Abstract
Current adeno-associated virus (AAV) gene therapy vectors package a transgene flanked by the terminal repeats (TRs) of AAV type 2 (AAV2). Although these vectors are replication deficient, wild-type (wt) AAV2 prevalent in the human population could lead to replication and packaging of a type 2 TR (TR2)-flanked transgene in trans during superinfection by a helper virus, leading to “mobilization” of the vector genome from treated cells. More importantly, it appears likely that the majority of currently characterized AAV serotypes as well as the majority of new novel isolates are capable of rescuing and replicating AAV2 vector templates. To investigate this possibility, we flanked a green fluorescent protein transgene with type 2 and, the most divergent AAV serotype, type 5 TRs (TR2 or TR5). Consistent with AAV clades, AAV5 specifically replicated TR5 vectors, while AAV2 and AAV6 replicated TR2-flanked vectors. To exploit this specificity, we created a TR5 vector production system for Cap1 to Cap5. Next, we showed that persisting recombinant AAV genomes flanked by TR2s or TR5s were mobilized in vitro after addition of the cognate AAV Rep (as well as Rep6 for TR2) and adenoviral helper. Finally, we showed that a cell line containing a stably integrated wt AAV2 genome resulted in mobilization of a TR2-flanked vector but not a TR5-flanked vector upon adenoviral superinfection. Based on these data and the relative prevalence of wt AAV serotypes in the population, we propose that TR5 vectors have a significantly lower risk of mobilization and should be considered for clinical use.
The adeno-associated virus (AAV) genome is a single-stranded DNA molecule flanked by two inverted terminal repeats (TRs) that are necessary for replication and packaging of the genome into viral capsids (2). AAV gene therapy vectors are typically designed from AAV serotype 2 (AAV2), the first and most thoroughly characterized AAV serotype. In tissue culture, the AAV2 TRs (TR2s) are involved in the establishment of latency through site-specific integration of wild-type (wt) AAV2 genomes into the host genome at the q arm of chromosome 19 by the viral replication proteins (Rep) (29). In addition, AAV TRs are capable of intermolecular recombination to integrate randomly into the host chromosome (21) or to form concatemeric episomes, the most common fate for TR-flanked genomes in nondividing cells (6, 23). When the host cell experiences environmental stress or coinfection with a helper virus, such as adenovirus (Ad) or herpesvirus, the latent AAV genome is “mobilized” to enter the lytic phase of its life cycle and produce new AAV virions (2).
Recombinant AAV (rAAV) vectors for gene therapy studies use AAV genomes in which the Rep and capsid (Cap) genes are replaced with a transgene expression cassette, leaving no part of the original AAV genome except the 145-bp TRs. As Rep is not present in rAAV vector constructs, the ability to drive specific integration into chromosome 19 is lost. Long-term persistence of rAAV occurs primarily through the formation of extrachromosomal episomes, accompanied by a low frequency of nonspecific integration events into the host chromosome (21, 23). These methods of persistence appear to be consistent between serotypes regardless of the TR (10, 36). The stability of episomal or chromosomally integrated rAAV genomes suggests that rAAV and wt AAV genomes could cohabit the same cell indefinitely. Moreover, helper virus superinfection would activate wt Rep and Cap expression, setting up conditions capable of rescuing, replicating, and encapsidating the rAAV genome in trans. The packaged rAAV genome could then be mobilized to nontarget cells or between individuals in the same manner as wt AAV.
The potential of rAAV mobilization has been demonstrated using molecular substrates. For example, the excision of the rAAV genome from a plasmid backbone used in rAAV production demonstrates the potential of the rAAV vector to be rescued from DNA in cis (27). Mobilization of wt AAV genomes from cells in culture has been demonstrated conclusively (2, 4) and mimicked to various degrees using rAAV (34). The closest example of AAV vector mobilization in vivo in a primate model was carried out by Afione and colleagues where, by using three different rescue assays, they observed AAV CFTR vector mobilization only when large doses of wt AAV were first seeded in the lower lung and then followed by vector and Ad superinfection (1).
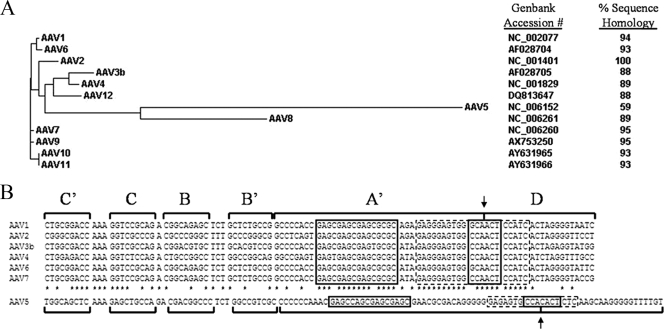
All of these studies point to the potential risk of rAAV mobilization. Because of the fact that AAV Rep proteins are largely nonspecific in their ability to recognize and act on the TRs of other serotypes and because numerous novel serotypes have been identified, the risk of rAAV vector mobilization seems to increase. For example, TR2 vectors can be replicated by AAV serotypes 1 to 4 and 6 (10, 11). This cross-compatibility exacerbates the potential of TR2-flanked rAAV vector mobilization since coinfection by a wide array of AAV serotypes could lead to replication and packaging of the transgene. Interestingly, AAV5 Rep (Rep5) cannot utilize TR2s as viral origins of replication, nor can Rep2 drive the replication of TR5 vectors (5, 18). The Rep proteins of AAV serotypes 1, 2, 3, 4, and 6 to 12 all share approximately 90% sequence homology within the N-terminal 200 amino acids of the large Rep proteins (Rep78 and Rep68), the portion of Rep responsible for binding and nicking the TR (8) (see Fig. 1A). Other recently identified simian AAVs also share 94% protein homology to Rep2 in this region. (30) In contrast, Rep5 shares only 58% sequence homology with AAV2 in this region. Additionally, TR5 has a cleavage site (AGTG/TGGC) and a predicted secondary structure distinct from those found in the TRs of other AAV serotypes, as well as an additional 30 nucleotides in overall length (5) (see Fig. 1B). The incompatibility of TR5s with the Reps of other serotypes offers the potential to create TR5 rAAV vectors with lower risk of mobilization. Since AAV5 is much less prevalent in the human population than are AAV2 or TR2-compatible serotypes (35 to 80% of adult humans are seropositive for AAV2 and 30% for AAV6, compared to 10 to 20% for AAV5) (13, 14, 20, 33), rAAV vectors flanked by TR5s should have a lower risk of mobilization.
