Abstract
The E1^E4 protein of human papillomavirus type 16 (HPV16) causes cytokeratin reorganization in the middle and upper epithelial layers and is thought to contribute to multiple facets of the virus life cycle. Although little is known as to how HPV16 E1^E4 (16E1^E4) functions are controlled following the first expression of this protein, the finding that low-risk E1^E4 proteins can be phosphorylated in vivo suggests an important role for kinases. Here, we show that 16E1^E4 is phosphorylated by cyclin-dependent kinase 1 (CDK1) and CDK2, extracellular signal-regulated kinase (ERK), protein kinase A (PKA), and PKC α, with CDK1/2 serine 32 and ERK threonine 57 phosphorylations representing the two primary events seen in cells in cycle. Interestingly, T57 phosphorylation was found to trigger a structural change in the 16E1^E4 protein that compacts the central fold region, leading to an increase in 16E1^E4 stability and overall abundance in the cell. When compared to wild-type 16E1^E4, a T57D phosphomimic was found to have greatly enhanced keratin-binding ability and an ability to modulate the binding of the unphosphorylated form, with keratin binding protecting the T57-phosphorylated form of 16E1^E4 from proteasomal degradation. In HPV16 genome-containing organotypic rafts, the T57-phosphorylated form was specifically detected in the intermediate cell layers, where productive infection occurs, suggesting that T57 phosphorylation may have a functional role at this stage of the viral life cycle. Interestingly, coexpression with 16E5 and ERK activation enhanced T57 phosphorylation, suggesting that E1^E4 and E5 may work together in vivo. Our data suggest a model in which the expression of 16E5 from the major E1^E4-E5 mRNA promotes T57 phosphorylation of E1^E4 and keratin binding, with dephosphorylation occurring following the switch to late poly(A) usage. Other forms of E1^E4, with alternative functional roles, may then increase in prevalence in the upper layers of the epithelium.
Human papillomaviruses (HPVs) are small, nonenveloped viruses with a double-stranded DNA genome of about 8 kb (27). They infect stratified epithelial cells and induce proliferative lesions ranging from benign warts to malignant carcinomas. More than 200 HPVs have so far been identified, with a subgroup of these causing cervical cancer (17, 18). HPV type 16 (HPV16) is the most prevalent of these high-risk HPV types and is responsible for over half of all cases of cervical cancer worldwide (44, 45, 57).
The HPV life cycle is tightly linked to epithelial differentiation. Infection begins in the basal epithelial cells, where the virus genome is maintained as an extrachromosomal episome at a low copy number. As the infected cells differentiate and migrate toward the epithelial surface, the productive stage of the virus life cycle begins and is marked by the expression of the E1^E4 protein and by the amplification of viral genomes. Infectious virions assemble in the upper epithelial layers and are shed from the epithelial surface with the cornified squames (19).
The E1^E4 protein is abundantly expressed in productive infections and coincides with the onset of viral genome amplification (9, 24, 42, 55). E1^E4-deficient mutants of several high-risk HPV types, as well as cottontail rabbit papillomavirus, show reduced levels of viral genome amplification, indicating an important role for E1^E4 during the productive stage of the papillomavirus life cycle (46, 48, 59, 60). E1^E4 is translated from a spliced mRNA, with the first five amino acids being encoded by the E1 open reading frame (23, 25, 47), and although the biological functions of E1^E4 have not been fully addressed, it has been shown previously that this protein associates with and reorganizes cytokeratin networks both in vivo and in vitro (22, 58). Like other high-risk E1^E4 proteins, HPV16 E1^E4 (16E1^E4) contains a conserved leucine cluster motif (LLXLL) near its N terminus that is important for cytokeratin association and a C-terminal domain that is involved in E4 multimerization and amyloid formation (20, 41, 52). The disruption of the keratin-binding motif of 16E1^E4 leads to a defect in viral DNA replication both in proliferating, poorly differentiated basal cells and in the intermediate and superficial layers, where the productive stage of the virus life cycle occurs (46). Several studies have demonstrated that the E1^E4 proteins of HPV1, HPV11, HPV16, and HPV18 can induce cell cycle arrest in G2/M, which may limit cell cycle progression and facilitate efficient replication during the productive cycle (12, 14, 15, 36). 16E1^E4 has been shown previously to colocalize with cyclin-dependent kinase 2 (CDK2)/cyclin A and CDK1/cyclin B and to sequester these proteins in the cytoplasm, to bind to proteins thought to be involved in RNA processing, and to associate with mitochondria, although the significance of these activities is still unclear (3, 13, 21, 51).
Our recent work has shown that the 16E1^E4 protein can be modified by N-terminal deletion, which removes the keratin-binding motif and stimulates the formation of E1^E4 amyloid fibers in the uppermost epithelial layers (41). The N terminus of 16E1^E4 plays an important role in maintaining the structure of the protein but is also involved in mitochondrial binding and in triggering efficient genome amplification. In the upper epithelial layers, the full-length 16E1^E4 protein associates with keratin filaments and causes their reorganization, which along with the disruption of cornified envelope formation is thought to damage the integrity of the cell and facilitate virus release. N-terminal cleavage is not the only posttranslational modification that E1^E4 undergoes, however, and for the low-risk HPV types, it appears that phosphorylation may also important. In benign lesions caused by HPV1, the extent of E1^E4 phosphorylation increases dramatically as infected cells migrate toward the epithelial surface, although the significance of this pattern is unknown (4, 33). Phosphorylation is also seen in the HPV11-infected genital epithelium and has been reported to affect the intracellular location of the E1^E4 protein (6). Despite these intriguing observations, our understanding of how phosphorylation affects E1^E4 structure and function is poorly developed, and there is currently no information as to whether such posttranslational modifications modulate high-risk E1^E4 structure and function during the virus life cycle. Given the observed effects of the high-risk E1^E4 proteins on cell proliferation, genome amplification, cell cycle progression, and the integrity of the cell, it is now important to know where the various modified forms are found.
In this study, we provide the first insight into the phosphorylation of the high-risk 16E1^E4 protein during epithelial differentiation and characterize the effect of phosphorylation on viral protein function. Four phosphorylation sites were identified, with extracellular signal-regulated kinase [ERK] phosphorylation of the residue at one of these sites, threonine 57, leading to a structural change in the central loop of the protein that greatly enhances keratin binding and the subsequent protection from proteasomal degradation. T57 phosphorylation correlated with the accumulation of the full-length protein in the cell and was apparent in the mid-epithelial layers of infected epithelial tissue following the activation of ERK. Our preliminary data suggest that E5, which is known to stimulate ERK and which is encoded on the E1^E4-E5 transcript, may modulate the activity of E1^E4.
MATERIALS AND METHODS
Cell culture.
SiHa is a cervical carcinoma cell line that constitutively expresses 16E6 and 16E7 (26). The SW13 cell line clone 2 (Cl2) was derived from a human adrenal cortex carcinoma which lacks the expression of any intermediate filament networks but contains intact microtubule and actin cytoskeleton networks (53). Clone T7K, derived from Cl2, expresses functional human keratin 8/18 networks. Both cell lines were a gift from Robert Evans, University of Colorado Health Sciences Center, Denver. These cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 1% penicillin and streptomycin. NIKS, an HPV-negative spontaneously immortalized human keratinocyte cell line (1), and primary human keratinocytes (PHK; Clonetics) were maintained at subconfluence on γ-irradiated J2 3T3 feeder cells in F medium with all supplements as described previously (30, 32). All cells were incubated at 37°C in a 5% CO2 environment. For λ phosphatase treatment, 106 SiHa cells infected with a recombinant adenovirus expressing 16E1^E4 (rAd16E1^E4) were lysed in 50 μl of 0.8% (vol/vol) Empigen. Twenty microliters of the lysate was incubated with 1× λ phosphatase buffer, 2 mM MnCl2, and 1 μl of λ phosphatase (New England Biolabs, United Kingdom) for 30 min at 30°C to allow protein dephosphorylation.
Plasmid construction, site-directed mutagenesis, and sequencing.
To generate the 16E5-expressing vector pBabe-16E5-puro, a 252-bp BamHI-EcoRI fragment containing the entire open reading frame for 16E5 was made by PCR using a plasmid, pMT3H16E5KC (a kind gift from Dan DiMaio, Department of Genetics, Yale University School of Medicine, CT), as a template and cloned into the unique BamHI-EcoRI sites in the pBabe-puro vector (43). The forward primer ATCGGGATCCCCACCATGACAAATCTTGATACTGC and the reverse primer ACTGGAATTCTTATGTAATTAAAAAGCG were used. The pET-28(b)+/16E1^E4 expression vector was made by cloning the 16E1^E4 BamHI-XhoI fragment into the pET-28(b)+ construct (Novagen, United Kingdom). The pMV11-16E1^E4 (15) and pET-28(b)+/16E1^E4 mutants were produced by PCR-based site-directed mutagenesis using the QuikChange site-directed mutagenesis kit according to the protocol of the manufacturer (Stratagene, United Kingdom). The primers used for the 16E1^E4 mutants are shown in Table 1.
TABLE 1.
Primers for mutagenesis
| Mutation | Forward primer | Reverse primer |
|---|---|---|
| S32A | CCATACCAAAGCCGGCGCCTTGGGCACCG | CGGTGCCCAAGGCGCCGGCTTTGGTATGG |
| S43A S44A | CGAAGAAACACAGACGACTAGCCGCCGACCAAGATCAGAGCCAG | CTGGCTCTGATCTTGGTCGGCGGCTAGTCGTCTGTGTTTCTTCG |
| S49A | CCAGCGACCAAGATCAGGCCCAGACACCGGAAACCCC | GGGGTTTCCGGTGTCTGGGCCTGATCTTGGTCGCTGG |
| T51A | CAAGATCAGAGCCAGGCACCGGAAACCCCTG | CAGGGGTTTCCGGTGCCTGGCTCTGATCTTG |
| T54A | CCAGACACCGGAAGCCCCTGCCACACC | GGTGTGGCAGGGGCTTCCGGTGTCTGG |
| T57A | CGGAAACCCCTGCCGCACCACTAAGTTGTTG | CAACAACTTAGTGGTGCGGCAGGGGTTTCCG |
| T57D | GACACCGGAAACCCCTGCCGACCCACTAAGTTGTTGCACAG | CTGTGCAACAACTTAGTGGGTCGGCAGGGGTTTCCGGTGTC |
All constructions were confirmed by DNA sequencing. The sequencing PCR was carried out using the BigDye Terminator cycle sequencing kit, version 1.1, according to the instructions of the manufacturer (Applied Biosystems, United Kingdom). Each reaction was performed in duplicate using the forward primer CTTGATCATATGGCTGATCCTGCAGCAGCAAC for 16E1^E4 and the forward and reverse primers described above for 16E5. The PCR fragments were sequenced using MegaBACE (GE Healthcare, United Kingdom).
Recombinant adenovirus infection and transfection with plasmids.
rAd16E1^E4 and recombinant adenovirus expressing β-galactosidase (rAdβ-Gal) have been described previously (15, 21). SiHa cells (5 × 105) were infected with rAd16E1^E4 or rAdβ-Gal at a multiplicity of infection of 100 and harvested 24 h postinfection unless otherwise stated. Transfection was performed using the Effectene transfection kit according to the protocol of the manufacturer (Qiagen, United Kingdom). For the 16E5-expressing cell line, SiHa cells were transfected with the 16E5-expressing vector pBabe-16E5-puro. A negative control cell line was also produced using pBabe-puro. Both cell lines were selected by puromycin. 16E5-positive clones were detected by reverse transcription-PCR (RT-PCR).
