Abstract
Varicella-zoster virus (VZV) is a human alphaherpesvirus that is highly cell associated in cell culture. Because cell-free virus yields are too low to permit the synchronous infections needed for time-resolved analyses, information is lacking about the sequence of events during the VZV replication cycle. To address this challenge, we differentially labeled VZV-infected inoculum cells (input) and uninfected (output) cells with fluorescent cell dyes or endocytosed nanogold particles and evaluated newly infected cells by confocal immunofluorescence or electron microscopy (EM) at the single-cell level at defined intervals. We demonstrated the spatiotemporal expression of six major VZV proteins, ORF61, IE62, IE63, ORF29, ORF23, and gE, representing all putative kinetic classes, for the first time. Newly synthesized ORF61, as well as IE62, the major VZV transactivator, appeared within 1 h, and they were targeted to different subnuclear compartments. The formation of VZV DNA replication compartments started between 4 and 6 h, involved recruitment of ORF29 to putative IE62 prereplication sites, and resulted in large globular nuclear compartments where newly synthesized viral DNA accumulated. Although considered a late protein, gE accumulated in the Golgi compartment at as early as 4 h. ORF23 capsid protein was present at 9 h. The assembly of viral nucleocapsids and mature enveloped VZ virions was detected by 9 to 12 h by time-resolved EM. Although syncytium formation is a hallmark of VZV infection, infection of neighboring cells did not require cell-cell fusion; its occurrence from 9 h is likely to amplify VZV replication. Our results define the productive cycle of VZV infection in a single cell as occurring in 9 to 12 h.
Varicella-zoster virus (VZV) is a ubiquitous human alphaherpesvirus that causes varicella (chickenpox) during primary infection, can establish latency in sensory ganglia, and may reactivate to cause herpes zoster (shingles) (11, 24). VZV is related to herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) and simian varicella virus and has a linear DNA genome of 125 kbp that has at least 70 open reading frames (ORFs) encoding known or predicted viral proteins (11).
Like those of other herpesviruses, VZV particles are presumed to enter cells by fusion of the virion envelope with the plasma membrane or by endocytosis followed by the transport of capsids and associated virion tegument proteins to the cell nucleus (11, 46). The major VZV transactivating protein, referred to as immediate-early 62 (IE62) is a tegument component, as are other VZV regulatory proteins, including IE4, ORF10, IE63, and the viral kinases ORF47 and ORF66 (11, 29, 30, 50). As has been demonstrated in cells infected with HSV and other herpesviruses, VZV gene transcription is believed to occur in a cascade that leads to the synthesis of viral proteins that are classified as immediate-early, early, and late, based on the time course of their expression after virus entry (11). Studies using VZV-infected cells to inoculate uninfected cells in conjunction with metabolic pulse-labeling of newly synthesized proteins and Western blot analysis have indicated that viral proteins are expressed by 4 to 6 h after infection (1, 2). However, because VZV is so highly cell associated in cultured cells, experiments that reveal the timing of gene transcription or the spatiotemporal characteristics of VZV protein expression in single cells within one infectious cycle have not been performed (11). Achievable titers of cell-free VZV are too low to permit synchronous infections of cultured cells, as is done to define the kinetics of viral mRNA and protein synthesis for HSV-1 and other herpesviruses (10, 13, 25, 26). Therefore, information is lacking, and there is some controversy about when and where VZV proteins are expressed in newly infected cells, how the assembly of VZV nuclear replication compartments is orchestrated, the time required to complete one infectious cycle, and the role of cell-cell fusion in VZV propagation, which is of interest, given the extensive syncytium formation that characterizes VZV replication (11, 12, 26, 52).
VZV experiments are usually done by adding an infected-cell inoculum of human fibroblasts or melanoma (MeWo) cells to a monolayer of uninfected cells. Initial events during replication are assessed by using low numbers of infected inoculum cells as a means to enrich for newly infected cells. Infection is then monitored for 24 to 72 h to demonstrate viral spread within the monolayer and to allow enough new VZV protein synthesis for detection by Western blotting, confocal microscopy, or other methods. Since VZV is not released into media, secondary plaque formation does not occur during the 72-h interval. Many important parameters of VZV genome replication, protein expression, and virus-host cell interactions have been defined by using this approach (11). However, these experimental conditions are not compatible with generating an accurate time-resolved analysis of events in the VZV replication cycle because the infected cells are a mixed population that reflect different stages of viral infection.
Overcoming the experimental challenges to studies of the VZV replication cycle requires a strategy that permits the use of high numbers of infected inoculum cells so that enough cells are infected to evaluate the earliest time points while allowing unequivocal discrimination of the inoculum and newly infected cells at sequential time points. To address these challenges, we used methods to label either the input cells (VZV-infected inoculum cells) or the output cells (uninfected cells) and investigated the VZV infectious cycle, including the VZV genome and viral protein synthesis, by confocal immunofluorescence (IF) or by standard electron microscopy (EM). Analyses using confocal microscopy were done by labeling VZV-infected inoculum cells with fluorescent cell dyes. In EM experiments, the cells in the uninfected monolayer were preincubated with protein A-gold (PA-gold) beads before being inoculated with VZV-infected cells, allowing their identification as newly infected cells based on detection of the PA-gold particles in the cytoplasm (38).
With these methods, we demonstrated the time course of expression of major VZV proteins of all putative kinetic classes, as well as their cellular localization patterns, for the first time. Among other observations, we found that ORF61, as well as IE62, is expressed very early, less than 1 h after infection, in human fibroblasts and that these proteins are targeted to different subnuclear compartments. We tracked the formation of VZV DNA replication compartments, showing the accumulation of newly synthesized viral DNA by 4 h after infection, and demonstrated the assembly of viral nucleocapsids and mature enveloped VZ virions by 9 to 12 h after infection. These experiments have also documented that VZV infection of neighboring cells does not require cell-cell fusion but that cell-cell fusion beginning at 9 h is likely to substantially amplify VZV replication.
MATERIALS AND METHODS
Cells and viruses.
VZV (rOka) was propagated in human embryonic lung fibroblast (HELF) cells by using standard methods (53). Inoculum titers were determined by infectious focus assay; inoculum titers of cells labeled with fluorescent dyes (see below) were determined after labeling (53).
Fluorescent cell tracker labeling.
For IF experiments, infected inoculum cells were labeled with green BODIPY CellTracker (Invitrogen) according to the manufacturer's instructions. Green BODIPY CellTracker labels the cytoplasm with bright green fluorescence that is retained within the labeled cells and is not released into the medium. In cell-cell fusion experiments, the uninfected output cells were also labeled, using orange CellTracker (CMRA; Invitrogen). Cell-cell fusion is demonstrated when the cytoplasmic mixing of red and green fluorescence produces yellow cytoplasmic fluorescence.
Cell labeling by endocytosis of PA-gold particles.
In EM experiments, the output cells were preincubated with PA-gold particles (CMC, Utrecht, The Netherlands) before unlabeled VZV-infected inoculum cells were added (38). The PA-gold was diluted 1:50 into growth medium and was added to HELF cells at 3 h before infection to allow endocytosis. PA-gold-labeled output cells were then washed several times with medium and chased for 45 min to remove PA-gold from the medium and to allow further uptake of surface-bound PA-gold. The PA-gold accumulated in lysosomes, where it served as a readily visible electron-dense marker to identify output cells by standard EM.
Time course experiments.
For IF time course experiments, uninfected HELF (output) cells were seeded onto sterile glass coverslips, grown to 90% confluence, and inoculated with green-BODIPY-CellTracker-labeled inoculum cells (input) at a ratio of 1 infected cell per 50 uninfected cells. Inoculum cells were allowed to settle on the monolayer during a 30-min incubation on ice. The coverslips were transferred to 37°C to initiate infection. Cells were fixed with 4% paraformaldehyde at 0 h, 2 h, 4 h, 6 h, 9 h, or 12 h; in some experiments, samples were also prepared at 30, 60, and 90 min, or 3 h, 8 h, 10 h, and 24 h after inoculation. At least three coverslips were examined at each time point in all experiments, and at least two independent time course experiments were performed to evaluate the kinetics of expression of each VZV protein. For time-resolved EM experiments, infected cells were fixed at 3 h, 6 h, 9 h, and 12 h.
Antibodies and IF staining.
Cells on coverslips were permeabilized for 15 min with 0.5% Triton X-100 in phosphate-buffered saline, washed, and blocked in digoxigenin (DIG) blocking solution (Roche) for 30 min. Primary antibodies were diluted in blocking solution, and incubation was generally for 1 h at room temperature in a humid chamber. The primary antibodies used included rabbit polyclonal antibodies against ORF61 generated in our laboratory (L. Wang, unpublished data), IE62 (a gift from Paul Kinchington, University of Pittsburgh), IE63 (a gift from William Ruyechan, University of Buffalo), ORF29 (a gift from S. Silverstein, Columbia University, New York), and ORF23 (9) and monoclonal antibodies against gE (Chemicon, Temecula, CA), anti-DIG (Roche), and anti-BrdU-biotin conjugate (Invitrogen). Secondary detection was done with Texas Red-conjugated (goat) anti-rabbit or anti-mouse antibodies (Jackson ImmunoResearch). Additional antibodies used for some double-staining experiments included polyclonal rabbit anti-promyelocytic leukemia (PML) (Santa Cruz Biotech), monoclonal mouse anti-PML (Santa Cruz Biotech) with secondary reagents, including (goat) anti-mouse-Alexa Fluor 488 (Invitrogen), (donkey) anti-rabbit-fluorescein isothiocyanate (FITC), (donkey) anti-rabbit or anti-mouse Alexa Fluor 647 (Invitrogen), and streptavidin-FITC and streptavidin-Texas Red (Jackson ImmunoResearch). Hoechst 22358 was used as a nuclear counterstain in all experiments.
