Abstract
The innate immune system recognizes nucleic acids during viral infection and stimulates cellular antiviral responses. Intracellular detection of RNA virus infection is mediated by the RNA helicases RIG-I (retinoic acid inducible gene I) and MDA-5, which recognize viral RNA and signal through the adaptor molecule MAVS (mitochondrial antiviral signaling) to stimulate the phosphorylation and activation of the transcription factors IRF3 (interferon regulatory factor 3) and IRF7. Once activated, IRF3 and IRF7 turn on the expression of type I interferons, such as beta interferon. Interestingly, unlike other signaling molecules identified in this pathway, MAVS contains a C-terminal transmembrane (TM) domain that is essential for both type I interferon induction and localization of MAVS to the mitochondrial outer membrane. However, the role the MAVS TM domain plays in signaling remains unclear. Here we report the identification of a function for the TM domain in mediating MAVS self-association. The activation of RIG-I/MDA-5 leads to the TM-dependent dimerization of the MAVS N-terminal caspase recruitment domain, thereby providing an interface for direct binding to and activation of the downstream effector TRAF3 (tumor necrosis factor receptor-associated factor 3). Our results reveal a role for MAVS self-association in antiviral innate immunity signaling and provide a molecular mechanism for downstream signal transduction.
The enhanced production of type I interferons is one of the earliest and most important host-protective cellular responses to virus infection (1, 9). Type I interferons, including beta interferon (IFN-β), are secreted cytokines that activate an early innate immune response and provide cellular antiviral protection (3). Together with other cytokines, type I interferons contribute to the establishment of adaptive immunity to virus infection. In most cell types, RIG-I (retinoic acid inducible gene I) and MDA-5 are thought to be the primary receptors that transduce signals to stimulate type I interferon production in response to infections by RNA viruses. RIG-I and MDA-5 each recognize distinct types of viral RNA, and both signal using a common intermediary protein called MAVS (mitochondrial antiviral signaling; also known as IPS-1/VISA/Cardif) (10). Signal transduction downstream of MAVS culminates in the activation of the IRF3/7 (interferon regulatory factors 3 and 7) and NF-κB transcription factors and the production of type I interferons and proinflammatory cytokines.
Although MAVS has been shown genetically and biochemically to be a critical downstream component of a signaling pathway connecting RIG-I and MDA-5 to the activation of IRF3/7 and NF-κB (10), how MAVS functions or is regulated remains enigmatic. MAVS is a mitochondrial outer membrane protein that contains a C-terminal transmembrane (TM) domain and an N-terminal caspase recruitment domain (CARD), both of which have been shown to be functionally important (10). Removal of the TM domain by artificial means or by proteolytic cleavage with the hepatitis C virus (HCV) NS3/4A protease prevents MAVS from localizing to the mitochondrial outer membrane and inducing IFN-β (20, 23, 28). Deletion of the MAVS CARD also prevents the induction of IFN-β (15, 23, 28, 30). Mutagenesis experiments suggest that the MAVS CARD is likely required for binding to a downstream effector (28). However, the identity of such a molecule has yet to be discerned.
The ubiquitin (Ub) ligase TRAF3 (tumor necrosis factor receptor-associated factor 3) has been implicated to play a role in RIG-I/MDA-5 signaling downstream of MAVS (24, 25). TRAF3 associates with MAVS, and TRAF3 ubiquitination is stimulated by the activation of RIG-I/MDA-5 signaling (16, 25). Here, we have uncovered an important role for MAVS self-association in RIG-I/MDA-5 signaling. We find that the TM domain is essential for MAVS to both self-associate and signal. Self-association of the MAVS N-terminal CARD allows for the direct binding and activation of TRAF3. Our data provide novel insight into how MAVS functions in antiviral innate immune signaling.
MATERIALS AND METHODS
Cell culture and transfections.
MAVS−/− mouse embryonic fibroblasts (MEFs) were a generous gift from Zhijian Chen and have been described previously (30). HEK293 and HEK 293T cells were from the ATCC. All cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 50 units penicillin/ml, and 50 μg streptomycin/ml (Invitrogen). Transient transfections in HEK293T cells and MEFs were performed using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Poly(I:C) was transfected at 4 μg/ml with Lipofectamine 2000. For luciferase assays, cells were cotransfected with pRL-TK Renilla luciferase (Promega) and luciferase activity was measured by a dual-luciferase reporter assay system (Promega), using Renilla luciferase as an internal control. Cells were lysed and measurements were taken 20 to 24 h following plasmid transfection.
Reagents.
Poly(I:C) was purchased from GE Healthcare Life Sciences. Rabbit anti-MAVS antibody was obtained from Bethyl. Mouse monoclonal hemagglutinin (HA) and rabbit polyclonal HA and AU1 antibodies were from Covance. FLAG M2 antibody was purchased from Sigma. PhosphoIRF3 (Ser 396) rabbit polyclonal antibodies were from Cell Signaling. AP1510 was obtained from ARIAD and used at 100 nM. Bis-maleimidohexane (BMH) cross-linker was purchased from Pierce.
Plasmids.
