Abstract
Human cytomegalovirus (HCMV) regulates NF-κB during infection by a variety of mechanisms. For example, the HCMV gene product, UL144, is known to activate NF-κB in a tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6)-dependent manner, causing the upregulation of the chemokine CCL22 (MDC). Viral UL144 is expressed from the UL/b′ region of the HCMV genome at early times postinfection and is a TNFR1-like homologue. Despite this homology to the TNFR1 receptor superfamily, UL144 does not bind to members of the TNF ligand superfamily. We show here that the upregulation of NF-κB by UL144 is dependent upon cellular tripartite motif 23 (TRIM23) protein. We propose a mechanism by which UL144 activates NF-κB through a direct interaction with the cellular protein TRIM23 in a complex containing TRAF6. In contrast, TRIM23 is not involved in conventional double-stranded RNA signaling via NF-κB. Therefore, we present a novel role for TRIM23 that is specific to UL144-mediated activation of NF-κB during the course of virus infection.
Human cytomegalovirus (HCMV) is a species-specific betaherpesvirus that causes severe disease in neonates and immunosuppressed individuals such as allograft transplant recipients and patients with AIDS (24). HCMV is the largest known human herpesvirus comprising 236 kb of double-stranded DNA, and a number of its genes are nonessential for replication in fibroblasts (8). UL144 is encoded in the 15-kb unique region of the genome termed UL/b′, which encodes at least 19 open reading frames (ORFs). UL/b′ is found in HCMV clinical isolates but has been lost in extensively passaged laboratory strains and is dispensable for growth in vitro (4). Evidence suggests that UL/b′ is important for viral infection in vivo since clinical isolate strains can replicate in SCID mice into which human tissues were implanted, whereas the laboratory strain AD169, lacking UL/b′, cannot (39). Consistent with this, roles for several UL-b′ proteins have been identified, and all of these suggest that genes encoded in UL/b′ play some role in the regulation of immunity or tissue tropism. For example, recent studies have shown that UL/b′ genes play a role in viral evasion of NK cell killing (UL141, UL142, and UL147) (4, 36, 40) and cell tropism (UL128, UL130, and UL131A) (4, 11, 16, 30).
We have previously described a role for UL144 in the upregulation of the chemokine CCL22 (also termed macrophage-derived chemokine [MDC]), a TH2 chemoattractant that may act to subvert the TH1 immune response (28). UL144 achieves this by activating NF-κB in a tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6)-dependent manner, similar to the mechanism used by Epstein-Barr virus (EBV)-encoded LMP-1 (23, 31). The mechanism of NF-κB activation by LMP-1 has been fairly well characterized. Expression of LMP-1 induces TRAF6 ubiquitination (33), and the subsequent activation of NF-κB has recently been shown to depend upon the Rab acceptor protein PRA1 (21). PRA1 dictates the perinuclear clumping of TRAF6 mediated by LMP-1. While the mechanism by which UL144 activates NF-κB via TRAF6 has not been characterized, as with LMP-1, TRAF6 has been found to clump in a perinuclear region during UL144-mediated activation of NF-κB (28). Here, we now show that UL144 binds directly to a Rab acceptor domain-containing protein, TRIM23, and this interaction may dictate the perinuclear association of TRAF6 with UL144. TRIM23 has been shown to have E3 ubiquitin ligase activity (38), and we demonstrate that this is essential for the upregulation of NF-κB by HCMV UL144.
MATERIALS AND METHODS
Cells and viruses.
Human fetal foreskin fibroblasts (HFFF) and U373 cells were maintained in Eagle minimal essential medium containing 10% fetal calf serum. Infections with HCMV AD169 and Toledo have been described previously (2, 22). Primary human monocyte-derived macrophages (MDM) were generated as described previously (26). Briefly, peripheral blood mononuclear cells were isolated by density gradient centrifugation, using Ficoll-Hypaque (Nycomed Pharma AS). Primary monocytes were obtained by adherence for 1.5 h at 37°C in 5% CO2. Following adherence, the monocytes were cultured in Iscove modified Dulbecco medium (Gibco-BRL) supplemented with 15% horse serum (Sera-Lab), 15% fetal calf serum, 2 mM l-glutamine (Gibco-BRL), 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 5 × 10−5 M hydrocortisone sodium succinate (Sigma) for 5 days at 37°C in 5% CO2. After 5 days, the cultures were stimulated with 10 ng of phorbol myristate acetate per ml overnight to induce differentiation to HCMV-permissive macrophages.
Plasmids and siRNAs.
Reporter plasmids containing the firefly luciferase gene under the control of NF-κB-driven expression, p(PRDII)5tk-lucter, and the constitutive β-galactosidase reporter plasmid pJATlac were provided by S. Goodbourn (St. George's Hospital Medical School, University of London, London, United Kingdom) and have been described previously (27, 28). pGL3 plasmids containing the wild-type MDC promoter driving the expression of the firefly luciferase protein (pGL3-MDC) has been described previously (23). Expression of the T7-tagged UL144 gene from the pCDNA3 vector has been described previously (28). UL144 antibody was kindly provided by C. Benedict (La Jolla Institute, La Jolla, CA). V-tagged TRIM23 was obtained from Hybridgenics, Paris, France. DNA encoding human ubiquitin was amplified from genomic DNA by using the KOD polymerase (Novagen) and cloned into pCMV5-HA plasmid by using the BamHI and NotI restriction sites. Vectors in which K48 or K63 were mutated to R (pCMV-HA Ub K48R/K63R) were created by using a QuikChange mutagenesis kit (Stratagene). All mutations were verified by DNA sequencing. Human TRAF6 was amplified from genomic DNA by using KOD polymerase and ligated into the BamHI/NotI sites of pCMV4-Flag. All small interfering RNAs (siRNAs) were obtained from Qiagen (HP Guaranteed). Wild-type TRAF6 and TRAF6 C70A were kindly provided by Felix Randow (Laboratory of Molecular Biology, Cambridge, United Kingdom) (3).