VZV DNA in situ hybridization.
The VZV-specific DNA probe was prepared by random priming, using a DIG DNA-labeling kit (Roche Diagnostics, Penzberg, Germany) as described previously (45). One milliliter of the hybridization mix contained 50% formamide, 10% dextran sulfate, 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate), 1× Denhardt's solution, 0.5 mg salmon sperm DNA, and 0.01 ml DIG-labeled VZV DNA probe. Cells on coverslips were permeabilized as for IF and treated with RNase A (1 mg/ml) for 20 min at 37°C. After being dehydrated with 70%, 90%, and 100% ethanol for 2 min each, coverslips were air dried and transferred onto 15-μl drops of preheated hybridization mix on a clean glass slide. Simultaneous heat denaturation of the probe and sample DNA was achieved by placing the slide on a heat block (98°C) for 10 min; slides were incubated at 37°C in a humid chamber for 12 h. Coverslips were then washed twice in 2× SSC and 0.1× SSC (10 min each) and blocked in DIG blocking solution (Roche) for 30 min. Sections were incubated with anti-DIG monoclonal antibody (Roche) for 1 h. Secondary detection was with anti-mouse DIG-conjugated antibody (Roche) and sheep anti-DIG-Fab fragments conjugated with rhodamine (Roche) for 1 h. When DNA-fluorescence in situ hybridization (FISH) was combined with BrdU or protein detection, the hybridized DNA probe was detected directly with rhodamine-conjugated anti-DIG-Fab fragments.
Drug inhibition experiments.
Uninfected cells grown on coverslips were either untreated or were pretreated by incubation with cycloheximide (100 μg/ml), actinomycin D (4 μg/ml), or phosphonoacetic acid (PAA; 0.5 mg/ml and 1 mg/ml) for 30 min at 37°C. All these drugs were purchased from Sigma-Aldrich. The uninfected output cells were then inoculated with green-labeled infected cells and incubated for 4 h at 37°C in the presence of the drugs at the same concentrations. The 4-h incubation was chosen to allow infection to proceed to a time at which newly synthesized IE62 or ORF61 would be detected easily, as determined in IF kinetics experiments. The cells were then fixed and stained for IE62 or ORF61.
Confocal microscopy.
IF analysis was performed with a Leica TCSSP2 confocal laser scanning microscope (Heidelberg, Germany) and a 63×/1.4 Plan Apochromat. Images were scanned at 1,024 × 1,024 pixels with at least 4× frame averaging and the pinhole adjusted to 1 airy unit. Brightness and contrast were adjusted using iPhoto (Apple) and Photoshop CS3 (Adobe). For each time point in the time course experiments, 50 fields with 30 to 50 output cells were scanned; this method allowed the analysis of the staining pattern for each VZV protein or for VZV DNA in at least 20 to 50 newly infected single cells for each time point.
Electron microscopy.
HELF cells were grown and infected on a 10-cm Petri dish. At specified time points, cells were trypsinized, gently pelleted, and fixed in 4% paraformaldehyde and 2% glutaraldehyde in phosphate buffer (0.1 M [pH 7.2]). Cells were postfixed with 1% osmium tetroxide (2 h) and incubated in 1% aqueous uranyl acetate overnight. The samples were dehydrated in a series of increasing ethanol concentrations followed by a final propylenoxide step. The samples were embedded in Embed 812 (Electron Microscopy Sciences, Fort Washington, PA). Ultrathin sections (60 nm) were prepared with a diamond knife (Diatome) and an ultramicrotome (Ultracut, Leica). Sections were stained with 3.5% aqueous uranyl acetate for 10 min and with 0.2% lead citrate for 3 min. The sections were analyzed using a Jeol 1230 transmission electron microscope at 80 kV, and digital photographs were taken with a Gatan Multiscan 701 digital camera. In the time-resolved EM studies, only cells that showed no PA-gold clusters in lysosomes were included in the morphological analysis.
RESULTS
Differentiation of cells newly infected with VZV from infected inoculum cells.
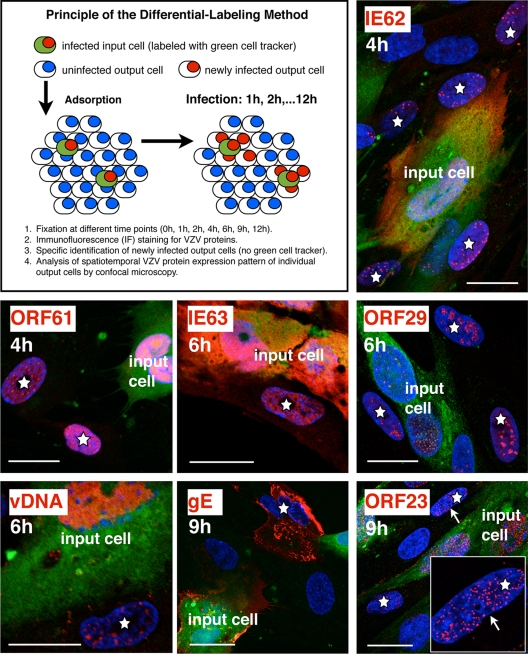
Initial experiments were done to evaluate the feasibility of using fluorescent-dye labeling of VZV-infected inoculum cells (input cells) to investigate the kinetics of expression of VZV proteins in newly infected HELF cells (output cells) (Fig. 1). Expression of ORF61, IE62, IE63, ORF29, gE, and ORF23 proteins was examined at 0, 2, 4, 6, 9, and 12 h after inoculation, using well-characterized antibodies to these proteins, and was compared to that of the uninfected HELF controls. The accumulation of VZV DNA over 2 to 12 h was followed by in situ hybridization, using a DIG-labeled probe. The technique allowed the discrimination of newly infected cells from inoculum cells at all time points, as illustrated by examples showing ORF61 and IE62 at 4 h, ORF29 and IE63 at 6 h, and gE and ORF23 at 9 h (Fig. 1). The cells identified as newly infected by the presence of VZV protein or DNA (red signal) and the absence of the green-fluorescence cell label were observed adjacent to or in close proximity to VZV-infected inoculum cells, as illustrated in a newly infected cell expressing IE62 at 4 h (Fig. 1, upper right panel). In many cases, several newly infected cells were observed adjacent to one infected inoculum cell.
FIG. 1.
Differentiation of cells newly infected with VZV from the infected-cell inoculum. The labeling method and selective analysis of newly infected (output) cells are outlined in the upper left panel. Examples of the spatiotemporal expression patterns of several viral proteins in newly infected cells (white stars) are shown. Unlabeled and uninfected (output) HELF cells were seeded on coverslips and infected with green-CellTracker-labeled and infected inoculum cells. The cells were then fixed at various time points and stained with specific antibodies for VZV proteins (red) IE62 and ORF61 (fixation at 4 h), IE63 and ORF29 (fixation at 6 h), or gE and ORF23 (fixation at 9 h) and Texas Red-conjugated secondary antibodies. Viral DNA (vDNA; red) was detected by DNA in situ hybridization (fixation at 6 h). The cell nuclei (blue) were counterstained with Hoechst 22358. The white arrow in the panel for ORF23 marks a newly infected cell nucleus that is also shown at higher magnification (inset). Scale bars are 20 μm.
Spatiotemporal expression of selected VZV proteins and viral genomic DNA in the first 12 h after infection.
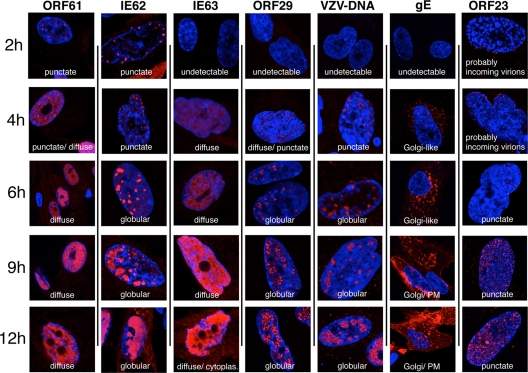
When the differential-labeling protocol and time course conditions had been established and staining with VZV-specific antibodies was optimized, this method was used to define the temporal pattern of expression of six major VZV proteins and the characteristics of their spatial locations within newly infected cells at intervals of 2, 4, 6, 9, and 12 h after inoculation (Fig. 2). The proteins included IE62 and IE63, which have been designated putative immediate-early proteins, based on the kinetics of expression of their HSV orthologs; the ORF61 gene product, which is related to HSV-1 ICP0; and ORF29, the single-stranded DNA binding protein. gE and the ORF23 capsid protein were evaluated as putative late gene products (3, 11, 14, 22, 31, 39, 41, 46). This analysis was done by staining for each protein separately to avoid potential cross-reactions and “bleed through”; fifty microscopic fields were examined for each time point, which allowed a single-cell assessment of protein expression in 20 to 50 newly infected cells at each time point after inoculation. Protein expression patterns within the newly infected cells at the same time point were quite similar during the interval before 6 h after infection. For example, all newly infected cells showed a similar punctate nuclear localization of IE62 at 4 h (Fig. 1, upper right panel; Fig. 2). Patterns of protein expression in the newly infected cells became more heterogeneous when examined at 6, 9, or 12 h. Most cells showed increased levels and changes in intracellular distribution of the VZV proteins at the later time points, but some cells showed patterns consistent with those observed at 2 h and 4 h. These observations are consistent with the expected continued transfer of virus from input cells into uninfected cells over the 12-h time course. However, because the pattern of expression had been established for each VZV protein in the analyses done at 2 h and 4 h, it was possible to differentiate the infected cells in the 6-to-12-h specimens that showed progression of infection from those that exhibited the pattern that was characteristic of early infection.