FIG. 1.
Rep and TR homology between AAV serotypes. (A) A phylogenetic tree of amino acids 1 to 200 of the large Rep proteins of AAV serotypes 1 to 12. The distance from the root of the tree represents relative amino acid substitutions. The GenBank accession number for each serotype is shown, as well as the percent sequence homology of the amino terminus of each serotype to that of AAV2. (B) A nucleotide alignment of the TRs of AAV serotypes 1 to 7. The complementary strands of the A′ and D loops are omitted. The stems of the TRs are indicated, as are the Rep binding element (large box), terminal resolution site (arrow), and predicted stem-loop involved in nicking (dashed boxes). Nucleotides conserved between serotypes 1 and 4 and serotypes 6 and 7 are indicated with asterisks.
Although rAAV vector mobilization requires three hit kinetics in an individual cell (e.g., infected by rAAV, wt AAV, and helper virus), the chances of mobilization outside a laboratory setting are largely unknown and the concept warrants further investigation. Possible solutions may enable researchers to avoid the problem of rAAV mobilization before it becomes one. To develop assays for the investigation of rAAV mobilization, we have designed and used novel vectors carrying green fluorescent protein (GFP) flanked by TR2s or TR5s (see Fig. 2A). We assayed the ability of Rep2, Rep5, or Rep6 to replicate our GFP vectors, confirming previous reports that only Rep5 is able to replicate TR5 vectors while Rep2 and Rep6 can drive the replication of TR2 vectors. To exploit the specificity of the Rep5-TR5 interaction, we created a TR5 rAAV production system for the packaging of TR5 vector genomes into Cap1 to Cap5, achieving similar titers and only slightly altered transduction efficiency in a capsid-specific manner relative to those of TR2 rAAV previously described (24). With the ability to produce TR5 rAAV vectors, we hypothesized that TR5 vectors would be at lower risk of vector mobilization due to relative frequencies of type 5 in the populace and the promiscuity of TR2s with respect to other AAV serotypes (13, 14, 20, 33). Here, we demonstrate that Rep-TR specificity for rAAV production is conserved for the mobilization of rAAV genomes, suggesting that only AAV5 could lead to the mobilization of a TR5-flanked vector. Thus, vector genomes produced using TR5s should possess a significant safety advantage over the TR2-flanked genomes currently in use.
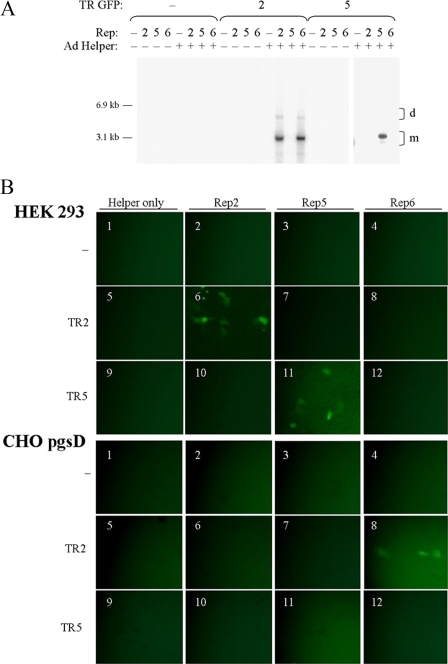
FIG. 2.
The Rep5-TR5 interaction is unique among the fully characterized AAV serotypes. (A) Vector constructs used in this study. GFP expression was driven by a CMV promoter and simian virus 40 poly(A) element. The neomycin cassette included the thymidine kinase promoter and the bovine growth hormone poly(A) element. TR2s or TR5s flanked the vectors. pTR5-eGFP contained an additional 500 bp ahead of the 3′ TR. (B) Southern blot of Hirt DNA comparing the abilities of Rep2Cap2, Rep5Cap2, and Rep6Cap6 to replicate TR2s or TR5-flanked vector genomes. Hirt DNA was isolated 48 h after transfection. DpnI cuts only the input plasmid, not the newly replicated AAV genomes. The two major replicative forms of AAV are indicated (m, double-stranded monomer; d, double-stranded dimer). Higher-order replicative forms are also visible. +, present; −, absent. (C) Transduction of HEK 293 cells (transducible by Cap2) or CHO pgsD cells (transducible by Cap6 or Cap2) with crude lysate from cells harvested 48 h after transfection of an Ad helper plasmid only or triple transfection of an Ad helper plasmid, TR2 or TR5 GFP, and either Rep2Cap2, Rep5Cap2, or Rep6Cap6. The numbers shown correspond to the lane of the gel in Fig. 2B.
MATERIALS AND METHODS
Plasmids.
