Abstract
Glycoprotein B (gB) homologs are conserved throughout the family Herpesviridae and appear to serve essential, universal functions, as well as specific functions unique to a particular herpesvirus. Genetic analysis is a powerful tool to analyze protein function, and while it has been possible to generate virus mutants, complementation of essential virus knockouts has been problematic. Human cytomegalovirus (HCMV) gB (UL55) plays an essential role in the replication cycle of the virus. To define the function(s) of gB in HCMV infection, the BAC system was used to generate a recombinant virus in which the UL55 gene was replaced with galK (pAD/CreΔUL55). UL55 deletions in the viral genome have been made before, demonstrating that UL55 is an essential gene. However, without being able to successfully complement the genetic defect, a phenotypic analysis of the mutant virus was impossible. We generated fibroblasts expressing HCMV gB that complement pAD/CreΔUL55 and produce infectious virions lacking the UL55 gene but containing wild-type gB on the virion surface (ΔUL55-gB HCMV). This is the first successful complementation of an HCMV mutant with a glycoprotein deleted. To characterize ΔUL55 infection in the absence of gB, noncomplementing cells were infected with ΔUL55-gB virus. All stages of gene expression were detected, and significant amounts of DNase-resistant viral DNA genomes, representing whole intact virions, were released into the infected cell supernatant. Gradient purification of these virions revealed they lacked gB but contained other viral structural proteins. The gB-null virions were able to attach to the cell surface similarly to wild-type gB-containing virions but were defective in virus entry and cell-to-cell spread. Glycoprotein B-null virions do, however, contain infectious DNA, as IE gene expression can be detected in fibroblasts following treatment of attached gB-null virions with a membrane fusion agent, polyethylene glycol. Taken together, our results indicate that gB is required for virus entry and cell-to-cell spread of the virus. However, HCMV gB is not absolutely required for virus attachment or assembly and egress from infected cells.
Human cytomegalovirus (HCMV), a ubiquitous, opportunistic pathogen, is a member of the Herpesviridae family of viruses (1, 44). Infection with HCMV is generally asymptomatic, but naïve or immunosuppressed individuals, such as neonates, AIDS patients, and transplant recipients, often manifest serious disease (1, 44). HCMV is a promiscuous pathogen, able to enter and initiate infection in almost every vertebrate cell type tested. Similarly, disease can occur in nearly every organ system and cell type in the human body (1, 44).
The ability of HCMV to enter such a wide variety of cell types requires the coordinated interaction of several virus-encoded envelope glycoproteins with multiple cell surface receptors (15). Herpesvirus entry, in general, is very complex and not well understood. For HCMV, the initial attachment step involves glycoprotein M (gM) and gB, both of which bind to heparan sulfate proteoglycans (HSPGs) on the cell surface (17, 33). The virus then quickly moves to a more stable docking interaction with one or more cellular receptors (15). It has recently been discovered that gB is able to interact with specific integrin heterodimers, a step that greatly enhances HCMV entry (21). It has also been reported that gB interacts with the epidermal growth factor receptor to mediate virus entry (62, 63), although our laboratory (28) and others (13) have not been able to confirm these findings. During the final phase of virus entry, the viral and cellular membranes fuse, releasing the tegument proteins and capsid into the cytoplasm. This fusion event is thought to require gB, as well as the gH/gL complex (6, 35, 46, 58, 59). It is possible that gB interaction with β1 integrins activates its fusogenic activity by some as yet unknown mechanism (21; A. L. Feire and T. Compton, unpublished data). Additionally, αVβ3 integrins were also recently identified as cellular receptors for gH (62) and are possibly required to activate its fusogenic activity, as well.
gB, an abundant glycoprotein on the virus envelope (61) and the most highly conserved glycoprotein of the human herpesviruses, plays a critical role in the HCMV entry process, as it is believed to be involved in the initial virion-tethering step and the second, more stable attachment step, as well as the fusion event itself (reviewed in reference 15). Evidence supporting an attachment role for gB includes the ability of soluble gB (gBs) to bind heparin, a soluble mimic of HSPGs (17, 34). Additionally, gBs manifests two-step binding kinetics to cells, in which the protein is initially dissociable with soluble heparin but quickly becomes resistant to removal by heparin washes. This suggests that gB moves from one receptor, presumably HSPGs, to another, likely cellular integrins, during the attachment process (7). Evidence supporting a role of cellular integrins in the HCMV entry process includes the disintegrin-like domain (DLD) of gB binding specifically to β1 integrins (21; Feire et al., unpublished).
Antibodies to HCMV gB, including those to the DLD and to specific integrin heterodimers, are able to efficiently neutralize virus entry at a postattachment stage, suggesting that gB and integrins are involved in the fusion event (23, 48; Feire et al., unpublished). Additionally, by comparison with other gB homologs, such as herpes simplex virus type 1 (HSV-1) gB, which has a well-defined role in virus entry, it is likely that HMCV gB also acts as a fusion mediator for the virus. Finally, gB contains a heptad repeat region, which is predicted to form an alpha-helical coiled coil, commonly found in many class I viral fusion proteins (41). Synthetic peptides that mimic the heptad repeat region of HCMV gB efficiently inhibit virus entry without interfering with virus attachment, presumably at the level of virus fusion (19, 41).
Aside from its role in virus entry and fusion, gB is also believed to be required for cell-to-cell spread of the virus (23, 46, 48). During plaque formation, virions are thought to spread directly through cellular contacts or across cellular junctions (reviewed in reference 30). Evidence for the role of gB in HCMV cell-to-cell spread is indirect and includes the ability of gB-neutralizing antibodies to prevent plaque formation in infected cells (23, 46, 48). However, gB proteins from other herpesviruses, including HSV-1 and HSV-2, pseudorabies virus (PRV), Epstein-Barr virus (EBV), and Kaposi's sarcoma-associated herpesvirus (KSHV), are also required for cell-to-cell spread (25, 45, 50, 52, 60).
gB proteins from a variety of herpesviruses have also been proposed to be necessary to mediate virion assembly and egress from infected cells. Herpesvirus egress is thought to involve an initial envelopment step at the inner nuclear membrane, followed by a de-envelopment step at the outer nuclear membrane and, last, a reenvelopment step at a Golgi apparatus-derived membrane before the virion-containing vesicle fuses with the plasma membrane to release the virion outside the cell (43). Presumably, the fusogenic activity of viral glycoproteins is required for the initially enveloped particles to fuse with the outer nuclear membrane to be released into the cytoplasm (reviewed in reference 43). PRV, EBV, and KSHV have all been reported to require gB for virion egress (37, 39, 49). However, reports on HSV-1, as well as another, conflicting report on PRV, suggest that gB is not absolutely required for egress from infected cells, although slight defects are seen with HSV-1 (11, 20, 52, 53). For HSV-1, it appears that either gB or gH on the virion envelope is sufficient to mediate virion egress; however, mutants lacking both glycoproteins are unable to be released into the cytoplasm (20).
Other than the fusogenic activity of HCMV gB, the protein also initiates multiple signaling events, even in the absence of other virion components (reviewed in reference 4). HCMV attachment and entry induce activation of innate immune responses, including the interferon response and inflammatory cytokine induction (reviewed in reference 4). gB has been demonstrated to bind Toll-like receptor 2, likely leading to the induction of inflammatory cytokines (5); however, the mechanism of activation of the interferon response is currently unknown. Additionally, the induction of cytoskeletal rearrangements and activation of focal adhesion kinase likely occur through gB binding to β1 integrins through its DLD and/or gH binding to β3 integrins during the entry process (21, 62; Feire, et al. unpublished).