FIG. 2.
The patterns of spatiotemporal expression of selected VZV immediate-early, early, and late proteins and viral genomic DNA. Fixation for this time course analysis was at 0 h (not shown), 2 h, 4 h, 6 h, 9 h, and 12 h after inoculation of unlabeled HELF cells with input cells labeled with green CellTracker. The cells were then stained with specific antibodies for VZV proteins (red, from left to right) ORF61, IE62, IE63, ORF29, gE, and ORF23 followed by Texas Red-conjugated secondary antibodies. VZV DNA was detected by DNA in situ hybridization (red). Cell nuclei were counterstained with Hoechst 22358. Ten fields of 30 to 50 output cells were scanned to determine the representative staining patterns in newly infected cells at each time point. Single representative cells or nuclei are shown. PM, plasma membrane; cytoplas., cytoplasm.
In these comparative kinetics experiments, ORF61 was detected at the earliest 2 h time point in a punctate nuclear distribution (Fig. 2). This pattern was maintained, but more punctae were observed by 4 h. The punctae appeared in a relatively uniform distribution in nuclei and did not show enrichment along the nuclear margins, in contrast to IE62. The punctae became progressively less distinct, so that the punctate pattern was almost completely replaced at 6 h by a diffuse nuclear expression that increased in intensity but remained nuclear up to 12 h.
Like ORF61, IE62 was detected at 2 h and, at 2 h and 4 h, appeared as distinct nuclear punctae, which were located predominantly along the inner nuclear rim (Fig. 2). In many newly infected cells at these time points, IE62 punctae were enriched at the nuclear rim closest to the adjacent VZV-infected input cell. By 4 h, the diameter of the punctae had increased to form globular structures. By 6 h, these globular IE62-expressing domains occupied up to 25% of the nuclear space, enlarging to an estimated 50% by 9 h. IE62 expression covered 75% to 90% of the nuclei by 12 h, but the IE62 nuclear domains retained their globular shape, with distinct boundaries, throughout the time course. Limited but discernible levels of IE62 appeared in the cytoplasm at 12 h.
IE63 became minimally detectable only at 4 h at low levels and was present in a diffuse distribution in the nuclei of newly infected cells (Fig. 2). By 6 h, the intensity of IE63 nuclear expression had increased markedly and continued to increase, generating extensive nuclear expression at 12 h. In contrast to ORF61 and IE62, nuclear IE63 remained diffuse without localizing to punctae or distinct globular domains. Weak cytoplasmic expression of IE63 was detectable by 9 h and had increased at 12 h.
ORF29, the single-stranded DNA binding protein, was first apparent by 4 h as a very weak diffuse nuclear signal along with a few (1 to 5) tiny but distinct punctae (Fig. 2). By 6 h, more and larger ORF29 punctae were visible and formed widely distributed clusters in the nuclei of infected cells. These clusters of ORF29 punctae formed a pattern with contours like the IE62 nuclear compartments and also increased in size over 9 to 12 h, suggesting that ORF29 and IE62 were present in the same nuclear domains.
Sufficient newly synthesized VZV DNA had accumulated to be detected unequivocally by 4 h and was localized in small punctae, usually near the nuclear rim (Fig. 2). By 6 h, VZV DNA was observed within globular domains that increased in size over 9 to 12 h, in a pattern similar to that in the IE62 and ORF29 nuclear compartments.
Although gE has been considered a late gene product, it was detectable by 4 h as a low intensity but distinct signal in a Golgi-compartment-like cytoplasmic distribution, and it increased at this location by 6 h. gE was not apparent on plasma membranes at 4 h or 6 h (Fig. 2). By 9 h, gE was present not only in Golgi-compartment regions but also in a patchy cytoplasmic distribution and as an intense signal on plasma membranes. This pattern was maintained at 12 h.
The ORF23 capsid protein, the ortholog of HSV VP26, was detectable in the nuclei of newly infected cells at all time points (Fig. 2). However, careful analysis using optical sections in different focal planes (Z stacks) revealed that tiny discrete punctae visible at 2 h and 4 h were probably incoming virions that had reached the outer envelope of the nuclear membrane but remained in the cytoplasm. Similar structures have been observed in studies of the transport of pseudorabies virus or HSV particles (17, 33). Optical sections through the middle of the nuclear volume did not reveal punctae within nuclei at these earliest time points. By 6 h, a few punctae were visible in optical sections through the center of some nuclei, consistent with the presence of newly synthesized ORF23 capsid protein and possibly of a few assembled nucleocapsids. At 9 h and 12 h, nuclei were filled with tiny but distinct ORF23 punctae. By 12 h, some ORF23 punctae had accumulated in spherical clusters within the nucleus.
ORF61 and IE62 proteins are targeted to different subnuclear compartments during very early infection.
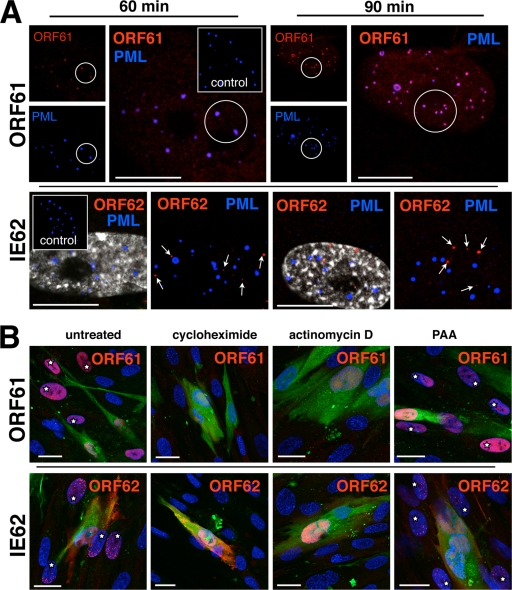
The HSV-1 proteins, ICP0 and ICP4, which are orthologs of ORF61 and IE62, associate with early viral replication compartments and colocalize or localize adjacent to nuclear bodies that express PML (18-20, 28, 35, 36, 49, 51). Therefore, in order to analyze the earliest events in the formation of VZV prereplication and replication compartments, ORF61 and IE62 expression, as well as their nuclear localization in relation to PML bodies, was examined in newly infected cells at 30, 60, or 90 min and at 2 h. ORF61 and IE62 were found in a few newly infected cells at 30 min (data not shown), and both were detected consistently by 60 or 90 min (Fig. 3A).
FIG. 3.
ORF61 and IE62 proteins are targeted to different subnuclear compartments and require active transcription and translation but not viral DNA synthesis for very early expression. (A) Unlabeled HELF cells were infected and fixed after 60 min (left panels) or 90 min (right panels). The cells were then double stained with either a combination of rabbit polyclonal anti-ORF61 antibody (upper panels, red) and anti-PML (blue; monoclonal mouse) or with rabbit polyclonal anti-IE62 (lower panels, red) and anti-PML (blue; monoclonal mouse). Secondary antibodies were donkey anti-rabbit Alexa Fluor 546 and donkey anti-mouse Alexa Fluor 647. Uninfected control cells (insets) were stained with the same antibody combinations. The small images in the ORF61 panels show single-PML (blue)- or ORF61 (red)-staining patterns that completely overlap (pink) in the larger overlay images. The two images shown in the IE62 panels (at 60 min or 90 min) depict the same nucleus. In the images on the left, nuclear staining (Hoechst 22358) is shown with the IE62 (red) and PML (blue) signals to clearly mark the nuclear areas. In the images on the right, the nuclear staining was left away for better visibility of the IE62 punctae (red; white arrows) that do not overlap with the PML signal (blue). Scale bars are 10 μm. (B) Uninfected HELF cells were pretreated for 30 min with cycloheximide, actinomycin D, or PAA or left untreated (control) and then inoculated with infected, untreated, green-labeled input cells. Incubation occurred in the presence of the same drugs and was stopped by fixation at 4 h. Cells were then stained for either ORF61 (red, upper panels) or IE62 (red, lower panels), and nuclei (blue) were counterstained with Hoechst 22358. Output cells adjacent to infected inoculum cells (green cytoplasmic staining plus red nuclear staining of ORF61 or IE62) were analyzed for nuclear expression of ORF61 or IE62 (red). Nuclear ORF61 or IE62 (red) is visible in several newly infected cells (white stars) in either the control (far left panels) or the PAA-treated cells (far right panels) but not in the cycloheximide- or actinomycin D-treated cells (middle panels). Scale bars are 20 μm.
HSV1 ICP0 has been reported to localize to PML nuclear bodies at very early times, before these nuclear structures are degraded (18, 51). When newly infected cells were stained for ORF61 and PML at 60 or 90 min, almost all ORF61 punctae were associated with PML punctae (Fig. 3A, upper panels). However, the colocalization of ORF61 with PML within individual PML bodies appeared in two patterns, which were either partial or complete (Fig. 3A, upper panels). Only the PML signal was detected in uninfected HELF controls (Fig. 3A, upper panels, inserts).