Gene expression, protein analysis, and primary antibodies.
For reporter gene assays, plasmids were transfected into HFFF or U373 cells with Lipofectamine 2000, using standard protocols and modifications described previously (28). TNF and double-stranded RNA (dsRNA) inductions have also been described previously (27). Lysates were prepared and analyzed for luciferase and β-galactosidase activities (26, 27, 28). For Western blotting, cells were harvested in Laemmli buffer and probed with anti-rabbit, anti-mouse, anti-goat, or anti-rat primary antibodies as described below, followed by horseradish peroxidase-conjugated secondary anti-mouse, anti-goat, anti-rat, or anti-rabbit immunoglobulins (Cell Signaling), and visualized with ECL reagent as described previously (28). The immunoprecipitation protocol has been described previously (28). IE86 was detected with a custom HCMV fluorescein isothiocyanate (FITC)-conjugated antibody without Evans blue (Chemicon International), UL144 with an anti-T7 tag mouse monoclonal antibody (Novagen) or an anti-UL144 rat monoclonal antibody and actin with rabbit polyclonal antibody (Cell Signaling). Flag tag and hemagglutinin (HA)-tagged primary antibodies were obtained from Cell Signaling, and antibodies to endogenous TRAF6 and TRIM23, as well as anti-V tag antibody, were from AbCam. The MDC enzyme-linked immunosorbent assay carried out in accordance with the manufacturer's protocol (RND Systems) on supernatants from macrophages derived from peripheral blood monocytes that were transfected as described using an electroporator with a macrophage Nucleofector kit (both from Amaxa, Inc.).
Immunofluorescence and FRET.
Infected and transfected cells were fixed and permeabilized for immunofluorescence confocal imaging as described previously (28, 29). The primary antibodies described in the previous section alongside isotype controls were used, followed by secondary antibodies (ensuring no species crossover with other secondaries or the primary antibodies) conjugated to FITC, Alexa Fluor 594, or Alexa Fluor 647 (Molecular Probes). Coverslips were mounted in Citifluor media and examined by using a Nikon microscope for confocal and fluorescence resonance energy transfer (FRET) analyses as described previously (29), using ImagePro WCIF ImageJ software (National Institutes of Health) and TCS NF/SP confocal software. The images were detected by using excitation wavelengths of 488, 568, and 647 nm.
RESULTS
UL144 targets canonical TAK1-mediated NF-κB signaling, but not via Pra1.
The LMP-1 protein of EBV has recently been shown to activate the canonical NF-κB pathway utilizing Pra1 as an accessory protein (21). Since both UL144 and LMP-1 activate NF-κB in a TRAF6-dependent manner, we tested whether Pra1 was also involved in UL144-mediated NF-κB signaling.
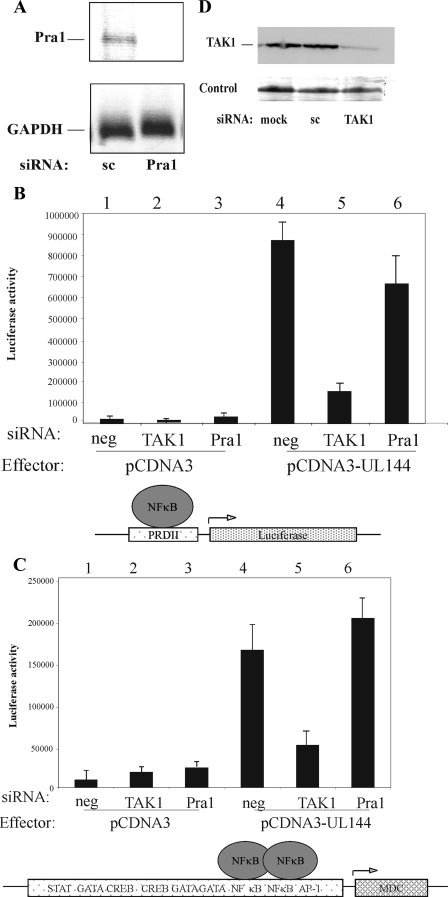
There is no antibody available for the detection of Pra1. Consequently, we tested the ability of Pra1-specific siRNA to abrogate levels of Pra1 mRNA by reverse transcription-PCR. Figure 1A shows this was effective after 48 h. However, the removal of Pra1 did not prevent UL144 from activating the PRDII NF-κB-responsive promoter or the MDC promoter driving the expression of luciferase (Fig. 1B and C, compare lanes 4 and 6).
FIG. 1.
TAK1 is required for UL144-mediated upregulation of MDC via NF-κB, but not Pra1. HFFF cells were transfected with p(PRDII)5tk-lucter or the −722-MDC-luciferase with pJATlac (B and C), and either a mammalian expression plasmid driving the overexpression of the HCMV protein UL144 or the control empty vector (B and C). Cells were then cotransfected with siRNAs: scramble control, to the cellular proteins TAK1 or Pra1 as indicated. Cells were transfected for 48 h, and cell extracts were prepared. TAK1 levels were analyzed by Western blotting (D), and Pra1 levels were analyzed by RT-PCR (A). Equal loading was verified by using Ponceau red staining (control). Luciferase and β-galactosidase activities were determined from cellular extracts, and relative expression values were calculated accordingly (expressed relative to the induced level of the vector-only sample = 1.0). The histograms present the results of three independent experiments, and standard deviations are indicated.
LMP-1 is also known to use TRAF6 to target canonical NF-κB signaling (31), which uses TAK1 as part of the pathway (19); therefore, this was used as an additional control in our experiments. To do this, we used siRNA to TAK1, a known essential component of the TNF-induced NF-κB signaling pathway (18, 19), to deplete cells of TAK1 and analyzed the ability of UL144 activate the NF-κB responsive promoters PRDII or MDC (Fig. 1). First, we tested the ability of our TAK1 siRNAs to deplete TAK1 from cells, which was effective by 48 h posttransfection (Fig. 1D). Furthermore, although the presence of a control siRNA had no effect on the ability of UL144 to stimulate an NF-κB-responsive promoter (Fig. 1B, compare lanes 1 and 4), the TAK1 siRNA prevented UL144 from activating the NF-κB responsive promoter, PRDII, or the endogenous MDC gene (Fig. 1B and C, compare lanes 2 and 5).