Expression constructs for epitope-tagged human RIG-I, MAVS, TBK1 (TANK-binding kinase 1), IRF3, and NS3/4A were constructed by PCR amplification and standard recombinant DNA techniques. FPK3 was amplified by PCR from pcDNA3-FLAG-IKKβ-FPK3 (11) and subcloned into pcDNA3. The authenticity of all constructs was confirmed by sequencing (UCLA sequencing core). Point mutations and deletion mutants were constructed by PCR mutagenesis (Stratagene). The AU1-Ub expression construct has been described previously (31). Cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) constructs were subcloned into pECFP-C1 and pEYFP-C1 (Clontech). The expression construct for MAVSΔTM includes residues 1 to 510. MAVS-TM was constructed by overlap extension PCR, fusing residues 1 to 510 of MAVS to residues 202 to 233 of Bcl-xL. MAVS CARD contains residues 1 to 98 of MAVS. MAVS CARDmt contains a substitution of alanine for tryptophan at residue 68. MAVS Q145N contains a substitution of asparagine for glutamine at residue 145 located in a TRAF2/3 consensus binding motif (25). MAVSΔTMx2 contains residues 1 to 468 of MAVS fused in tandem to itself. RIG-IΔRD contains residues 1 to 734 of RIG-I. pLUC-IFN-β, pLUC-PRD(III-I)3, and pLUC-PRD(II)2 have been described previously (7). GAL4-IRF3 and GAL4-IRF7 were constructed by inserting residues 66 to 427 of IRF3 and residues 81 to 503 of IRF7, respectively, into pPFA-dbd (Stratagene).
Immunoblotting and immunoprecipitations.
For all coimmunoprecipitation assays except for that represented in Fig. 1D, HEK293T cells were washed once with phosphate-buffered saline (PBS) (Invitrogen) and then lysed in EBC150 lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% Nonidet P-40, 50 mM NaF, 0.1 mM Na3VO4, 1 mM dithiothreitol). For the coimmunoprecipitation assay represented in Fig. 1D, cells were lysed in Triton X-100 lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 30 mM NaF, 2 mM sodium pyrophosphate, 0.1 mM Na3VO4, 1 mM dithiothreitol). All lysis buffers were supplemented with complete EDTA-free protease inhibitor cocktail (Roche). Ubiquitination of immunoprecipitated TRAF3 was performed as described previously (31).
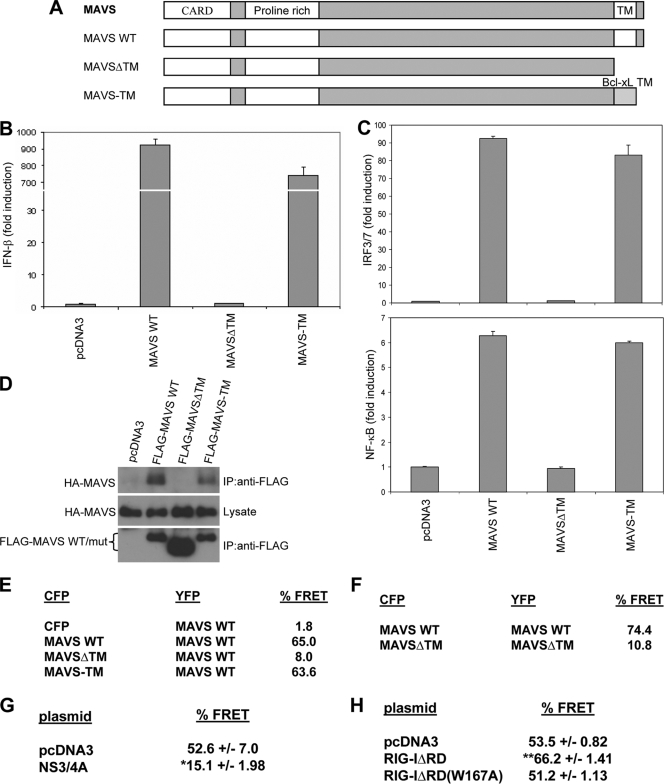
FIG. 1.
The TM domain is required for MAVS self-association. (A) Schematic representation of MAVS mutant constructs. WT, wild type. (B) Luciferase reporter assay for HEK293T cells cotransfected with wild-type or mutant MAVS and an IFN-β promoter reporter construct, pLUC-IFN-β. (C) Luciferase reporter assay for HEK293T cells cotransfected with wild-type or mutant MAVS and the pLUC-PRD(III-I)3 or pLUC-PRD(II)2 reporter construct. (D) Coimmunoprecipitation assay with lysates prepared from HEK293T cells cotransfected with FLAG-tagged wild-type or mutant (mut) MAVS and HA-tagged wild-type MAVS. Anti-FLAG immunoprecipitates (IP) or lysates were immunoblotted for HA- or FLAG-tagged MAVS constructs. (E, F) FRET analysis of MAVS−/− MEFs cotransfected with YFP, YFP-tagged wild-type MAVS, or YFP-MAVSΔTM and either CFP, CFP-tagged MAVS, or CFP-MAVSΔTM constructs. The percentage of cells that coexpressed both CFP and YFP and displayed FRET was measured by flow cytometry. (G, H) FRET analysis of MAVS−/− MEFs transfected with CFP-MAVS and YFP-MAVS along with NS3/4A, RIG-IΔRD, or RIG-IΔRD(W167A). The percentage of cells that expressed both CFP and YFP and displayed FRET was measured (mean ± standard deviation). Samples were analyzed in triplicate. *, P values of <0.01 for comparison with pcDNA3 (n = 2); **, P values of <0.0001 for comparison with pcDNA3 (n = 3).
Flow cytometry and FRET analysis.