RNA isolation and RT-PCR.
Total RNA was extracted using the RNase Easy minikit according to the protocol of the manufacturer (Qiagen, United Kingdom). One microliter of the extracted RNA was added to 25 pmol of oligo(dT) 12- to 18-mer primer (Invitrogen, United Kingdom), 0.5 μl of 10 mM deoxynucleoside triphosphate, and 4.5 μl double-distilled water, and the mixture was incubated at 65°C for 5 min and then placed on ice for 1 min. RT was carried out by adding 2 μl of first-strand buffer (Invitrogen, United Kingdom), 0.5 μl of 0.1 M dithiothreitol, 0.5 μl of RNasin (Promega, United Kingdom), and 0.5 μl of SuperScript III reverse transcriptase (Invitrogen, United Kingdom) and incubating the mixture at 50°C for 90 min and then at 70°C for 15 min. The PCR program was 94°C for 3 min, 94°C for 45 s, 55°C for 45 s, and 72°C for 30 s, with 29 cycles. 16E5 was detected using the primers described above.
His-tagged protein preparation and in vitro kinase assays.
C-terminally hexahistidine-tagged 16E1^E4 with a leucine-glutamic acid linker (His-16E1^E4) was expressed from the pET-28(b)+/16E1^E4 expression vector in BL21 Star cells (Invitrogen, United Kingdom) as described by the manufacturer. Cells were grown at 37°C to an optical density at 600 nm of 0.6 in the presence of 50 μg/ml kanamycin before being induced by the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Growth was allowed to continue at 30°C for a further 3 h before the cells were pelleted and lysed by sonication in pH 7.0 urea buffer (8 M urea, 0.01 M Tris, 0.1 M NaH2PO4, and 25 mM complete protease inhibitor). The His-tagged proteins were bound to preequilibrated Talon metal affinity resin (BD Biosciences, United Kingdom) and washed with 10 bed volumes of the pH 7.0 urea buffer at 4°C. The His-16E1^E4 was eluted using pH 4.3 urea buffer (8 M urea, 0.01 M Tris, 0.1 M NaH2PO4) and refolded by dialyzing into phosphate-buffered saline (containing 2 mM dithiothreitol) overnight at 4°C using Slide-A-Lyzer mini-dialysis units with a 5,000-Da cutoff (Pierce, United Kingdom). Radioactive in vitro kinase assays were carried out using protein substrate His-16E1^E4, [γ-32P]ATP, and recombinant kinases according to the manufacturers' instructions. The kinases used were CaMKII, CDK1/cyclin B, CDK2/cyclin A, CKII, and ERK (New England Biolabs, United Kingdom), protein kinase A (PKA; Promega, United Kingdom), and PKC α (Calbiochem, United Kingdom). The phosphorylated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Phosphorylation was analyzed using a Storm860 phosphorimager (GE Healthcare, United Kingdom) and ImageQuant 5.0 software. When required for mass spectrometry, the kinase reactions were carried out as described above but nonradioactive ATP was used.
Two-dimensional (2D) gel electrophoresis.
Isoelectric focusing (IEF) and SDS-PAGE were carried out according to the instructions of the supply manufacturer (GE Healthcare, United Kingdom). For the first dimension, 1 to 2 μg of protein per immobilized pH gradient (IPG) strip was loaded and focused in the Multiphor II system (GE Healthcare, United Kingdom). For the second dimension, 15% acrylamide gels were used. The IPG strips were applied to the tops of the SDS-PAGE gels, and electrophoresis was performed for 1.5 h at 150 V. The pHs at different points of the IPG strip were calculated using charts (catalogue no. 18114060; GE Healthcare, United Kingdom).
Fractionation, coimmunoprecipitation, and Western blotting.
Cell fractionation (into soluble and insoluble fractions) was performed as described previously (58). For coimmunoprecipitation, 3 × 106 cells were pelleted, resuspended in 500 μl of Empigen buffer (1% [vol/vol] Empigen and 25 mM complete protease inhibitor in phosphate-buffered saline), and incubated on ice for 30 min before being spun. Fifty microliters of the supernatant was added to 150 μl of Empigen buffer, and the mixture was incubated for 1 h at 4°C with 2 μl of the anti-keratin 8/18 monoclonal antibody L2A1 (a kind gift from Bishr Omary, Palo Alto VA Medical Center and Stanford University School of Medicine, CA). Complexes were pulled down on protein G-Sepharose beads (GE Healthcare, United Kingdom) and incubated with 400 μl of NP-40 buffer containing 0.4 μg of refolded wild-type (WT) or mutant His-16E1^E4 at 4°C for 2 h. After being washed, the beads were resuspended in 25 μl of 2× Laemmli's buffer. For Western blotting, the samples were separated on SDS-15% or 10% (for active ERK) polyacrylamide gels and transferred onto Immobilon membranes (Millipore, United Kingdom) or Protran membranes (for detecting active ERK; Schleicher and Schuell Bioscience, Geneflow Ltd., United Kingdom). The primary antibodies used were anti-16E1^E4 rabbit polyclonal antibody N-Term (24), monoclonal antibody TGV402 (against 16E1^E4) (21), anti-keratin 8/18 rabbit polyclonal antibody 8592 (a kind gift from Bishr Omary), anti-active ERK (Promega, United Kingdom, or Invitrogen, United Kingdom), and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) clone 374 (Chemicon), followed by anti-rabbit (NA9340) or anti-mouse (NA931) horseradish peroxidase conjugates (Amersham Pharmacia Biotech). Western blots were developed using an ECL kit (Amersham Pharmacia Biotech).
Ferguson plot.
Ferguson plot analysis was used to distinguish a change in the size or shape of a protein from a change in the charge (29, 35). Extracts from rAd16E1^E4-infected SiHa cells were separated by SDS-PAGE with 12, 15, and 20% acrylamide concentrations. The log Rm values, where Rm is the electrophoretic mobility calculated as the distance from the top of the gel to the dye front, are plotted against the acrylamide concentrations.
FACS.
SiHa cells (5 × 107 to 7 × 107) infected with rAd16E1^E4 were fixed with 70% ethanol and stained with Alexa 488-conjugated anti-16E1^E4 antibody TVG 405 (24). Fluorescence-activated cell sorting (FACS) was performed using a DakoCytomation high-speed cell sorter (Dako UK Ltd).
Immunofluorescence and immunohistochemistry.
Immunofluorescence and immunohistochemistry analyses were performed as described previously (58). The antibodies used were anti-keratin 8/18 rabbit polyclonal antibody 8592, anti-16E1^E4 antibody TVG 405 conjugated to Alexa 488, and anti-active ERK antibody (Cell Signaling).
Peptide synthesis.
The T57-phosphorylated peptide for antibody production (PETPATPPLSC, where TP is phosphorylated T57) and the peptides for tryptophan fluorescence energy transfer measurements (T57-phosphorylated and unphosphorylated forms of amino acids 24 to 61 of 16E1^E4) were synthesized using the FastMoc solid-phase peptide synthesis method, purified by C18 reverse-phase high-performance liquid chromatography, and then freeze-dried (56). Molecular weights were confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry.
Generation of polyclonal antibody specific for T57-phosphorylated 16E1^E4.
For generating the polyclonal antibody specific for T57-phosphorylated 16E1^E4, the peptide PETPATPPLSC was conjugated with a maleimide-containing carrier protein, mariculture keyhole limpet hemocyanin, via the cysteine's free sulfhydryl (SH) group by using the Imject maleimide-activated mariculture keyhole limpet hemocyanin kit according to the protocol of the manufacturer (Pierce, United Kingdom). The efficiency of conjugation was detected by cysteine assays using Ellman's reagent (Pierce, United Kingdom). Harlan Sera-Lab (Loughborough, United Kingdom) produced the polyclonal antibody.
Fluorescence measurements.
Fluorescence measurements were made on a SPEX FluoroMax fluorimeter. Total intensity measurements were made using 295-nm excitation with emission scanned from 300 to 450 nm. To monitor the conformational changes induced upon the phosphorylation of 16E1^E4 by ERK, fluorescence emission spectra were collected at regular intervals during an in vitro ERK phosphorylation reaction. MIANS [2-(4′-maleimidylanilino)naphthalene-6-sulfonic acid, sodium salt; Molecular Probes] labeling of the 16E1^E4 peptides (amino acids 24 to 61) was carried out by reacting the peptides with a fivefold excess of the probe and separating the labeled peptide from the unlabeled probe on a PD10 column. Energy transfer from tryptophan to the MIANS label, observed at 280 nm, was studied by recording the excitation spectra of the labeled peptides and comparing them to that of the MIANS alone.
Generation of HPV16 genome-containing NIKS cell lines and organotypic raft cultures.
The plasmid containing HPV16 DNA was digested with BamHI to release the HPV16 genome from the vector. The linearized HPV16 DNA was recircularized and recovered as described previously (38). Twenty-four hours after cotransfection with recovered DNA and a blasticidin-resistant plasmid (pcDNA6; Invitrogen), the NIKS cells were selected with 6 μg/ml of blasticidin for 5 to 7 days. After being screened by PCR and Southern blotting, HPV16-positive clones were grown in organotypic raft cultures as described previously (38).
RESULTS
16E1^E4 is phosphorylated by CDK1/2, ERK, PKA, and PKC α, with ERK phosphorylation stimulating a gel shift.