Plasmids were constructed by subcloning AAV5 replication genes (Rep5) and AAV1 to AAV4 capsid genes (Cap1-4) into a Bluescript backbone (see Fig. 3A). The Rep5 fragment was amplified from the plasmid AAV5-2 (gift from R. M. Kotin, hereafter referred to as pRep5Cap5) via PCR using the forward primer rep-5 F1 5′-CGAGCTCGGCGCGTATGAGTTCTCGC-3′ and the reverse primer rep-5 R1 5′-GACTACTCGCTTTATTTACTGTTC-3′, which added an upstream SacI restriction site. The Cap2 fragment was amplified from the plasmid pXR2 (24), using forward primer 5′-ATGGCTGCCGATGGTTATCTTC-3′ and reverse primer 5′-TGGTGATGACTCTGTCGCCC-3′, which included a NotI restriction site downstream from Cap2. The Rep5 and Cap2 fragments were ligated back into the pXR2 plasmid via the SacI and NotI restriction sites, in a triple-fragment ligation, to make pRep5Cap2. The pRep5Cap2 plasmid was used as a template for construction of the remaining plasmids. The Cap1, Cap3, and Cap4 fragments were amplified from plasmids pXR1, pXR3, and pXR4, respectively, (24). The pXR plasmid series contain rep2 or chimeric replication genes with the cap genes from AAV serotypes that correspond to the plasmid number (24). The forward primers were pxr1/3 F (5′-ATGGCTCGGCATCCTTATCTTC-3′) and pxr4F (5′-ATGGCTGCTGACGGTTACC-3′), and the reverse primers were pxr1/3 R (5′-CTATGACCATGATTACGCCAAGC-3′) and pxr4 R (5′-CAGCTATGACCATGATTACGC-3′). The amplified region included a NotI restriction site downstream from the capsid genes. The Rep5 and Cap fragments were ligated into the pRep5Cap2 backbone via the PpuMI and NotI restriction sites. Rep5Cap1-4 plasmids were confirmed by restriction site analysis and DNA sequencing. Vector plasmids included the transgene enhanced GFP (eGFP) preceded by the cytomegalovirus (CMV) promoter and followed by the simian virus 40 polyadenylation [poly(A)] signal, flanked by AAV2 TR (TR2-eGFP) or AAV5 TR (TR5-eGFP) (see Fig. 2A). A neomycin cassette lies downstream, driven by a thymidine kinase promoter and the bovine growth hormone poly(A) signal. Note that the TR5-eGFP plasmid has an additional 500-bp insert upstream of the 3′ TR in order to distinguish it from TR2-eGFP on a gel. The pRep6Cap6 plasmid was obtained from David Russell (26).
FIG. 3.
Creation and characterization of Rep5 helper constructs for Cap1 to Cap5. (A) The AAV5 Rep and AAV1 to AAV4 Cap genes were subcloned into pXR2, a non-TR-containing plasmid (see Materials and Methods for details). These new helper plasmids were used to package TR5 vectors into Cap1 to Cap5. (B) Southern blot using a GFP-specific probe of Hirt DNA extracted from HEK 293 cells transfected with pRep5Cap1-5, an Ad helper plasmid, and TR5-GFP or TR2-GFP. The two replicative forms of the vector are indicated. (C) Cell lysate from triple transfections described above were used to infect naïve HEK 293 cells. Cells infected with lysate from TR5-eGFP vector transfection of capsid serotypes 1 to 5 were positive for GFP (panels 1 to 5 corresponding to AAV1 to AAV5). HEK 293 cells infected with lysate from TR2-eGFP vector transfection of capsid serotypes 1 to 5 did not express GFP (panel 6, AAV1; representative of AAV2 to AAV5). (D) Graph comparing the titers achieved in the production of TR2 versus TR5 rAAV. Titers of samples were determined in duplicate by quantitative PCR. Standard errors are indicated. (E) Graph comparing the transducing units per vector genome of rAAV TR2 versus TR5 vectors. Note that values on the y axis are multiplied by 1 × 10−7. Virus was serially diluted and used to infect cells. GFP-positive cells were quantitated and transducing units per microliter calculated before conversion to transducing units per vector genome. Samples were measured in duplicate, and standard errors are indicated.
Cell culture.
HEK 293 and Cos1 cell lines were originally obtained from the American Type Culture Collection (ATCC). Detroit 5 (D5) and Detroit 6 (D6) human bone marrow cells were a gift from K. Berns. The D5 cell line was originally cultured as a clone of wt AAV2 latently infected D6 cells (4) and contained wt AAV2 DNA integrated into the chromosomal DNA. All cells were maintained at 37°C with 5% CO2 saturation in media supplemented with 10% fetal bovine serum (Sigma) and 20 units/ml penicillin-streptomycin. HEK 293, Cos1, D5, and D6 cells were cultured in Dulbecco modified Eagle medium, while CHO pgsD-677 cells were grown in Ham F-12 medium. CHO pgsD-677 cells were obtained from the ATCC (Manassas, VA) (32).
Production of rAAV.
Approximately 8 × 106 HEK 293 cells were triple transfected with 5 μg Rep- and Cap-encoding plasmid (pRep5Cap1-5 or pXR1-5), 15 μg XX680 (Ad helper plasmid), and 5 μg of either TR5-eGFP or TR2-eGFP plasmid transgene vector. These constructs were mixed with 50 μl of 2.5 M CaCl2, 440 μl double-distilled H2O, and 500 μl 2× HEPES, and then added dropwise to the HEK 293 cells. At 48 h posttransfection, cells were harvested for the collection of cell lysate, Hirt DNA, and protein. Virus particles were collected from cell lysates produced from triple freezing and thawing of cells and centrifugation to remove cell debris.
Hirt DNA analysis.
At 48 h posttransfection, DNA from transfected cells was isolated using the Hirt method previously described (15). Fifteen micrograms of each DNA sample was digested with the DpnI enzyme, and both digested and undigested DNA were separated by electrophoresis on a 0.8% agarose gel. Following transfer to a nylon membrane, the Southern blot was probed with either a 735-bp GFP fragment or a fragment containing the Rep2 and Cap2 open reading frames (ORFs). The GFP probe was obtained from the digestion of a TR-eGFP plasmid with the AgeI and NotI enzymes. The Rep2Cap2 fragment was isolated from the digestion of pXR2 with PvuII.
In vitro transduction assay.
Lysate from each transfection was tested for infectivity to ascertain virus production. A total of 1 × 105 HEK 293 or Cos1 cells was seeded in each well of 48-well plates. Cos1 and CHO pgsD cells were used to assay AAV4 and AAV6, respectively, since they transduce HEK 293 cells at a low efficiency. Original cell lysates harvested from transfections were diluted to a range from 1 × 10−1 to 1 × 10−8. One hundred microliters of each serial dilution in addition to Ad (multiplicity of infection [MOI] of 5) were added to cells. Each infection was done in duplicate. At 24 h postinfection, GFP-positive cells were visualized. Under ×200 magnification, the number of GFP-positive cells per field was counted. Ten fields per well were counted, and the average number of GFP-positive cells/field was determined. This number was multiplied by the number of fields per well and then divided by the amount of lysate added to each well (as given by the dilution factor) to determine the number of transducing units (TU) per μl of cell lysate.