Forty-eight hours after MAVS−/− MEFs were transfected with expression constructs for CFP and YFP proteins, cells were washed once with PBS (Invitrogen), trypsinized, and resuspended in regular growth medium. All cytometric data were collected using FACS Diva (BD Biosciences). A 405-nm laser line at 50 milliwatts and a 488-nm laser line at 20 milliwatts were used to excite CFP and YFP, respectively. CFP signals were collected using a 450/50-nm band-pass filter, and YFP signals were collected using a 560/50-nm band-pass filter. Förster resonance energy transfer (FRET) signals were measured by excitation with the 405-nm laser line and collection with the 560/50-nm band-pass filter. For vesicular stomatitis virus-green fluorescent protein (GFP) infection measurements, HEK293 cells were transfected with expression constructs and 48 h later were infected at a multiplicity of infection of 0.0001. AP1510 was added prior to infection where indicated. For infections, cells in 12-well tissue culture plates were washed once with PBS (Invitrogen) and incubated with 0.5 ml serum-free DMEM containing virus. After 1 h, 0.5 ml of DMEM supplemented with 20% fetal bovine serum, 50 units penicillin/ml, and 50 μg streptomycin/ml was added. Cells were analyzed for GFP signal by flow cytometry 24 h following infection.
Cross-linking.
Mitochondria were isolated from HEK293T cells by using a mitochondrion isolation kit according to the manufacturer's instructions (Pierce). Cells were transfected with poly(I:C) or mock treated for 8 h prior to mitochondrion isolation. Purified mitochondria were resuspended in 30 μl of PBS containing dimethyl sulfoxide or 5 mM BMH at room temperature for 30 min. Cross-linking was quenched by the addition of sodium dodecyl sulfate (SDS) sample buffer, and proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotted with anti-MAVS antibody.
Measurement of IFN-β.
IFN-β enzyme-linked immunosorbent assays were performed according to the manufacturers instructions (PBL). Supernatants were collected 36 h later, following plasmid transfection of HEK293 cells. AP1510 was added 12 h prior to analysis where indicated.
Yeast two-hybrid interaction.
MAVS and TRAF3 cDNAs were cloned into the pGBKT7 bait and pGADT7 prey vectors (Clontech). Constructs were transformed into AH109 (Clontech), and multiple colonies grown on minimal medium lacking leucine and tryptophan were streaked out onto minimal medium lacking leucine and tryptophan or leucine, tryptophan, adenine, and histidine (Clontech) and incubated at 30°C overnight.
Statistical analysis.
The statistical significance of any differences in cytokine concentration or virus infection was determined by Student's one-sided t test.
RESULTS
The MAVS TM domain mediates self-association.
To study the function of the MAVS TM domain, we first made expression constructs for a deletion mutant of MAVS lacking its C-terminal TM domain (MAVSΔTM) as well as a mutant version of MAVS with its TM domain replaced with the analogous domain from Bcl-xL (MAVS-TM) (Fig. 1A). Similar constructs were used previously to demonstrate that the TM domain of Bcl-xL could functionally substitute for the TM domain of MAVS in inducing IFN-β (28). Consistent with these previous results, we found that wild-type MAVS and MAVS-TM both activated an Ifnb promoter construct whereas MAVSΔTM was defective (28) (Fig. 1B). Similar results were found when the IRF3/7-responsive pLUC-PRD(III-I)3 or NF-κB-responsive pLUC-PRD(II)2 reporter construct was used, suggesting that the TM domain is required for MAVS to activate both IRF3/7 and NF-κB transcription factors (Fig. 1C).
We next examined whether the MAVS TM domain might be important for self-association. Previously, studies have shown a role for the TM domains of some proteins in mediating homodimeric interactions (4, 5, 13, 18, 19, 22, 29). We investigated first whether wild-type MAVS could form homooligomers in vivo. In coimmunoprecipitation assays, we found that wild-type MAVS could self-associate (Fig. 1D). To confirm these results, we used FRET analysis to examine for protein-protein interactions. We transfected MAVS−/− MEFs with both CFP- and YFP-tagged MAVS and used flow cytometry to detect FRET signals among those cells that expressed both proteins. As a negative control, we transfected MEFs with CFP and YFP-MAVS. The YFP-MAVS fusion protein was functional because it was able to rescue the sensitivity of these cells to transfection of the synthetic double-stranded-RNA analog poly(I)-poly(C) [poly(I:C)] (see Fig. S1 in the supplemental material). We found that a significant portion of cells expressing CFP-MAVS and YFP-MAVS, but not CFP and YFP-MAVS, displayed a FRET signal (65% versus 1.8%), demonstrating that MAVS homooligomerizes in live cells (Fig. 1E).
To test the importance of the MAVS TM domain in self-interactions in vivo, we first examined the abilities of wild-type MAVS, MAVSΔTM, and MAVS-TM to coimmunoprecipitate with MAVS. Unlike wild-type and MAVS-TM, MAVSΔTM failed to associate with MAVS (Fig. 1D). Furthermore, in MAVS−/− MEFs coexpressing CFP-MAVSΔTM with YFP-MAVS, the percentage of cells displaying a FRET signal was significantly lower than when CFP-MAVS or CFP-MAVS-TM was coexpressed with YFP-MAVS (8% versus 65% or 63.6%, respectively) (Fig. 1E). These results suggest that a TM domain is required for MAVS proteins to associate with wild-type, full-length MAVS.
Since the removal of the TM domain prevents MAVS from localizing to the mitochondria where wild-type MAVS resides, we also tested whether MAVSΔTM could itself self-associate (28). We found that CFP-MAVSΔTM and YFP-MAVSΔTM were defective in binding to one another in MAVS−/− MEFs (Fig. 1F). Thus, the TM domain of MAVS is necessary for its ability to self-associate. Interestingly, given the increased percentage of FRET signals in cells coexpressing CFP-MAVSΔTM with YFP-MAVS compared to the level for CFP and YFP-MAVS (8% versus 1.8%), there may be a lower affinity self-association domain distinct from the TM domain (Fig. 1E).