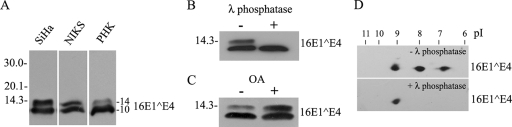
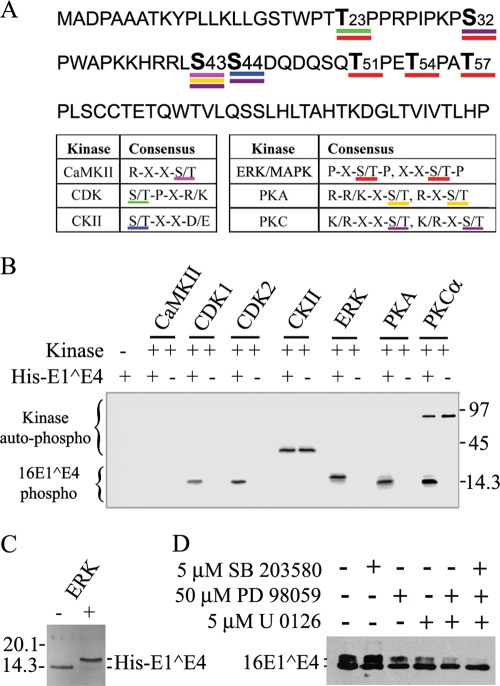
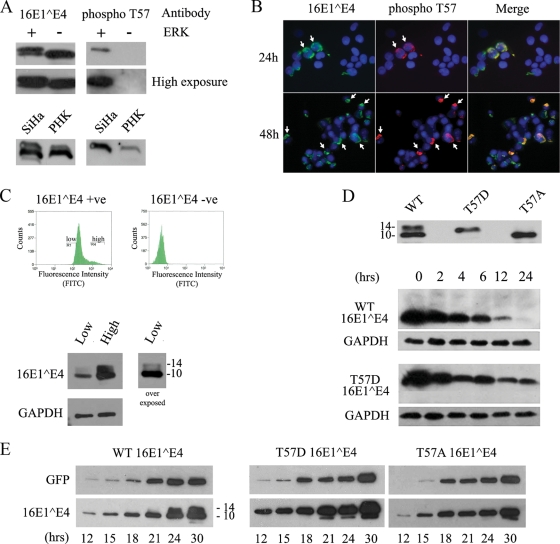
When 16E1^E4 was expressed in SiHa, NIKS, or PHK cells by infection with rAd16E1^E4 for 24 h, the protein migrated as two bands corresponding to molecular masses of 10 and 14 kDa (Fig. 1A). Treatment with λ phosphatase removed the 14-kDa band (Fig. 1B), while okadaic acid (OA), a protein serine/threonine phosphatase PP2A inhibitor, greatly enhanced it (Fig. 1C). It appears from this result that the upper band is a serine- or threonine-phosphorylated form of 16E1^E4. 2D gel electrophoresis allowed the 16E1^E4 protein to be resolved into three prominent isoelectric variants with pI values of approximately 9.2, 8.0, and 6.7, with only the most basic spot of pI 9.2 being visible following λ phosphatase treatment (Fig. 1D). When taken together, these results indicate that 16E1^E4 has multiple phosphorylation sites and can be either singly or multiply phosphorylated in monolayer cell cultures. This conclusion prompted us to examine the sites of 16E1^E4 phosphorylation and the specific kinases involved. By using NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/), ScanProsite (http://ca.expasy.org/tools/scanprosite), and protein kinase consensus site analysis, the16E1^E4 protein was found to have putative sites for CDK1/2, CKII, CaMKII, ERK/mitogen-activated protein kinase (MAPK), PKA, and PKC (Fig. 2A). In vitro kinase assays using His-16E1^E4 confirmed 16E1^E4 to be a target for CDK1, CDK2, ERK, PKA, and PKC α (Fig. 2B). No phosphorylation was apparent following incubation with CaMKII or CKII. Interestingly, ERK phosphorylation stimulated a gel shift similar to that seen following the expression of 16E1^E4 in cells. Unphosphorylated His-16E1^E4 has a molecular mass of 14.5 kDa, with ERK-phosphorylated His-16E1^E4 migrating at approximately 16 kDa (Fig. 2C).
FIG. 1.
16E1^E4 is phosphorylated in cell culture. (A) Extracts from rAd16E1^E4-infected SiHa, NIKS, and PHK cells were separated by SDS-PAGE and probed using anti-16E1^E4 antibody. The 16E1^E4 protein migrated as two bands corresponding to molecular masses of 10 and 14 kDa. Molecular mass standards (in kilodaltons) are shown to the left, and the molecular masses of 16E1^E4 bands (in kilodaltons) are shown to the right. (B) SiHa cells were infected for 24 h with rAd16E1^E4, and the cell extracts were treated with or without λ phosphatase and then analyzed by Western blotting with anti-16E1^E4 antibody. λ phosphatase treatment removed the 14-kDa band. +, with; −, without. (C) SiHa cells were infected for 24 h with rAd16E1^E4 and then treated with OA at 1 μg/ml or methanol only (as a control) for 1 h prior to harvest. The cell extracts were analyzed by Western blotting with anti-16E1^E4 antibody. OA treatment enhanced the 14-kDa band. (D) SiHa cells were infected for 24 h with rAd16E1^E4, and the cell extracts were treated with or without λ phosphatase, separated in the first dimension by IEF and in the second dimension by SDS-PAGE, and probed with anti-16E1^E4 antibody TGV402. Note that the two phosphorylated variants with pI values of 8.0 and 6.7 were removed after λ phosphatase treatment.
FIG. 2.
16E1^E4 protein is phosphorylated by multiple kinases, with ERK stimulating a gel shift. (A) Putative phosphorylation sites of 16E1^E4 based on common kinase consensus sites are shown. (B) His-16E1^E4 was used as a substrate in kinase assays with [γ-32P]ATP and CDK1, CDK2, CKII, ERK, PKA, or PKC α. Samples were separated by SDS-PAGE, and then phosphorylation was detected using a phosphorimager. His-16E1^E4 was phosphorylated by CDK1, CDK2, ERK, PKA, and PKC α. Autophosphorylation (auto-phospho) of the kinase is apparent in the CKII and PKC α lanes. Molecular mass standards (in kilodaltons) are shown to the right. 16E1^E4 phospho, phosphorylated 16E1^E4; +, present; −, absent. (C) His-16E1^E4 was incubated with or without ERK under kinase assay conditions and then analyzed by silver staining. 16E1^E4 phosphorylation by ERK caused a gel shift. Molecular mass standards (in kilodaltons) are shown to the left. (D) rAd16E1^E4-infected SiHa cells were incubated with the kinase inhibitors SB203580, PD98059, and U0126 (separately or in combination) 6 h postinfection. Cells were harvested 24 h postinfection, and extracts were analyzed by Western blotting with anti-16E1^E4 antibody. The MEK inhibitors PD98059 and/or U0126 reduced the intensity of the upper 16E1^E4 band.
To examine whether ERK could be responsible for the gel shift seen when 16E1^E4 was expressed in cells, kinase inhibitors were used. The inhibitors SB203580 for p38 MAPK and PD98059 and U0126 for MEK1 and MEK2 (upstream activators of ERK1/2) were added to the cell culture medium 6 h postinfection of SiHa cells with rAd16E1^E4, with dimethyl sulfoxide serving as a control. Cell extracts were analyzed by Western blotting after 24 h, and the results showed that the MEK inhibitors, but not the p38 inhibitor, significantly reduced the intensity of the upper 16E1^E4 band compared with the negative control (Fig. 2D). This finding suggests that phosphorylation by ERK, but not p38 MAPK, is most likely to be responsible for the gel shift.
ERK-threonine 57 and CDK1/CDK2-serine 32 represent the two predominant 16E1^E4 phosphorylation events in monolayer cell cultures.
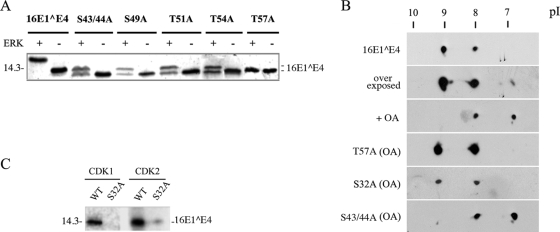
In order to examine how phosphorylation may affect 16E1^E4 structure and its possible functions, we wished to confirm the specific amino acid residues involved. For this purpose, His-16E1^E4 was purified and phosphorylated in vitro with the kinases described above (CDK1, CDK2, ERK, PKA, and PKC α) prior to analysis by matrix-assisted laser desorption ionization-time of flight mass spectrometry or nanospray quadrupole ion trap mass spectrometry coupled with collision-induced fragmentation. These techniques confirmed CDK1 phosphorylation at serine 32 (13) and revealed ERK phosphorylation to occur within a fragment encompassing residues 35 to 59 (see Fig. S1 and S2 in the supplemental material). To map this site more precisely, site-directed mutagenesis was carried out. S32 and each serine or threonine within the region of residues 35 to 59 were replaced with an alanine to yield the His-16E1^E4 mutants S32A, S43/44A, S49A, T51A, T54A, and T57A. Although mutations in this region altered the migration patterns of the His-tagged proteins relative to that of the WT form, when each mutant was tested in ERK kinase assays, only T57A had lost the ability to be phosphorylated (Fig. 3A), suggesting that ERK phosphorylation at T57 is most probably responsible for the effect on protein migration. To confirm that T57 is phosphorylated in the cell, 2D gel electrophoresis was used. Although the gel shift associated with E1^E4 phosphorylation was not apparent following the IEF step, it was clear that the two primary phosphorylation states in cell culture involve S32 and T57 (Fig. 3B). OA elevated the levels of both the singly and multiply phosphorylated forms of 16E1^E4, indicating that (as might be expected) 16E1^E4 phosphorylation is dynamic. Even in the presence of OA, however, both the 16E1^E4 mutants T57A and S32A (as a control) (13) migrated as a singly phosphorylated form, consistent with the loss of one of the two primary phosphorylation sites in each case, with S32A showing reduction of phosphorylation by both CDK1 and CDK2 (Fig. 3C). Interestingly, the T57 phosphorylation site lies in a negatively charged region on one side of the flexible loop within 16E1^E4, whereas the S32 site is in an adjacent region with an overall positive charge (see Fig. 4) (41). The S43/44A 16E1^E4 mutant, which has lost a putative PKA site and a PKC site, showed no change following 2D gel electrophoresis, suggesting that the residues at these sites are not phosphorylated in monolayer cell cultures (Fig. 3B).
FIG. 3.
The ERK-mediated gel shift is triggered by the phosphorylation of the threonine residue at consensus site position 57. (A) The His-16E^E4 WT and mutants S43/44A, S49A, T51A, T54A, and T57A were used in nonradioactive kinase assays and detected by SDS-PAGE and silver staining. Only 16E1^E4 T57A (with a mutation in the ERK/MAPK consensus site) failed to show the gel shift. A molecular mass standard (in kilodaltons) is shown to the left. +, present; −, absent. (B) 2D SDS-PAGE and Western blotting show that in the cells transfected with WT or mutant 16E1^E4, WT 16E1^E4 exists as an unphosphorylated protein (pI 9.2) and a singly phosphorylated form (pI 8). A minor multiply phosphorylated species (pI 6.7) increases in abundance in the presence of OA. In the presence of OA (PP2A inhibition), the T57A and S32A mutants, but not mutant S43/44A, failed to produce the second main spot. (C) His-16E1^E4 WT and S32A proteins were analyzed by nonradioactive kinase assays and detected by SDS-PAGE and silver staining. S32A phosphorylation by CDK1 was abolished, and that by CDK2 was clearly reduced. A molecular mass standard (in kilodaltons) is shown to the left.
FIG. 4.