Quantitative PCR for virus titer.
The number of virus particles per microliter was determined for each virus preparation. Ten microliters of partially purified cell lysate (see “Production of rAAV” above) was added to 90 μl DNase I solution (10 μg DNase I, 10 mM Tris [pH 7.5], 10 mM MgCl2, 2 mM CaCl2) and incubated for 1 h at 37°C and stopped with the addition of 6 μl of 0.5 M EDTA. Then, 120 μl of proteinase K solution (1 M NaCl, 1% Sarkosyl, 20 μg proteinase K) was added, and each sample was incubated for 2 h at 55°C. Each sample was heated at 95°C for 10 min, diluted 1:500 or 1:5,000, and used directly as a template for quantitative PCR.
For quantitation of DNase-resistant viral genomes in the sample, 2 μl were used as a template in a 10 μl real-time PCR. The LightCycler FastStart DNA master SYBR green I kit (Roche catalog number 12239264001) was used by following the manufacturer's instructions, using 3 mM MgCl2 and 500 nM of each primer. All reactions were carried out on a Roche diagnostic real-time PCR LightCycler 2.0 kit. For each reaction, the cycling parameters were as follows: 10 min at 95°C; 5 cycles at 95°C for 15 s, 64°C for 5 s, and 72°C for 15 s; 5 cycles at 95°C for 15 s, 62°C for 5 s, and 72°C for 15 s; 40 cycles at 95°C for 15 s, 60°C for 5 s, and 72°C for 15 s. At the end of each run, a melting curve analysis was performed, in which the PCR products were annealed at 72°C and the temperature was gradually raised to 99°C. In all cases, the PCR products melted in a narrow temperature range, indicating a pure PCR product without detectable nonspecific amplification. The following primers were used: QGFP1-F (5′-AGCAGCACGACTTCTTCAACTCC-3′) and QGFP1-R (5′-TGTAGTTGTACTCCAGCTTGTGCC-3′). A plasmid containing GFP, pTR2-GFP, was used to generate the standard curve.
Dot blot analysis.
Dot blot analyses were performed using 10 μl of purified cell lysate from the various transduction assays, as described previously (8). The samples were blotted onto a nylon membrane, using a dot blot manifold, and UV cross-linked at 60 mJ (UV Stratalinker 1800; Stratagene). The Southern blot was hybridized to a radiolabeled GFP probe (see “Hirt DNA analysis” above) and exposed to film or visualized using a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
DNA alignment and generation of phylogenetic trees.
ClustalW2 was used for all DNA alignments and to generate phylogenetic trees (17). The amino-terminal 200 amino acids of the large Rep proteins from AAV1 to AAV12 were used to generate a phylogenetic tree by the PHYLIP method. This program also calculated the percent homology between the Rep proteins of different serotypes.
Creation and mobilization of AAV genomes from HEK 293 stable GFP-expressing cell lines.
A total of 1 × 105 HEK 293 cells were infected with 10,000 vector genomes/cell of TR2- or TR5-eGFP rAAV and were then cultured for 18 days. Cells (approximately 1% GFP positive) were then trypsinized and subjected to flow sorting with a Beckman-Coulter Dako MoFlo cell sorter to isolate GFP-positive cells. A total of 1,500 GFP-positive cells were pooled into one well of a 96-well plate and allowed to propagate. GFP expression was monitored to ensure a homogeneous GFP-positive population in which the transgene was maintained. AAV helper plasmids were transfected into these cells in 10-cm dishes and incubated for 48 h prior to Hirt DNA extraction and preparation of crude lysate.
TR5 and TR2 vector mobilization assay.
D6 and D5 cell lines were infected under four different conditions. A 10-cm plate of each cell line, approximately 75% confluent, was incubated overnight at 37°C in 5% CO2 saturation with 50 μl AAV2 TR2-eGFP or AAV2 TR5-eGFP cell lysate. Cells were then washed four times with 10 ml Dulbecco modified Eagle medium and coinfected with Ad (MOI of 10). Control plates were infected with only AAV2 TR2-eGFP or only Ad. At 96 h postinfection, cell lysate and Hirt DNA from each infection were harvested as described above. Hirt DNA was separated by electrophoresis on a 0.8% agarose gel and transferred to a nylon membrane. The blot was probed for GFP as described above. The membrane was stripped by boiling with 1% sodium dodecyl sulfate. The membrane was rehybridized with a probe specific for Rep2 and Cap2. Additionally, cell lysates from each infection and Ad (MOI of 5) were used to infect 1 × 105 fresh HEK 293 cells. Cells positive for GFP were visualized 24 h postinfection.
RESULTS
Rep5-TR5 interaction is unique among fully characterized AAV serotypes.
As previously reported, Rep1 to Rep4 and Rep6 are unable to catalyze replication of TR5-flanked genomes, while Rep5 is unable to catalyze the replication with vectors flanked by TR1 to TR4 and TR6 (10, 11). Due to high sequence homology, it is likely that AAV7 to AAV12 are also compatible with TR2s and not TR5s (Fig. 1A). In order to demonstrate the specificity of the Rep5-TR5 interaction, two GFP vectors were utilized (Fig. 2A). These constructs, TR2-eGFP and TR5-eGFP, are flanked by either TR2s or TR5s and express GFP from a CMV promoter. The rAAV vectors were transfected into HEK 293 cells along with an Ad helper plasmid (pXX680) and either Rep2Cap2, Rep5Cap2, or Rep6Cap6. Rep6 was included to confirm the cross-compatibility of AAV2 and AAV6 replication, as well as to underscore the ability of other naturally occurring serotypes to replicate TR2 vectors. After 48 h, cells were harvested for Hirt DNA and crude lysate. Hirt DNA (15) was analyzed by Southern blotting with a probe for the GFP ORF. A DpnI digestion was performed to remove transfected methylated plasmid DNA, but not unmethylated genomes which had been replicated in the cell. The results validated Rep-TR specificity for the serotypes used, with Rep2 and Rep6 driving replication of only TR2 vectors and Rep5 driving replication of only the TR5 vector (Fig. 2B). Crude lysate from these cells was used to transduce HEK 293 cells (highly transducible by Cap2) and CHO pgsD cells (transducible by Cap6) (Fig. 2C). Specificity in the production of rAAV vectors followed the same pattern as that of replication, with Rep2 and Rep6 each able to produce infectious TR2 rAAV particles and Rep5 able to produce only infectious TR5 rAAV particles.