The HCV NS3/4A protease is known to inhibit RIG-I/MDA-5 signaling by cleaving off the TM domain from MAVS (20, 23). Consistent with a requirement for the TM domain of MAVS for self-association, we found that expression of NS3/4A interfered with the interaction between CFP-MAVS and YFP-MAVS in cells (Fig. 1G). These data together indicate the importance of the TM domain of MAVS for its ability to self-associate in vivo.
Stimulation of MAVS self-association by active RIG-I.
Next, we examined whether MAVS self-association may be regulated by the activation of RIG-I. We tested the effect of expressing RIG-IΔRD, a constitutively active form of RIG-I that is missing its C-terminal repressor domain (RD) (26), on MAVS self-association as measured by FRET. In MAVS−/− MEFs expressing both CFP-MAVS and YFP-MAVS, the percentage of cells that displayed a FRET signal was enhanced upon coexpression of RIG-IΔRD (Fig. 1H). In contrast, an inactive mutant version of RIG-IΔRD carrying an alanine substitution at a conserved tryptophan residue in the second CARD had no effect (Fig. 1H; also see Fig. S2 in the supplemental material). Thus, active RIG-I can enhance MAVS self-association.
Forced oligomerization or dimerization of MAVS can overcome the requirement for a TM domain.
Since the TM domain is important for both MAVS signaling and the ability of MAVS to self-associate, we sought to test whether this domain could be replaced functionally by a heterologous self-association domain. As such, we replaced the TM domain of MAVS with a tag containing three tandemly repeated dimerization domains of FPK (FPK3) to create MAVSΔTM-FPK3. FPK is a modified version of human FKBP12, an FK506 binding protein. Proteins containing FPK can be induced to dimerize by the addition of the cell-permeable ligand AP1510 (21). We found that fusion of FPK3 to MAVSΔTM allowed MAVSΔTM to partially regain the ability to induce the Ifnb promoter and IFN-β production in the presence of a dimerizer (Fig. 2A and B).
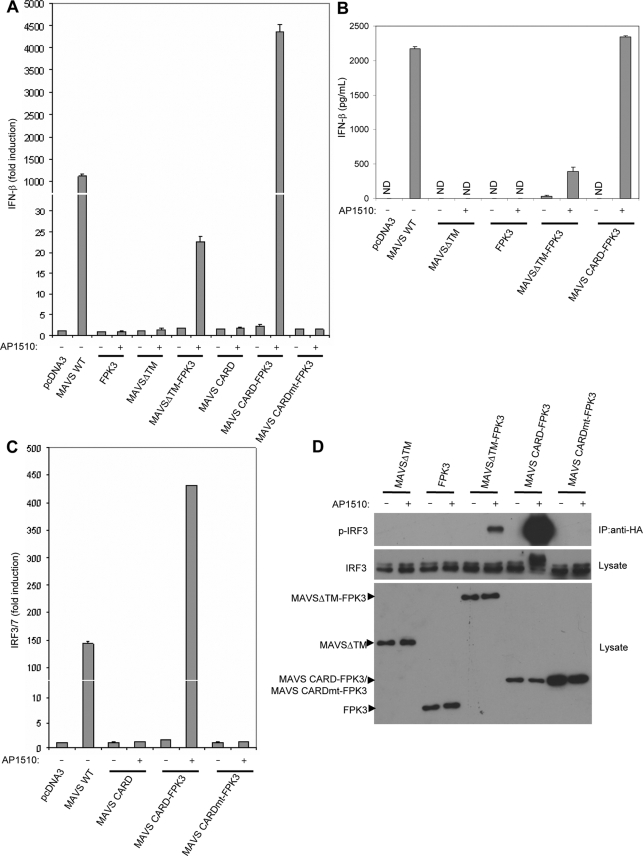
FIG. 2.