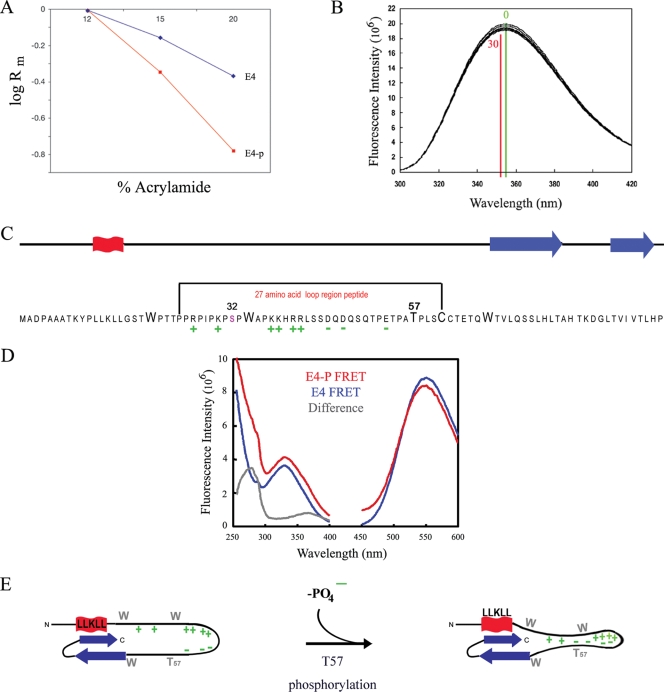
T57 phosphorylation triggers 16E1^E4 structural change. (A) Dependence of electrophoretic mobilities (Rm) of the 16E1^E4 phosphorylated (E4-p) and unphosphorylated (E4) species on the acrylamide concentration in SDS-PAGE (Ferguson plot). (B) Intrinsic fluorescence upon the phosphorylation of His-16E1^E4 by ERK was monitored. The fluorescence maxima prior to phosphorylation (time, 0 min; green) and upon the completion of phosphorylation (time, 30 min; red) are indicated on the plot. The decrease in fluorescence intensity and the red shift of the spectrum accompanying phosphorylation are consistent with the tryptophan residues becoming more buried in a hydrophobic environment. (C) The 16E1^E4 protein has elements of secondary structure at its C and N termini (red, α helix; blue, β sheet). T57 is located in a highly charged unstructured region of the protein, and its phosphorylation contributes to the polarization of this region. The effect of T57 phosphorylation on this unstructured region of the protein was assessed using a 27-amino-acid peptide as indicated. (D) The effect of T57 phosphorylation on the structure of the peptide (indicated in panel C) was monitored by recording fluorescent resonance energy transfer (FRET) between the tryptophan residue and a fluorescence acceptor molecule on the terminal cysteine. The excitation (250- to 400-nm) and emission (450- to 600-nm) spectra of the phosphorylated (E4-P) and unphosphorylated (E4) peptides are shown. The peak at 280 nm in the difference spectrum reveals that fluorescence transfer is observed only in the phosphorylated peptide, which indicates that T57 phosphorylation compacts the peptide. (E) Schematic summarizing the effect of T57 phosphorylation by ERK on the structure of 16E1^E4. Upon phosphorylation, the addition of a negative charge on T57 increases the charge polarization in the loop region of the protein and, as indicated by the findings of the tryptophan fluorescence studies, results in additional restraints in this region.
Threonine 57 phosphorylation by ERK drives a structural change within 16E1^E4 that increases the negative charge and compacts the central loop of the protein.
The slower migration of the T57-phosphorylated form of 16E1^E4 than of the unphosphorylated form on SDS-polyacrylamide gels may be due to a change in the electrostatic charge of 16E1^E4 (resulting in altered SDS binding) or a conformational change in the protein structure. To distinguish between these two possibilities, we looked at the migration of the upper (T57-phosphorylated) and lower (non-T57-phosphorylated) bands of 16E1^E4 (from rAd16E1^E4-infected SiHa cell extracts) as a function of the acrylamide concentration in SDS-PAGE (Fig. 4A). With increasing acrylamide concentrations, the decrease in the mobility of the upper band was much more pronounced than that in the mobility of the lower band. If the different mobility states of the upper and lower bands of 16E1^E4 were due simply to charge, then they would be independent of the acrylamide concentration. These data indicate that T57 phosphorylation of 16E1^E4 by ERK causes a structural change which increases the effective Stokes radius of 16E1^E4. To determine if this conformational change occurs under more physiological conditions, we have studied the intrinsic tryptophan fluorescence of 16E1^E4. Tryptophan fluorescence was monitored during His-16E1^E4 phosphorylation by ERK in vitro to determine if the phosphorylation-induced conformational change affects the tryptophan residues. The decrease in total tryptophan fluorescence, combined with a red shift in the spectrum upon phosphorylation (Fig. 4B), indicates that the tryptophan residues become more buried upon phosphorylation, suggesting a more compact structure with a decreased Stokes radius. The tryptophan residues and T57 are located within an unstructured and highly charged region of E1^E4 (Fig. 4C), which in the context of the folded protein forms a loop linking the secondary structure elements at the N and C termini (41). The charge polarization of this region is thought to contribute to the maintenance of the loop structure in the folded protein (41). It is anticipated that phosphorylation at T57 may enhance this effect by reinforcing the charge differential, thereby forming a more compact structure. This prediction would explain the observation that the tryptophan residues are more buried in the phosphorylated protein. To assess the effect of phosphorylation on this highly charged loop region of the protein, we used two peptides corresponding to the T57-phosphorylated and unphosphorylated forms of residues 24 to 61 of 16E1^E4, as indicated in Fig. 4C. The C-terminal cysteine (C61 in the context of the full-length protein) of each peptide was labeled with a fluorescence acceptor (MIANS), and the contribution of the tryptophan residue (W34 in the context of the full-length protein) to the fluorescence excitation spectrum of this label was measured (Fig. 4D). The difference excitation spectrum reveals a peak at 280 nm for the phosphorylated peptide, indicating that the tryptophan (W34) and the MIANS label (C61) are in closer proximity in the phosphorylated form of the peptide than in the unphosphorylated form. Taken together, these data indicate that the introduction of a negative charge on T57 by ERK phosphorylation reinforces restraints in the loop region of the protein, forming a more compact structure (Fig. 4E). This structural change explains the retarded migration of ERK-phosphorylated E1^E4 on SDS-PAGE gels and suggests a way in which phosphorylation may affect E1^E4 function and stability, as discussed below.
16E1^E4 T57 phosphorylation is associated with increased protein stability and the accumulation of full-length 16E1^E4 protein in the cell.
The observation that T57 phosphorylation can affect 16E1^E4 structure prompted us to examine the functional consequences of this modification. To investigate this issue, an antipeptide antibody specific for the T57-phosphorylated form of 16E1^E4 was prepared in rabbits (Fig. 5A, upper panel) and used to probe 16E1^E4 expressed in SiHa or PHK cells by Western blotting. This antibody picked up only the slower-migrating band of 16E1^E4, confirming that the slower-migrating band is the T57-phosphorylated form of 16E1^E4. When the antibody was used for immunofluorescence analysis of SiHa cells expressing 16E1^E4, cells were fixed for 5 min in 5% formaldehyde and stained either directly (Fig. 5B) or after epitope exposure by microwave treatment. Analysis of the immunofluorescence results suggested that in each case, the anti-phospho-T57 antibody stained only a subset of 16E1^E4-expressing cells and that the stained cells typically showed the most extensive and/or intense staining with TVG 405 (Fig. 5B). As immunofluorescence intensity is difficult to quantify visually, we decided to examine this pattern further using a FACS approach. To do this, cells expressing high and low levels of E1^E4 were separated and analyzed by Western blotting. As shown in Fig. 5C, the population with high-level staining separated by FACS did indeed show a higher level of E1^E4 expression than the population with low-level staining. More importantly, however, the ratio of the T57-phosphorylated (slower-migrating) form to the non-T57-phosphorylated form was increased in cells containing the more abundant 16E1^E4 (Fig. 5C). One interpretation of this finding is that T57 phosphorylation affects the stability of the full-length protein, causing the accumulation of the 16E1^E4 protein in the cell following the activation of ERK. To test this hypothesis, two 16E1^E4 mutants, T57A and T57D (a T57 phosphomimic), were used. When expressed in SiHa cells, 16E1^E4 T57D migrated more slowly than the T57A form, with the two mutants showing a migration difference similar to that seen following ERK phosphorylation of the WT 16E1^E4 protein (Fig. 5D, top panel). To examine the effect of T57 phosphorylation on protein stability, 16E1^E4-expressing cells were treated with cycloheximide to inhibit protein synthesis 24 h posttransfection and the soluble fraction, which contains soluble keratin-associated E4, and the insoluble fraction, which contains keratin network-bound E4 (58), were analyzed by Western blotting over a 24-h period. No significant difference among the WT and mutant proteins in the insoluble fraction was found (data not shown), whereas in the soluble fraction, T57D appeared to be more stable than the WT and T57A relative to GAPDH loading controls and could still be detected in the cell 24 h after the initiation of the block (Fig. 5D). WT 16E1^E4 and the T57A mutant (data not shown) were consistently absent at this time point in repeated experiments, indicating a role of ERK phosphorylation in regulating 16E1^E4 stability in the cell. To look at this issue further, the rate of 16E1^E4 accumulation following the first synthesis of the protein in the cell was also examined. 16E1^E4 (WT, T57D, or T57A) was expressed following cotransfection with a green fluorescent protein (GFP) marker as a transfection control, and the relative abundance of total E1^E4 between 12 and 36 h posttransfection was measured. The overall trend was similar to that seen in the cycloheximide experiments with regard to protein stability and abundance, with T57D 16E1^E4 accumulating to higher levels than WT 16E1^E4 and the T57A mutant at early time points. When taken together, these results suggest that T57 phosphorylation may increase the accumulation of the full-length protein in the cell and reduce its turnover, both of which would increase the level of full-length E1^E4 protein.
FIG. 5.
T57 phosphorylation increases the stability and abundance of full-length 16E1^E4 in the cell. (A) His-16E1^E4 was incubated at 30°C in the presence (+) or absence (−) of ERK for 1 h and analyzed by Western blotting using antibodies to 16E1^E4 or to T57-phosphorylated 16E1^E4 (phospho T57). High- and low-level exposures of these blots are shown, demonstrating that the phosphospecific antibody did not detect unphosphorylated E1^E4 (upper panels). SiHa or PHK cells were infected with rAd16E1^E4 for 24 h, and cell lysates were analyzed by Western blotting using antibodies to 16E1^E4 or to T57-phosphorylated 16E1^E4. The antibody to T57-phosphorylated 16E1^E4 detects only the slower-migrating band, confirming that this band is the T57-phosphorylated form of 16E1^E4 (lower panels). (B) SiHa cells were infected with rAd16E1^E4 for 24 or 48 h, fixed with 5% formaldehyde, and triple stained with antibodies to total 16E1^E4 (green) and T57-phosphorylated 16E1^E4 (red) and with DAPI (4′,6-diamidino-2-phenylindole; blue). T57-phosphorylated 16E1^E4 appeared only in cells containing abundant E1^E4 (shown by arrows). Images were captured using a 20× objective. (C) SiHa cells were infected with rAd16E1^E4 for 24 h, and then the SiHa cells expressing high and low levels of E1^E4 were separated by FACS and analyzed by Western blotting with antibodies against 16E1^E4 and GAPDH (as a loading control). Fluorescence intensity correlated with protein abundance and the presence of the T57-phosphorylated (slower-migrating) E1^E4 form. The molecular masses of 16E^E4 forms (in kilodaltons) are shown to the right of the lower panel. +ve, positive; −ve, negative; FITC, fluorescein isothiocyanate. (D, top panel) SiHa cells were transfected with a 16E1^E4 WT-, T57A-, or T57D-expressing plasmid for 24 h prior to SDS-PAGE and Western blotting to reveal the different migration patterns. The molecular masses of 16E1^E4 forms (in kilodaltons) are shown to the left. (Bottom panels) SiHa cells were transfected with a 16E1^E4 WT- or T57D-expressing plasmid for 24 h before being treated with 40 μg/ml cycloheximide for 0, 2, 4, 6, 12, or 24 h prior to harvest. Equal volumes of the soluble protein fraction were analyzed by Western blotting to show the increased stability of 16E1^E4 T57D relative to those of the non-T57-phosphorylated WT 16E1^E4 and the GAPDH loading controls. The results shown are typical of results from triplicate experiments. (E) SiHa cells were cotransfected with a plasmid expressing either T57A, T57D, or WT 16E1^E4 and the GFP-expressing plasmid pXJ-GFP (as a transfection control). Cells were harvested at multiple time points posttransfection, as indicated. Total extracts were run on SDS-polyacrylamide gels and analyzed with antibodies against GFP and 16E1^E4 to show the accumulation of 16E1^E4 in the cell. The time course shows the more rapid accumulation of the T57D (phosphomimic) form than of the WT 16E1^E4 (which lacks a T57-phosphorylated form before the 24-h time point) and the 16E1^E4 T57A mutant (which lacks the T57 phosphorylation site). The results shown are typical of results from triplicate experiments. The molecular masses of 16E1^E4 forms (in kilodaltons) are shown to the right (left panel).