In contrast to ORF61, IE62 (Fig. 3A) did not colocalize with PML nuclear bodies or was adjacent to these structures in newly infected cells at 60 or 90 min after inoculation (Fig. 3A, lower panels). The nuclear localization of IE62 and PML at 60 and 90 min was also examined in cells in which nuclei were stained with Hoechst. PML domains were clearly visible, but no association with IE62 punctae was observed. This finding was further documented in 80 infected cell nuclei examined at 2 h; these nuclei contained 1,039 punctae that expressed IE62, but only 30 (2.9%) showed colocalization with any of 930 PML bodies.
ORF61 and IE62 require active transcription and translation but not viral DNA synthesis for very early detection.
In order to determine whether ORF61 and IE62 detected at 60 and 90 min represented newly synthesized proteins or had reached the nuclei in association with entering capsids, their expression was analyzed in cells treated with drugs that inhibit transcription (actinomycin D), translation (cycloheximide), or viral DNA replication (PAA) and compared with untreated VZV-infected HELF cells. Both ORF61 and IE62 were detected at 4 h in newly infected cells adjacent to green-labeled inoculum cells with or without PAA treatment (Fig. 3B), and their intracellular localization was similar to that observed in the time course experiments (Fig. 1 and 2). The size and morphology of the nuclear compartments that expressed IE62 were not altered in PAA-treated cells at 4 h. PAA concentrations of either 0.5 mg/ml (data not shown) or 1 mg/ml did not reduce the expression of either ORF61 or IE62. However, no ORF61 or IE62 was detected in cells treated with actinomycin D or cycloheximide. None of the punctate signals seen at 60 or 90 min in the absence of these drugs were detected in cells adjacent to the inoculum cells, indicating that ORF61 and IE62 detected at these times represented newly synthesized VZV proteins (Fig. 3A).
The ORF29 single-stranded DNA binding protein is targeted to nuclear domains that express IE62 and transform into viral replication compartments.
HSV-1 ICP4, which is related to IE62, and ICP8, the single-stranded DNA binding protein, are involved in the formation and function of replication compartments in infected cell nuclei (15, 44). Analyses of the stages in their formation indicate a stepwise recruitment of ICP8 and other viral replication proteins to ICP4 in generating active replication compartments (7, 15, 32). To explore the timing and define the steps in the creation of VZV replication compartments, the spatiotemporal expression of IE62 and ORF29 was analyzed, along with the localization of nascent VZV DNA, detected by BrdU pulse-labeling, and of accumulated VZV DNA, detected by DNA-FISH. The 2- to 12-h kinetics experiments showed that IE62, ORF29, and viral DNA accumulated in globular nuclear domains over time (Fig. 2). These experiments also revealed that both viral DNA and ORF29 were detectable unequivocally only at about 4 h after infection, whereas IE62 was detectable within 1 h (Fig. 2 and 3). These observations suggested that ORF29 might be recruited to nuclear compartments containing IE62 to form active viral replication sites, where VZV genomic DNA accumulated.
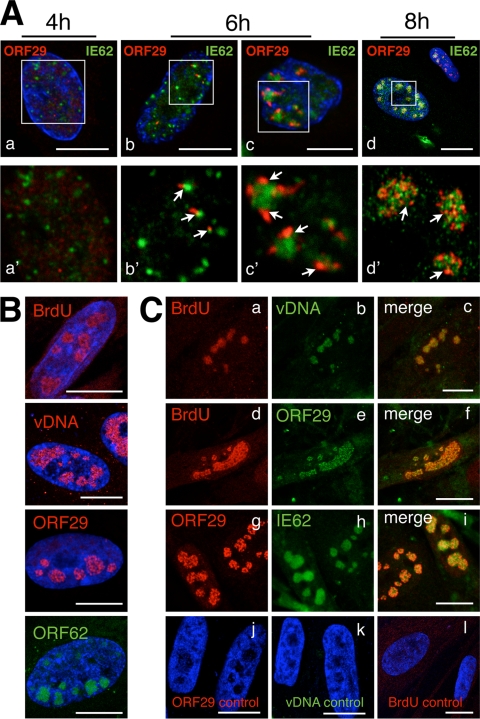
IE62 punctae were prominent at 4 h, whereas ORF29 expression was limited and diffuse at this time point (Fig. 4A, panels a and a'), confirming the observations shown in Fig. 2. By 6 h, single ORF29 punctae (Fig. 4A, panels b and b') or clusters of ORF29 punctae (Fig. 4A, panels c and c') that were clearly associated with the periphery of small or more globular IE62 compartments (Fig. 4A, panels b' and c') were observed. However, ORF29 was only partially colocalized with IE62. By 8 h, both ORF29 and IE62 were present within globular compartments, where they remained in a pattern of partial colocalization (Fig. 4A, panel d').
FIG. 4.
The single-stranded DNA binding protein ORF29 is targeted to IE62 nuclear domains that transform into viral replication compartments. (A) HELF cells were seeded on glass coverslips and infected with green-labeled inoculum cells for 4 h, 6 h, or 8 h and were double stained with anti-IE62 (green; mouse monoclonal antibody) and anti-ORF29 (red; polyclonal rabbit antibody) followed by secondary anti-mouse Alexa Fluor 488 or anti-rabbit Texas Red-conjugated secondary antibodies. Nuclei (upper panels a through d; blue) were counterstained with Hoechst 22358. Newly infected cells were identified by the absence of green cytoplasmic staining and were analyzed for IE62 and ORF29 expression by confocal microscopy. The upper panels (a through d) show an overview of the nuclei with merged blue, green, and red channels. Areas of the same nuclei (within the white squares in the upper panels) are shown at higher magnification in lower panels a' through d' (for better visualization of red and green signals, no blue channels are shown). White arrows point to red ORF29 punctae associated with green-stained IE62 nuclear domains. Scale bars are 10 μm. (B) HELF cells were seeded and infected and then additionally treated at 12 h after infection with a 30-min pulse of 0.1 mM BrdU (in medium) before being fixed at 12.5 h. Cells were then stained with anti-ORF29 (red) or -ORF62 (green; polyclonal antibody) and Texas Red (ORF29)- or FITC (IE62)-conjugated secondary antibodies, respectively, processed for DNA in situ hybridization (red; DIG-labeled VZV DNA probe), or stained for incorporated BrdU (red; biotinylated anti-BrdU monoclonal antibody and streptavidin-Texas Red conjugate for secondary detection). Scale bars are 10 μm. Each of these single stainings reveals similarly shaped globular nuclear compartments. (C) Cells from the same experiment as for panel B were double stained for either BrdU (a, red) with viral DNA (vDNA; b, green), BrdU (d, red) with ORF29 (e, green), or ORF29 (g, red) with IE62 (h, green). The merged channels are shown in panels c, f, and i. Control stainings for ORF29, viral DNA, and BrdU are shown in panels j, k, and l. Scale bars are 10 μm.
Whether viral DNA replication was associated within these globular compartments that expressed IE62 and ORF29 was then determined by labeling nascent VZV DNA with BrdU and by detecting accumulated VZV DNA, using DNA-FISH. Newly infected cells were examined at 12 h after infection, because staining protocols to detect incorporated BrdU or DNA-FISH together with immunofluorescence for viral proteins were usually much less sensitive than those to detect VZV DNA or proteins separately. As shown in Fig. 2, larger and more intensely stained nuclear IE62, ORF29, or VZV DNA domains were present at 12 h than at 6 h, making experiments using BrdU detection or DNA-FISH in combination with antibodies to detect VZV proteins more robust and easier to interpret at this time point.
At 12 h, cells were pulse-labeled with BrdU for 30 min before being fixed. First, single stainings for incorporated BrdU, VZV DNA, ORF29, or IE62 were performed (Fig. 4B). As expected from the time course experiments, each single staining revealed large globular nuclear compartments that were of similar size and shape, suggesting that incorporated BrdU, VZV DNA, ORF29, and IE62 resided in the same nuclear domains at this time point. To confirm this observation, double staining of newly infected cells from the same experiment were performed (Fig. 4C). Incorporated BrdU and VZV DNA signals exhibited colocalization, demonstrating that nascent VZV DNA was within the same compartment as accumulated viral DNA (Fig. 4C, panels a through c). To demonstrate that ORF29 was present with nascent viral DNA, infected cells were stained for incorporated BrdU and ORF29, which showed colocalization (Fig. 4C, panels d through f). Dual staining for ORF29 and IE62 confirmed that both proteins resided in the same nuclear compartments at 12 h (Fig. 4C, panels g through i). Control staining for ORF29 and VZV DNA in uninfected cells or for BrdU in untreated cells demonstrated the specificity of the staining protocols (Fig. 4C, panels j through l).
The harsh conditions of the BrdU-labeling and DNA-FISH protocols prevented the combined visualization of VZV DNA with ORF29 or IE62 at 4 h or 8 h. Nevertheless, these results suggest that IE62 and ORF29 accumulated together with nascent and accumulated VZV DNA within the same nuclear domains and that these domains represent VZV replication compartments. Both ORF29 and VZV DNA signals were weak at 4 h but were clearly recognizable at 6 h in single-staining experiments (Fig. 2), indicating that the first active VZV replication compartments form between 4 and 6 h in newly infected HELF.
VZV nucleocapsid formation and plasma membrane expression of glycoprotein gE.