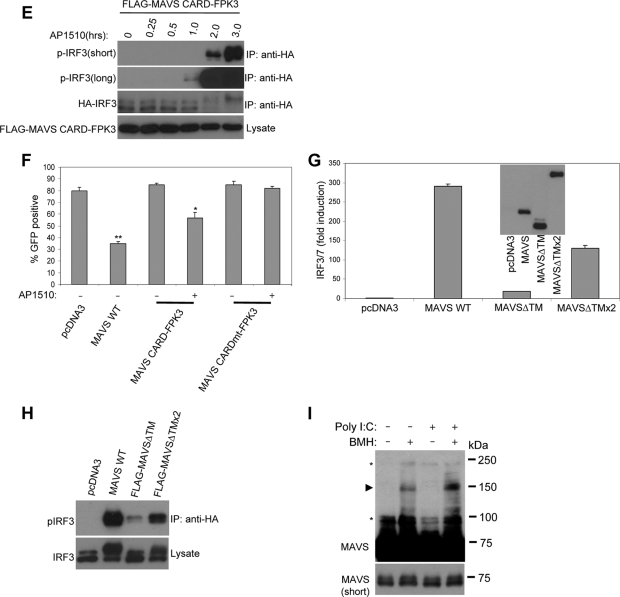
Restoration of MAVS signaling in the absence of a TM domain by induced self-association. (A) Luciferase reporter assay for HEK293T cells transfected with wild-type (WT) or mutant MAVS and an IFN-β reporter, pLUC-IFN-β. AP1510 was added 12 h prior to cell lysis. (B) Human IFN-β enzyme-linked immunosorbent assays at 36 h with tissue culture supernatants of HEK293 cells transfected with wild-type MAVS or mutant MAVS constructs. Samples were prepared in duplicate. The concentration of IFN-β is shown (mean ± standard deviation). AP1510 was added 12 h prior to analysis. ND, not detectable. (C) Luciferase reporter assay for HEK293T cells transfected with wild-type or mutant MAVS and pLuc-PRD(III-I)3. AP1510 was added 12 h prior to cell lysis. (D) Immunoblotting for phosphorylated IRF3 (Ser 396) from anti-HA immunoprecipitates (IP) prepared from lysates of HEK293T cells transfected with FLAG-tagged MAVS, FPK3, or MAVS-FPK3 fusions along with HA-IRF3. Lysates were immunoblotted for HA-IRF3 and FLAG-tagged proteins. (E) Immunoblotting for phosphorylated IRF3 (Ser 396) from anti-HA immunoprecipitates in lysates prepared from HEK293T cells transfected with FLAG-tagged MAVS CARD-FPK3. AP1510 was added prior to lysis for various times. Images representing both short and long exposures are shown. Anti-HA immunoprecipitates were also immunoblotted for HA-IRF3. Lysates were immunoblotted for MAVS CARD-FPK3. (F) Monitoring for VSV-GFP replication by flow cytometry with infected HEK293 cells transiently transfected with MAVS constructs. The percentage of cells positive for GFP is indicated (mean ± standard deviation). Virus infection was performed 48 h following transfection. AP1510 was added 12 h prior to infection. Regular culture medium was added after infection. *, P values of <0.001 for comparison with pcDNA3 (n = 3); **, P values of <0.00001 for comparison with pcDNA3 (n = 3). (G) Luciferase reporter assay for HEK293T cells cotransfected with FLAG-tagged wild-type MAVS, MAVSΔTM, or MAVSΔTMx2 and an IRF3/7 reporter, pLuc-PRD(III-I)3. Lysates were immunoblotted for FLAG-tagged proteins. (H) Immunoblotting for phosphorylated IRF3 (Ser 396) and phosphorylated IκBα (Ser 32) from anti-HA immunoprecipitates in lysates prepared from HEK293T cells transfected with FLAG-tagged wild-type MAVS, MAVSΔTM, or MAVSΔTMx2 along with HA-IRF3. Immunoprecipitates were also immunoblotted for HA-IRF3. (I) Cross-linking of presumptive MAVS homodimer from purified mitochondria isolated from HEK293T cells transfected with poly(I:C). Mitochondria were incubated with vehicle (dimethyl sulfoxide) or BMH, and total proteins were separated by SDS-PAGE and probed for endogenous MAVS. The arrowhead indicates 150-kDa presumptive MAVS homodimeric species. Asterisks indicate nonspecific reactive bands. The bottom panel shows an image representing a short exposure, demonstrating monomeric, 75-kDa full-length MAVS.
Previously, a fusion protein containing only the MAVS CARD joined to the TM domain was shown to be sufficient to activate the Ifnb promoter (28). Notably, we found that fusion of FPK3 to the MAVS CARD gave the MAVS CARD the ability to activate the Ifnb promoter and induce IFN-β production as well as wild-type MAVS in the presence of a dimerizer (Fig. 2A and B). MAVS CARD-FPK3 was able to activate IRF3/7 (Fig. 2C). The specific importance of the CARD was shown by the fact that a mutant version with an alanine substitution at a conserved tryptophan residue in the CARD (CARDmt) was functionally inactive (Fig. 2A and C).
MAVSΔTM-FPK3 and MAVS CARD-FPK3 could both also induce phosphorylation of IRF3 on Ser 396, an essential phosphoacceptor site directly targeted by TBK1 (6, 27) (Fig. 2D). The induction of IRF3 phosphorylation by MAVS CARD-FPK3 was detectable within as early as 1 hour after the addition of a dimerizer (Fig. 2E). Along with the ability to induce IFN-β, MAVS CARD-FPK3 was also able to function as an antiviral protein, like wild-type MAVS (Fig. 2F). These data suggest that enforced oligomerization of MAVS can overcome the requirement of a TM domain for antiviral signaling and that oligomerization of the CARD is sufficient.
Because FPK3 has multiple surfaces for binding to AP1510, higher-order oligomers in addition to dimers may form. To test whether dimerization was sufficient to restore the function of a MAVS protein without a TM domain, we created a tandem repeat of MAVSΔTM with two copies fused end to end (MAVSΔTMx2). This fusion displayed an enhanced ability to phosphorylate IRF3 and activate IRF3/7 (Fig. 2G and H; also see Fig. S3A in the supplemental material). Fusion of two copies of only the MAVS CARD could also partially restore function (see Fig. S3B in the supplemental material). Thus, dimerization of MAVS can restore its signaling properties in the absence of a TM domain.
Dimerization of endogenous MAVS in response to pathway activation.
In order to examine whether endogenous MAVS forms oligomers in response to RIG-I/MDA-5 activation, we isolated mitochondria from cells and treated them with the chemical cross-linker BMH. Total proteins were analyzed by SDS-PAGE, followed by immunoblotting with a MAVS antibody. To stimulate RIG-I/MDA-5 signaling, we transfected cells with poly(I:C), a double-stranded-RNA analog that stimulates MDA-5- and MAVS-dependent interferon production (8, 14, 17, 30). We found that when cells were transfected with poly(I:C), a 150-kDa-molecular-mass immunoreactive band was strongly induced in the presence of a cross-linker, a result consistent with the formation of MAVS homodimers (Fig. 2I). Interestingly, mock-treated cells displayed a slight amount of homodimer formation, a level that may be insufficient to activate downstream signaling. These results suggest that a fraction of endogenous MAVS forms homodimers in vivo in response to the activation of RIG-I/MDA-5 signaling.