16E1^E4 T57 phosphorylation stimulates enhanced keratin binding, which prevents intracellular proteasomal degradation.
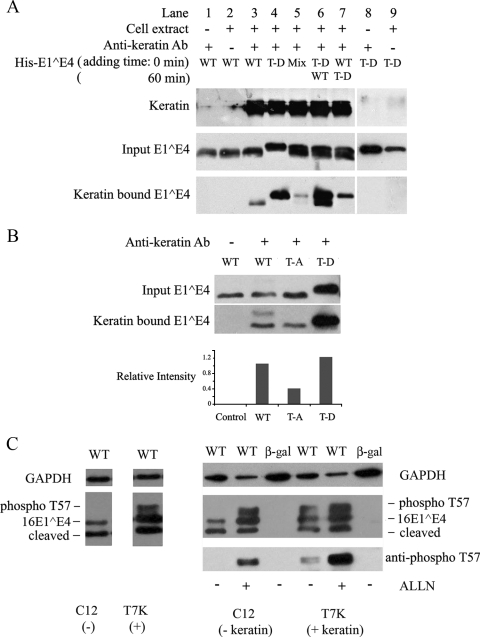
E4 stability can be enhanced by N-terminal loss, which stimulates the formation of amyloid fibers in the upper epithelial layers of differentiating cells (41). In SiHa cells and monolayer NIKS cells (unlike differentiating rafts and Cos7 cells), however, N-terminal cleavage is very limited and not generally observed except when 16E1^E4 becomes abundant (Fig. 5E, late time points). In fact, T57 phosphorylation appears to actively inhibit N-terminal cleavage in in vitro assays (P. McIntosh and S. Hinz, unpublished data). Given that T57 phosphorylation appears to mediate the N-terminal cleavage of the full-length 16E1^E4 protein and that the slower-migrating (ERK-phosphorylated) form is present only in the insoluble cellular fraction (22, 58), we suspected that the observed effects on stability may reflect changes in keratin binding. To examine this possibility, an in vitro keratin-binding assay was used. Keratins 8 and 18 were first immunoprecipitated from SiHa cells by using the monoclonal antibody L2A1 and protein G-Sepharose beads (Fig. 6A). The keratin-bound beads were then incubated with unphosphorylated WT His-16E1^E4 or the phosphomimic T57D His-16E1^E4. The bound proteins were detected by SDS-PAGE and Western blotting using the anti-16E1^E4 antibody TVG402. As shown in Fig. 6A, both His-16E1^E4 proteins (the WT and T57D) could associate with the immunoprecipitated keratins but the T57D mutant bound much more strongly (lanes 3 and 4). The results shown are typical of those from replicate experiments. Interestingly, when keratin-bound beads were incubated with premixed WT (unphosphorylated) His-16E1^E4 and T57D His-16E1^E4, the level of binding of T57D to keratin was much lower than that of unmixed T57D and only a very faint WT His-16E1^E4 band was detected (Fig. 6A, lane 5). When immunoprecipitated keratins 8 and 18 were incubated with T57D His-16E1^E4 first and then WT His-16E1^E4 was added 1 h later, both T57D and WT His-16E1^E4 proteins associated strongly with keratin (Fig. 6A, lane 6). In contrast, when keratins 8 and 18 were incubated with WT His-16E1^E4 first and T57D was added 1 h later, only the T57D band, with lower intensity than that observed when keratins were incubated with T57D first, was seen (Fig. 6A, lane 7). These results suggest that T57 phosphorylation may facilitate the binding of 16E1^E4 to keratins and the formation of a tripartite complex that includes the non-T57-phosphorylated form. Unphosphorylated 16E1^E4 binds keratin less well than phosphorylated 16E1^E4 and, under some circumstances (Fig. 6A, lanes 5 and 7), can suppress the binding of the T57D phosphomimic. To examine the association in the cell, coimmunoprecipitation was carried out using cells expressing WT 16E1^E4, 16E1^E4 T57A, or T57D (Fig. 6B). The levels of the different E1^E4 forms present in the input tracks reflect the relative abundances of the different E4 forms in the cell (as described above). To examine whether these 16E1^E4 variants do indeed show differential keratin binding, an anti-keratin 8/18 antibody was used to coimmunoprecipitate keratins along with the associated E1^E4 proteins. Relative to the input levels, 16E1^E4 T57D associated most effectively, with 16E1^E4 T57A associating less well (Fig. 6B). Interestingly, the slower-migrating band shown in Fig. 5A and confirmed to be the T57-phosphorylated form of 16E1^E4 was enriched in the keratin-bound pool from the soluble fraction of cells expressing WT 16E1^E4 (Fig. 6B, second lane), which is not normally seen in this fraction, in accordance with the enhanced keratin-binding ability outlined herein. This phosphorylated form may stimulate the binding of the unphosphorylated form, as suggested by the results shown in Fig. 6A (lane 6). The T57A (phosphorylation-deficient) mutant indicates what might happen when only the pure unphosphorylated form of 16E1^E4 is present (Fig. 6B, third lane). To establish whether the increased 16E1^E4 stability associated with T57 phosphorylation reflects the enhanced ability to bind to keratins, WT 16E1^E4 was expressed in the isogenic cell line pair Cl2 and T7K; these cell lines differ from each other only in the presence of a cytokeratin network (Fig. 6C). In Cl2 cells, which lack a cytokeratin network, no T57-phosphorylated form of 16E1^E4 was present, whereas in the T7K line (which contains a keratin 8/18 network), the T57-phosphorylated form was detectable. The role of keratins in stabilizing the T57-phosphorylated form was apparent following the addition of the proteasome inhibitor acetyl-leucyl-leucyl-norleucinal (ALLN), which restored the T57-phosphorylated form in C12 cells to levels similar to those seen in the T7K cell line. Proteasome inhibition also increased the level of the non-T57-phosphorylated 16E1^E4 slightly, although not as dramatically as that of the T57-phosphorylated form, but had no effect on the N-terminally cleaved form which is detectable in these cells. This result is compatible with the findings in a recent report (41) suggesting that this form is present as stable amyloid fibers that are no longer accessible to proteasomal degradation.
FIG. 6.
16E1^E4 T57D binds keratin more strongly than the 16E1^E4 WT and is protected from proteasomal degradation. (A) Keratins immunoprecipitated from Empigen-soluble fractions of SiHa cells were incubated with 0.4 μg of the His-16E1^E4 WT, His-16E1^E4 T57D (T-D), or a mixture of the two forms. Control immunoprecipitations were carried out in the absence of either cell extract (lane 8) or antikeratin antibody (lane 9). For lanes 6 and 7, T57D and WT His-16E1^E4, respectively, were preincubated with keratin (time point, 0 min) and then the WT (lane 6) and T57D (lane 7) were added 1 h later (time point, 60 min). All samples were analyzed by Western blotting using anti-16E1^E4 antibody and antikeratin antibody (Ab). The T57D form of 16E1^E4 showed greatly enhanced keratin binding compared to that of the unphosphorylated WT 16E1^E4 protein. +, present; −, absent. (B) Empigen-soluble extracts from WT, T57A (T-A), or T57D E1^E4-expressing SiHa cells were immunoprecipitated with a mouse monoclonal antikeratin antibody (L2A1) and analyzed by Western blotting with rabbit polyclonal antibodies against 16E1^E4. T57D was more effectively immunoprecipitated than T57A 16E1^E4 and was enriched compared to the input. The relative intensities of keratin-bound 16E1^E4 WT, T57A, and T57D proteins versus those of the respective inputs are shown in the histogram. (C) An isogenic cell line pair either expressing (T7K) or not expressing (Cl2) keratins (8/18) were infected with rAd16E1^E4 for 24 h. E1^E4 expression was examined by Western blotting, with GAPDH levels being used as a loading control (left panel). The T57-phosphorylated form of 16E1^E4 (phospho T57) is observed only in the keratin-containing cells. The right panel shows results for cells infected with either rAdE1^E4 (WT) or rAdβ-Gal (β-gal) in the presence of the proteasome inhibitor ALLN (10 μM; Calbiochem) for 16 h preharvest. The phosphorylated form of 16E1^E4 was restored in the keratin-negative cell line Cl2 (right panel, second lane) and enhanced in the keratin-positive cell line T7K (right panel, fifth lane) following ALLN treatment.
T57-phosphorylated 16E1^E4 and activated ERK are observed in the intermediate layers in HPV16 genome-containing raft tissues.
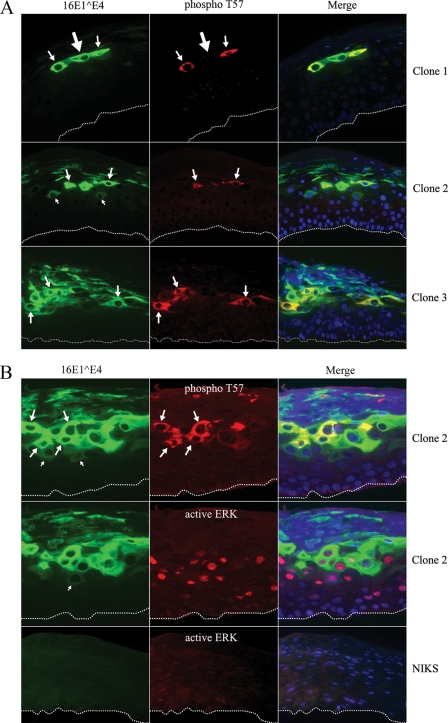
The proposed role of ERK-mediated phosphorylation of 16E1^E4 in facilitating effective keratin binding through structural modification predicts that during the papillomavirus life cycle, T57 phosphorylation would occur soon after E1^E4 synthesis in cells that contain active ERK. As 16E1^E4 is known to be first expressed in cells that are still in cycle, we wished to examine whether this is indeed the case. To investigate this prediction, E4 phosphorylation in HPV16-containing rafts was evaluated by immunofluorescence analysis using phosphospecific antibodies and by Western blotting using cell extracts prepared from raft material. 2D gel electrophoresis showed that 16E1^E4, like that in monolayer cells, undergoes at least two predominant phosphorylation events during epithelial differentiation. Western blotting showed the T57-phosphorylated form to be only a minor species, however, suggesting that the protein may be only transiently phosphorylated at T57 during productive infection (data not shown). This possibility was supported by the findings of immunofluorescence experiments, which revealed T57 phosphorylation to be restricted to a narrow strip of cells in the intermediate epithelial layers of HPV16 rafts (Fig. 7A). By using TVG 405 and polyclonal 16E1^E4 antibodies, the E4 protein can be seen first as a poorly staining cytoplasmic gene product (Fig. 7). T57 phosphorylation occurs in the epithelial layers immediately above the cells with this poorly staining product (Fig. 7) and appears to correlate with 16E1^E4 accumulation as the cell migrates toward the epithelial surface. The ability to detect the T57 phosphoepitope does not depend on E4 abundance, however (Fig. 7A, top panels), and the T57-phosphorylated form of 16E1^E4 did not appear to persist to the epithelial surface (Fig. 7A, middle and bottom panels), suggesting a possible role for phosphatases as seen in monolayer cell cultures. Abundant E1^E4 expression appears to persist following the loss of phospho-T57 staining, but it is not known at present whether this expression is that of N-terminally cleaved E1^E4 or whether the 16E1^E4 protein is additionally phosphorylated by CDK or PKC (during progression into G2 and during differentiation) as might be expected. To examine the timing of ERK activation and 16E1^E4 phosphorylation in HPV16 rafts, adjacent sections were double stained with TVG 405 and anti-16E1^E4 phospho-T57 or with TVG 405 and an antibody that recognizes activated ERK (anti-active ERK). As expected, neither E1^E4 nor active ERK was apparent in rafts that did not contain HPV16 (Fig. 7B, bottom panels), indicating the importance of HPV infection in activating ERK during epithelial differentiation. Although activated ERK had a predominantly nuclear distribution, shown in Fig. 7B, it is known that ERK is activated in the cytoplasm and then shuttled to the nucleus. Activated ERK has been shown previously to phosphorylate cytoplasmic, membranous, and nuclear substrates (31, 54, 61). As shown in Fig. 7B, activated ERK appeared to precede the first appearance of E1^E4 by one or two cell layers. During epithelial differentiation, it seems that 16E1^E4 first becomes apparent in cells that stain positively for ERK, with T57 phosphorylation becoming apparent in the cell layers immediately above. Not every ERK-positive cell appears to support E1^E4 expression, however, and in those that do, the extent of phospho-T57 staining diminishes rapidly following the loss of active ERK. This staining pattern is compatible with the hypothesis that ERK activation and the effects on viral proteins such as E1^E4 may be confined to a short period of time as the cell moves into S phase during genome amplification. The loss of active ERK and staining for 16E1^E4 phospho-T57 supports the idea that the cells then progress through G2 and succumb to differentiation.