Genetic analysis is a powerful tool to determine functional roles in the context of viral infections. The recent development of the bacterial artificial chromosome (BAC) system has proven to be a great asset for reverse genetics studies; however, like all mutagenesis systems, it is limited. Essential gene mutants cannot be isolated and propagated unless the mutation is able to be efficiently complemented. Until now, all attempts to complement a gB deletion virus, or any essential HCMV glycoprotein knockout, have been unsuccessful. To further define the role of gB in the HCMV life cycle, we constructed a BAC plasmid containing a replacement of the UL55 gene, which encodes gB, with a galactose operon gene, galK (pAD/CreΔUL55). This ΔUL55 BAC was transfected into gB-expressing normal human dermal fibroblasts (NHDF-gB), which compensated for the loss of the UL55 gene, generating infectious virus with a deletion of the UL55 gene in the viral genome but pseudotyped with wild-type gB on the virion surface (ΔUL55-gB HCMV). When noncomplementing NHDF were infected with ΔUL55-gB HCMV, all stages of gene expression were detected and significant quantities of DNase-resistant viral genomes were released into the supernatant, indicating that viral egress occurs in the absence of gB. These ΔUL55 gB-null virions were gradient purified and demonstrated to contain other virion structural proteins, including gH, and tegument proteins pp150, pp71, and pp28. Using gB-null virions, we found HCMV could attach to cells in the absence of gB, as well as wild-type gB-containing virions, but the virus was completely noninfectious, confirming that gB is required for virus-cell fusion. These gB-null virions could, however, be chemically forced to fuse with NHDF by treatment with polyethylene glycol (PEG), which artificially induces the fusion of nearby membranes, leading to immediate-early (IE) gene expression. Additionally, the ΔUL55-gB virus was not able to spread from cell to cell in an NHDF plaque overlay assay, indicating that gB is also required for cell-to-cell spread of the virus. These genetic data complement prior approaches and demonstrates that gB mediates entry, but not egress. Additionally, this is the first report of a successful complementation of an HCMV glycoprotein mutant and establishes the ability to examine essential glycoprotein mutants in the context of a viral infection using a complementation system.
MATERIALS AND METHODS
Cell lines and viruses.
NHDF were purchased from Clonetics (San Diego, CA). NHDF and 293T cells were grown at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT), 1% l-glutamine (Invitrogen), and 1% penicillin-streptomycin-amphotericin B (BioWhittaker, Walkersville, MD). HCMV strain AD169, AD169 BAC, and pAD/Cre (66), which was a gift from Thomas Shenk (Princeton University) and is the wild-type parent of pAD/CreΔUL55 and pAD/CreΔUL55R, were all propagated in NHDF as previously described (16).
Antibodies.
Mouse monoclonal gB antibodies 27-78 and 58-15 were generous gifts from William J. Britt (University of Alabama—Birmingham) and have been previously described (3, 9, 56). Mouse monoclonal antibody 1203, which recognizes HCMV IE genes (IE1-72 and IE2-86) was purchased from the Rumbaugh-Goodwin Institute for Cancer Research, Inc. (Plantation, FL). Mouse monoclonal antibody UL44 (ICP36) was purchased from Virusys Corporation (Sykesville, MD). Rabbit polyclonal gH antibody 6824 was previously described (14). Mouse monoclonal antibodies to pp71 (2H10-9), pp28 (CMV 157), and pp150 (CMV 127) were a kind gift from Robert F. Kalejta and have been previously described (31, 47). Goat anti-mouse and goat anti-rabbit horseradish peroxidase secondary antibodies were purchased from Pierce Biotechnology (Rockford, IL).
Retrovirus production and transduction.
The gB retroviral vector pCMMP-UL55-IRES-GFP was previously described (36). Vector encoding murine leukemia virus Gag-Pol proteins, pMD-gagpol, and vector encoding vesicular stomatitis virus gG, pMD-G, were gifts from Richard C. Mulligan (Harvard Medical School). Recombinant retrovirus was generated as previously described (36). Briefly, 293T cells were transfected using the calcium phosphate method in 100-mm plates. Retrovirus was collected at 48 and 72 h posttransfection and filtered through 45-μm filters, and stocks were frozen at −80°C. For retroviral delivery of the UL55 gene to cells, recombinant retrovirus was incubated overnight with subconfluent low-passage NHDF in the presence of 8 μg/ml Polybrene in a minimal amount of DMEM containing 10% FBS. The retrovirus was removed and replaced with DMEM containing 10% FBS. Upon reaching confluence, cells were passaged before undergoing a second round of transduction. Cells were not selected with drug resistance, sorting, or clonal selection, but a mixed population containing various degrees of stable gB expression was used for all experiments. Cells were not used past passage 20, whereupon freshly transduced, low-passage cells were prepared.
Construction of pAD/CreΔUL55 and pAD/CreΔUL55R.
The UL55 gene of pAD/Cre (66) was deleted by replacing it with galK using the Red recombination method as described previously (64). pGalK, a gift from Neal Copeland (National Cancer Institute), was used to generate a recombination selection cassette by amplification using PCR primers to galK that contain homology arms to the UL55 gene (primer pair A) (Table 1). Following PCR, the recombination cassette was digested with DpnI (New England Biolabs, Ipswich, MA) and gel purified. A second round of PCR was conducted, followed by ethanol precipitation and resuspension in water. The recombination cassette was electroporated into bacterial strain SW102, a gift from Neal Copeland, which harbored pAD/Cre and had been heat shocked at 42°C for 20 min to induce Red recombination proteins. Recombinants were selected for the ability to use galactose as a carbon source on minimal medium and were verified by streaking them on MacConkey galactose plates. To generate a revertant, a wild-type UL55 recombination cassette was generated by PCR using primer pair B (Table 1). The recombination cassette was electroporated into SW102 bacteria harboring pAD/CreΔUL55 that had been heat shocked as described above. Recombinants that reverted to wild type were selected for their resistance to 2-deoxygalactose in minimal medium containing glycerol as a carbon source.
TABLE 1.
PCR primers
| Primer pair | Primer name | Sequencea | Product length (bp)/fragment generated |
|---|---|---|---|
| A | gB51-100galKF | 5′-CTGTCTGGGTGCTGCGGTTTCCTCTTCTAGTACTTCCCATGCAATTCTTCCTGTTGACAATTAATCATCG-3′ | 1,331/galK recombination cassette |
| gB2622-71galKR | 5′-TGCGATGTCGCAGCCGGTCTAGCAGGTTAGGCTTCTGTCCCTTGTCCTGCTCAGCACTGTCCTGCTCCTT-3′ | ||
| B | gB1RecF | 5′-GGATCTGGTGCCTGGTAGTCTGC-3′ | 2,701/UL55 recombination cassette |
| gB2RecR | 5′-TCTTCTTCGTCGGAGTCTTTCAA-3′ | ||
| C | UL56F | 5′-CTCCTCCTCGACTGCGGGTGTTTCC-3′ | 2,806/UL56-UL55 or 1,526/UL56-galK-UL55 fragment |
| UL55R | 5′-TCTTCTTCGTCGGAGTCTTTCAA-3′ | ||
| D | UL55SoF | 5′-ACGCCGACAAGTTTTTC-3′ | 598/UL55 Southern probe |
| UL55SoR | 5′-CCGCGCAACGTGTCATAGGTG-3′ | ||
| E | galKSoF | 5′-GTTCTGCCTGCGCGATTGATTAT-3′ | 599/galK Southern probe |
| galKSoR | 5′-AGGGCTGGCTGCTGGAAGAAAC-3′ |
Italics indicate gB sequence; boldface indicates galK sequence.