Therefore, it appears that UL144 activates NF-κB via TRAF6 using the canonical TAK1 pathway. However, unlike LMP-1 of EBV, it does not use the Pra1 accessory protein to do this.
TRIM23 complexes with UL144.
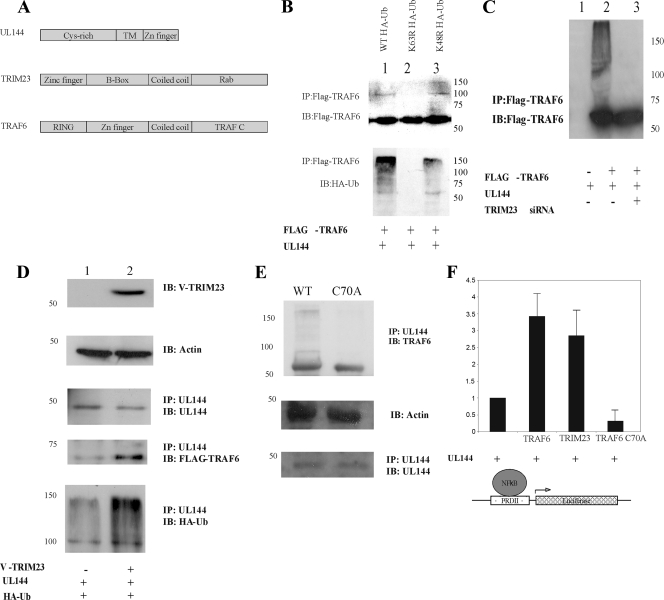
Previously, we have shown that UL144 complexes with TRAF6 in cells (28), and our observations thus far suggest that TAK1, but not Pra1, is important for UL144-mediated activation of NF-κB. Therefore, we next carried out a yeast-two-hybrid screen to identify whether TRAF6 and/or TAK1 interacted “directly” with UL144 and also to identify any other cellular proteins that may interact with UL144. Although this screen did not detect TRAF6 or TAK1 using the intracellular domain of UL144, it did identify six interacting clones. DNA sequencing showed one of these to be TRIM23 (the interaction domains are summarized in Fig. 5A).
FIG. 5.
UL144 activates NF-κB through TRIM23-mediated TRAF6 autoubiquitination. (A) The domains of TRIM23 and TRAF6 are summarized. HFFF cells were transfected with either a mammalian expression plasmid driving the overexpression of the cellular protein TRAF6 (Flag tagged), TRAF6 C70A, or TRIM23 (V tagged). HCMV protein UL144, ubiquitin (HA tagged), or the control empty vector with or without or without control or TRIM23 siRNA are as indicated (B, C, D, and E). Cells were transfected for 48 h, and cell extracts prepared either directly by lysis in Laemmli buffer or for immunoprecipitation (IP). Samples were analyzed by Western blotting (IB) with the indicated antibodies (B, C, D, and E). Alternatively, HFFF cells were transfected with p(PRDII)5tk-lucter and pJATlac (A and B) and a plasmid driving the overexpression of the HCMV protein UL144, with plasmids expressing either empty vector, wild-type (WT) TRIM23, TRAF6, or TRAF6 C70A. Cells were transfected for 48 h, and cell extracts were prepared. The luciferase and β-galactosidase activities were determined from cellular extracts, and the relative expression values were calculated accordingly (expressed relative to the induced level of the UL144-only sample = 1.0).
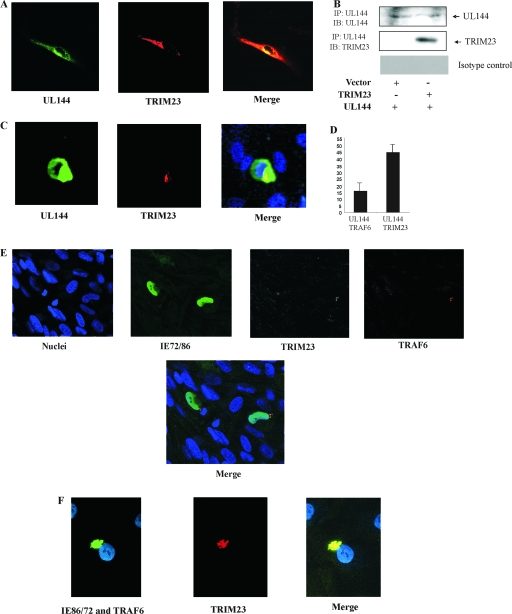
To confirm that TRIM23 and UL144 interact in vivo, we tested for colocalization in immunofluorescence studies. Figure 2A shows the localization of UL144 and TRIM23 in the HCMV-permissive cell line, HFFF. Cells were transfected for the overexpression of UL144 (red) and TRIM23 (green). As observed previously, UL144 is predominantly perinuclear but also visible throughout the cytoplasm, and TRIM23 is also predominantly cytoplasmic; the two proteins colocalize (merge). Furthermore, the ability of TRIM23 and UL144 to complex in HFFF cells was demonstrated by immunoprecipitation and Western blotting (Fig. 2B).
FIG. 2.