TRAF3 binding to MAVS requires the TM domain and self-association.
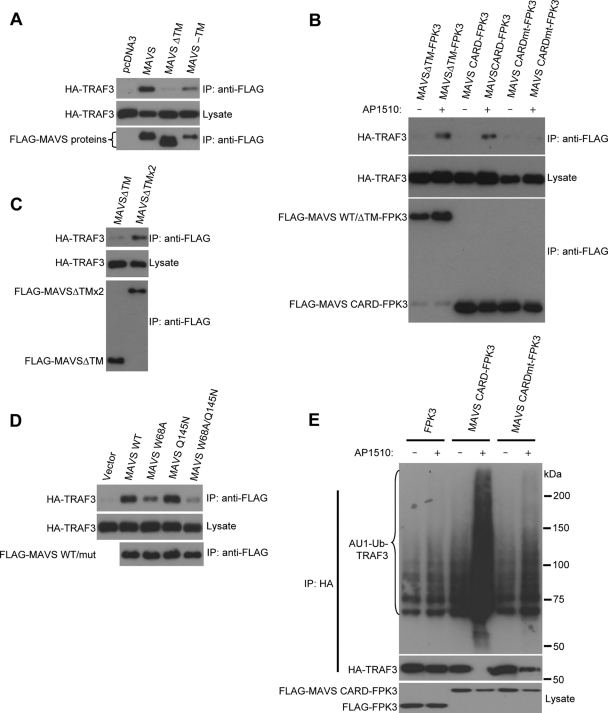
Given that TRAF3 was previously identified as an important downstream target of MAVS in RIG-I/MDA-5 signaling (25), we were interested to see whether the TM domain of MAVS was necessary for the interaction between MAVS and TRAF3. We found that wild-type MAVS and MAVS-TM, but not MAVSΔTM, coimmunoprecipitated with TRAF3, indicating that the TM domain was important for TRAF3 binding (Fig. 3A). To see if the TM-dependent binding of TRAF3 to MAVS was due to the inability of MAVSΔTM to self-associate, we tested whether TRAF3 might be able to bind to MAVSΔTM when induced to self-associate through FPK. Indeed, MAVSΔTM-FPK3 regained affinity for TRAF3 in the presence of a dimerizer (Fig. 3B). In addition, MAVSΔTMx2 displayed enhanced binding to TRAF3, demonstrating that MAVS dimerization was sufficient to partially restore TRAF3 binding (Fig. 3C). We also found that TRAF3 bound MAVS in a yeast two-hybrid interaction assay, suggesting they likely interact directly (see Fig. S4 in the supplemental material). Thus, TRAF3 binding to MAVS is dependent on a TM domain, at least in part due to the ability of the TM domain to confer on MAVS the ability to self-associate.
FIG. 3.
Self-association of MAVS allows for TRAF3 binding. (A) Coimmunoprecipitation assay with lysates prepared from HEK293T cells cotransfected with FLAG-tagged MAVS constructs together with HA-tagged TRAF3. Anti-FLAG-immunoprecipitates (IP) were immunoblotted for HA-TRAF3 or FLAG-MAVS proteins. Lysates were immunoblotted for HA-TRAF3. (B) Coimmunoprecipitation assay with lysates prepared from HEK293T cells cotransfected with FLAG-tagged MAVS constructs together with HA-tagged TRAF3. Anti-FLAG immunoprecipitates were immunoblotted for HA-TRAF3 or FLAG-MAVS proteins. Lysates were immunoblotted for HA-TRAF3. AP1510 was added 12 h prior to cell lysis. WT, wild type. (C) Coimmunoprecipitation assay with lysates prepared from HEK293T cells cotransfected with FLAG-MAVSΔTM or FLAG-MAVSΔTMx2 together with HA-tagged TRAF3. Anti-FLAG-immunoprecipitates were immunoblotted for HA-TRAF3 or FLAG-MAVS proteins. Lysates were immunoblotted for HA-TRAF3. (D) Coimmunoprecipitation assay with lysates prepared from HEK293T cells cotransfected with FLAG-tagged MAVS constructs together with HA-tagged TRAF3. Anti-FLAG immunoprecipitates were immunoblotted for HA-TRAF3 or FLAG-MAVS proteins. Lysates were immunoblotted for HA-TRAF3. mut, mutant. (E) Immunoblotting for Ub-conjugated TRAF3 from HEK293T cells cotransfected with FLAG-tagged FPK3, MAVS CARD-FPK3, or MAVS CARDmt-FPK3 along with AU1-Ub and HA-TRAF3. Anti-HA immunoprecipitates were immunoblotted for AU1-Ub conjugates or HA-TRAF3. Lysates were immunoblotted for FLAG-tagged proteins. AP1510 was added 12 h prior to cell lysis.
Self-associated MAVS requires an intact CARD to signal.