Creation and characterization of Rep5 helper constructs for Cap1-5.
As the prevalence of AAV5 in the human population is lower than that of AAV2 or AAV6, and considering the unique specificity between Rep5 and TR5, we decided to create an rAAV TR5 production system for transcapsidation into Cap1-5 that is similar to a previously described system for AAV type 2 (24). In order to confirm the efficacy of TR5 vectors with respect to that of existing TR2 vectors, Rep5 helper constructs were created (Fig. 3A). This new system for producing virus vectors utilizes triple transfection with AAV helper plasmids containing the AAV5 Rep gene and one of the AAV1 to AAV5 Cap genes (pRep5Cap1-5), a reporter transgene plasmid with GFP flanked by TR5s, and an Ad helper plasmid (XX680). HEK 293 cells were transfected with pRep5Cap1-5, an Ad helper plasmid, and TR2 or TR5 eGFP. Analyses of Hirt DNA extracted from these cells showed that Rep5 functioned properly, generating the expected DpnI-resistant AAV monomer and dimer replication intermediates when delivered with TR5-eGFP but not the TR2-eGFP vector (Fig. 3B). Additionally, cell lysate harvested from each transfection was tested for infectivity to ascertain the system's ability to produce functional recombinant virus. HEK 293 or Cos1 cells were exposed to lysate from the TR5 transfections and assayed for GFP expression at 24 h postinfection. Lysates carrying capsid-specific sequences (types 1 to 5) all produced GFP-positive cells when TR5 was complemented with Rep5 expression plasmids during vector production (Fig. 3C, panels 1 to 5). Cells exposed to lysate from TR2 transfections in the presence of Rep5 proteins were negative for transgene expression (Fig. 3C, panel 6).
Having developed a functional AAV capsid production system using TR5s, we sought to compare it to current Rep2-TR2 production yields. The protocol designed by our lab (24) for the production of transcapsidated rAAV2 (i.e., using helper plasmids that contain AAV Rep2 and serotype 1 to 5 capsid genes, a reporter transgene [GFP] in a TR2 vector cassette, and an Ad helper plasmid) was used as a comparison to evaluate the new Rep5-TR5 system. Cell lysate was harvested from each transfection and assayed for virus production by titer determination using quantitative PCR to determine the number of virus particles per unit volume of lysate (Fig. 3D). The measurements obtained from the dot blot analysis are comparable for TR2 and TR5 vectors when packaged in AAV serotype 1 to 4 capsids. Interestingly, TR2 vector titers were noticeably reduced when the vectors were packaged in an AAV5 capsid, possibly due to the evolutionary divergence of AAV5 with respect to the other serotypes (Fig. 1A). Both TR5 and TR2 vector production systems were also assayed for infectivity as measured by the number of transducing viral units per vector genome (Fig. 3E). A total of 1 × 105 HEK 293 cells were exposed to serial dilutions of cell lysate from each transfection, and the number of resulting transgene-positive cells was used to calculate the number of transducing units per microliter (TU/μl) of cell lysate. This was then divided by the viral titer (vg/μl) to yield the transducing units per vector genome (TU/vg). TR5 vectors displayed a minor drop in transduction efficiency compared to that of TR2 vectors, ranging from 5- to 10-fold in Cap1 to Cap3, while TR5 vectors performed better in Cap5. These data demonstrate the transduction potential of each individual vector genome, again suggesting that due to evolutionary divergence, TR5 vectors may have slightly better transduction potential when encapsidated in an AAV5 capsid. Transduction of the HEK 293 cells by Cap4 was below the detection threshold of the assay. The results indicate that while the yields of TR5 rAAV vector production are equivalent to the widely utilized TR2 system, there may be a capsid-specific effect on the ability of these vectors to transduce cells efficiently.
AAV genomes conferring long-term transgene expression in cultured cells can be rescued, replicated, and packaged.
During infection, AAV genomes not degraded have two fates: episomal formation or chromosomal integration (6, 21). While wt AAV2 has been shown in tissue culture cells to integrate into the human chromosome in a site-specific fashion (4, 29), it has been demonstrated that Rep is required for this form of latency (2). Ideally, rAAV vectors should be delivered in the absence of Rep, wherein numerous studies have determined that rAAV genomes remain episomal, typically circularizing or forming into concatemers as a mechanism of vector persistence (7, 31, 35). Thus, the infrequent event of integration by rAAV genomes is not site specific (21). Regardless of the method of molecular persistence, AAV genomes are able to excise themselves from the chromosome or episome upon Ad superinfection to enter the lytic phase of the AAV life cycle, suggesting that wt AAV persists in a conservative manner, keeping at least one TR sequence intact (28).
To test whether persisting rAAV vectors could be rescued after infection in the absence of site-specific integration, HEK 293 cells were infected with 10,000 vector genomes/cell of either TR2 or TR5 GFP virus, as determined by dot blot analysis. After 18 days, GFP cells were sorted and pooled to approximate a population of cells infected by the vector. While we did not confirm that these vectors had integrated into the host chromosome, we did confirm that after sorting, GFP remained in 100% of the cells for longer than 2 months. Rep2Cap2, Rep5Cap2, or Rep6Cap6 and an Ad helper plasmid were transfected into the mock, TR2-, or TR5-containing cell lines, and both Hirt DNA and crude lysate were isolated. Figure 4A reveals that both TR2 and TR5 genomes were capable of being rescued and replicated (lanes 6, 8, and 11). Specificity remained consistent for these vectors, with both Rep2 and Rep6 able to rescue and replicate TR2s and only Rep5 able to replicate TR5s. The pTR5-eGFP panel was exposed longer than were the TR2 or mock cell lines due to the small amount of replicated vector DNA isolated from these cells.