Recombinant BACs were purified using a Spin Doctor BAC Prep Kit (Gerard Biotech, Oxford, OH). To verify correct recombination, the UL56-UL55 genes were amplified by PCR using primer pair C (Table 1). BACs were digested with EcoRI (New England Biolabs), followed by agarose gel electrophoresis. Southern blotting detected UL55 and galK genes in recombinant BAC digests. Southern blot probes were generated using the PCR primer pair D and E (Table 1) for the UL55 and galK genes, respectively. The probes were labeled using a HybQUEST Complete (DNP) System Kit purchased from Mirus Bio Corporation (Madison, WI), and Southern blotting was performed according to the manufacturer's instructions.
Electroporation of BAC plasmids and propagation of infectious ΔUL55-gB HCMV.
To generate virus from the BAC plasmids, 15 μl of purified pAD/Cre, pAD/CreΔUL55, or pAD/CreΔUL55R was mixed with 1 μg of HCMV pp71-expressing plasmid (pCGN-HA-pp71 [31]) and 5 × 106 NHDF or NHDF-gB in a total volume of 265 μl in serum-free DMEM and electroporated in a Gene Pulser with 4-mm-gap cuvettes (Bio-Rad) with settings of 260 V and 960 μF. Following electroporation, cells were seeded in 10% serum-containing medium in 100-mm plates and were supplied with fresh serum-containing medium the next day. When confluence was reached, the cells were passaged 1:2. Approximately 3 weeks postelectroporation, the cells were stained with crystal violet to visualize plaque formation or were incubated until an 80 to 100% cytopathic effect was reached. Virus-containing supernatant was collected at that point.
Growth curves.
For single-step growth curves, 1.5 × 105 NHDF or NHDF-gB/well in a 12-well plate were infected at a multiplicity of infection (MOI) of 5 PFU/cell. Infected cell supernatants were collected at 24-hour intervals for 6 days. For multistep growth curves, 1.5 × 105 cells/well in a 12-well plate were infected with virus at an MOI of 0.01 PFU/cell. Infected cell supernatants were collected at 3-day intervals for 21 days. The cell supernatants were stored at −80°C until the end of the experiment. Cellular debris was removed by centrifugation, and the virus titer was determined by plaque assay. Infections were done in triplicate for each time point except for the ΔUL55-gB growth curve, which was done in duplicate.
SDS-PAGE and immunoblotting.
Cells were harvested in phosphate-buffered saline (PBS) containing 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) (Sigma, St. Louis, MO). The protein concentration was determined with a Bio-Rad (Hercules, CA) protein assay kit using bovine immunoglobulin G as a standard. Equivalent amounts of total protein were boiled in sodium dodecyl sulfate (SDS) gel loading buffer and subjected to SDS-10% polyacrylamide gel electrophoresis (PAGE), followed by Western immunoblotting.
Real-time PCR.
DNA was extracted from virus supernatant or infected cell lysate using the QIAamp Mini Elute Virus Spin Kit (Qiagen, Valencia, CA). The DNA was eluted in 50 μl nuclease-free water (Ambion, Austin, TX). Viral genomes were quantitated using the primer pair pp549s and pp812as and UL83 6-carboxyfluorescein-6-carboxytetramethylrhodamine probe as previously described (22). Unknown sample values were determined based on a standard curve of known UL83 copy numbers using pCGN-HA-pp65 (2). PCR mixtures contained 5, 2.5, or 1.25 μl of extracted DNA; 50 nM primers and probe; 12.5 μl Taq-Man Universal PCR Master Mix (Applied Biosystems, Foster City, CA); and nuclease-free water to a total volume of 25 μl. Real-time PCR was run on an ABI 7900HT, and data were analyzed using the SDS 2.2.1 program.
Immunofluorescence.
For gB detection following transduction, NHDF plated on coverslips in 12-well plates were fixed in 3% paraformaldehyde (PFA) in PBS and analyzed by immunofluorescence using antibody 27-78. To detect IE gene expression following HCMV infection, 1.5 × 105 cells/well in a 12-well plate were infected with HCMV, and 24 hours postinfection, the cells were fixed in 3% PFA and analyzed for IE gene expression by immunofluorescence using antibody 1203. For the IE-spreading assay, cells were electroporated with HCMV BAC plasmids as described above. Seven days postelectroporation, the cells were passaged and plated on coverslips in 12-well plates. One week later, the cells were fixed in 3% PFA and analyzed for IE gene expression using indirect immunofluorescence with antibody 1203. Alexa Fluor goat anti-mouse 594 secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Molecular Probes (Eugene, OR).
Virion egress assays.
NHDF were infected with AD169 or ΔUL55-gB virus, or NHDF-gB were infected with ΔUL55-gB virus, at an MOI of 1.0 PFU/ml. Six days postinfection, infected cell supernatant was collected, cellular debris was removed by centrifugation, and for real-time PCR, any contaminating DNA was removed by DNase I (Roche Applied Science, Indianapolis, IN) digestion at 37°C for 30 min. DNA from the supernatant was extracted and analyzed to detect viral genomes by real-time PCR as described above. To purify released virions, the infected cell supernatant was concentrated by pelleting it over a 20% sorbitol cushion, followed by banding in a 20 to 70% sorbitol step gradient. The virus-containing band was isolated and analyzed by SDS-PAGE and immunoblotted to detect gB (58-15), gH, pp28, pp71, and pp150 as described above.
Attachment assay.
AD169, ΔUL55-gB, or ΔUL55 gB-null HCMV with or without soluble heparin (100 μg/ml; Sigma, St. Louis, Mo) was incubated with NHDF in 12-well plates with equivalent genome copy numbers as determined by real-time PCR at 4°C. The virus was allowed to attach at 4°C for 1 hour, followed by three washes with serum-free DMEM. The cells were lysed in 1% CHAPS in PBS, and DNA was extracted and analyzed by real-time PCR to detect viral genomes as described previously.
PEG assay.
AD169, ΔUL55-gB, or gB-null virus was allowed to bind and enter cells for 1 h at 37°C. The virus was removed, and the cells were washed three times in serum-free DMEM or in decreasing concentrations of PEG 8000 (50%, 25%, and 12.5%) in serum-free DMEM. DMEM containing 2% FBS was added to the cells after they were washed, and the cells were returned to 37°C. Twenty-four hours postinfection, the cells were fixed, and IE gene expression was detected by indirect immunofluorescence as described previously.
Plaque overlay.
NHDF in a six-well plate were infected with 10-fold dilutions of ΔUL55-gB HCMV for 2 hours at 37°C. The cells were washed with PBS and overlaid with 1% agarose in Eagle minimum essential medium. Two days postinfection, 2% FBS-containing DMEM was added to the cells. The medium was changed every 3 days until plaques formed, and the cells were stained with crystal violet fixing reagent (0.07% crystal violet, 5.55% formaldehyde, 0.5× PBS).
RESULTS
Construction of pAD/CreΔUL55 and pAD/CreΔUL55R.
The pAD/CreΔUL55 BAC was created using the recombineering system (64) to replace an approximately 2.5-kb region of the UL55 gB gene with a 1.2-kb galK gene in the wild-type HCMV AD169 BAC (pAD/Cre) (66), as demonstrated in Fig. 1A. PCR primer pair A (Table 1) was used to generate the galK recombination cassette, which contained 50 bp on the 5′ and 3′ ends homologous to the UL55 gB gene. The 5′ end of the recombination cassette corresponded to nucleotide positions 51 to 100 of UL55, while the 3′ end corresponded to nucleotide positions 2622 to 2671. The recombination cassette therefore consisted of the galK gene flanked on either side with 50 bp of UL55 gene sequence (Fig. 1A).