TRIM23 colocalizes with UL144. HFFF cells were transfected with UL144 (A and B), T7-tagged UL144 (C), and/or TRIM23 (V tagged) (A and B) as indicated for 48 h and then fixed and permeabilized. (E and F) Alternatively, HFFF cells were infected with TB40E for 48 h before being fixed and permeabilized. UL144 was detected with rat monoclonal anti-UL144 antibody, followed by Alexa Fluor 594-conjugated donkey anti-rat antibody staining (A) or mouse monoclonal anti-T7-FITC antibody (C). TRIM23 was detected by using mouse monoclonal anti-V tag antibody, followed by Alexa Fluor 645-conjugated goat anti-mouse antibody staining (A), or the endogenous TRIM23 gene was detected with rabbit anti-TRIM23 antibody, followed by TRITC (tetramethyl rhodamine isothiocyanate) donkey anti-rabbit antibody staining (C, D, E, and F). Endogenous TRAF6 was detected with goat anti-TRAF6 antibody, followed by swine anti-goat-Alexa Fluor 647 or swine anti-goat-FITC (E and F, respectively). Nuclei were stained with Hoechst stain (C and E). Infected cells were detected by using mouse monoclonal antibody to IE72/86, which was either FITC conjugated (E) or followed by Alexa Fluor 647-conjugated anti-mouse antibody (F). Cells were visualized by confocal microscopy (A, C, E, and F) and FRET (C and D). The FRET was quantitated by using WCIF ImageJ (D). For immunoprecipitation (B) cells were lysed and immunoprecipitated with anti-UL144 monoclonal antibody or an isotype-matched control and immunoblotted with anti-V5 tag for the detection of TRIM23 or anti-UL144 rat monoclonal antibody to detect UL144.
We were also able to detect endogenous TRIM23 (Fig. 2C), which colocalized with expressed UL144 and, to assess this interaction, FRET analysis was used. These data were analyzed by using ImageJ software, and the percentage of FRET was determined (Fig. 2D). Interestingly, the ratio of FRET between TRIM23 and UL144 was 46%, which is considerably higher than that observed between UL144 and TRAF6 (28), potentially indicating a closer interaction between TRIM23 and UL144 (Fig. 2E). From these results it appears that UL144 colocalizes and interacts with both TRIM23 and TRAF6 and, consistent with this, we were also able to confirm that TRIM23 and TRAF6 colocalize in the context of virus infection (Fig. 2E and F). TRIM23 and TRAF6 were only found to colocalize in the perinuclei of infected (green) and not in those of uninfected cells (Fig. 2E). Red and green colocalization was also demonstrated for TRAF6 and TRIM23 in infected cells (Fig. 2F). Together, these data show that TRIM23 and UL144 complex in both overexpression analyses and in the context of virus infection, indicating that UL144 and TRAF6 may interact in a complex mediated by TRIM23.
TRIM23 is critical for the upregulation of NF-κB by UL144.
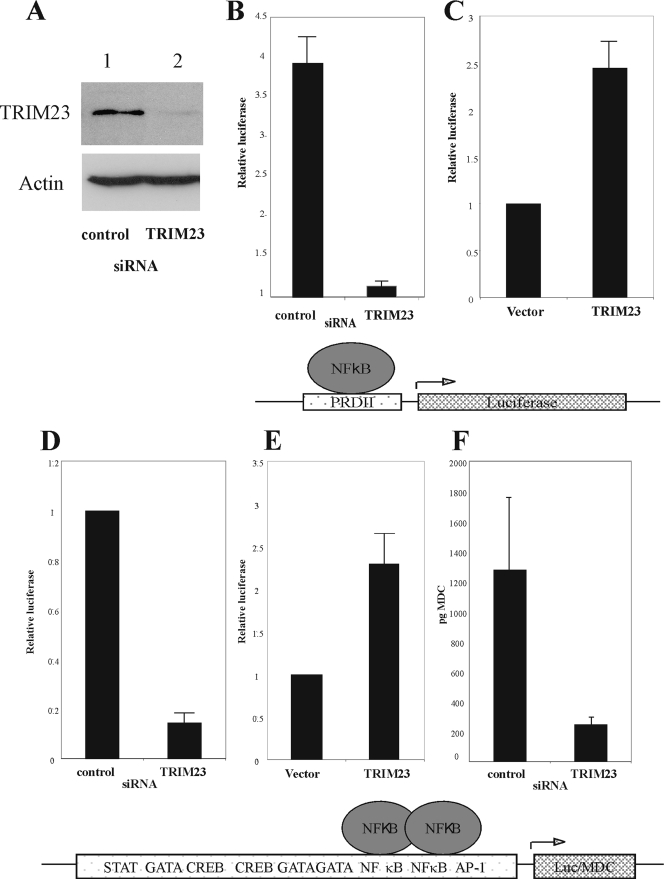
To determine the functional relevance of the interaction between UL144 and TRIM23, we examined whether knockdown of TRIM23 with siRNAs would have any effect on NF-κB signaling by UL144. To address this, we generated siRNAs to TRIM23 and tested their ability to reduce TRIM23 levels in cells. Compared to a control siRNA, TRIM23 siRNA specifically reduced the levels of TRIM23 (Fig. 3A, compare lanes 1 and 2). Next, luciferase reporter assays were used to determine the effect of the presence or absence of TRIM23 on the ability of UL144 to stimulate an NF-κB-only driven promoter (Fig. 3B) or the MDC promoter, shown previously to be upregulated by UL144 and clinical isolate strains of HCMV (28) (Fig. 3C). Figure 3B and D demonstrate that the removal of TRIM23 from the cellular environment abrogated the ability of UL144 to activate the NF-κB-specific (B) or the MDC (D) promoter. Furthermore, overexpression of TRIM23 stimulates UL144-mediated activation of NF-κB to either promoter (Fig. 3C and E) and, when monocyte derived macrophages were transfected with UL144 and siRNA to TRIM23, there was a decrease in MDC secretion compared to cells cotransfected with a control siRNA (Fig. 3F). Therefore, TRIM23 is an essential cofactor for upregulating UL144-mediated NF-κB signaling.
FIG. 3.
TRIM23 is required for NF-κB upregulation by UL144. HFFFs (A to E) or peripheral blood mononuclear cell macrophages (F) were transfected with p(PRDII)5tk-lucter (B and C) or pGL3-MDC-luc (D and E) and pJATlac (B to D), with either a mammalian expression plasmid driving the overexpression of the HCMV protein UL144 (B to F) or the cellular protein TRIM23 or the control empty vector (A, B, and D). In panels B, D, and F, cells were cotransfected with siRNAs: either a scramble control or to TRIM23. Cells were transfected for 48 h, and cell extracts were prepared. Cell lysates were either prepared for Western blotting (A), or the luciferase and β-galactosidase activities were determined from cellular extracts; the relative expression values were calculated accordingly (expressed relative to the induced level of the vector-only sample = 1.0). Histograms present the results of three independent experiments, with the standard deviations indicated as error bars (B to E).