FIG. 7.
T57 of 16E1^E4 is phosphorylated in the intermediate layers in HPV16-containing raft tissue. (A) Organotypic raft cultures from three different HPV16-containing NIKS cell lines (clones 1, 2, and 3) were triple stained for total 16E1^E4 (green), T57-phosphorylated 16E1^E4 (phospho T57; red), and DNA (blue). The cells showing weak cytoplasmic staining of 16E1^E4 are indicated by the small arrows. The cells showing T57 phosphorylation correlating with 16E1^E4 accumulation are indicated by the medium-sized arrows. The cell with abundant 16E1^E4 staining but no detectable T57-phosphorylated E1^E4 is indicated by the large arrow. The dotted lines indicate the position of the basal layer. The images were taken using a 20× objective. (B) Raft cultures from HPV16-containing NIKS cell line clone 2 or HPV16-negative NIKS cells were triple stained with antibodies against total 16E1^E4 (green), T57-phosphorylated 16E1^E4 (red in the upper row), or active ERK (red in the middle and bottom rows) and DNA (blue). The cells showing weak cytoplasmic staining of 16E1^E4 are indicated by the small arrows. The cells showing T57 phosphorylation correlating with 16E1^E4 accumulation are indicated by the medium-sized arrows. The dotted lines indicate the position of the basal layer. The images were taken using a 20× objective.
16E5 enhances ERK-mediated phosphorylation of 16E1^E4 in epithelial cells.
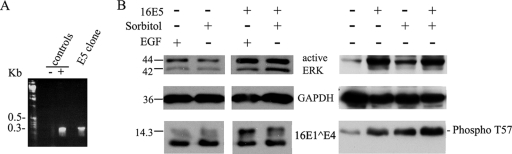
It appears from the above data that HPV16 infection activates ERK in differentiating epithelial cells. Although this effect may be due to E6 and E7, it is generally thought that the expression of these proteins during HPV16 infection extends throughout the epithelial layers rather than just the differentiating layers. The expression of E5 is, in contrast, linked to that of E1^E4 during productive infection, as the two proteins are encoded by the same abundant bicistronic transcript (5, 25). One of the best-characterized biological effects of 16E5 is ERK activation (10, 11), prompting us to consider whether 16E5 could affect 16E1^E4 phosphorylation. To test this possibility, a 16E5-expressing SiHa cell line was established by transfection with an E5-expressing plasmid, pBabe-16E5-puro. By RT-PCR, E5-encoding mRNA was detected in this cell line but not in the SiHa cells transfected with an empty vector (Fig. 8A). 16E5 has been shown previously to activate ERK in the presence of epidermal growth factor (EGF) or sorbitol (10, 11). Therefore, 24 h postinfection with rAd16E1^E4, SiHa cells were incubated with 10 ng/ml EGF or 0.5 M sorbitol for 10 min. Cell extracts were analyzed by Western blotting with anti-active ERK antibody and antibodies against 16E1^E4 or GAPDH (Fig. 8B). Levels of active ERK in the E5-positive cells were higher than those in the E5-negative cells. T57 phosphorylation of 16E1^E4 was also increased in the E5-positive cells, as shown by an increase in the intensity of the upper band in relation to that of the lower band in the Western blot (Fig. 8B, left panel). Interestingly, when examining the ERK activation and T57 phosphorylation of 16E1^E4 in 16E5- and 16E1^E4-coexpressing cells in the absence of sorbitol or EGF treatment, we found that the presence of 16E5 strongly increased ERK activity and stimulated T57 phosphorylation of 16E1^E4 (Fig. 8B, right panel). The results of these preliminary experiments suggest a potential mechanism by which 16E1^E4 phosphorylation is driven following the activation of the p670 promoter and the expression of E1^E4-E5 mRNA.
FIG. 8.
Effect of 16E5 on ERK and 16E1^E4 phosphorylation. (A) RT-PCR was carried out with a 16E5-expressing SiHa cell line to detect the presence of E5-encoding mRNA. SiHa cells transfected with empty plasmid were used as the negative control. For the positive control, the plasmid pMT3H16E5KC was used as the template for the PCR. (B) E5-positive or E5-negative SiHa cells were infected with rAd16E1^E4 for 24 h and incubated with 10 ng/ml EGF or 0.5 M sorbitol for 10 min prior to harvest. Cell extracts with (+) or without (−) EGF or sorbitol treatment were analyzed by Western blotting with antibodies against active ERK, GAPDH, 16E1^E4, or T57-phosphorylated 16E1^E4 (phospho T57). Molecular mass standards (in kilodaltons) are shown to the left.
DISCUSSION
Although phosphorylation has been shown previously to be important in regulating the functions of HPV proteins (2, 4, 6, 33, 37, 39, 40), little is known as to how such modifications influence events during productive infection in epithelial cells. Phosphoepitope antibodies were used here to establish the timing of 16E1^E4 threonine 57 phosphorylation by ERK during the infectious cycle, while biophysical and functional analyses were used to demonstrate a role for this posttranslational modification in compacting 16E1^E4 structure and enhancing protein stability and keratin binding. The activation of ERK and the phosphorylation of E1^E4 at T57 occur at a defined stage during the HPV16 productive cycle in the mid-epithelial layers, where genome amplification is also triggered (14, 42). It is interesting, therefore, that E1 is also a target of ERK, with phosphorylation stimulating E1 nuclear accumulation and enhancing viral replication (16, 62).
During productive papillomavirus infection, the 16E1^E4 protein is expressed from a transcriptional unit located downstream of the differentiation-dependent p670 promoter, which also directs the expression of transcripts encoding E1, E2, and E5. The E1 and E1^E4 transcripts share their first six codons (49), with splice site selection regulating the relative abundances of the two messages. This organizational strategy has previously suggested a link between the functions of E1 and E1^E4, which is supported by the findings of recent studies showing that E1^E4 loss compromises the efficiency of genome amplification (46, 48, 59, 60). The changes in 16E1^E4 structure and keratin binding mediated by ERK are likely to complement the ERK-mediated changes in E1 (and possibly also E2) during genome amplification and to contribute to the reorganization of keratin structure that ultimately results from 16E1^E4 accumulation. The observation that both E1 and E1^E4 can be functionally modified by ERK suggests that kinase activation may coordinate multiple effects in the virus life cycle (13; this study). For both proteins (E1 and E1^E4), ERK and CDK1/2 appear to be important regulatory kinases (13; this study).
In this study, ERK was found to phosphorylate the 16E1^E4 threonine residue at position 57, a site that is distinct from the serine 32 CDK1/2 site (Fig. 3) (13). In cells in cycle, it appears that these are in fact the only obvious phosphorylation sites in 16E1^E4. The phosphorylation sites of PKA and PKC remain to be determined, with PKC being expected to modify E1^E4 during epithelial differentiation (exact residues could not be identified from our mass spectrometry experiments) (data not shown). Only ERK phosphorylation was found to affect 16E1^E4 migration in SDS-PAGE, however, and lead to a structural change. Threonine 57 is located in the loop region of 16E1^E4 (41) that is rich in polarized amino acid residues (Fig. 4C). In the folded form of the protein (41), S32 is predicted to be in close proximity to T57 and to be contained within a region of positive charge that is thought to contribute to the stability of the 16E1^E4 loop structure (41). T57 phosphorylation increases the negative charge on the already negatively charged side of the loop and acts to increase the compactness of the structure, whereas phosphorylation among the positively charged amino acids (e.g., at residue S32) would be expected to have an opposite effect and to lead to a less constrained loop structure. These observations suggest a general strategy, supported by the experimental data, by which phosphorylation can affect 16E1^E4 structure and perhaps the structure of other E1^E4 proteins given that similar polarized regions are widely conserved among E1^E4 sequences. Our preliminary data obtained using antibodies to the 16E1^E4 S32 phosphoepitope suggest that S32 phosphorylation follows T57 phosphorylation as the epithelial cells migrate toward the epithelial surface (A. Kennedy, unpublished data), which is compatible with cell cycle progression from an S-phase-like to a G2-like environment as genome amplification proceeds (12).
Such a structural change may cause increased or more stabilized exposure of the keratin-binding motif (LLXLL) (Fig. 4E), explaining the ability of the T57-phosphorylated form of 16E1^E4 to associate with cytokeratin. The results of our immunoprecipitation experiments do indeed support this hypothesis; the phosphomimic, T57D, bound more strongly to immunoprecipitated keratins than the unphosphorylated form (Fig. 6A and B). The fact that the T57-phosphorylated form (i.e., the slower-migrating species) appears only in the keratin-associated fraction of E1^E4-expressing cells (58) supports the observation presented herein that T57 phosphorylation plays an important role in regulating the association of E1^E4 with keratin.
Our observation that T57-phosphorylated E1^E4 is detected predominantly in cells having high levels of total E1^E4 (Fig. 5B and C), together with the increased half-life and quick accumulation of the 16E1^E4 phosphomimic T57D (Fig. 5D and E), suggests that T57 phosphorylation serves to enhance the stability of the full-length 16E1^E4 protein. This increased stability appears to be due to enhanced keratin binding. When human fibroblast cells with or without keratin 8/18 were transfected with a plasmid expressing 16E1^E4, a much greater number of 16E1^E4 positively stained cells were seen among the keratin 8/18-expressing cells than among the keratin 8/18-negative cells (data not shown). Furthermore, the slower-migrating 16E1^E4 species was present only in the cells expressing keratins, consistent with the idea of stabilization through association with the cellular keratin network (Fig. 6C). Treatment with the proteasome inhibitor ALLN rescued the levels of T57-phosphorylated 16E1^E4 in keratin 8/18-negative cells, indicating that in the absence of keratins, ERK phosphorylation of 16E1^E4 can occur but that the phosphorylated protein does not accumulate. Although the significance of the keratin-binding activity of 16E1^E4 in the context of the viral life cycle is still not fully understood, mutations in the keratin-binding motif of 16E1^E4 have been shown previously to severely reduce viral DNA amplification (46). It appears from results of the work presented herein that keratin binding may in fact be a regulated process involving the phosphorylation of E1^E4 by ERK.