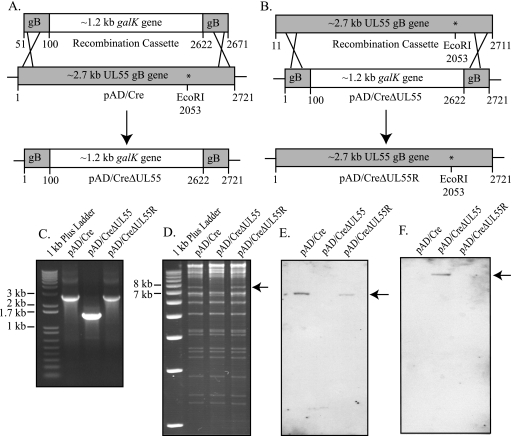
FIG. 1.
Recombination replacing the HCMV UL55 gB gene with galK in pAD/Cre. (A) The galK recombination cassette was PCR amplified using primer pair A (Table 1), adding 50 bp of flanking gB sequence to either end of the galK gene. The ∼2.7-kb UL55 gB gene of pAD/Cre was replaced via recombination with the ∼1.2-kb galK gene, generating pAD/CreΔUL55. Recombinants were selected with chloramphenicol on minimal-medium plates containing galactose. (B) To revert pAD/CreΔUL55, a 2.7-kb wild-type UL55 recombination cassette was recombined into pAD/CreΔUL55, generating pAD/CreΔUL55R. Recombinants were selected with chloramphenicol on minimal-medium plates containing deoxygalactose. Correct recombination for all BACs was confirmed via PCR (C), EcoRI digestion (D), and Southern blotting (E and F). (C) PCR primer pair C (Table 1) corresponds to UL56 and to the 3′ end of UL55 that was not replaced during recombination generating 2.8- or 1.5-kb PCR fragments in pAD/Cre and pAD/CreΔUL55R or pAD/CreΔUL55, respectively. (D) The loss of an EcoRI site in UL55 following recombination leads to the disappearance of a 7.3-kb fragment (arrow), which is restored in pAD/CreΔUL55R. Southern blot probes to UL55 (E) or galK (F) detected these genes (arrows) in pAD/Cre and pAD/CreΔUL55R or pAD/CreΔUL55, respectively.
To generate pAD/CreΔUL55, Escherichia coli strain SW102, which harbored pAD/Cre, was heat shocked at 42°C to express the phage recombination proteins (64), followed by electroporation of the galK recombination cassette. Following recovery at 32°C and plating on galactose minimal-medium plates, colonies were screened by plating them on MacConkey-galactose minimal-medium plates. Bright-red colonies were picked and screened by colony-direct PCR using primer pair C (Table 1) to the UL55 and UL56 genes to confirm correct insertion of the galK gene and generation of pAD/CreΔUL55. Wild-type HCMV BAC template generates a product of 2,803 bp, while an HCMV BAC in which UL55 has been replaced by galK generates a product of 1,517 bp (Fig. 1C).
A revertant pAD/CreΔUL55 (pAD/CreΔUL55R) was also generated via recombineering (Fig. 1B). First, a wild-type UL55 recombination cassette whose 5′ and 3′ ends corresponded to the remaining UL55 sequence in pAD/CreΔUL55 was PCR amplified using primer pair A (Table 1). This recombination cassette was electroporated into E. coli SW102, which had been heat shocked at 42°C to express the phage recombination proteins (64) and harbored pAD/CreΔUL55. Following recovery at 32°C and plating on M63-2-deoxy-galactose minimal-medium plates, colonies were screened by colony-direct PCR using primer pair D (Table 1) to the UL55 and UL56 genes to confirm correct insertion as described above (Fig. 1C).
Correct recombination for all BACs was confirmed by EcoRI restriction digestion of pAD/Cre, pAD/CreΔUL55, and pAD/CreΔUL55R (Fig. 1D), followed by Southern blot analysis (Fig. 1E and F). Replacement of the UL55 gene by galK leads to a loss of an EcoRI site. EcoRI digestion of wild-type pAD/Cre generates a 7.3-kb fragment, which is lost when the UL55 gene is replaced with the galK gene, but this fragment reappears in the revertant BAC plasmid (Fig. 1D). When the EcoRI BAC digests were probed with the 598-bp DNP-labeled UL55 probe, a 6.6-kb fragment was detected in the pAD/Cre and pAD/CreΔUL55R lanes (Fig. 1E). When the EcoRI BAC digests were probed with the 599-bp DNP-labeled galK probe, a 12.6-kbp fragment was detected in the pAD/CreΔUL55 lane only (Fig. 1F).
Generation of virus from BAC plasmids.
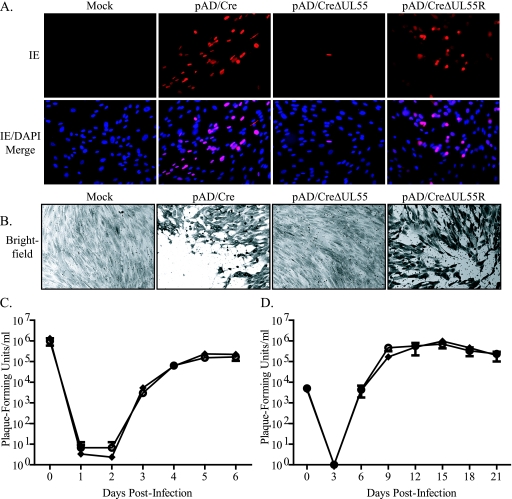
To produce virus from the BAC plasmids, NHDF were electroporated with serum-free DMEM (mock), pAD/Cre, pAD/CreΔUL55, or pAD/CreΔUL55R. Cells were passaged approximately 7 days postelectroporation, when the cells reached confluence. To detect infected cells, IE gene expression was visualized by indirect immunofluorescence 14 days postelectroporation (Fig. 2A). The results demonstrated that in the absence of gB, transfection of viral genomes can express IE, but infection does not spread compared to wild-type or revertant BACs. Approximately 3 weeks postelectroporation, cells were stained with crystal violet fixing reagent to visualize plaque formation (Fig. 2B), and the absence of plaques following pAD/CreΔUL55 electroporation confirmed an inability to spread and implied that infectious particles were not produced.
FIG. 2.
Generation and growth kinetics analysis of HCMV from electroporation of BAC plasmids into fibroblasts. NHDF were electroporated in the absence of a BAC plasmid (Mock) or with pAD/Cre, pAD/CreΔUL55, or pAD/CreΔUL55R. At 14 days postelectroporation, IE gene expression was detected via indirect immunofluorescence (A), and 1 week later, the cells were stained with crystal violet to detect plaque formation (B). NHDF were infected at an MOI of 5 (C) or 0.01 (D) with virus generated from electroporation of pAD/Cre (⧫) or pAD/CreΔUL55R (○). Virus-containing supernatant was removed and replaced with fresh medium supplemented with 2% FBS. At time zero and various times postinfection, supernatant was collected and titrated on NHDF. The values represent averages and standard deviations from three separate infections during one experiment.