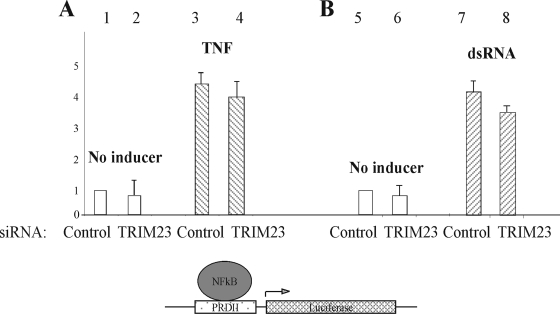
TRIM23 does not mediate conventional NF-κB signaling by TNF or dsRNA.
Although UL144 does not bind members of the TNF ligand superfamily, it is a homologue of the TNFR superfamily and signals via TRAF6 (28). Consequently, we tested whether TRIM23 plays any role in conventional TNF signaling in the absence of UL144. Figure 4 shows that in the presence of a control siRNA, TNF causes the stimulation of the NF-κB responsive promoter, PRDII (Fig. 4, compare lanes 1 and 3). However, removal of TRIM23 from the cellular environment with TRIM23-specific siRNA did not prevent induction of the PRDII promoter by TNF (Fig. 4A, compare lanes 3 and 4). Therefore, despite being a homologue of the TNFR superfamily, UL144 uses an alternative pathway to canonical TNF-mediated signaling involving TRIM23. To further test whether TRIM23 plays a role in virally stimulated NF-κB signaling, the ability of cells to respond to dsRNA (a known viral stimulator of NF-κB) was tested in the presence or absence of TRIM23 (Fig. 4B). Again, the absence of TRIM23 did not prevent cells from activating the NF-κB-driven luciferase expression in response to dsRNA (Fig. 4B, compare lanes 1 and 3 to lanes 2 and 4). Thus, in contrast to NF-κB activation by UL144, the upregulation of NF-κB by TNF or dsRNA is not prevented by the absence of TRIM23.
FIG. 4.
TRIM23 is not required for NF-κB activation via dsRNA or TNF. U373 cells were transfected with p(PRDII)5tk-lucter and pJATlac (A and B) and either a plasmid driving the overexpression of the HCMV protein UL144 or the control empty vector. Cells were cotransfected with siRNAs: either a scramble control or to TRIM23 as indicated. Cells were transfected for 48 h, followed by induction with dsRNA (A) or TNF (B), and then cell extracts were prepared. The luciferase and β-galactosidase activities were determined from cellular extracts, and relative expression values were calculated accordingly (expressed relative to the induced level of the vector-only sample = 1.0). Histograms present the results of two independent experiments (A and B).
UL144 mediates K63-linked TRAF6 ubiquitination in a TRIM23-dependent manner.
To address possible mechanisms by which UL144, TRIM23, and TRAF6 may mediate NF-κB signaling, the interaction between UL144 and TRIM23 was analyzed. Yeast two-hybrid binding assays showed that, when the UL144 zinc finger-binding domain was used as bait, a cDNA expressing the N-terminal TRIM23 zinc finger domain was identified as a positive interacting partner (data not shown, but the domains of UL144, TRIM23, and TRAF6 are summarized in Fig. 5A). TRIM23 also contains a coiled-coil or multimerization domain, which is similar to the TRAF6 multimerization domain. Multimerization and autoubiquitination of TRAF6 initiates a signaling cascade culminating in the activation of NF-κB signaling. Other viruses also cause TRAF6 multimerization to mediate NF-κB upregulation, such as human T-cell leukemia virus, which does so via the Tax protein (another coiled-coil motif-containing protein) (43).
Therefore, we tested whether TRIM23 and UL144 regulate NF-κB-mediated signaling via TRAF6 ubiquitination. TRAF6 could be detected by immunoprecipitation from cells which had been transfected to overexpress Flag-tagged TRAF6, HA-ubiquitin, and UL144 (Fig. 5B, lanes 1 to 3). Higher-molecular-weight products were weakly visible when TRAF6 was immunoprecipitated (Fig. 5B, lane 1, top panel), and when this was blotted for cotransfected HA-tagged ubiquitin, the immunoprecipitated TRAF6 was shown to be ubiquitinated (Fig. 5B, lane 1, bottom panel). Since TRAF6 signaling has been shown to require K63 and not K48 ubiquitination, we tested which type of TRAF6 ubiquitination was being mediated by UL144 (Fig. 5B, lanes 2 and 3). Cells were cotransfected with HA-tagged ubiquitin, K63R, or K48R mutants. These mutants have the indicated lysines mutated to arginine, thus preventing ubiquitin linkages at these residues. When cells that had been transfected with UL144, TRAF6 (Flag tagged), and HA-tagged ubiquitin mutants were immunoprecipitated with anti-Flag, we detected TRAF6 protein, but higher-molecular-weight species of TRAF6 were only detected using the K48R and not the K63R-HA-tagged ubiquitin molecules (Fig. 5B, lanes 2 and 3). Consistent with these higher-molecular-weight species being due to ubiquitination, reblotting with an anti-HA antibody (which only detects ubiquitinated products) showed that ubiquitinated TRAF6 was only detected when the K48R and not the K63R mutant was cotransfected (Fig. 5B, lanes 2 and 3, bottom panel), indicating that K63-linked TRAF6 ubiquitination is induced by UL144.
To test whether the ubiquitination of TRAF6 observed in Fig. 5B is due to the presence of TRIM23, siRNA analysis was performed to look at the ability of TRAF6 to become ubiquitinated in the presence of UL144. Figure 5C shows that when TRIM23 was removed from the cellular environment, UL144 was no longer able to induce ubiquitination of TRAF6 (compare lanes 2 and 3).