When expressed in monolayer SiHa cells, 16E1^E4 is phosphorylated at T57 and/or S32. The phosphorylation of T57 is mediated by the MEK-ERK pathway and does not appear to involve p38 MAPK (Fig. 2D). CDK1 and CDK2 phosphorylate 16E1^E4 primarily at serine 32 (Fig. 3) (13). Interestingly, when 16E1^E4 expressed in SiHa cells was analyzed by IEF and mass spectrometry, phosphorylated S32 was not detected in the monophosphorylated form of 16E1^E4 (P. Das, unpublished data). It is therefore possible that, in SiHa cells, T57 is preferentially phosphorylated over S32, perhaps because ERK is more active than CDK1/2 in these cells. ERK pathways can be activated by high-risk HPV E6 proteins, including 16E6 (7), and it is known that SiHa cells contain an integrated HPV16 sequence and constitutively express E6 and E7 (26). Indeed, in organotypic rafts generated with HaCaT cells transfected with 16E6, the levels of phospho-ERK1/2 were higher in 16E6-expressing cells than in 16E6-negative cells (7). 16E7 was recently shown to associate with the phosphatase PP2A and prevent PKB/Akt dephosphorylation (50). Levels of phosphorylated 16E1^E4 may be preserved by this activity of E7, since the treatment of 16E1^E4-expressing SiHa cells with OA, an inhibitor of PP2A, increased levels of the 16E1^E4 slower-migrating band (Fig. 1C). Since PP2A can also dephosphorylate active ERK (63), E7 may both protect 16E1^E4 from dephosphorylation and inhibit the deactivation of active ERK.
During epithelial differentiation of cells containing episomal genomes, ERK activation occurred in the middle layers of the HPV16-containing raft tissue (Fig. 7B) but not in the control (HPV16-negative NIKS raft tissue), with the increase in the T57-phosphorylated form of E1^E4 being closely linked to the presence of active ERK. It would be interesting to see whether 16E6 and/or 16E7 activates the ERK pathway in vivo, thus affecting 16E1^E4 phosphorylation and function. In this study, another HPV16-encoded protein, E5, was found to enhance 16E1^E4 phosphorylation. E5 is a highly hydrophobic membrane-bound protein that is associated with the Golgi apparatus, endoplasmic reticulum, and perinuclear membrane (8). It has been shown previously to activate ERK by EGF receptor-dependent and -independent mechanisms (10, 11, 34). We found that the level of T57 phosphorylation was higher when 16E1^E4 was expressed in a 16E5-expressing cell line than when it was expressed in a 16E5-negative cell line. This finding correlated with an increase in activated ERK (Fig. 8). E5 proteins may therefore have an impact on E1^E4 activity in vivo by increasing the levels of endogenous active ERK. Indeed, a 31E5 knockout genome appeared to have an effect on 31E1^E4 levels. The expression of 31E1^E4 protein in an organotypic raft culture of cells transfected with E5 mutant HPV31 was remarkably reduced compared to that in WT transfectants, suggesting cooperation between E1^E4 and E5 in the viral life cycle (28). 31E1^E4 has an ERK consensus site homologous to T57 of 16E1^E4, and it would be interesting to analyze the extent of 31E1^E4 phosphorylation in the presence or absence of 31E5. Although the loss of 16E5 did not seem to cause a decrease in 16E1^E4 levels in HPV16-containing rafts (32), the results of this study demonstrate the ability of 16E5 to enhance the phosphorylation of 16E1^E4 at T57, which increases protein stability and the affinity of the protein for keratin (Fig. 5, 6, and 8).
When considered alongside existing knowledge, the data presented herein suggest a putative model by which ERK phosphorylation can regulate 16E1^E4 activity during the viral life cycle (Fig. 9). As HPV-infected epithelial cells leave the basal layer and migrate toward the epithelial surface, the differentiation-dependent promoter becomes upregulated, driving the production of the E1^E4-E5 mRNA, along with transcripts encoding E1 and E2, which also contain the E5 open reading frame as their second or third exon. The increase in the abundance of the 16E5 protein is predicted to enhance the activation of the MEK-ERK pathway, which facilitates 16E1^E4 phosphorylation at T57. As shown in this study, T57 phosphorylation stimulates a structural change which enhances 16E1^E4 cytokeratin binding and protein stability. 16E6 and 16E7 are likely to contribute to this effect by activating MEK-ERK and/or inhibiting protein phosphatase PP2A. The modified functions of T57-phosphorylated 16E1^E4 provide at least one explanation for the initial accumulation of the full-length protein in the cell, and it may be that established functions of 16E1^E4, such as cyclin/CDK sequestration and G2 arrest, enhance genome amplification by maintaining a cellular environment conducive to viral replication. The switch from early to late polyadenylation site usage means that the synthesis of E1^E4-E5 transcripts is replaced by that of E1^E4-L1 transcripts, and it is likely that ERK activity would be reduced under these conditions. Our preliminary data suggest that the eventual loss of activated ERK in the upper epithelium and subsequent T57 dephosphorylation (Fig. 7) may facilitate N-terminal cleavage and the formation of E4 amyloid. Other phosphorylated or dephosphorylated forms of 16E1^E4 may then become more prevalent and play alternate roles during the later stages of infection.
FIG. 9.
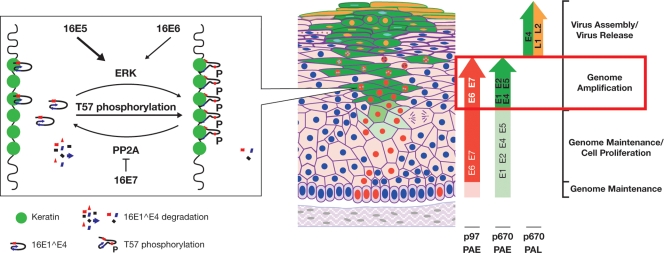
Putative model for E4 association with keratin following T57 phosphorylation during the viral life cycle. T57 phosphorylation of 16E1^E4 by ERK can be activated by 16E5, along with E6 and E7, and triggers a structural change which enhances 16E1^E4 keratin binding and stability. This process is illustrated graphically in the leftmost box, with a pointer to indicate where this step appears to occur in the model of productive infection (19) previously described by Middleton et al. (42) (right). Following the activation of the p670 promoter during epithelial differentiation, the E1^E4-E5 mRNA is upregulated (indicated by the vertical green arrow on the right). 16E5 expression can activate the MEK-ERK pathway, facilitating the phosphorylation of ERK targets such as 16E1^E4. The phosphorylation of 16E1^E4 at T57 induces a structural change that enhances 16E1^E4-keratin association and protein stability. 16E1^E4 accumulation may contribute to the late stages of the virus life cycle, possibly through association with CDK or the reorganization of the keratin network. In the upper epithelial layers, the E5, E6, and E7 proteins are no longer expressed and E1^E4 expression is directed from the E1^E4-L1 mRNA, leading to a loss of ERK activity and subsequent T57 dephosphorylation. PAE, early polyadenylation site; PAL, late polyadenylaion site.
Supplementary Material
Acknowledgments
We thank Bishr Omary for anti-keratin 8/18 monoclonal antibody L2A1 and anti-keratin 8/18 rabbit polyclonal antibody 8592, D. DiMaio for plasmid pMT3H16E5KC, Robert Evans for SW13 cell line Cl2 and clone T7K, and Jonathan Stoye for his support and encouragement during the course of this work.
This work was supported by the United Kingdom Medical Research Council.