In all experiments, the revertant had a phenotype identical to that of the wild type, indicating that the loss of UL55 was the source of the no-plaque phenotype for pADCreΔUL55. To confirm that the wild type and revertant were similar, single-step (MOI, 0.01 PFU/cell) (Fig. 2C) and multistep (MOI, 5.0 PFU/cell) (Fig. 2D) growth curves were conducted on NHDF. Infectious virus generated from electroporation of wild-type pAD/Cre and pAD/CreΔUL55R was used to infect NHDF at an MOI of either 0.01 or 5.0 PFU/cell. Infected cell supernatants were collected at various times postinfection and plaqued on NHDF to determine the titer of virus released from infected cells. As demonstrated in Fig. 2C and D, the revertant virus displayed growth kinetics similar to those of the wild-type virus, suggesting that pAD/CreΔUL55 is wild type except for the loss of the UL55 gene.
Generation of an NHDF line expressing HCMV gB that complements pAD/CreΔUL55.
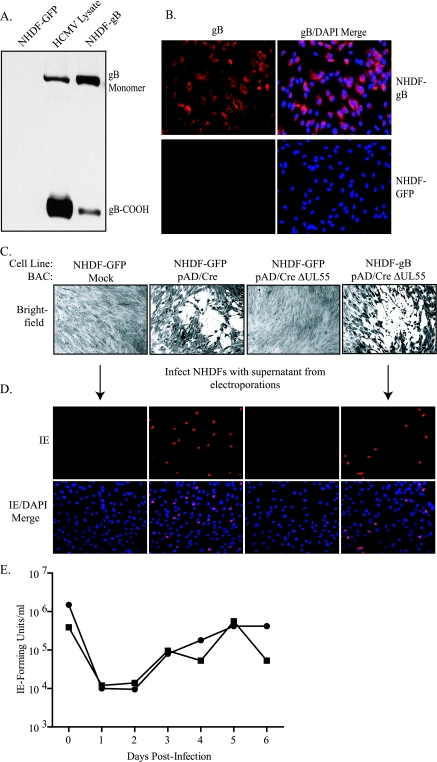
gB is an essential gene product in the life cycle of HCMV, as deletion of the UL55 gene generates noninfectious virus, as demonstrated previously (18, 27, 65) and in Fig. 2A and B. Therefore, to propagate pAD/CreΔUL55 and generate HCMV ΔUL55 virus containing wild-type gB on the virion surface (ΔUL55-gB HCMV), HCMV permissive cells that constitutively expressed gB were generated. A retroviral transduction and expression system that had been used previously to examine cell-cell fusion induced by HCMV glycoproteins (36) was utilized to generate the gB-expressing cell line. Retrovirus containing pCMMP-UL55-IRES-GFP expression vector was produced and used to transduce NHDF, leading to the expression of HCMV gB (NHDF-gB) (Fig. 3). Retrovirus containing the empty vector pCMMP-IRES-GFP was also generated and used to transduce NHDF (NHDF-GFP) as a control. As demonstrated in Fig. 3A, retroviral transduction can be used to express gB to levels similar to those seen in HCMV-infected cells (lane HCMV Lysate). Additionally, retroviral transduction can be used to express gB in most cells without selection, as demonstrated by detection of gB via indirect immunofluorescence (Fig. 3B). Cells were transduced two times, and expression levels were examined 1 week posttransduction. Although the cells were stably transduced, expression of gB decreased over time; therefore, new NHDF-gB were generated after approximately 20 passages.
FIG. 3.
gB expression in NHDF and complementation of pAD/CreΔUL55. (A) Following transduction of NHDF with recombinant (-GFP or -gB) retrovirus, cells were collected and lysates were analyzed by SDS-PAGE, along with HCMV AD169-infected cell lysate (HCMV Lysate). The gel was transferred to nitrocellulose and immunoblotted to detect gB using antibody 58-15. (B) Transduced cells were plated on coverslips, and gB was detected via indirect immunofluorescence using antibody 27-78. (C) NHDF-GFP or NHDF-gB were electroporated in the absence of BAC plasmid or with pAD/Cre or pAD/CreΔUL55. Approximately 3 weeks postelectroporation, the cells were stained with crystal violet to visualize plaque formation. (D) NHDF on coverslips were infected with supernatant from the BAC electroporations, and 24 h postinfection, IE gene expression was detected via indirect immunofluorescence. (E) Single-step growth curve comparing ΔUL55-gB virus (▪) replication on NHDF-gB to HCMV revertant virus (•) on NHDF-gB. The cells were infected at an MOI of 5.0. At various times postinfection, cells and supernatants were collected and titrated on NHDF-gB or -GFP. The values represent averages from duplicate infections in one experiment.
To determine if pAD/CreΔUL55 could be complemented by NHDF-gB, NHDF-GFP were mock electroporated or pAD/Cre or pAD/CreΔUL55 was electroporated into either NHDF-GFP or NHDF-gB. Approximately 10 days postelectroporation, cytopathic effects caused by virus replication were readily visualized following pAD/Cre or pAD/CreΔUL55 electroporation into NHDF-gB only. At 3 weeks postelectroporation, the cells were stained with crystal violet so plaques could be more easily visualized (Fig. 3C). Cell supernatants were collected approximately 4 weeks postelectroporation and used to infect new NHDF. IE gene expression was detected from only those electroporations in which plaque formation had been visible. These data indicate that infectious virus was generated and complementation of pAD/CreΔUL55 was successful (Fig. 3D). To quantitate the growth kinetics of the ΔUL55 virus, one-step growth curves were performed comparing revertant HCMV to the ΔUL55-gB infection in complementing NHDF-gB (Fig. 3E). As the ΔUL55 virus is not infectious in the absence of gB, no growth curves were conducted under noncomplementing conditions.
Characterization of HCMV gene expression and virion production by ΔUL55 virus.
Virus-containing supernatant from electroporation of pAD/CreΔUL55 into NHDF-gB was used to infect new NHDF-gB, generating an infectious ΔUL55-gB HCMV stock. It was expected that in the first round of replication, ΔUL55-gB infection would result in normal stages of gene expression and a defect would not be evident until the second round of replication, when either virions did not assemble and/or egress properly in the absence of gB or gB-null virions were produced and could not infect new cells. We next sought to measure the stages of gene expression following ΔUL55-gB infection into noncomplementing NHDF. Infected cell lysates were collected, analyzed by SDS-PAGE, and immunoblotted to detect IE (IE-1 and IE-2), early (UL44), and late (pp28) gene expression. As demonstrated in Fig. 4A, ΔUL55 infection proceeded normally, even in noncomplementing cells, as would be expected during the first round of replication for ΔUL55 HCMV. However, when the infection was allowed to proceed, plaque formation did not occur following ΔUL55-gB infection in noncomplementing NHDF (see Fig. 6), similar to that seen following electroporation. Lack of plaque formation could be the result of a failure to assemble or release viral particles (an egress defect) or the release of viral particles that are noninfectious (an entry defect).
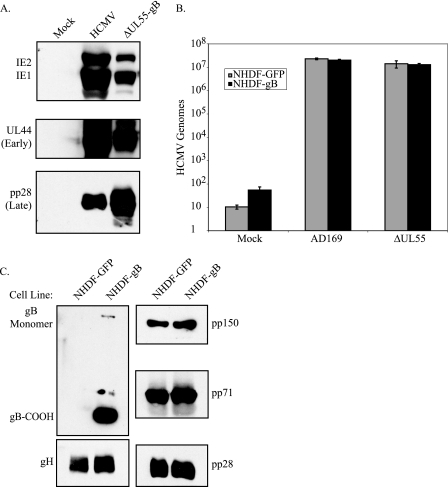
FIG. 4.