To analyze the role that TRIM23 played in UL144-mediated TRAF6 ubiquitination, further coimmunoprecipitations were carried out (Fig. 5D). UL144, TRAF6 (Flag-tagged), and HA-ubiquitin were overexpressed in cells also expressing V-tagged TRIM23 (Fig. 5D, lane 2) or empty vector control (Fig. 5D, lane 1). The V-tagged TRIM23 was detected in the appropriate controls (Fig. 5D, lane 2, top panel), and UL144 was effectively immunoprecipitated from cells (Fig. 5D, lanes 1 and 2, third panel). TRAF6 was coimmunoprecipitated more effectively by TRIM23 overexpression than by cells expressing only endogenous levels of TRIM23 (Fig. 5D, fourth panel, compare lanes 1 and 2), and the amount of ubiquitinated protein associated with UL144 was also enhanced (Fig. 5D, bottom panel, compare lanes 1 and 2). Input controls (second panel) show equal loading.
To address whether the association of TRIM23 and TRAF6 causes TRAF6 autoubiquitination or whether it is the known E3 ligase activity of TRIM23 that causes the ubiquitination of TRAF6, a TRAF6 mutant devoid of autoubiquitination activity but still capable of being ubiquitinated was obtained which has a point mutation C70A, located in the RING finger domain of TRAF6 (3). Although both the wild-type and C70A forms of TRAF6 were able to associate with UL144 (Fig. 5E, top and bottom panels), higher-molecular-weight products were only observed in the presence of wild-type TRAF6, but not in the presence of C70A TRAF6, arguing that it is TRAF6 autoubiquitination that is required for UL144-mediated activation.
Then, to analyze the functional effects of this TRAF6 mutant on UL144-dependent NF-κB activation, a luciferase reporter assay was carried out (Fig. 5F). UL144 alone stimulates the NF-κB-responsive promoter driving the expression of luciferase. However, if wild-type TRIM23 or TRAF6 is overexpressed, UL144-mediated NF-κB activation is enhanced (Fig. 5F). However, when the TRAF6 mutant C70A, which lacks autoubiquitination ability, is overexpressed, UL144-mediated activation of NF-κB is substantially reduced, suggesting that this mutant actually acts as a dominant-negative and that the autoubiquitination activity of TRAF6 is critical for UL144-mediated NF-κB activation.
Taken together, these data suggest that UL144 complexes with TRIM23, which in turn causes K63-linked autoubiquitination of TRAF6, leading to NF-κB signaling to the MDC promoter.
DISCUSSION
The regulation of NF-κB during the course of herpesvirus infection appears to be very tightly controlled, with viral functions modulating the extent of NF-κB activation at different times of infection. Alphaherpesviruses, such as herpes simplex virus, stimulate NF-κB (1, 15, 25), which can inhibit apoptosis (12-14), and gammaherpesviruses such as EBV are known to stimulate NF-κB by the LMP-1 protein using a Pra1-dependent mechanism to achieve continual cell proliferation (21). Betaherpesviruses, such as HCMV, appear to target NF-κB signaling differentially throughout the course of infection (5, 6, 7, 44). For instance, it is well established that NF-κB is induced by HCMV binding to the outside of cells. However, this induction of NF-κB activity is downregulated at immediate-early times of infection by HCMV IE86 (35). At later times of infection, however, NF-κB is activated again by UL144, even in the presence of IE86 (26, 28). The NF-κB regulation appears, therefore, to be carefully tailored by HCMV for its specific needs during the course of infection. Interestingly, HCMV also ensures that virus-specific regulation of NF-κB dominates in the infected cell is not compromised by cell-specific signals. For instance, the TNFR, a potent NF-κB inducer is downregulated from the surface of HCMV-infected cells (2), presumably ensuring that any cell-specific signals do not compromise virus-specific signaling of NF-κB.
NF-κB targeting is clearly important at some time or other during the course of infection with all herpesvirus subfamilies. For example, herpes simplex virus type 1 encodes protein ICP27 that stimulates persistent NF-κB signaling by targeting the p65 subunit of NF-κB (17). Both the EBV LMP-1 protein and the HCMV UL144 protein upregulate NF-κB by targeting TRAF6; however, the exact mechanism by which this is achieved is different. Liu et al. (21) have shown that, in addition to the requirement for TRAF6, LMP-1 signaling is dependent on the direct interaction of LMP-1 with the prenylated Rab acceptor protein 1. In contrast, our data show that Pra1 is dispensable for UL144-mediated NF-κB upregulation and that UL144 complexes with TRIM23 and TRAF6 to mediate signaling. Consequently, although the exact mechanism by which NF-κB is activated by UL144 and LMP-1 is different between these viruses, they both upregulate NF-κB and mediate CCL22 secretion by similar but not identical mechanisms. It is interesting that Pra1 and TRIM23 share some domain homology in that both have “Rab” domains (Fig. 5A), although the relevance of this domain for any function in NF-κB activation remains to be determined. The use of these two different signaling intermediates (Pra1 and TRIM23) may reflect that NF-κB stimulation by UL144 of HCMV is transient, unlike LMP-1 of EBV, which constantly activates NF-κB, probably as part of its stimulation of continual cell proliferation. Thus, Pra1-mediated activation of NF-κB through LMP-1 may be less responsive to “stop-start” signaling than TRIM-23-mediated activation of NF-κB through UL144.
The necessity for TAK1 in LMP-1 mediated NF-κB signaling remains controversial (37, 41). These discrepancies may be due to the cell types and viral strains used for the experiments. In our analysis, activation of NF-κB by UL144 clearly requires TAK1; lack of either TAK1 or TRIM23 prevents UL144 from stimulating NF-κB responsive promoters.