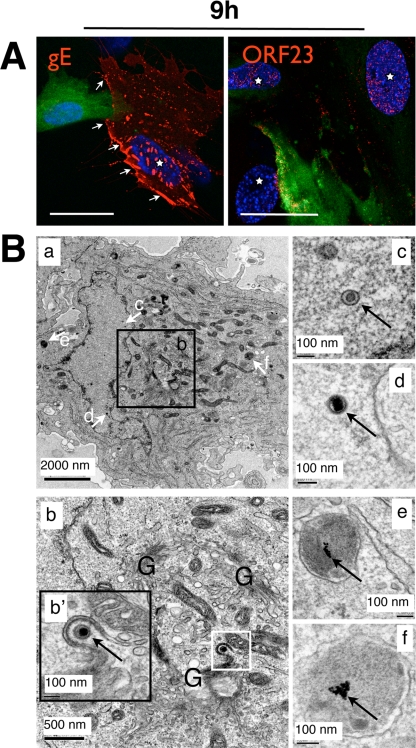
Having observed that VZV DNA replication occurs well before 9 h in VZV-infected HELF cells, we analyzed later events in the VZV replication cycle, including nucleocapsid assembly, glycoprotein expression on plasma membranes, and virion envelopment and egress. The kinetics experiments revealed that gE was expressed as early as 4 h and accumulated in a Golgi-compartment-like pattern in the cytoplasm from 4 to 6 h (Fig. 2). However, gE association with plasma membranes was minimal at the early times compared to 9 h, when it was expressed extensively on plasma membranes as well as in the cytoplasm (Fig. 2 and 5A). These experiments also revealed that distinct ORF23 punctae were very rare within the nucleus at 6 h but had become abundant by 9 h (Fig. 2 and 5A). However, confocal microscopy did not reveal whether ORF23 is associated with assembled nucleocapsids.
FIG. 5.
VZV nucleocapsids and surface expression of glycoprotein gE are detectable at 9 h after infection. (A) Uninfected HELF cells were inoculated with green-labeled inoculum cells and fixed after 9 h. Unpermeabilized cells were stained with anti-gE (left panel, red; monoclonal antibody) and a secondary anti-mouse Texas Red antibody. White arrows mark areas of plasma membrane-expressed gE protein (red). Other cells were permeabilized and stained with rabbit polyclonal anti-ORF23 (right panel, red) and a secondary anti-rabbit Texas Red-conjugated antibody. Newly infected cells are marked with a white star. Scale bars are 30 μm. (B) Time-resolved standard EM analysis of newly infected HELF cells. (a) Overview of an infected HELF cell 9 h after inoculation. The Golgi-compartment area (b, white square) is shown at higher magnification in the inset panel b', which shows a nucleocapsid (arrow) undergoing envelopment with a Golgi-compartment-derived membrane. The white arrows labeled c through f in the overview image point to structures that are shown at much higher magnification in the images on the right (c through f). (c and d) Nuclear viral nucleocapsids are indicated by arrows. (e and f) Lysosomes with clusters of internalized 10-nm gold particles (arrows) that were used to discriminate newly infected cells from infected inoculum cells are shown.
EM experiments were done to assess whether VZV nucleocapsid assembly, nuclear egress of virions into the cytoplasm, and secondary envelopment of cytoplasmic virions had occurred by 9 h after infection. In order to distinguish between inoculum cells and newly infected cells, the uninfected cells were labeled by fluid-phase endocytosis of PA-gold particles before infection. The accumulation of the electron-dense 10-nm PA-gold particles within lysosomes differentiated the output cells from VZV-infected input cells. Assessment of VZV-induced changes in the ultrathin sections of these samples was done by examining only these labeled cells. Cells within multinucleated syncytia were excluded from this analysis because fusion of an infected input cell with a labeled output cell could result in the presence of virion structures that were made in the inoculum cells rather than in the newly infected cell. Only cells that had a single nucleus and a large area of cytoplasm with at least two lysosomes containing electron-dense PA-gold clusters were evaluated.
EM analysis at 2 h and 6 h did not reveal morphological changes in the PA-gold-labeled cells compared to unlabeled cells, indicating that the PA-gold particles did not alter cellular structures. No evidence of VZV infection was detectable, although small numbers of incoming virions would not be likely to be observed in these ultrathin sections. Importantly, PA-gold particles were not present on cell surfaces, between cells, or in the heavily infected inoculum cells at these early time points, showing that PA-gold particles were not transferred from labeled output cells to VZV-infected inoculum cells, which would interfere with interpretation of the results. By 9 h, ultrathin sections revealed VZV nucleocapsids in the nuclei of newly infected cells (Fig. 5B, panels a, c, and d) that were identified by PA-gold clusters in lysosomes (Fig. 5B, panels a, e, and f). Several virions were also present in the cytoplasm adjacent to Golgi-compartment-derived membranes (Fig. 5B, panel b, inset b'). The apparent density of nucleocapsids in the EM sections was less than might be expected, given the extent of ORF23 expression observed at 9 h by confocal analysis, but it is explained by the evaluation of a much smaller cell volume in ultrathin sections (60 nm) than in a confocal optical section (500 to 1,000 nm). Although nucleocapsids and cytoplasmic virions were observed, cytoplasmic vacuoles containing numerous enveloped VZ virions were essentially absent at 9 h after infection, as were extracellular virions.
The kinetics of cell-cell fusion after VZV infection.
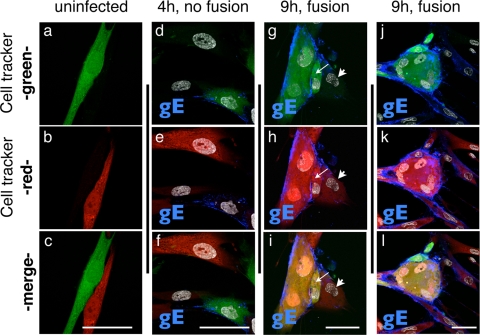
The time course experiments showed that gE was expressed in a Golgi-compartment-like pattern as early as 4 h and was enriched on plasma membranes by 9 h. Since these experiments relied on distinguishing inoculum cells by their green-fluorescent-dye labeling, only newly infected cells that had not fused with inoculum cells were analyzed. These conditions demonstrated that very early expression of VZV proteins, e.g., ORF61 and IE62, nascent VZV DNA synthesis, and expression of later proteins, e.g., gE and ORF23, could occur in the absence of cell-cell fusion. However, cell-cell fusion and the formation of multinucleated cells is a hallmark of VZV replication in cultured cells, as well as in the pathogenesis of VZV skin infection (11, 12). Therefore, to examine this characteristic of VZV replication, fluorescent-dye labeling was used to determine the time course of cell-cell fusion in VZV-infected HELF cells. VZV-infected and mock-infected cells were labeled with green BODIPY CellTracker, and uninfected output cells were labeled with orange CellTracker (CMRA). The cell populations were washed thoroughly with medium before the inoculum cells were added to the uninfected-cell monolayer. As expected, control experiments demonstrated that the two cell populations could be distinguished by confocal microscopy without “bleed-through” of the red fluorescence into the green channel or vice versa (Fig. 6a through c). In order to follow cell-cell fusion events, cells were also stained with anti-gE antibody and a secondary Alexa Fluor 647 (far red)-conjugated antibody and with nuclear Hoechst 22358 stain to facilitate the detection of syncytia (Fig. 6d through l).
FIG. 6.
Demonstration of cell-cell fusion events at 9 h after VZV infection. HELF cells were seeded on glass coverslips and stained with orange CellTracker (CMRA), washed, and then inoculated with green-CellTracker-labeled uninfected HELF cells (a through c) or infected HELF cells (d through l). Cells were fixed at 4 h (d through f) or 9 h (g through i and j through l) after infection and stained for gE protein (blue; monoclonal antibody and secondary anti-mouse Alexa Fluor 647 conjugate). Nuclei were stained with Hoechst 22358 (d through l, gray). Cells were analyzed for the mixing of green and red fluorescent signals in the cytoplasm that would indicate fusion of newly infected cells with an input cell. The thin white arrows in panels g through i point to a cell (red and green cytoplasm) that seems to have partially fused with fused input cells. The adjacent output cell (arrowhead) has not yet fused (only red fluorescence). Scale bars are 50 μm.
Multinucleated cells that exhibited only green fluorescence were detected at each time point, consistent with the presence of fused cells in the inoculum (data not shown). No clear evidence of cell-cell fusion between green- and red-labeled cells was detected from 2 to 6 h. In an example at 4 h, an infected inoculum cell was adjacent to output cells that were either not yet infected or did not yet express gE (Fig. 6d through f). In this case, distinct red or green cytoplasmic staining was apparent with no evidence of a yellow-orange color that would indicate cytoplasmic mixing. However, by 9 h, many syncytia with yellow fluorescence were observed, indicating fusion of inoculum and output cells (Fig. 6g through i and j through l). Some cells adjacent to a syncytium showed some evidence of red/green cytoplasmic mixing but still seemed to be partially separated, suggesting that only partial fusion had occurred (Fig. 6g through i). However, other cells adjacent to a syncytium exhibited only red fluorescence, indicating that no fusion had occurred (Fig. 6g through i and k through l). These experiments indicate that cell-cell fusion is usually a relatively late event in the VZV replication cycle, becoming prominent at 9 h, and that it is not necessary for the spread of VZV to adjacent uninfected cells.
Detection of VZV nucleocapsids, enveloped virions in cytoplasmic vacuoles, and extracellular enveloped virions.