MAVS has a TRAF2/3 interaction motif (TIM) that can bind to TRAF3 in vitro (25), but the functional significance of this motif has yet to be explored. In contrast, the N-terminal CARD has been shown to be important for signaling, but its target remains unclear (15, 23, 28, 30). In order to investigate whether the TIM was required for MAVS signaling, we compared the signaling abilities of wild-type MAVS and two missense mutants. One mutant carries an alanine substitution at a conserved tryptophan residue in the CARD (W68A), and another has an asparagine substitution at a conserved glutamine residue in the TIM (Q145N). Using expression constructs for these MAVS mutants, we found that the CARD but not the TIM of MAVS was critical for the full activation of IRF3/7 (see Fig. S5A in the supplemental material). Also, the phosphorylation of IRF3 was dependent on the MAVS CARD but not the TIM (see Fig. S5B in the supplemental material). When the activity of MAVSΔTM was restored by oligomerization through FPK3, the CARD but not the TIM was also important for the ability to induce IRF3 phosphorylation (see Fig. S5C in the supplemental material). Thus, an intact CARD, but not a TIM, is required for MAVS signaling.
The MAVS CARD mediates TRAF3 binding and activation.
To see whether the CARD or TIM was important for TRAF3 association, we performed coimmunoprecipitation experiments. We found that TRAF3 binding to MAVS was diminished significantly by mutation of the CARD but not the TIM (Fig. 3D). The contribution of the MAVS TIM to TRAF3 binding was detectable only when both CARD and TIM mutations were combined. Furthermore, the MAVS CARD alone fused to FPK3, but not a mutant CARD, could bind to TRAF3 in the presence of a dimerizer, suggesting that oligomerization of the CARD alone was sufficient for binding (Fig. 3B).
Ubiquitination of TRAF3 has previously been shown to occur following virus infection and may be an indication of the activation of its Ub ligase activity (16). We found that oligomerization of the MAVS CARD but not an inactive mutant version was able to potently stimulate TRAF3 ubiquitination (Fig. 3E). These data suggest that upon self-association through the TM domain, MAVS CARD appears to directly recognize and activate TRAF3 as a downstream effector.
DISCUSSION
It is well established that the helicases RIG-I and MDA-5, as well as the adaptor molecule MAVS, play important roles in signaling pathways that contribute to the production of type I interferons in response to RNA virus infection. However, the molecular mechanisms that govern these innate immune responses remain unclear. Previous data had suggested that the C-terminal TM domain and the N-terminal CARD of MAVS were functionally important for downstream signaling, but their exact roles were unclear (28). Here, we uncover a role for the TM domain in mediating the self-association of MAVS and consequently the ability of the MAVS CARD to recognize the downstream effector TRAF3.
Our data support a role for the TM domain of MAVS in mediating self-association. A recent paper demonstrated that removal of the TM domain of MAVS impairs the ability of MAVS to oligomerize (2). Our results are in agreement with these findings and establish a critical functional role for the TM in self-association through experiments involving fusion to a heterologous dimerization domain and artificial dimerization. We also find evidence that dimerization of endogenous MAVS occurs in vivo and is enhanced in response to pathway activation. Furthermore, we provide an explanation for the functional importance of dimerization of MAVS, namely, in directly associating with and activating the downstream effector TRAF3. Our studies find that the MAVS N-terminal CARD is important for TRAF3 interaction, thus providing a signaling role for this motif not previously identified. There have been several previous examples of proteins whose homodimerization is mediated by their TM domains (4, 5, 13, 18, 19, 22, 29). Since the signaling ability of a TM domain-lacking mutant can be rescued by enforced dimerization, mitochondrial localization does not appear to be required for downstream signaling to effectors per se. Our observation that the HCV NS3/4A protease inhibits MAVS self-association provides a molecular explanation of how HCV antagonizes innate immune signaling. Also, given that the Bcl-xL TM domain can functionally replace that of MAVS, it would interesting to see if the TM domain of Bcl-xL also mediates its self-association.
We propose a model in which the binding of the RNA helicases RIG-I and MDA-5 to their specific RNA ligands induces conformational changes that expose their N-terminal CARDs for binding to MAVS. Binding of RIG-I and MDA-5 to MAVS through CARD-CARD homotypic interactions leads in some way to the stabilization and accumulation of “active” MAVS homodimers. These “active” homodimers are able to directly complex with the Ub ligase TRAF3 and activate its E3 ligase activity. Active TRAF3, in turn, may mediate the recruitment and activation of TBK1 through scaffold molecules, such as NEMO and TANK, through an unknown mechanism. Of note, since TRAF3 is not required for NF-κB activation (24) and the TM domain of MAVS is important for activating NF-κB in addition to IRF3/7 (Fig. 1C), self-association of MAVS is likely important for targeting at least one additional downstream molecule. It will be interesting to see how dimerization of MAVS relates to the function of STING/MITA, a recently uncovered new molecule in this signaling pathway (12, 32). It remains unclear whether STING/MITA interacts directly with either MAVS or RIG-I/MDA-5.
In summary, our results identify an essential role for self-association in the function of MAVS and account for the importance of its TM domain for signaling. Our findings provide new insight in the search for potential therapeutic targets for the control of viral immunity. In addition, our data predict that antiviral innate immune responses can be stimulated or impaired by inducing or impeding MAVS self-association, respectively.
Supplementary Material
Acknowledgments
We greatly appreciate the assistance of Zhijian Chen, Kun-Liang Guan, Kate Fitzgerald, and Genhong Cheng, who provided reagents. We also thank Tammy Phung and Ingrid Schmid for help with flow cytometric experiments.
This work was supported by grants from the National Institutes of Dental and Craniofacial Research (NIDCR) to C.-Y.W. E.D.T. was supported in part by a UCLA T32 Oral Health-Scientist Training Fellowship.