Footnotes
Published ahead of print on 11 February 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allen-Hoffmann, B. L., S. J. Schlosser, C. A. Ivarie, C. A. Sattler, L. F. Meisner, and S. L. O'Connor. 2000. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J. Investig. Dermatol. 114444-455. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa, M. S., C. Edmonds, C. Fisher, J. T. Schiller, D. R. Lowy, and K. H. Vousden. 1990. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and Sv40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation. EMBO J. 9153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, I., A. Martin, and S. Roberts. 2007. The E1^E4 protein of human papillomavirus interacts with the serine-arginine-specific protein kinase SRPK1. J. Virol. 815437-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitburd, F., O. Croissant, and G. Orth. 1987. Expression of human papillomavirus type-1 E4 gene products in warts, p. 115-122. In B. M. Steinberg, J. L. Brandsma, and L. B. Taichman (ed.), Papillomaviruses, vol. 5. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- 5.Brown, D. R., T. L. McClowry, R. A. Sidner, K. H. Fife, and J. T. Bryan. 1998. Expression of the human papillomavirus type 11 E5A protein from the E1E4,E5 transcript. Intervirology 4147-54. [DOI] [PubMed] [Google Scholar]
- 6.Bryan, J. T., A. Han, K. H. Fife, and D. R. Brown. 2000. The human papillomavirus type 11 E1E4 protein is phosphorylated in genital epithelium. Virology 268430-439. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti, O., K. Veeraraghavalu, V. Tergaonkar, Y. Liu, E. J. Androphy, M. A. Stanley, and S. Krishna. 2004. Human papillomavirus type 16 E6 amino acid 83 variants enhance E6-mediated MAPK signaling and differentially regulate tumorigenesis by notch signaling and oncogenic Ras. J. Virol. 785934-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad, M., V. J. Bubb, and R. Schlegel. 1993. The human papillomavirus type 6 and 16 E5 proteins are membrane-associated proteins which associate with the 16-kilodalton pore-forming protein. J. Virol. 676170-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crum, C. P., S. Barber, M. Symbula, K. Snyder, A. M. Saleh, and J. K. Roche. 1990. Coexpression of the human papillomavirus type 16 E4 and L1 open reading frames in early cervical neoplasia. Virology 178238-246. [DOI] [PubMed] [Google Scholar]
- 10.Crusius, K., E. Auvinen, and A. Alonso. 1997. Enhancement of EGF- and PMA-mediated MAP kinase activation in cells expressing the human papillomavirus type 16 E5 protein. Oncogene 151437-1444. [DOI] [PubMed] [Google Scholar]
- 11.Crusius, K., I. Rodriguez, and A. Alonso. 2000. The human papillomavirus type 16 E5 protein modulates ERK1/2 and p38 MAP kinase activation by an EGFR-independent process in stressed human keratinocytes. Virus Genes 2065-69. [DOI] [PubMed] [Google Scholar]
- 12.Davy, C., and J. Doorbar. 2007. G2/M cell cycle arrest in the life cycle of viruses. Virology 368219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davy, C. E., M. Ayub, D. J. Jackson, P. Das, P. McIntosh, and J. Doorbar. 2006. HPV16 E1-E4 protein is phosphorylated by Cdk2/cyclin A and relocalizes this complex to the cytoplasm. Virology 349230-244. [DOI] [PubMed] [Google Scholar]
- 14.Davy, C. E., D. J. Jackson, K. Raj, W. L. Peh, S. A. Southern, P. Das, R. Sorathia, P. Laskey, K. Middleton, T. Nakahara, Q. Wang, P. J. Masterson, P. F. Lambert, S. Cuthill, J. B. Millar, and J. Doorbar. 2005. Human papillomavirus type 16 E1^E4-induced G2 arrest is associated with cytoplasmic retention of active Cdk1/cyclin B1 complexes. J. Virol. 793998-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davy, C. E., D. J. Jackson, Q. Wang, K. Raj, P. J. Masterson, N. F. Fenner, S. Southern, S. Cuthill, J. B. Millar, and J. Doorbar. 2002. Identification of a G2 arrest domain in the E1^E4 protein of human papillomavirus type 16. J. Virol. 769806-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng, W., B. Y. Lin, G. Jin, C. G. Wheeler, T. Ma, J. W. Harper, T. R. Broker, and L. T. Chow. 2004. Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J. Virol. 7813954-13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Villiers, E. M. 1999. Human papillomavirus. Introduction. Semin. Cancer Biol. 9377. [DOI] [PubMed] [Google Scholar]
- 18.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 32417-27. [DOI] [PubMed] [Google Scholar]
- 19.Doorbar, J. 2005. The papillomavirus life cycle. J. Clin. Virol. 32(Suppl. 1)S7-S15. [DOI] [PubMed] [Google Scholar]
- 20.Doorbar, J., and G. Myers. 1996. The E4 protein, p. 58-80. In H. D. G. Myers and J. Icenogelet (ed.), Human papillomaviruses, vol. III. Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 21.Doorbar, J., R. C. Elston, S. Napthine, K. Raj, E. Medcalf, D. Jackson, N. Coleman, H. M. Griffin, P. Masterson, S. Stacey, Y. Mengistu, and J. Dunlop. 2000. The E1^E4 protein of human papillomavirus type 16 associates with a putative RNA helicase through sequences in its C terminus. J. Virol. 7410081-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352824-827. [DOI] [PubMed] [Google Scholar]
- 23.Doorbar, J., H. S. Evans, I. Coneron, L. V. Crawford, and P. H. Gallimore. 1988. Analysis of HPV-1 E4 gene expression using epitope-defined antibodies. EMBO J. 7825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doorbar, J., C. Foo, N. Coleman, L. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 23840-52. [DOI] [PubMed] [Google Scholar]
- 25.Doorbar, J., A. Parton, K. Hartley, L. Banks, T. Crook, M. Stanley, and L. Crawford. 1990. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology 178254-262. [DOI] [PubMed] [Google Scholar]
- 26.el Awady, M. K., J. B. Kaplan, S. J. O'Brien, and R. D. Burk. 1987. Molecular analysis of integrated human papillomavirus 16 sequences in the cervical cancer cell line SiHa. Virology 159389-398. [DOI] [PubMed] [Google Scholar]
- 27.Favre, M., N. Ramoz, and G. Orth. 1997. Human papillomaviruses: general features. Clin. Dermatol. 15181-198. [DOI] [PubMed] [Google Scholar]
- 28.Fehrmann, F., D. J. Klumpp, and L. A. Laimins. 2003. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J. Virol. 772819-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferguson, K. A. 1964. Starch-gel electrophoresis—application to the classification of pituitary proteins and polypeptides. Metabolism 13(Suppl.)985-1002. [DOI] [PubMed] [Google Scholar]
- 30.Flores, E. R., and P. F. Lambert. 1997. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 717167-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furuno, T., N. Hirashima, S. Onizawa, N. Sagiya, and M. Nakanishi. 2001. Nuclear shuttling of mitogen-activated protein (MAP) kinase (extracellular signal-regulated kinase (ERK) 2) was dynamically controlled by MAP/ERK kinase after antigen stimulation in RBL-2H3 cells. J. Immunol. 1664416-4421. [DOI] [PubMed] [Google Scholar]
- 32.Genther, S. M., S. Sterling, S. Duensing, K. Munger, C. Sattler, and P. F. Lambert. 2003. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J. Virol. 772832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grand, R. J., J. Doorbar, K. J. Smith, I. Coneron, and P. H. Gallimore. 1989. Phosphorylation of the human papillomavirus type 1 E4 proteins in vivo and in vitro. Virology 170201-213. [DOI] [PubMed] [Google Scholar]
- 34.Gu, Z., and G. Matlashewski. 1995. Effect of human papillomavirus type 16 oncogenes on MAP kinase activity. J. Virol. 698051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedrick, J. L., and A. J. Smith. 1968. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch. Biochem. Biophys. 126155-164. [DOI] [PubMed] [Google Scholar]
- 36.Knight, G. L., J. R. Grainger, P. H. Gallimore, and S. Roberts. 2004. Cooperation between different forms of the human papillomavirus type 1 E4 protein to block cell cycle progression and cellular DNA synthesis. J. Virol. 7813920-13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhne, C., D. Gardiol, C. Guarnaccia, H. Amenitsch, and L. Banks. 2000. Differential regulation of human papillomavirus E6 by protein kinase A: conditional degradation of human discs large protein by oncogenic E6. Oncogene 195884-5891. [DOI] [PubMed] [Google Scholar]
- 38.Lambert, P. F., M. A. Ozbun, A. Collins, S. Holmgren, D. Lee, and T. Nakahara. 2005. Using an immortalized cell line to study the HPV life cycle in organotypic “raft” cultures. Methods Mol. Med. 119141-155. [DOI] [PubMed] [Google Scholar]
- 39.Liang, Y. J., H. S. Chang, C. Y. Wang, and W. C. Yu. 2008. DYRK1A stabilizes HPV16E7 oncoprotein through phosphorylation of the threonine 5 and threonine 7 residues. Int. J. Biochem. Cell Biol. 402431-2441. [DOI] [PubMed] [Google Scholar]
- 40.Massimi, P., D. Pim, C. Kuhne, and L. Banks. 2001. Regulation of the human papillomavirus oncoproteins by differential phosphorylation. Mol. Cell. Biochem. 227137-144. [PubMed] [Google Scholar]
- 41.McIntosh, P. B., S. R. Martin, D. J. Jackson, J. Khan, E. R. Isaacson, L. Calder, K. Raj, H. M. Griffin, Q. Wang, P. Laskey, J. F. Eccleston, and J. Doorbar. 2008. Structural analysis reveals an amyloid form of the human papillomavirus type 16 E1-E4 protein and provides a molecular basis for its accumulation. J. Virol. 828196-8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Middleton, K., W. Peh, S. Southern, H. Griffin, K. Sotlar, T. Nakahara, A. El-Sherif, L. Morris, R. Seth, M. Hibma, D. Jenkins, P. Lambert, N. Coleman, and J. Doorbar. 2003. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J. Virol. 7710186-10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 183587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munoz, N., and F. X. Bosch. 1997. Cervical cancer and human papillomavirus: epidemiological evidence and perspectives for prevention. Salud Publica Mex. 39274-282. [DOI] [PubMed] [Google Scholar]
- 45.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348518-527. [DOI] [PubMed] [Google Scholar]
- 46.Nakahara, T., W. L. Peh, J. Doorbar, D. Lee, and P. F. Lambert. 2005. Human papillomavirus type 16 E1^E4 contributes to multiple facets of the papillomavirus life cycle. J. Virol. 7913150-13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nasseri, M., R. Hirochika, T. R. Broker, and L. T. Chow. 1987. A human papilloma virus type 11 transcript encoding an E1-E4 protein. Virology 159433-439. [DOI] [PubMed] [Google Scholar]
- 48.Peh, W. L., J. L. Brandsma, N. D. Christensen, N. M. Cladel, X. Wu, and J. Doorbar. 2004. The viral E4 protein is required for the completion of the cottontail rabbit papillomavirus productive cycle in vivo. J. Virol. 782142-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peh, W. L., K. Middleton, N. Christensen, P. Nicholls, K. Egawa, K. Sotlar, J. Brandsma, A. Percival, J. Lewis, W. J. Liu, and J. Doorbar. 2002. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 7610401-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pim, D., P. Massimi, S. M. Dilworth, and L. Banks. 2005. Activation of the protein kinase B pathway by the HPV-16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene 247830-7838. [DOI] [PubMed] [Google Scholar]
- 51.Raj, K., S. Berguerand, S. Southern, J. Doorbar, and P. Beard. 2004. E1^E4 protein of human papillomavirus type 16 associates with mitochondria. J. Virol. 787199-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts, S., I. Ashmole, L. J. Gibson, S. M. Rookes, G. J. Barton, and P. H. Gallimore. 1994. Mutational analysis of human papillomavirus E4 proteins: identification of structural features important in the formation of cytoplasmic E4/cytokeratin networks in epithelial cells. J. Virol. 686432-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schweitzer, S. C., M. W. Klymkowsky, R. M. Bellin, R. M. Robson, Y. Capetanaki, and R. M. Evans. 2001. Paranemin and the organization of desmin filament networks. J. Cell Sci. 1141079-1089. [DOI] [PubMed] [Google Scholar]
- 54.Shaul, Y. D., and R. Seger. 2007. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim. Biophys. Acta 17731213-1226. [DOI] [PubMed] [Google Scholar]
- 55.Stoler, M. H., S. M. Wolinsky, A. Whitbeck, T. R. Broker, and L. T. Chow. 1989. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology 172331-340. [DOI] [PubMed] [Google Scholar]
- 56.Tiburu, E. K., P. C. Dave, J. F. Vanlerberghe, T. B. Cardon, R. E. Minto, and G. A. Lorigan. 2003. An improved synthetic and purification procedure for the hydrophobic segment of the transmembrane peptide phospholamban. Anal. Biochem. 318146-151. [DOI] [PubMed] [Google Scholar]
- 57.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 18912-19. [DOI] [PubMed] [Google Scholar]
- 58.Wang, Q., H. Griffin, S. Southern, D. Jackson, A. Martin, P. McIntosh, C. Davy, P. J. Masterson, P. A. Walker, P. Laskey, M. B. Omary, and J. Doorbar. 2004. Functional analysis of the human papillomavirus type 16 E1^E4 protein provides a mechanism for in vivo and in vitro keratin filament reorganization. J. Virol. 78821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson, R., F. Fehrmann, and L. A. Laimins. 2005. Role of the E1^E4 protein in the differentiation-dependent life cycle of human papillomavirus type 31. J. Virol. 796732-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson, R., G. B. Ryan, G. L. Knight, L. A. Laimins, and S. Roberts. 2007. The full-length E1E4 protein of human papillomavirus type 18 modulates differentiation-dependent viral DNA amplification and late gene expression. Virology 362453-460. [DOI] [PubMed] [Google Scholar]
- 61.Yoon, S., and R. Seger. 2006. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 2421-44. [DOI] [PubMed] [Google Scholar]
- 62.Yu, J. H., B. Y. Lin, W. Deng, T. R. Broker, and L. T. Chow. 2007. Mitogen-activated protein kinases activate the nuclear localization sequence of human papillomavirus type 11 E1 DNA helicase to promote efficient nuclear import. J. Virol. 815066-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, B., Z. X. Wang, Y. Zhao, D. L. Brautigan, and Z. Y. Zhang. 2002. The specificity of extracellular signal-regulated kinase 2 dephosphorylation by protein phosphatases. J. Biol. Chem. 27731818-31825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.