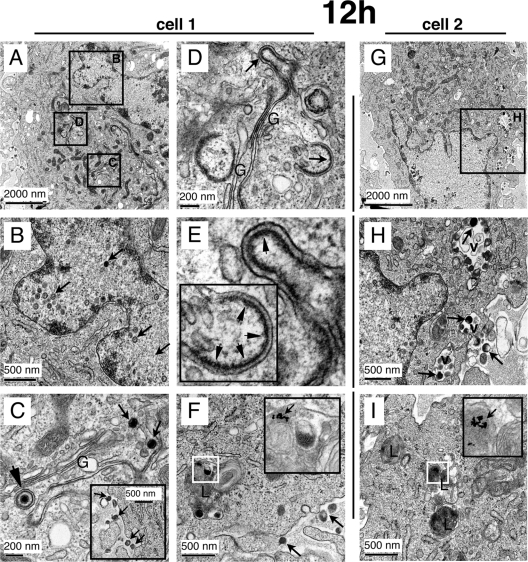
VZV gE and the ORF23 capsid protein were expressed extensively by 9 h after infection, as determined by confocal analysis (Fig. 2 and 5A), and time-resolved EM revealed that nucleocapsids and cytoplasmic virions were present in newly infected cells by 9 h (Fig. 5B). However, since few virions in cytoplasmic transport vacuoles or extracellular virions were detected at 9 h, further EM analyses of newly infected cells were done at 12 h, again assessing only cells with a single nucleus and endocytosed PA-gold particles within at least two lysosomes. Figure 7 shows detailed ultrastructural images of two newly infected cells at 12 h. Cell 1, shown at lower magnification in Fig. 7A, has numerous nucleocapsids within the nucleus (Fig. 7B), cytoplasmic virions in the Golgi-compartment area (Fig. 7C), and modified Golgi-compartment membranes with putative tegument protein (Fig. 7D and E), all within the same cell. Extracellular virions were also associated with this cell (Fig. 7C, inset).
FIG. 7.
Detection of VZV nucleocapsids, intracellular enveloped virions in cytoplasmic vacuoles, and extracellular enveloped virions at 12 h after infection. Areas of two cells (A through F, cell 1; G through I, cell 2) that were fixed after 12 h of infection are shown at the ultrastructural level after processing for time-resolved standard EM. (A) Overview of cell 1 with black squares indicating the areas that are shown in panels B through D at a higher magnification. (B) Demonstration of many nucleocapsids (arrows) in the nucleus. (C) Stack of Golgi cisternae (G) with adjacent nucleocapsids (small arrows) and one enveloped nucleocapsid (arrowhead). The inset shows extracellular virions associated with the plasma membrane of cell 1. (D and E) Another Golgi stack (G) in the same cells with modified Golgi cisternae (D, arrows) and enrichment of electron-dense putative tegument proteins (E, arrowheads) at the concave sides of these cisternae. (F) A multilamellar lysosome (L) is shown with a cluster of internalized 10-nm gold particles that was used to identify cell 1 as a newly infected cell (see Materials and Methods). (G) Overview of cell 2. The black square marks an area shown at higher magnification in panel H. (H) A nucleus with nucleocapsids (left side) is visible, and three cytoplasmic vacuoles (v) that contain VZV particles (arrows) can be seen. An extracellular virion is also marked (right side, arrow). (I) Three lysosomes are marked (L), one of which harbors a cluster of 10-nm gold particles (white square) that are clearly visible at a higher magnification (arrow in inset).
Cell 2 (Fig. 7G) has several nucleocapsids within the nucleus (Fig. 7H) and enveloped virions within cytoplasmic vacuoles (Fig. 7H). Both cells contained 10-nm PA-gold particles within lysosomes, as shown in Fig. 7F and I. Thus, by 12 h after infection, VZV-infected cells exhibited an abundance of nucleocapsids, extensively modified Golgi-compartment membranes, cytoplasmic vacuoles with many virions with secondary envelopment, and completely enveloped virions in the cytoplasm, along with numerous extracellular virions on plasma membranes.
DISCUSSION
In this study, the kinetics of major events during the first 12 h of the VZV replication cycle was established, using novel strategies to differentiate virus-infected inoculum cells from newly infected cells in time-resolved confocal IF and EM analyses of individual human fibroblasts after VZV entry. Since VZV is highly cell associated, studies of its replication are done with infected-cell or low-titer, cell-free virus inocula that cannot produce uniform, synchronous infections (11). In confocal microscopy experiments, this obstacle to evaluating the spatiotemporal expression of VZV proteins and viral DNA synthesis was overcome by labeling VZV-infected inoculum cells with fluorescent dyes that are nontoxic and do not diffuse into the medium. These fluorescent dyes are commonly employed to trace cell lineages over intervals of hours up to several days in developmental biology experiments; the characterization of entosis, a form of nonapoptotic cell death, is a recent application of the method (43).
In our investigations of VZV replication, green fluorescence labeling of inoculum cells and detection of viral proteins and DNA with Texas Red-conjugated reagents proved to be the most effective. Even though the inoculum cells were heavily infected, the green CellTracker dye was not released, and the labeled cells retained their infectivity for uninfected fibroblasts. Any effect of the fluorescent dye on the VZV replication cycle was excluded because the cell tracer was not available to be taken up by newly infected cells. This approach allowed an elucidation of the kinetics of synthesis and intracellular localization of VZV proteins and genomic DNA and provided a semiquantitative and reproducible assessment of events in VZV replication based on examining a defined number of individual infected cells at each time point. The method also permitted studies of different VZV proteins in parallel at each interval, the effects of drugs that inhibit various stages of replication, and associations between viral proteins, VZV DNA, and cell proteins in samples from the same assay.
As a complementary strategy for examining VZ virion assembly, intracellular trafficking, and egress in a time-resolved analysis of events at the ultrastructural level, the uninfected cell population was labeled with PA-gold particles by fluid-phase endocytosis before inoculation and EM analysis (38). The newly infected cells, identifiable by the presence of gold particles within lysosomes, showed no ultrastructural differences from unlabeled cells and remained susceptible to productive VZV infection. Both strategies should be applicable for investigating the replication cycle of other cell-associated viruses. Differential labeling of inoculum and newly infected cells with fluorescent dyes can also be used for sorting by flow cytometry or selective harvest of newly infected cells by laser capture at specific intervals, either of which may facilitate large-scale, time-resolved profiling of virus and host cell gene transcription and of the virus or host cell proteome for VZV or other viruses. In addition, the two cell-labeling techniques could be combined in multiscale correlative IF/EM experiments to examine rare events in virus-infected cells (45).
Events during the first 12 h after VZV infection were defined by tracking the spatiotemporal expression of IE62 and IE63, tegument proteins that regulate VZV transcription; OR61, a poorly characterized regulatory protein; ORF29, the VZV single-stranded DNA binding protein; gE, the major VZV glycoprotein; and ORF23, a nucleocapsid protein and ortholog of HSV VP26 (11). These experiments were done in parallel with ultrastructural examination of virion assembly and egress. Of these six VZV proteins, only ORF61 and IE62 were detected at the earliest time point and were newly synthesized within 1 h. VZV DNA accumulated with IE62 in nuclear domains as early as 4 h, and these domains enlarged to occupy 75 to 90% of the nuclear compartment by 12 h. VZV gE and ORF23 expression was extensive by 9 h, and consistent with these observations, nucleocapsids and cytoplasmic virions in the Golgi-compartment region were detected at 9 h. By 12 h, newly infected cells contained many nucleocapsids, enveloped capsids in the Golgi compartment, many VZ virions with secondary envelopes in intracellular vacuoles, and extracellular enveloped virions associated with plasma membranes. These experiments demonstrated that completion of the VZV replication cycle within individual cells, leading to the release of progeny virions, takes only 9 to 12 h in human fibroblasts. The time course established with these six VZV proteins provides a point of reference for grouping other VZV proteins with regard to the timing of their synthesis after viral entry. In addition, this information about the normal VZV replication cycle will be useful in defining disruptions resulting from mutations of the VZV genome, revealing when the functions of the mutated gene or promoter region are important in the course of the VZV replication cycle. These observations also confirm that VZV replication is comparatively slower than that of HSV-1, which produces new genomic DNA within 2 h after infection. Large replication compartments are formed in HSV-infected cells by 6 h, compared to between 9 and 12 h in VZV-infected cells (8, 15).
IE62, the predominant VZ virion tegument protein, is predicted to be an important immediate-early protein because it functions as the most potent and promiscuous transactivator of VZV gene promoters, including its own, and is essential for VZV replication (47, 48). The functions of ORF61 are much less well defined, although it also has regulatory activity in transient expression experiments (42). Using drug inhibition protocols and spatiotemporal analyses of individual newly infected cells, we demonstrated synthesis and nuclear translocation of both ORF61 and IE62 within 1 h after VZV infection. Of interest, ORF61 and IE62 were targeted to different nuclear subcompartments. ORF61 was consistently associated with ND10 bodies, identified by PML expression, whereas IE62 was associated with fewer than 3% of PML-expressing nuclear domains. The strong association of ORF61 with PML is consistent with observations about ICP0, which is its HSV-1 ortholog (40, 41). ICP0 localization to ND10 bodies and its ubiquitin ligase function is essential for their molecular rearrangement and degradation in HSV-1-infected cells (6, 18). ORF61 has an N-terminal ring finger domain that shares approximately 26% amino acid identity with the ICP0 ring finger motif, and ORF61 complements HSV-1-ICP0 deletion mutants (11, 40, 41). These observations suggest that ORF61 may compensate for the absence of ICP0-mediated degradation of ND10 bodies in cells infected with these mutants. Whether ORF61 shares the other functions of ICP0 that are necessary to inhibit the host cell antiviral response during initial replication is not known.