Footnotes
Published ahead of print on 4 February 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124783-801. [DOI] [PubMed] [Google Scholar]
- 2.Baril, M., M. E. Racine, F. Penin, and D. Lamarre. 2009. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J. Virol. 831299-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden, E. C., G. C. Sen, G. Uze, R. H. Silverman, R. M. Ransohoff, G. R. Foster, and G. R. Stark. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6975-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin, C. N., J. N. Sachs, and D. M. Engelman. 2005. Transmembrane homodimerization of receptor-like protein tyrosine phosphatases. FEBS Lett. 5793855-3858. [DOI] [PubMed] [Google Scholar]
- 5.Choi, S., E. Lee, S. Kwon, H. Park, J. Y. Yi, S. Kim, I. O. Han, Y. Yun, and E. S. Oh. 2005. Transmembrane domain-induced oligomerization is crucial for the functions of syndecan-2 and syndecan-4. J. Biol. Chem. 28042573-42579. [DOI] [PubMed] [Google Scholar]
- 6.Clément, J.-F., A. Bibeau-Poirier, S.-P. Gravel, N. Grandvaux, E. Bonneil, P. Thibault, S. Meloche, and M. J. Servant. 2008. Phosphorylation of IRF-3 on Ser 339 generates a hyperactive form of IRF-3 through regulation of dimerization and CBP association. J. Virol. 823984-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4491-496. [DOI] [PubMed] [Google Scholar]
- 8.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 1038459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiscott, J. 2007. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 18483-490. [DOI] [PubMed] [Google Scholar]
- 10.Hiscott, J., R. Lin, P. Nakhaei, and S. Paz. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 1253-56. [DOI] [PubMed] [Google Scholar]
- 11.Inohara, N., T. Koseki, J. Lin, L. del Peso, P. C. Lucas, F. F. Chen, Y. Ogura, and G. Nunez. 2000. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 27527823-27831. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa, H., and G. N. Barber. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, G., J. den Hertog, and T. Hunter. 2000. Receptor-like protein tyrosine phosphatase α homodimerizes on the cell surface. Mol. Cell. Biol. 205917-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441101-105. [DOI] [PubMed] [Google Scholar]
- 15.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6981-988. [DOI] [PubMed] [Google Scholar]
- 16.Kayagaki, N., Q. Phung, S. Chan, R. Chaudhari, C. Quan, K. M. O'Rourke, M. Eby, E. Pietras, G. Cheng, J. F. Bazan, Z. Zhang, D. Arnott, and V. M. Dixit. 2007. DUBA: a deubiquitinase that regulates type I interferon production. Science 3181628-1632. [DOI] [PubMed] [Google Scholar]
- 17.Kumar, H., T. Kawai, H. Kato, S. Sato, K. Takahashi, C. Coban, M. Yamamoto, S. Uematsu, K. J. Ishii, O. Takeuchi, and S. Akira. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 2031795-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemmon, M. A., J. M. Flanagan, H. R. Treutlein, J. Zhang, and D. M. Engelman. 1992. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry 3112719-12725. [DOI] [PubMed] [Google Scholar]
- 19.Li, R., R. Gorelik, V. Nanda, P. B. Law, J. D. Lear, W. F. DeGrado, and J. S. Bennett. 2004. Dimerization of the transmembrane domain of Integrin alphaIIb subunit in cell membranes. J. Biol. Chem. 27926666-26673. [DOI] [PubMed] [Google Scholar]
- 20.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 10217717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacCorkle, R. A., K. W. Freeman, and D. M. Spencer. 1998. Synthetic activation of caspases: artificial death switches. Proc. Natl. Acad. Sci. USA 953655-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendrola, J. M., M. B. Berger, M. C. King, and M. A. Lemmon. 2002. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 2774704-4712. [DOI] [PubMed] [Google Scholar]
- 23.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 4371167-1172. [DOI] [PubMed] [Google Scholar]
- 24.Oganesyan, G., S. K. Saha, B. Guo, J. Q. He, A. Shahangian, B. Zarnegar, A. Perry, and G. Cheng. 2006. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439208-211. [DOI] [PubMed] [Google Scholar]
- 25.Saha, S. K., E. M. Pietras, J. Q. He, J. R. Kang, S. Y. Liu, G. Oganesyan, A. Shahangian, B. Zarnegar, T. L. Shiba, Y. Wang, and G. Cheng. 2006. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 253257-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito, T., R. Hirai, Y. M. Loo, D. Owen, C. L. Johnson, S. C. Sinha, S. Akira, T. Fujita, and M. Gale, Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 104582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 2789441-9447. [DOI] [PubMed] [Google Scholar]
- 28.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122669-682. [DOI] [PubMed] [Google Scholar]
- 29.Sulistijo, E. S., T. M. Jaszewski, and K. R. MacKenzie. 2003. Sequence-specific dimerization of the transmembrane domain of the “BH3-only” protein BNIP3 in membranes and detergent. J. Biol. Chem. 27851950-51956. [DOI] [PubMed] [Google Scholar]
- 30.Sun, Q., L. Sun, H. H. Liu, X. Chen, R. B. Seth, J. Forman, and Z. J. Chen. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24633-642. [DOI] [PubMed] [Google Scholar]
- 31.Tang, E. D., C. Y. Wang, Y. Xiong, and K. L. Guan. 2003. A role for NF-kappaB essential modifier/IkappaB kinase-gamma (NEMO/IKKgamma) ubiquitination in the activation of the IkappaB kinase complex by tumor necrosis factor-alpha. J. Biol. Chem. 27837297-37305. [DOI] [PubMed] [Google Scholar]
- 32.Zhong, B., Y. Yang, S. Li, Y. Y. Wang, Y. Li, F. Diao, C. Lei, X. He, L. Zhang, P. Tien, and H. B. Shu. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29538-550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.