HCMV gB is not required for virion egress. (A) Infectious AD169 HCMV or ΔUL55-gB virus generated from propagation on NHDF-gB and purified by sorbitol cushioning was used to infect noncomplementing NHDF. Infected cell lysate was collected 6 days postinfection and analyzed by immunoblotting to detect IE, early (UL44), and late (pp28) gene expression during a single round of virus replication. (B) NHDF-GFP or NHDF-gB were mock infected or infected with HCMV AD169 or ΔUL55-gB at an MOI of 1 PFU/cell. Six days postinfection, the supernatant was collected and cell debris was removed by centrifugation, the supernatant was DNase I treated, and HCMV DNA was quantitated by real-time PCR to determine the number of DNase-resistant genomes that were released into the supernatant following infection. The error bars indicate standard deviations for three experiments. (C) Infected cell supernatants from ΔUL55-gB infection of NHDF-GFP or NHDF-gB were concentrated over a 20% sorbitol cushion, followed by gradient purification in a 20 to 70% sorbitol step gradient. Virus bands were extracted, and HCMV structural proteins, including gB, gH, pp150, pp71, and pp28, were detected by Western immunoblotting.
FIG. 6.
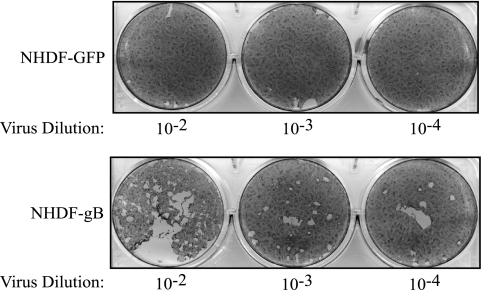
gB is required for cell-to-cell spread. NHDF-GFP or NHDF-gB were infected with dilutions of HCMV ΔUL55-gB virus. The virus inoculum was removed, and the cells were overlaid with plaquing medium. Approximately 3 weeks postinfection, the cells were stained with crystal violet to visualize plaque formation.
As viral glycoproteins are known to be required for both steps, we first asked whether HCMV gB is required for virion maturation, assembly, and/or egress in infected cells. For herpesviruses, the role of gB in virion egress appears to vary between subfamilies, and more data are needed to complete several of these studies. To determine if HCMV gB is required for virion assembly and egress, we asked if viral particles could be produced in the absence of gB. NHDF-GFP or NHDF-gB were infected with ΔUL55-gB virus or HCMV AD169 at an MOI of 1.0 PFU/cell, and infected cell supernatants were collected, followed by DNase I treatment to remove exogenous DNA not protected within virions and quantitation using real-time PCR. As demonstrated in Fig. 4B, DNase-resistant genomes produced from ΔUL55-gB virus infection in noncomplementing NHDF-GFP were found at levels similar to that of AD169 infection in NHDF-GFP or ΔUL55-gB infection in NHDF-gB, suggesting that gB has no evident role in virion egress from infected cells.
To confirm these results, infected cell supernatant from ΔUL55-gB infection in NHDF-GFP or NHDF-gB was concentrated over a 20% sorbitol cushion, followed by gradient purification in a 20% to 70% sorbitol step gradient. Virus bands were collected, analyzed by SDS-PAGE, and immunoblotted to detect virion structural proteins, including gB and gH and tegument proteins pp150, pp71, and pp28. As demonstrated in Fig. 4C, gB is present in virions only when the ΔUL55 virus is passaged in complementing NHDF-gB. However, when ΔUL55-gB virus is passaged one time in noncomplementing NHDF-GFP, gB-null virus can be generated. These virions lack gB but contain other virion structural proteins (gH, pp150, pp71, and pp28), suggesting that in the absence of gB, HCMV particles are produced that are physically indistinguishable from the wild type by all criteria analyzed, except for the presence of gB. A more complete proteomics and electron microscopy analysis of gB-null virions will be the subject of future studies.
Characterization of HCMV virion attachment and fusion in the absence of gB.
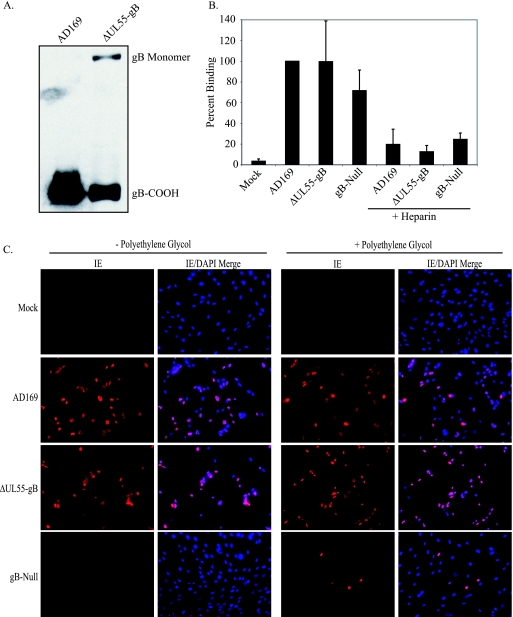
gB is thought to play a role in multiple stages of HCMV entry, including attachment to cells during the first step of initial tethering to HSPGs, as well as the second, more stable heparin-resistant attachment step, possibly through binding one or more cellular receptors (7, 17, 21, 33, 62). gB is also likely to be required for virion fusion (23, 48). gB homologs from other herpesviruses have been demonstrated to be involved in virion attachment, although none are absolutely required (10, 38, 40), likely because other glycoproteins contain redundant attachment functions that are sufficient to compensate in the absence of gB. However, the fusogenic activity of gB does appear to be required for all herpesviruses tested, with the possible exception of EBV, which incorporates only minimal amounts of gB into the virion (24). To determine if the inability of gB-null HCMV to infect cells is a defect at the level of virion attachment or fusion, similar numbers of AD169 and ΔUL55-gB virions were first analyzed by SDS-PAGE and immunoblotting to determine the level of gB incorporation into virions (Fig. 5A). Similar genome copy numbers of AD169, ΔUL55-gB, or gB-null virus were incubated with NHDF at 4°C. Virus was added alone or incubated with soluble heparin, which served as a positive control for attachment, prior to addition to cells. Serum-free DMEM alone was used as a mock-treated control. Following attachment, cell lysates were collected and HCMV DNA copy numbers were determined by real-time PCR to determine the number of genome-containing particles that attached to cells. As demonstrated in Fig. 5B, although the gB-null ΔUL55 virus may have a slight defect in attachment to cells compared to gB-containing virus, gB is not absolutely required for virion attachment. Additionally, incubation of both gB-null and gB-containing virus with soluble heparin inhibited virion attachment by approximately 90%, as expected.
FIG. 5.
gB-null HCMV can attach to cells and contains infectious HCMV DNA. (A) gB from equivalent numbers of purified HCMV AD169 and ΔUL55 gB virions was analyzed by immunoblotting. (B) NHDF were treated with AD169, ΔUL55-gB, or ΔUL55 gB-null virus or virus pretreated with soluble heparin at 4°C for 1 h. The cells were washed three times with serum-free DMEM, the cell lysate was collected, DNA was extracted, and the viral genomes were quantitated by real-time PCR. The graph is representative of one of three similar experiments. The error bars represent standard deviations from triplicate samples in one experiment. (C) NHDF were infected with HCMV AD169, ΔUL55-gB, or ΔUL55 gB-null virus. The supernatant was removed, and the cells were washed three times with serum-free DMEM or serum-free DMEM containing decreasing concentrations of PEG (50, 25, and 12.5%). DMEM supplemented with 2% FBS was added back to the cells, and 24 h postinfection, IE gene expression was detected by indirect immunofluorescence.