At the earliest time points, IE62 tended to be enriched at the inner nuclear rim in the newly infected cell that was nearest to an infected inoculum cell. While IE62 has important differences, both IE62 and HSV1 ICP4 have DNA binding domains and the capacity to interact with cellular transactivators to regulate transcription from viral gene promoters (21, 46, 47). Functionally, IE62 can complement HSV-1 mutants that lack ICP4 (22). Since ICP4 has been demonstrated to mark the location of HSV-1 prereplication compartments (19, 20, 32), it is likely that the first detection of IE62 punctae at the inner nuclear rim adjacent to an attached VZV-infected input cell represented its localization with incoming VZV genomes. As noted, IE62 showed essentially no localization with PML nuclear bodies during the first 2 h after VZV entry. In contrast, although earlier reports differ, recent studies indicate that ICP4 associated with these structures within 30 min after HSV-1 entry (32).
The time-resolved assessment of IE62, ORF29, the single-stranded DNA binding protein, and newly synthesized genomic DNA provides the first information that addresses the formation of VZV replication compartments at very early time points. IE62, ORF29, and VZV DNA began to colocalize in putative replication compartments between 4 to 6 h after infection. However, ND10 components, represented by PML, did not seem to be deposited in these locations within infected cell nuclei at these early time points (data not shown). The stepwise assembly of active HSV-1 replication compartments from prereplicative sites has been demonstrated by tracking the intracellular localization of ICP4; ICP8, which is the HSV-1 single-stranded DNA binding protein; and incorporated BrdU as a marker for nascent viral DNA (7, 8, 15, 32, 44, 46). Although delayed compared to that for HSV-1-infected cells, a similar sequence of events was observed in VZV-infected cells in that IE62 was first observed at less than 4 h in nuclear punctae that did not yet contain ORF29, the VZV single-stranded DNA binding protein, and probably represented prereplicative sites. ORF29 was first detectable at low levels in a diffuse nuclear distribution at 4 h. IE62-expressing domains grew into larger globular nuclear structures and ORF29 began to appear at these sites from 4 to 6 h after infection. When evaluated at 4 h and 8 h, small ORF29 punctae were localized specifically and exclusively at the margins of IE62 punctae, and small, although somewhat more globular IE62 sites, followed at later times by a distribution together with IE62 in larger globular compartments. For comparison, HSV-1 ICP4 and ICP8 colocalization was observed at 3 h, and small ICP8 punctae were associated with the periphery of ICP4-positive domains; colocalization of the helicase/primase and viral polymerase complexes was also shown (32). Similarly, ORF29 seems to be recruited to sites where IE62 has accumulated and that may serve as platforms for the assembly of proteins required for VZV replication. Although we did not analyze VZV helicase/primase or polymerase proteins, their recruitment would be expected, since the basic replication machinery is predicted to be highly conserved among herpesviruses (46).
The VZV ORF63 gene product is a tegument phosphoprotein with some regulatory functions, e.g., enhancing IE62 transactivation of some VZV promoters. It is related to the HSV-1 US1.5 protein, which is expressed colinearly with ICP22 (3). IE63 has been designated an immediate-early protein based on its presumed kinetics (16). However, IE63 nuclear expression was detectable only as a weak, diffuse signal at 4 to 6 h, which was substantially later than that of IE62 and ORF61. A difference in the sensitivity of IE63 detection is unlikely, because the anti-IE63 antibody is a high-potency reagent (34) and the intensity of the IE63 signal showed a rapid increase when evaluated between 6 to 8 h after infection. However, because IE63 was diffusely expressed and was not enriched in discrete compartments like the IE62 signal, earlier expression of IE63 might escape detection. IE63 expression remained diffuse throughout the nuclei, suggesting that this protein may be present within IE62-positive replication compartments but is not specifically enriched at these sites even though IE63 binds to IE62 by residues in the IE63 N terminus (3). IE63 was predominantly nuclear throughout the 9- to 12-h VZV replication cycle and remained so up to 24 h, but IE63 expression in the cytoplasm increased beginning at 9 h. This extensive cytoplasmic expression of IE63 at later times may be important for its role as an antiapoptotic factor, as has been demonstrated in cultured neurons (27).
VZV gE is the most abundant glycoprotein in infected cells and is a prominent envelope glycoprotein. In contrast to other alphaherpesviruses which replicate in the absence of gE, VZV requires gE for viral replication (11, 39). While it has been considered to be a late gene product, our time course experiments revealed that gE synthesis occurred quite early in newly infected cells in a pattern similar to that described for HSV gB (4, 5). gE appeared as a weak but readily detectable signal in a Golgi-compartment-like distribution as early as 4 h, progressing significantly in intensity and extent at this site by 6 h. This early gE expression may serve to rearrange the Golgi compartment and prepare Golgi-compartment-derived membranes for VZ virions exiting the nucleus by accumulating tegument proteins such as IE62 on membrane-bound gE (11, 23, 54). Because gE was absent from the plasma membrane and did not show a punctate endosome-like cytoplasmic pattern at early time points, it is unlikely that we detected endocytosed gE. ORF23, which is a VZV capsid protein related to HSV-1 VP26, became enriched within nuclei at 9 h, suggesting that viral nucleocapsids could become assembled for release from the nucleus and envelopment in the premodified Golgi-compartment area by 6 to 9 h after infection. In contrast to its Golgi-compartment localization, substantial gE expression on plasma membranes was evident only at 9 h.
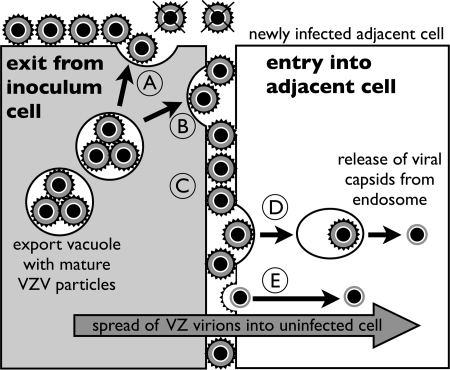
While syncytium formation is a hallmark of VZV replication in cultured cells, our analyses of the VZV replication cycle were done by tracking events during the infection of individual cells that were located next to an infected inoculum cell but had no evidence of green-fluorescent-dye transfer into the cytoplasm. These experiments suggested that VZV infection was initiated without cell-cell fusion and could be attributed to the transfer of extracellular virions on surfaces of inoculum cells, as visualized by EM. This explanation was supported by the observation that newly infected cells were consistently found adjacent to inoculum cells. The inoculum cells were also washed extensively to eliminate any unabsorbed fluorescent dye, which also removed the original culture medium and any cell-free VZ virions it may have contained. Furthermore, it is known that secondary plaques attributable to virus release do not appear in VZV-infected monolayers; in addition, extracellular VZ virions are quite labile, whereas enveloped VZV particles are abundant on the surfaces of heavily infected fibroblasts and are also present in intracellular vacuoles near the plasma membranes (11). When the inoculum cell was attached in proximity to the uninfected cell, the space between the plasma membranes appeared to be minimal, therefore placing surface virions next to the uninfected cell membrane and potential entry receptors of the uninfected cell and limiting the dispersal of virions. These conditions would favor entry of cell-associated extracellular virus particles without requiring cell-cell fusion (Fig. 8). Of interest, VZV gE may promote these conditions, since gE enhances the formation of tight junctions between plasma membranes (37).
FIG. 8.
Model of the cell-to-cell spread of VZV before cell fusion in vitro. Mature VZV particles are transported in an export vacuole to the plasma membrane of an infected inoculum cell (gray cell on left). (A) VZV particles released by exocytosis adhere to the plasma membrane at the interface with culture media. Infectious VZV particles are not released into the media or rapidly lose infectivity, since secondary plaques do not form in VZV-infected monolayers. (B) Some VZV particles are released from the inoculum cell in close proximity to the plasma membrane of the adjacent uninfected cell (white cell on right). (C) Envelope glycoproteins of extracellular VZV particles at these sites have an increased probability of binding to cell surface receptors on the plasma membrane of the uninfected cell. Entry into the adjacent cell occurs by endocytosis (D) or direct entry (E), which is followed by the release of partially tegumented capsids into the cytoplasm of the newly infected cell. Our model predicts that the spread of cell-associated extracelluar VZ virions in vitro is determined by the proximity of infected and uninfected cells and does not require cell fusion.
Although VZV infection occurred immediately after the exposure of uninfected cells to the infected cell inoculum without the formation of polykaryocytes, the kinetics experiments done using two cell dyes with different emission wavelengths (red and green fluorescence) to label inoculum and uninfected cells revealed that abundant cell-cell fusion and syncytium formation occurred by 9 h after infection. Some earlier fusion may have been undetected if a syncytium consisting of several green-labeled inoculum cells had fused with one red-labeled output cell, diluting the red fluorescence signal. In any case, while viral entry into uninfected cells occurs without it, cell-cell fusion at 9 h should amplify VZV replication substantially, since this time point coincides with the detection of enveloped cytoplasmic virions and precedes their appearance in large numbers on the surfaces of plasma membranes, which became prominent at 12 h.
In summary, this kinetic analysis of the VZV replication cycle in individual fibroblasts demonstrated the spatiotemporal expression of six VZV proteins, newly synthesized viral DNA, and virion morphogenesis. The earliest event after infection was new IE62 and ORF61 synthesis within 1 h. VZV DNA replication was observed at 4 h, and virion assembly and envelopment were detected at 9 h. Our results reveal that one complete VZV replication cycle leading to a new generation of infectious VZV particles takes between 9 and 12 h.
Acknowledgments
This work was supported by NIH grants (AI053846, AI20459, and CA049605) to A.M.A. and by a postdoctoral fellowship to M.R. from the Deutsche Forschungsgemeinschaft (DFG, Germany; Geschaeftszeichen RE2716/1-1).