FIG. 4.
AAV genomes conferring long-term transgene expression in cultured cells can be rescued, replicated, and packaged. (A) HEK 293 cells were infected with either TR2 or TR5 eGFP vectors and passaged 18 days before cells still expressing GFP were sorted and pooled. AAV helper plasmids (Rep2Cap2, Rep5Cap2, Rep6Cap6) in the presence (+) or absence (−) of Ad helper plasmid were then added to the assay for the ability of persistent AAV genomes to be rescued and undergo replication in the presence or absence of Ad helper plasmid. Hirt DNA was isolated and assayed via Southern blotting with a probe for the GFP ORF. The two major replicative forms of the vector genomes are indicated. Larger replicative forms are also visible. The TR5 with helper plasmid panel of the blot was subjected to a longer exposure in order to visualize the replicating vector genomes. The two major replicative forms of AAV are indicated (m, double-stranded monomer; d, double-stranded dimer). (B) Mobilized genomes were assayed for infectivity by transducing HEK 293 or CHO pgsD cells with crude lysate from the cells described for panel A (control, TR2, or TR5 persisting vector genomes transfected with the helper plasmids described) 48 h after addition of helper plasmids (all transfections for lysate used in Fig. 4B included an Ad helper plasmid).
To determine if these rescued, replicating genomes could be encapsidated and mobilized to naïve cells, lysate was added to HEK 293 or CHO pgsD cells. Figure 4B shows that rescued genomes were encapsidated and that persisting rAAV genomes can be mobilized into previously nontransduced cells (HEK 293, panels 6 and 11, and CHO pgsD, panel 8). Predictably, the transduction profile of the mobilized TR2 or TR5 vector genomes was dependent on the capsid into which they were packaged (Cap2 or Cap6), highlighting the potential danger of TR2 vector mobilization being driven by a range of wt AAV serotypes.
TR2 vectors, but not TR5, are mobilized by latent wt AAV2 and Ad coinfection.
AAV2 has been consistently demonstrated to be the most prevalent natural AAV serotype in the human population (13, 14, 20, 33); thus, there is a strong likelihood that a high percentage of human individuals harbor a latent AAV2 infection (22). For that reason, we obtained D5 cells demonstrated to contain a latent wt AAV2 infection in order to model the potential for TR2 or TR5 vector mobilization upon rAAV and Ad infection. The D5 cell line contains the wt AAV2 genome, which is stably integrated at chromosome 19 and is rescuable upon infection by helper virus (4, 29). The parental line, D6, is negative for wt AAV and was used as a control. Cell lysate containing TR2- or TR5-flanked rAAV GFP genomes encapsidated into Cap2 was harvested from triple-plasmid transfections of HEK 293 cells. Each type of lysate was used to infect D5 and D6 cells. After 24 h of incubation, cells were washed and coinfected with Ad helper virus. Control plates were exposed to lysate containing either AAV2 TR2-eGFP only or Ad only. Transgene GFP expression was observed in all cells receiving the original cell lysate, confirming the infectivity of AAV2 TR2-eGFP and AAV2 TR5-eGFP in D5 and D6 cells (data not shown). Hirt DNA analysis of infected cells revealed rescue of latent AAV2 genes, in the form of AAV2 replication intermediates, in D5 cells infected with Ad (Fig. 5A). As expected, D6 cells without latent AAV or D5 cells without Ad did not show AAV2 replication intermediates (Fig. 5A). In addition, rescued latent wt AAV2 genomes were able to complement rAAV vector genomes when assayed by Southern blot analysis. For example, replication intermediates were observed in D5 cells exposed to the TR2 vector, while no vector replication was observed in the cells exposed to the TR5 vector (Fig. 5B). Longer exposure revealed minor TR5 vector signal in D5 cells, comparable to background levels of TR5 and TR2 vector signals found in D6 Hirt DNA, indicating a lack of replication in the presence of Ad.
FIG. 5.
TR2 but not TR5 vectors are mobilized by latent wt AAV2 and Ad coinfection. D6 (latent AAV2 [−]) or D5 (latent AAV2 [+]) cell lines were infected with rAAV2 from crude lysate containing TR2 or TR5-eGFP in the presence or absence of Ad. Hirt DNA was isolated and analyzed by Southern blotting 48 h postinfection, using either an AAV2 (A) or GFP (B) probe. The two major replicative forms of AAV are indicated (m, double-stranded monomer; d, double-stranded dimer). Larger replicative forms are also visible. A size marker is denoted. (C) GFP expression was visualized in HEK 293 cells after crude lysate was added from either D5 or D6 cells infected with the vectors indicated above the panels. The numbers refer to the gel lanes in panel B. +, present; −, absent.
Lysate taken from TR2 or TR5 eGFP vector-infected D5 and D6 cells was used to infect naïve HEK 293 cells (Fig. 5C). HEK 293 cells given lysate taken from D5 cells exposed to the AAV2/TR2-eGFP vector and Ad were positive for GFP (Fig. 5C, panel 6), demonstrating that latent wt AAV2 was able to provide Rep and Cap in trans to mobilize the TR2-flanked GFP vector. In contrast, HEK 293 cells given lysate from D5 cells infected with the TR5-eGFP vector did not express GFP (Fig. 5C, panel 7), demonstrating that the latent AAV2 was not able to mobilize the TR5-flanked GFP vector. As expected, control lysate from D6 cells did not produce infectious GFP vectors using either TR, and infection of D5 cells without Ad helper virus or without TR2-eGFP vector did not produce infectious GFP vectors (Fig. 5C, panels 1 to 5).
DISCUSSION
For nearly 30 years, it has been known that a chromosomally integrated AAV genome could be rescued and replicated in order for AAV to enter the lytic phase of its life cycle (4). Ever since AAV was first cloned into a recombinant plasmid, we have known that the viral or vector genome could be rescued and replicated from exogenous DNA (27). Over 10 years ago, the first evidence for in vitro mobilization of an AAV vector transgene was presented (1), describing a potential scheme of vector mobilization between individuals. Yet, to date, no work has been published on preventing AAV vector mobilization, nor has the ever-increasing quantity of AAV vectors used for clinical trials been adapted to address this concern.