TRIM family proteins have attracted much attention recently, particularly because of their roles in virus infection. Examples include human immunodeficiency virus targets: TRIM5α, TRIM1, TRIM18, and TRIM19 contain SPRY domain, which have been shown to be critical for human immunodeficiency virus type 1 restriction (42). TRIM19 (also known as PML) has also been shown to play a role in both alpha- and betaherpesvirus restriction (9, 34). TRIM family proteins have also been implicated in various immune response signaling pathways. For instance, TRIM25 has been shown to play a role in the regulation of IRF3 and the induction of IFN by enhancing the ubiquitination of RIG-1 (10), and TRIM30 has been shown to negatively target the NF-κB induction in response to Toll-like receptors by targeting TAB2 and TAB3 for degradation (32). Here, we report that TRIM23, like TRIM25, enhances K63-linked ubiquitination. However, the target molecule for TRIM23 is TRAF6, whereas TRIM25 targets RIG-1. To our knowledge, this is a novel, thus-far-unreported, function of the TRIM23 protein. Thus, we propose that TRIM23 mediates TRAF6 autoubiquitination in the presence of UL144, resulting in the virally controlled activation of NF-κB stimulation at early times of HCMV infection. Furthermore, our studies show that TRIM23 is not involved in the ubiquitination of TRAF6 mediated by TNF or dsRNA signaling (Fig. 4). Nevertheless, it is likely that TRIM23 plays another role in the cellular regulation of TRAF6 ubiquitination, although the stimuli for this are yet to be ascertained.
Acknowledgments
We thank Christopher Benedict for the monoclonal antibody to UL144 and for reading the manuscript.
This study was funded by the Wellcome Trust (grant RG35737).
Footnotes
Published ahead of print on 28 January 2009.
REFERENCES
- 1.Amici, C., G. Belardo, A. Rossi, and M. G. Santoro. 2001. Activation of IκB kinase by herpes simplex virus type 1: a novel target for anti-herpetic therapy. J. Biol. Chem. 27628759-28766. [DOI] [PubMed] [Google Scholar]
- 2.Baillie, J., D. A. Sahlender, and J. H. Sinclair. 2003. Human cytomegalovirus infection inhibits tumor necrosis factor (TNF) alpha signaling by targeting the 55-kilodalton TNF receptor. J. Virol. 777007-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloor, S., G. Ryzhakov, S. Wagner, P. J. Butler, D. L. Smith, R. Krumbach, I. Dikic, and F. Randow. 2008. Signal processing by its coiled coil zipper domain activates IKKγ. Proc. Natl. Acad. Sci. USA 1051279-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 7078-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, G., E. R. Bivins-Smith, M. S. Smith, and A. D. Yurochko. 2008. Transcriptome analysis of NF-κB- and phosphatidylinositol 3-kinase-regulated genes in human cytomegalovirus-infected monocytes. J. Virol. 821040-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMeritt, I. B., L. E. Milford, and A. D. Yurochko. 2004. Activation of the NF-κB pathway in human cytomegalovirus-infected cells is necessary for efficient transactivation of the major immediate-early promoter. J. Virol. 784498-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMeritt, I. B., J. P. Podduturi, A. M. Tilley, M. T. Nogalski, and A. D. Yurochko. 2006. Prolonged activation of NF-κB by human cytomegalovirus promotes efficient viral replication and late gene expression. Virology 34615-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn, W. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 10014223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, R. D., C. Parada, P. Gripon, H. Sirma, and A. Orr. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 822661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gack, M. U., Y. C. Shin, C. H. Joo, T. Urano, C. Liang, L. Sun, O. Takeuchi, S. Akira, Z. Chen, S. Inoue, and J. U. Jung. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446916-920. [DOI] [PubMed] [Google Scholar]
- 11.Gerna, G., E. Percivalle, D. Lilleri, L. Lozza, C. Fornara, G. Hahn, F. Baldanti, and M. G. Revello. 2005. Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131-128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J. Gen. Virol. 86275-284. [DOI] [PubMed] [Google Scholar]
- 12.Goodkin, M. L., S. Epstein, P. A. Asbell, and J. A. Blaho. 2007. Nuclear translocation of NF-κB precedes apoptotic poly(ADP-ribose) polymerase cleavage during productive HSV-1 replication in corneal epithelial cells. Investig. Ophthalmol. Vis Sci. 484980-4988. [DOI] [PubMed] [Google Scholar]
- 13.Goodkin, M. L., E. R. Morton, and J. A. Blaho. 2004. Herpes simplex virus infection and apoptosis. Int. Rev. Immunol. 23141-172. [DOI] [PubMed] [Google Scholar]
- 14.Goodkin, M. L., A. T. Ting, and J. A. Blaho. 2003. NF-κB is required for apoptosis prevention during herpes simplex virus type 1 infection. J. Virol. 777261-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory, D., D. Hargett, D. Holmes, E. Money, and S. L. Bachenheimer. 2004. Efficient replication by herpes simplex virus type 1 involves activation of the IκB kinase-IκB-p65 pathway. J. Virol. 7813582-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 7810023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hargett, D., S. Rice, and S. L. Bachenheimer. 2006. Herpes simplex virus type 1 ICP27-dependent activation of NF-κB. J. Virol. 8010565-10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishitani, T., G. Takaesu, J. Ninomiya-Tsuji, H. Shibuya, R. B. Gaynor, and K. Matsumoto. 2003. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 226277-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson-Bernitsas, D. G., H. Ichikawa, Y. Takada, J. N. Myers, X. L. Lin, B. G. Darnay, M. M. Chaturvedi, and B. B. Aggarwal. 2007. Evidence that the TNF-TNFR1-TRADD-TRAF2-RIP-TAK1-IKK pathway mediates constitutive NF-κB activation and proliferation in human head and neck squamous cell carcinoma. Oncogene 261385-1397. [DOI] [PubMed] [Google Scholar]