Sample preparation, sectioning, and IF-EM analyses were performed in the CSIF/Stanford University Medical School.
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Asano, Y., and M. Takahashi. 1979. Studies on the polypeptides of varicella-zoster (V-Z) virus. 1. Detection of varicella-zoster virus polypeptides in infected cells. Biken J. 2281-89. [PubMed] [Google Scholar]
- 2.Asano, Y., and M. Takahashi. 1980. Studies on the polypeptides of varicella-zoster (V-Z) virus. II. Syntheses of viral polypeptides in infected cells. Biken J. 2395-106. [PubMed] [Google Scholar]
- 3.Baiker, A., C. Bagowski, H. Ito, M. Sommer, L. Zerboni, K. Fabel, J. Hay, W. Ruyechan, and A. M. Arvin. 2004. The immediate-early 63 protein of varicella-zoster virus: analysis of functional domains required for replication in vitro and for T-cell and skin tropism in the SCIDhu model in vivo. J. Virol. 781181-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beitia Ortiz de Zarate, I., L. Cantero-Aguilar, M. Longo, C. Berlioz-Torrent, and F. Rozenberg. 2007. Contribution of endocytic motifs in the cytoplasmic tail of herpes simplex virus type 1 glycoprotein B to virus replication and cell-cell fusion. J. Virol. 8113889-13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beitia Ortiz de Zarate, I., K. Kaelin, and F. Rozenberg. 2004. Effects of mutations in the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B on intracellular transport and infectivity. J. Virol. 781540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkham, J., D. M. Coen, C. B. Hwang, and S. K. Weller. 2001. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J. Virol. 752353-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkham, J., D. M. Coen, and S. K. Weller. 1998. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 7210100-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri, V., M. Sommer, J. Rajamani, L. Zerboni, and A. M. Arvin. 2008. Functions of varicella-zoster virus ORF23 capsid protein in viral replication and the pathogenesis of skin Infection. J. Virol. 8210231-10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J. J., Z. Zhu, A. A. Gershon, and M. D. Gershon. 2004. Mannose 6-phosphate receptor dependence of varicella zoster virus infection in vitro and in the epidermis during varicella and zoster. Cell 119915-926. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, J., S. Straus, and A. Arvin (ed.). 2007. Varicella-zoster virus replication, pathogenesis, and management, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA.
- 12.Cole, N. L., and C. Grose. 2003. Membrane fusion mediated by herpesvirus glycoproteins: the paradigm of varicella-zoster virus. Rev. Med. Virol. 13207-222. [DOI] [PubMed] [Google Scholar]
- 13.Cook, M. L., and J. G. Stevens. 1968. Labile coat: reason for noninfectious cell-free varicella-zoster virus in culture. J. Virol. 21458-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 671759-1816. [DOI] [PubMed] [Google Scholar]
- 15.Debrus, S., C. Sadzot-Delvaux, A. F. Nikkels, J. Piette, and B. Rentier. 1995. Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J. Virol. 693240-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55857-868. [DOI] [PubMed] [Google Scholar]
- 17.Döhner, K., K. Radtke, S. Schmidt, and B. Sodeik. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 808211-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 135062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett, R. D., G. Sourvinos, C. Leiper, J. B. Clements, and A. Orr. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 781903-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 773680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faber, S. W., and K. W. Wilcox. 1986. Association of the herpes simplex virus regulatory protein ICP4 with specific nucleotide sequences in DNA. Nucleic Acids Res. 146067-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felser, J. M., P. R. Kinchington, G. Inchauspe, S. E. Straus, and J. M. Ostrove. 1988. Cell lines containing varicella-zoster virus open reading frame 62 and expressing the “IE” 175 protein complement ICP4 mutants of herpes simplex virus type 1. J. Virol. 622076-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 686372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilden, D., R. Mahalingam, S. Deitch, and R. Cohrs (ed.). 2006. Varicella-zoster virus neuropathogenesis and latency. Caister Academic Press, Norwich, United Kingdom.
- 25.Harper, D. R., N. Mathieu, and J. Mullarkey. 1998. High-titre, cryostable cell-free varicella zoster virus. Arch. Virol. 1431163-1170. [DOI] [PubMed] [Google Scholar]
- 26.Harson, R., and C. Grose. 1995. Egress of varicella-zoster virus from the melanoma cell: a tropism for the melanocyte. J. Virol. 694994-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hood, C., A. L. Cunningham, B. Slobedman, A. M. Arvin, M. H. Sommer, P. R. Kinchington, and A. Abendroth. 2006. Varicella-zoster virus ORF63 inhibits apoptosis of primary human neurons. J. Virol. 801025-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinchington, P. R., D. Bookey, and S. E. Turse. 1995. The transcriptional regulatory proteins encoded by varicella-zoster virus open reading frames (ORFs) 4 and 63, but not ORF 61, are associated with purified virus particles. J. Virol. 694274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinchington, P. R., J. K. Hougland, A. M. Arvin, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J. Virol. 66359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinchington, P. R., G. Inchauspe, J. H. Subak-Sharpe, F. Robey, J. Hay, and W. T. Ruyechan. 1988. Identification and characterization of a varicella-zoster virus DNA-binding protein by using antisera directed against a predicted synthetic oligopeptide. J. Virol. 62802-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livingston, C. M., N. A. DeLuca, D. E. Wilkinson, and S. K. Weller. 2008. Oligomerization of ICP4 and rearrangement of heat shock proteins may be important for herpes simplex virus type 1 prereplicative site formation. J. Virol. 826324-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyman, M. G., B. Feierbach, D. Curanovic, M. Bisher, and L. W. Enquist. 2007. Pseudorabies virus Us9 directs axonal sorting of viral capsids. J. Virol. 8111363-11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch, J. M., T. K. Kenyon, C. Grose, J. Hay, and W. T. Ruyechan. 2002. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology 30271-82. [DOI] [PubMed] [Google Scholar]
- 35.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 742679-2690. [DOI] [PubMed] [Google Scholar]
- 36.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 21767-75. [DOI] [PubMed] [Google Scholar]
- 37.Mo, C., E. E. Schneeberger, and A. M. Arvin. 2000. Glycoprotein E of varicella-zoster virus enhances cell-cell contact in polarized epithelial cells. J. Virol. 7411377-11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Möbius, W., E. van Donselaar, Y. Ohno-Iwashita, Y. Shimada, H. F. Heijnen, J. W. Slot, and H. J. Geuze. 2003. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic 4222-231. [DOI] [PubMed] [Google Scholar]
- 39.Montalvo, E. A., R. T. Parmley, and C. Grose. 1985. Structural analysis of the varicella-zoster virus gp98-gp62 complex: posttranslational addition of N-linked and O-linked oligosaccharide moieties. J. Virol. 53761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moriuchi, H., M. Moriuchi, and J. I. Cohen. 1994. The RING finger domain of the varicella-zoster virus open reading frame 61 protein is required for its transregulatory functions. Virology 205238-246. [DOI] [PubMed] [Google Scholar]
- 41.Moriuchi, H., M. Moriuchi, H. A. Smith, S. E. Straus, and J. I. Cohen. 1992. Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. J. Virol. 667303-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moriuchi, H., M. Moriuchi, S. E. Straus, and J. I. Cohen. 1993. Varicella-zoster virus (VZV) open reading frame 61 protein transactivates VZV gene promoters and enhances the infectivity of VZV DNA. J. Virol. 674290-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Overholtzer, M., A. A. Mailleux, G. Mouneimne, G. Normand, S. J. Schnitt, R. W. King, E. S. Cibas, and J. S. Brugge. 2007. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131966-979. [DOI] [PubMed] [Google Scholar]
- 44.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36857-868. [DOI] [PubMed] [Google Scholar]
- 45.Reichelt, M., L. Zerboni, and A. M. Arvin. 2008. Mechanisms of varicella-zoster virus neuropathogenesis in human dorsal root ganglia. J. Virol. 823971-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA.
- 47.Sato, B., H. Ito, S. Hinchliffe, M. H. Sommer, L. Zerboni, and A. M. Arvin. 2003. Mutational analysis of open reading frames 62 and 71, encoding the varicella-zoster virus immediate-early transactivating protein, IE62, and effects on replication in vitro and in skin xenografts in the SCID-hu mouse in vivo. J. Virol. 775607-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato, B., M. Sommer, H. Ito, and A. M. Arvin. 2003. Requirement of varicella-zoster virus immediate-early 4 protein for viral replication. J. Virol. 7712369-12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sourvinos, G., and R. D. Everett. 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 214989-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson, D., K. L. Colman, and A. J. Davison. 1994. Characterization of the putative protein kinases specified by varicella-zoster virus genes 47 and 66. J. Gen. Virol. 75317-326. [DOI] [PubMed] [Google Scholar]
- 51.Tavalai, N., and T. Stamminger. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta. [DOI] [PubMed]
- 52.Yamanishi, K., Y. Matsunaga, T. Ogino, M. Takahashi, and A. Takamizawa. 1980. Virus replication and localization of varicella-zoster virus antigens in human embryonic fibroblast cells infected with cell-free virus. Infect. Immun. 28536-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zerboni, L., C. C. Ku, C. D. Jones, J. L. Zehnder, and A. M. Arvin. 2005. Varicella-zoster virus infection of human dorsal root ganglia in vivo. Proc. Natl. Acad. Sci. USA 1026490-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu, Z., M. D. Gershon, Y. Hao, R. T. Ambron, C. A. Gabel, and A. A. Gershon. 1995. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J. Virol. 697951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]