AAV serotypes have been discovered historically as a contaminating factor in Ad preparations (25). AAV, having evolved to require Ad coinfection, must persist in this manner in order to propagate itself. While the true risk of AAV vector mobilization is not known, there is a possibility that rAAV vectors with wt AAV TRs could constitute an AAV-associated virus, one which replicates in cells where productive Ad and AAV infections are taking place. This could lead to the spread of recombinant genetic material to healthy individuals.
More likely, however, is vector mobilization within an individual. An enormous amount of effort has gone into the creation of novel AAV capsids with specific tissue tropism (16, 19). This can be especially important when tissue-specific promoters are unavailable or incompatible with AAV therapeutics. However, these elegant experiments to deliver vectors to specific targets become meaningless if the vector genomes are mobilized by wt AAV into other body tissues by wt capsids.
Here, we have presented a means to reduce the risk of AAV vector mobilization. The two most prevalent human AAV serotypes (AAV2 and AAV6) both have the ability to replicate the TR2-flanked vectors currently used in AAV clinical trials (10). A less widespread AAV serotype, AAV5, has a unique Rep-TR interaction, making it the only human serotype able to replicate TR5-flanked vectors (5, 10, 18). This replicative specificity, as well as the relative abundance of these serotypes in the population (AAV5 is >4-fold less abundant than AAV2) (13, 14, 20, 33), led us to hypothesize that TR5-flanked vector genomes have a significantly reduced risk of vector mobilization.
To test this hypothesis, we created a TR5-based vector production system similar to the TR2-based system currently used to produce rAAV (Fig. 3A). This system worked well, exclusively packaging TR5-flanked vectors into Cap1 to Cap5 while yielding viral titers and transduction efficiency comparable to those yielded by the current TR2 vector production system. While our TR5 vectors may have shown a minor inherent decrease in transduction efficiency (potentially in a capsid-specific manner), optimization of our system may eliminate this disparity. A similar comparison between TR2 and TR5 vectors in vivo showed no such bias, (10), suggesting that the differential may be due to the sensitivity of our in vitro system or otherwise restricted to our assay.
In order to confirm that Rep-TR replicative specificity extended to vector mobilization, we adopted two cell culture assays. While the transformed cells used for these assays had the potential to behave differently from the primary cell types AAV vectors would encounter in vivo, we reasoned that Rep-TR specificity would remain consistent regardless of cell type. That said, demonstrating vector mobilization in primary cells remains an important step in establishing the potential danger to future gene therapy candidates.
We first created cell lines containing stably persisting TR2- or TR5-flanked vector genomes. While we did not determine whether these genomes were integrated into the host chromosome, the persistence of GFP signal in 100% of these cells 2 months after sorting suggests chromosomal integration. However, the possibility that they are persisting in some other manner lends only credence to the mobilization assay we have developed, as such genomes may recapitulate any number of modes of persistence in vivo. We next demonstrated that these persisting rAAV genomes could be rescued and replicated upon the transfection of AAV helper plasmids (Fig. 4A). These genomes were also encapsidated and able to transduce naïve cells (Fig. 4B). Predictably, the cell/tissue tropism of these mobilized genomes was dependent on the capsid into which they were mobilized.
Slightly different levels of replication were detected for pTR2-eGFP vectors in the presence of Rep2 or Rep6 (Fig. 2B). While this may suggest that Rep6 replicates TR2s with higher fidelity than does Rep2, it is more likely that more Rep6 protein was produced by the plasmid constructs. Western blot analyses were not performed due to the lack of a suitable Rep6 antibody. Interestingly, our TR5 vector was rescued with lower fidelity from 293 cells than was our TR2 vector (Fig. 4A). We have confirmed by Western blotting that our Rep2 and Rep5 constructs produce equal amounts of protein (data not shown) and that they drive similar amounts of vector genome replication (Fig. 2B). As such, the decreased rescue of TR5 genomes is most likely due to the inability of a subset of the TR5-flanked GFP vectors in this population to be rescued due to deletions of the integrated or concatemerized TRs as seen with AAV2 latent genomes (37). It is possible, however, that persisting TR5 genomes may be somewhat refractory to rescue, and further experiments may be required to definitively answer this question.
In addition, we showed that in cells with latent wt AAV2 infection, introduction of a TR2 vector and subsequent superinfection by Ad resulted in replication of the wt genome and the TR2-flanked transgene (Fig. 5A and B) and led to the production of infectious rAAV particles (Fig. 5C). These results demonstrated that latent wt AAV2 plus Ad reconstituted the replication-deficient TR2 vector system, allowing for mobilization of transgene vectors. Once again, AAV2 was unable to replicate or mobilize a TR5-flanked genome (Fig. 5B, lane 7), underlining the potential of our TR5-based system to decrease AAV vector mobilization due to relative AAV2, AAV5, and AAV6 prevalence in the population. While inclusion of a cell line harboring a latent AAV5 genome would have been ideal for this study, there are no reports of AAV5 integrating site-specifically into the human chromosome. Thus, any cell line harboring an AAV5 genome should be recapitulated by our mobilization system, shown in Fig. 4, by which a persisting TR5-flanked genome is rescued and replicated.
It is impossible to quantify the degree of safety TR5-based vectors would add to AAV clinical applications. Based on the exclusivity of the Rep5-TR5 interaction, as well as the small degree of AAV5 in the population compared to AAV2 and AAV6, we can only postulate that TR5 vectors possess a significantly lower risk of spreading after rAAV administration. While TR5-based vectors may be markedly safer, they are not a solution. However, at the moment, TR5-flanked vectors offer a safer means to ensure that AAV vectors remain where they are intended to be.
Acknowledgments
We thank Nathan Allen for his contributions, Robert Kotin for providing AAV5 TRs, David Russell for providing AAV6 Rep and Cap, Tal Kafri for helpful discussions, Matt Hirsch for discussions and critical reading of this work, and the reviewers for their helpful comments.