- 20.Kaspari, M., N. Tavalai, T. Stamminger, A. Zimmermann, R. Schilf, and E. Bogner. 2008. Proteasome inhibitor MG132 blocks viral DNA replication and assembly of human cytomegalovirus. FEBS Lett. 582666-672. [DOI] [PubMed] [Google Scholar]
- 21.Liu, H., C. C. Wu, and Y. S. Chang. 2006. PRA1 promotes the intracellular trafficking and NF-κB signaling of EBV latent membrane protein 1. EMBO J. 254120-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy, E., I. Rigoutsos, T. Shubuya, and T. E. Shenk. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. USA 10013585-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama, T., K. Hieshima, D. Nagakubo, E. Sato, M. Nakayama, K. Kawa, and O. Yoshie. 2004. Selective induction of Th2-attracting chemokines CCL17 and CCL22 in human B cells by latent membrane protein 1 of Epstein-Barr virus. J. Virol. 781665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pass, R. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 25.Patel, A., J. Hanson, T. I. McLean, J. Olgiate, M. Hilton, W. E. Miller, and S. L. Bachenheimer. 1998. Herpes simplex type 1 induction of persistent NF-κB nuclear translocation increases the efficiency of virus replication. Virology 247212-222. [DOI] [PubMed] [Google Scholar]
- 26.Poole, E., E. Atkins, T. Nakayama, O. Yoshie, I. Groves, A. Alcami, and J. Sinclair. 2008. NF-κB-mediated activation of the chemokine CCL22 by the product of the human cytomegalovirus gene UL144 escapes regulation by viral IE86. J. Virol. 824250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 30333-46. [DOI] [PubMed] [Google Scholar]
- 28.Poole, E., C. King, J. Sinclair, and A. Alcami. 2006. The UL144 gene product of human cytomegalovirus activates NF-κB via a TRAF6-dependent mechanism. EMBO J. 254390-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole, E., P. Strappe, H. P. Mok, R. Hicks, and A. M. L. Lever. 2005. HIV-1 gag-RNA interaction occurs at a perinuclear/centrosomal site: analysis by confocal microscopy and FRET. Traffic 61-15. [DOI] [PubMed] [Google Scholar]
- 30.Prichard, M. N., M. E. Penfold, G. M. Duke, R. R. Spaete, and G. W. Kemble. 2001. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 11191-200. [DOI] [PubMed] [Google Scholar]
- 31.Schultheiss, U., S. Puschner, E. Kremmer, T. W. Mak, H. Engelmann, W. Hammerschmidt, and A. Kieser. 2001. TRAF6 is a critical mediator of signal transduction by the viral oncogene latent membrane protein 1. EMBO J. 205678-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi, M., W. Deng, E. Bi, K. Mao, Y. Ji, G. Lin, X. Wu, Z. Tao, Z. Li, X. Cai, S. Sun, C. Xiang, and B. Sun. 2008. TRIM30α negatively regulates TLR-mediated NF-κB activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 9369-377. [DOI] [PubMed] [Google Scholar]
- 33.Song, Y.-L., K.-Y. Jen, V. Soni, E. Kieff, and E. Cahir-McFarland. 2006. IL-1 receptor-associated kinase 1 is critical for latent membrane protein 1-induced p65/RelA serine 536 phosphorylation and NF-κB activation. Proc. Natl. Acad. Sci. USA 1032689-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tavalai, N., P. Papior, S. Rechter, and T. Stamminger. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82126-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, R. T., and W. A. Bresnahan. 2006. Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFκB-dependent gene expression. J. Virol. 8010763-10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomasec, P., E. C. Wang, A. J. Davison, B. Vojtesek, M. Armstrong, C. Griffin, B. P. McSharry, R. J. Morris, S. Llewellyn-Lacey, C. Rickards, A. Nomoto, C. Sinzger, and G. W. Wilkinson. 2005. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat. Immunol. 6181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uemura, N., T. Kajino, H. Sanjo, S. Sato, S. Akira, K. Matsumoto, and J. Ninomiya-Tsuji. 2006. TAK1 is a component of the Epstein-Barr virus LMP1 complex and is essential for activation of JNK but not of NF-κB. J. Biol. Chem. 2817863-7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vichi, A., D. M. Payne, G. Pacheco-Rodriguez, J. Moss, and M. Vaughan. 2005. E3 ubiquitin ligase activity of the trifunctional ARD1 (ADP-ribosylation factor domain protein 1). Proc. Natl. Acad. Sci. USA 1021945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, W., S. L. Taylor, S. A. Leisenfelder, R. Morton, J. F. Moffat, S. Smirnov, and H. Zhu. 2005. Human cytomegalovirus genes in the 15-kilobase region are required for viral replication in implanted human tissues in SCID mice. J. Virol. 792115-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wills, M. R., O. Ashiru, M. B. Reeves, G. Okecha, J. Trowsdale, P. Tomasec, G. W. Wilkinson, J. Sinclair, and J. G. Sissons. 2005. Human cytomegalovirus encodes an MHC class-I-like molecule (UL142) that functions to inhibit NK cell lysis. J. Immunol. 1757457-7465. [DOI] [PubMed] [Google Scholar]
- 41.Wu, L., H. Nakano, and Z. Wu. 2006. The C-terminal activating region 2 of the Epstein-Barr virus-encoded latent membrane protein 1 activates NF-κB through TRAF6 and TAK1. J. Biol. Chem. 2812162-2169. [DOI] [PubMed] [Google Scholar]
- 42.Yap, M. W., M. P. Dodding, and J. P. Stoye. 2006. Trim-cyclophilin A fusion proteins can restrict human immunodeficiency virus type 1 infection at two distinct phases in the viral life cycle. J. Virol. 804061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, Q., Y. Minoda, R. Yoshida, H. Yoshida, H. Iha, T. Kobayashi, A. Yoshimura, and G. Takaesu. 2008. HTLV-1 Tax-mediated TAK1 activation involves TAB2 adapter protein. Biochem. Biophys. Res. Commun. 365189-194. [DOI] [PubMed] [Google Scholar]
- 44.Yurochko, A. D. 2008. Human cytomegalovirus modulation of signal transduction. Curr. Top. Microbiol. Immunol. 325205-220. [DOI] [PubMed] [Google Scholar]