Our data demonstrate that while productive infection requires gB, egress and attachment do not. This implies that the major role of gB may be during fusion. To determine if gB-null virions could deliver viral genomes to the nucleus if their fusion to cell membranes were artificially forced, NHDF were incubated with HCMV AD169, ΔUL55-gB virus, or gB-null virus, followed by washing with serum-free DMEM or serum-free DMEM containing decreasing concentrations (50, 25, and 12.5%) of PEG, a membrane-fusing agent (32). Twenty-four hours postinfection, IE gene expression was detected via indirect immunofluorescence. As shown in Fig. 5C, gene expression was detected only without PEG treatment for AD169 or ΔUL55-gB virus, but not gB-null virus, as demonstrated previously. However, we could restore IE gene expression with the gB-null virus by treatment with PEG, indicating that the gB-null virions are capable of mediating gene expression in the absence of gB if they can be artificially forced to fuse. This is also further evidence that these virions are capable of attaching to cells in the absence of gB.
gB is required for cell-to-cell spread of HCMV infection.
Plaque assays with gB-neutralizing antibodies have suggested that HCMV gB is required for cell-to-cell spread of the virus, similar to other well-studied herpesviruses (46). However, cell-to-cell fusion assays have indicated that gB alone is not sufficient to mediate cell-to-cell fusion, nor does it seem to enhance cell-to-cell fusion mediated by gH/gL (36). To determine if gB is required for cell-to-cell spread of HCMV, ΔUL55-gB virus was used to infect either noncomplementing NHDF-GFP or complementing NHDF-gB. Following infection, the cells were overlaid with agarose, and approximately 2 weeks postinfection, the cells were stained to visualize plaque formation. As demonstrated in Fig. 6, plaque formation occurred for ΔUL55-gB virus only when complementing NHDF-gB were infected, indicating that gB is required for cell-to-cell spread of the virus.
DISCUSSION
gB is the most highly conserved envelope protein throughout the herpesvirus family, which suggests that it plays an important role in the life cycle of these viruses. Deletion mutants from alpha-, beta-, and gammaherpesviruses, including HSV-1, HSV-2, PRV, Marek's disease virus, HCMV, EBV, and KSHV, have demonstrated that gB is essential for virus replication (12, 18, 26, 27, 37, 42, 52, 54, 55, 57, 65). With the development of the BAC system, herpesvirus mutants can be generated very efficiently; however, until now, the ability to complement any HCMV glycoprotein deletion mutant has been lacking, which has greatly limited the study of lethal gB mutants in the context of a viral infection. Here, we have described the development of a gB complementation system that allowed us to characterize the phenotype of a gB-null mutant. It will also allow the study of a variety of gB mutants that may otherwise be difficult to examine in the context of the virus. Because gB is essential for virus replication, mutations in the UL55 gene often result in a no-plaque phenotype when the BAC system is used. We found that a ΔUL55 BAC plasmid could be efficiently propagated in gB-expressing fibroblasts, generating ΔUL55 virus pseudotyped with wild-type gB (ΔUL55-gB).
A single-step growth curve was used to compare ΔUL55-gB replication to revertant virus replication in gB-expressing cells (Fig. 3E), demonstrating that replication is fairly similar in the two viruses. The ΔUL55-gB virus titer did appear to drop late in infection compared to that of revertant virus; however, as the NHDF-gB represent a range of gB expression, varying from no expression to high levels of expression, it is not surprising that complementation is not 100%. Further experimentation with high versus low NHDF-gB expressors may enhance complementation.
Following infection of noncomplementing fibroblasts by ΔUL55-gB HCMV, we saw levels of gene expression similar to those in a wild-type HCMV infection, indicating that the cells were at least initially infected. This was not surprising, as we expected the block in virus replication to occur late in infection, when virions cannot be assembled or released in the absence of gB, or in the second round of replication, when gB-null virions are generated but cannot reenter cells.
By passaging ΔUL55-gB HCMV one time in noncomplementing NHDF, we were able to generate gB-null virions. We used real-time PCR to detect DNase-resistant viral genomes that were released into the supernatant following ΔUL55-gB HCMV infection of normal fibroblasts. Presumably, these DNase-resistant genomes represented intact gB-null virions. To test this hypothesis, these gB-null, DNase-resistant genome-containing particles were gradient purified, and we found that virion structural proteins other than gB were present, including gH and various tegument proteins. These data suggest that HCMV gB is not absolutely required for virion egress, as may also be the case for alphaherpesviruses, such as PRV and HSV-1 (11, 52, 53). For HSV-1, it appears that gB and gH together are absolutely required for virion egress, suggesting that they have redundant roles, allowing one to compensate for the other in either one's absence (20). However, for other herpesviruses, such as KSHV, EBV, and a second, conflicting report on PRV, gB does appear to be absolutely required for virion egress, as enveloped virions lacking gB could not be isolated (37, 39, 49). Therefore, it appears that although gB is the most highly conserved glycoprotein in the herpesvirus family, functional differences do exist from one subfamily to another, and perhaps even within subfamilies.
By comparison with other herpesviruses, such as HSV-1, it is thought that HCMV gB likely plays a role in the virus entry process, including attachment and fusion events. Supporting data for HCMV include interaction between gB and cell surface HSPGs (17, 33), suggesting that gB plays a role in the initial virion-tethering step. The ability of gB to interact with β1 integrins through its DLD also suggests that gB may play a role in the secondary attachment and/or fusion step, as well (21; Feire et al., unpublished). Generation of a gB-null virus allowed us to directly test the requirement for gB in HCMV attachment. Using a quantitative real-time PCR-based assay, we demonstrated that HCMV gB is not required for virus attachment, as gB-null virions are capable of binding to the cell surface similarly to gB-containing virions, although a small defect in virion binding may exist in the absence of gB. This initial attachment of gB-null virions can be inhibited, similarly to wild-type virions, by incubation with soluble heparin, as also occurs for wild-type virus, likely because gM also binds HSPGs (17, 33). Given our results, gM is possibly the major envelope protein involved in the initial virion attachment to HSPGs, as it has also been demonstrated to have significant heparin binding capabilities (61); however, until a gM-null virus is generated, it is not known whether gB will be able to compensate for the loss of gM during virion attachment. It is possible that gB may play a nonessential accessory role that may or may not be sufficient to mediate virion attachment in the absence of gM. Similar results have also been found for other herpesviruses, suggesting that while all gB homologs possess some initial attachment capabilities, this attachment is not absolutely required for virus entry, as other glycoproteins can compensate for the loss of gB (10, 29, 38, 40). It is likely that for all herpesviruses the various glycoproteins have redundant functions and are capable of compensating in another's absence; for HCMV, both gB and gM appear to participate in virion adsorption.
Additionally, although these gB-null virions that were generated were not able to infect NHDF, they could be artificially induced to fuse using PEG, leading to the detection of IE gene expression. Although this assay is inefficient and it is difficult to increase the efficiency, as increasing PEG concentrations or incubation times leads to significant cell death, these data do indicate that the gB-null virions are intact and able to attach to cells and contain active tegument proteins, as well as infectious HCMV DNA. They are not, however, inherently fusogenic. gB is thought to have a fusogenic role in the HCMV life cycle during both virus-cell and cell-to-cell fusion events. Evidence for this includes neutralizing antibodies to gB that inhibit virus entry without affecting virus attachment, even when applied after attachment has already occurred (23, 48). Additionally, plaque assays in the presence of gB-neutralizing antibodies also suggest that gB is required for cell-to-cell spread of the virus (6, 8). However, it is possible that these antibodies interfere with another virion molecule required for fusion, although this is unlikely given the well-demonstrated fusogenic role of gB in virus entry for other herpesviruses (24, 51). Mutations in the cytoplasmic tail of HCMV gB have been demonstrated to affect its fusogenic activity; however, these experiments are usually performed in neuronal glioblastoma cells in which spontaneous fusion of non-gB-expressing cells can make interpretation difficult (58, 59). Finally, gB is not sufficient to mediate or enhance cell-to-cell fusion in fibroblasts in a virus-free system (36).
