver, and P. L. Bergquist. 1993. Some characteristics of a proteinase from a thermophilic Bacillus sp. expressed in Escherichia coli: comparison with the native enzyme and its processing in E. coli and in vitro. Appl. Environ. Microbiol. 59:1168-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plummer, T. H., Jr., A. L. Tarention, and C. R. Hauer. 1995. Novel specific O-glycosylation of secreted Flavobacterium meningosepticum proteins. J. Biol. Chem. 270:13192-13196. [DOI] [PubMed] [Google Scholar]
- 24.Rawlings, N. D., and A. J. Barrett. 2004. Introduction: metallopeptides and their clans, p. 231-268. In A. J. Barrett, N. D. Rawlings, and J. F. Woessner (ed.), Handbook of proteolytic enzymes, 2nd ed. Elsevier Press, London, United Kingdom.
- 25.Rawlings, N. D., F. R. Morton, and A. J. Barrett. 2006. MEROPS: the peptidase database. Nucleic Acids Res. 34(Database Issue):D270-D272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinhold, B. B., C. R. Hauer, T. H. Plummer, and V. Reinhold. 1995. Detailed structural analysis of a novel specific O-linked glycan from the prokaryote Flavobacterium meningosepticum. J. Biol. Chem. 270:13197-13203. [DOI] [PubMed] [Google Scholar]
- 27.Ryvnyak, V. V., and O. F. Dulgieru. 2003. Elastase involvement in extracellular and intracellular collagen degradation during postpartum involution of the uterus. Bull. Exp. Biol. Med. 136:206-208. [DOI] [PubMed] [Google Scholar]
- 28.Sachar, L. A., K. K. Winter, N. Sicher, and S. Frankel. 1955. Photometric method for estimation of elastase activity. Proc. Soc. Exp. Biol. Med. 90:323-326. [DOI] [PubMed] [Google Scholar]
- 29.Tarention, A. L., G. Quinones, B. G. Grimwood, C. R. Hauer, and T. H. Plummer, Jr. 1995. Molecular cloning and sequence analysis of flavastacin: an O-glycosylated prokaryotic zinc metalloendopeptidase. Arch. Biochem. Biophys. 319:281-285. [DOI] [PubMed] [Google Scholar]
- 30.Tarention, A. L. 2004. Flavastacin, p. 631-632. In A. J. Barrett, N. D. Rawling, and J. F. Woessner (ed.), Handbook of proteolytic enzymes, 2nd ed. Elsevier Press, London, United Kingdom.
- 31.Tsai, Y.-C., R.-Y. Juang, S.-F. Lin, S.-W. Chen, M. Yamasaki, and G. Tamura. 1988. Production and further characterization of an alkaline elastase produced by alkalophilic Bacillus strain Ya-B. Appl. Environ. Microbiol. 54:3156-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Usha, R., and T. Ramasami. 2002. Effect of hydrogen-bond-breaking reagent (urea) on the dimensional stability of rat tail tendon (RTT) collagen fiber. J. Appl. Polym. Sci. 84:975-982. [Google Scholar]
- 33.Zhang, X.-Y., Y.-J. Zhang, X.-L. Chen, Q.-L. Qin, D.-L. Zhao, T.-G. Li, H.-Y. Dang, and Y.-Z. Zhang. 2008. Myroides profundi sp. nov., isolated from deep-sea sediment of the southern Okinawa Trough. FEMS Microbiol. Lett. 287:108-112. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, Y., X, Liu, C. M. Sköld, H. Wang, T. Kohyama, F.-Q. Wen, R. F. Ertl, and S. I. Rennard. 2001. Collaborative interactions between neutrophil elastase and metalloproteinases in extracellular matrix degradation in three-dimensional collagen gels. Respir. Res. 2:300-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.