This work was supported by NIH grants GM0529299, HL066973, HL051818, and AI072176 awarded to R.J.S. and by NIAID grant AI007419 awarded to F.C.H.
Footnotes
Published ahead of print on 11 February 2009.
REFERENCES
- 1.Afione, S. A., C. K. Conrad, W. G. Kearns, S. Chunduru, R. Adams, T. C. Reynolds, W. B. Guggino, G. R. Cutting, B. J. Carter, and T. R. Flotte. 1996. In vivo model of adeno-associated virus vector persistence and rescue. J. Virol. 703235-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berns, K. I., and C. R. Parrish. 2007. Parvoviridae, p. 2437-2477. In D. M. Knipe. P. M. Howley, et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 3.Reference deleted.
- 4.Cheung, A. K., M. D. Hoggan, W. W. Hauswirth, and K. I. Berns. 1980. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J. Virol. 33739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiorini, J. A., S. Afione, and R. M. Kotin. 1999. Adeno-associated virus (AAV) type 5 Rep protein cleaves a unique terminal resolution site compared with other AAV serotypes. J. Virol. 734293-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, V. W., D. M. McCarty, and R. J. Samulski. 2006. Host cell DNA repair pathways in adeno-associated viral genome processing. J. Virol. 8010346-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, V. W., R. J. Samulski, and D. M. McCarty. 2005. Effects of adeno-associated virus DNA hairpin structure on recombination. J. Virol. 796801-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, M. D., J. Wu, and R. A. Owens. 2000. Mutational analysis of adeno-associated virus type 2 Rep68 protein endonuclease activity on partially single-stranded substrates. J. Virol. 742936-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reference deleted.
- 10.Grimm, D., K. Pandey, H. Nakai, T. A. Storm, and M. A. Kay. 2006. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 80426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimm, D., M. A. Kay, and J. A. Kleinschmidt. 2003. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol. Ther. 7839-850. [DOI] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Halbert, C. L., A. D. Miller, S. Mcnamara, J. Emerson, R. L. Gibson, B. Ramsey, and M. L. Aitken. 2006. Prevalence of neutralizing antibodies against adeno-associated virus (AAV) types 2, 5, and 6 in cystic fibrosis and normal populations: implications for gene therapy using AAV vectors. Hum. Gene Ther. 17440-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildinger, M., A. Auricchio, G. Gao, L. Wang, N. Chirmule, and J. M. Wilson. 2001. Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J. Virol. 756199-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26365-369. [DOI] [PubMed] [Google Scholar]
- 16.Koerber, J. T., J.-H. Jang, and D. V. Schaffer. 2008. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol. Ther. 161703-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. ClustalW and ClustalX version 2. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 18.Li, C., and R. J. Samulski. 2005. Serotype-specific replicating AAV helper constructs increase recombinant AAV type 2 vector production. Virology 33510-21. [DOI] [PubMed] [Google Scholar]
- 19.Li, W., A. Asokan, Z. Wu, T. Van Dyke, N. Diprimio, et al. 2008. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol. Ther. 161252-1260. [DOI] [PubMed] [Google Scholar]
- 20.Mayor, H. D., S. Drake, J. Stahmann, and D. M. Mumford. 1976. Antibodies to adeno-associated satellite virus and herpes simplex in sera from cancer patients and normal adults. Am. J. Obstet. Gynecol. 126100-104. [DOI] [PubMed] [Google Scholar]
- 21.McCarty, D. M., S. M. Young, Jr., and R. J. Samulski. 2004. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu. Rev. Genet. 38819-845. [DOI] [PubMed] [Google Scholar]
- 22.Mehrle, S., V. Rohde, and J. R. Schlehofer. 2004. Evidence of chromosomal integration of AAV DNA in human testis tissue. Virus Genes 2861-69. [DOI] [PubMed] [Google Scholar]
- 23.Penaud-Budloo, M., C. Le Guiner, A. Nowrouzi, A. Toromanoff, Y. Chérel, P. Chenuaud, M. Schmidt, C. von Kalle, F. Rolling, P. Moullier, and R. O. Snyder. 2008. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J. Virol. 827875-7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose, J. A., M. D. Hoggan, and A. J. Shatkin. 1966. Nucleic acid from an adeno-associated virus: chemical and physical studies. Proc. Natl. Acad. Sci. USA 5686-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samulski, R. J., K. I. Berns, M. Tan, and N. Muzyczka. 1982. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. USA 792077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samulski, R. J., A. Srivastava, K. I. Berns, and N. Muzyczka. 1983. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell 33135-143. [DOI] [PubMed] [Google Scholar]
- 29.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 103941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt, M., E. Grot, P. Cervenka, S. Wainer, C. Buck, and J. A. Chiorini. 2006. Identification and characterization of novel adeno-associated virus isolates in ATCC virus stocks. J. Virol. 805082-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnepp, B. C., K. R. Clark, D. L. Klemanski, C. A. Pacak, and P. R. Johnson. 2003. Genetic fate of recombinant adeno-associated virus vector genomes in muscle. J. Virol. 773495-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 721438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tobiasch, E., M. Rabreau, K. Geletneky, S. Laruë-Charlus, F. Severin, N. Becker, and J. R. Schlehofer. 1994. Detection of adeno-associated virus DNA in human genital tissue and in material from spontaneous abortion. J. Med. Virol. 44215-222. [DOI] [PubMed] [Google Scholar]
- 34.Tratschin, J. D., I. L. Miller, M. G. Smith, and B. J. Carter. 1985. Adeno-associated virus vector for high-frequency integration, expression, and rescue of genes in mammalian cells. Mol. Cell. Biol. 53251-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao, X., J. Li, and R. J. Samulski. 1996. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J. Virol. 708098-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan, Z., R. Zak, Y. Zhang, and J. F. Engelhardt. 2005. Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J. Virol. 79364-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, J., W. Zhou, Y. Zhang, T. Zidon, T. Ritchie, and J. F. Engelhardt. 1999. Concatemerization of adeno-associated virus circular genomes occurs through intermolecular recombination. J. Virol. 739468-9477. [DOI] [PMC free article] [PubMed] [Google Scholar]