Until now, direct evidence for the role of gB in virus-cell and cell-to-cell fusion has been lacking. Our data suggest that gB is required for virus-cell entry, as we can infect normal fibroblasts with ΔUL55-gB HCMV and can detect all stages of gene expression leading to the release of gB-null virions into the supernatant. These virions are able to attach to cells, but they are blocked at the second round of replication, at the level of virus entry, demonstrating that gB is required for the fusion of the viral and cellular membranes. Additionally, gB also appears to be required for cell-to-cell spread of the virus, as infection of normal fibroblasts with ΔUL55-gB HCMV does not lead to the spread of initially infected cells and plaque formation, which supports the conclusions from earlier studies using gB-neutralizing antibodies (23, 48).
In summary, our data confirm a role for gB in virus entry and cell-to-cell spread but not in attachment or egress. The ΔUL55 virus generated can be used as a tool to study HCMV gB mutations, even those conferring a lethal phenotype, in the context of a viral infection. Hopefully, these studies will also pave the way for the generation of complementation systems for other glycoprotein mutants, including gM and gH. Additionally, the ability to generate gB-null virions that attach to cells will allow a variety of studies of the signaling aspects of HCMV and gB to be conducted, including the induction of innate immune responses and integrin signaling events early in infection.
Acknowledgments
We thank Thomas Shenk for pAD/Cre, Robert F. Kalejta for HCMV antibodies, William J. Britt for gB antibodies, Richard C. Mulligan for pMD-gagpol and pMD-VSV-G, and Neal Copeland for the bacterial strain SW102 and pGalK.
This work was supported by NIH grant R01-AI034998.
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Alford, C. A. and W. J. Britt. 1993. Cytomegalovirus, p. 227-255. In B. Roizman, R. J. Whitley, and C. Lopez (ed.), Human herpesviruses. Raven Press, New York, NY.
- 2.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 714400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billstrom, M. A., and W. J. Britt. 1995. Postoligomerization folding of human cytomegalovirus glycoprotein B: identification of folding intermediates and importance of disulfide bonding. J. Virol. 697015-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehme, K. W., and T. Compton. 2006. Virus entry and activation of innate immunity, p. 111-130. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister, Norfolk, United Kingdom.
- 5.Boehme, K. W., M. Guerrero, and T. Compton. 2006. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J. Immunol. 1777094-7102. [DOI] [PubMed] [Google Scholar]
- 6.Bold, S., M. Ohlin, W. Garten, and K. Radsak. 1996. Structural domains involved in human cytomegalovirus glycoprotein B-mediated cell-cell fusion. J. Gen. Virol. 772297-2302. [DOI] [PubMed] [Google Scholar]
- 7.Boyle, K. A., and T. Compton. 1998. Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. J. Virol. 721826-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britt, W. J. 1984. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology 135369-378. [DOI] [PubMed] [Google Scholar]
- 9.Britt, W. J., and L. G. Vugler. 1992. Oligomerization of the human cytomegalovirus major envelope glycoprotein complex gB (gp55-116). J. Virol. 666747-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 622596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai, W. Z., S. Person, S. C. Warner, J. H. Zhou, and N. A. DeLuca. 1987. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J. Virol. 61714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheshenko, N., and B. C. Herold. 2002. Glycoprotein B plays a predominant role in mediating herpes simplex virus type 2 attachment and is required for entry and cell-to-cell spread. J. Gen. Virol. 832247-2255. [DOI] [PubMed] [Google Scholar]
- 13.Cobbs, C. S., L. Soroceanu, S. Denham, W. Zhang, W. J. Britt, R. Pieper, and M. H. Kraus. 2007. Human cytomegalovirus induces cellular tyrosine kinase signaling and promotes glioma cell invasiveness. J. Neurooncol. 85271-280. [DOI] [PubMed] [Google Scholar]
- 14.Compton, T. 1993. An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J. Virol. 673644-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Compton, T. 2004. Receptors and immune sensors: the complex entry path of human cytomegalovirus. Trends Cell Biol. 145-8. [DOI] [PubMed] [Google Scholar]
- 16.Compton, T., R. R. Nepomuceno, and D. M. Nowlin. 1992. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 191387-395. [DOI] [PubMed] [Google Scholar]
- 17.Compton, T., D. M. Nowlin, and N. R. Cooper. 1993. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 193834-841. [DOI] [PubMed] [Google Scholar]
- 18.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 10014223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.English, E. P., R. S. Chumanov, S. H. Gellman, and T. Compton. 2006. Rational development of beta-peptide inhibitors of human cytomegalovirus entry. J. Biol. Chem. 2812661-2667. [DOI] [PubMed] [Google Scholar]
- 20.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA 10410187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feire, A. L., H. Koss, and T. Compton. 2004. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. USA 10115470-15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gault, E., Y. Michel, A. Dehee, C. Belabani, J. C. Nicolas, and A. Garbarg-Chenon. 2001. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 39772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gicklhorn, D., M. Eickmann, G. Meyer, M. Ohlin, and K. Radsak. 2003. Differential effects of glycoprotein B epitope-specific antibodies on human cytomegalovirus-induced cell-cell fusion. J. Gen. Virol. 841859-1862. [DOI] [PubMed] [Google Scholar]
- 24.Gong, M., and E. Kieff. 1990. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, gp350/220 and gp110. J. Virol. 641507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290106-114. [DOI] [PubMed] [Google Scholar]
- 26.Herrold, R. E., A. Marchini, S. Fruehling, and R. Longnecker. 1996. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J. Virol. 702049-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 747720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isaacson, M. K., A. L. Feire, and T. Compton. 2007. Epidermal growth factor receptor is not required for human cytomegalovirus entry or signaling. J. Virol. 816241-6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacquet, A., M. Haumont, D. Chellun, M. Massaer, F. Tufaro, A. Bollen, and P. Jacobs. 1998. The varicella zoster virus glycoprotein B (gB) plays a role in virus binding to cell surface heparan sulfate proteoglycans. Virus Res. 53197-207. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 761-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalejta, R. F., J. T. Bechtel, and T. Shenk. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 231885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao, K. N., and M. R. Michayluk. 1974. A method for high-frequency intergeneric fusion of plant protoplasts. Planta 115355-367. [DOI] [PubMed] [Google Scholar]
- 33.Kari, B., and R. Gehrz. 1992. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J. Virol. 661761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kari, B., and R. Gehrz. 1993. Structure, composition and heparin binding properties of a human cytomegalovirus glycoprotein complex designated gC-II. J. Gen. Virol. 74255-264. [DOI] [PubMed] [Google Scholar]
- 35.Keay, S., and B. Baldwin. 1991. Anti-idiotype antibodies that mimic gp86 of human cytomegalovirus inhibit viral fusion but not attachment. J. Virol. 655124-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinzler, E. R., and T. Compton. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 797827-7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnan, H. H., N. Sharma-Walia, L. Zeng, S. J. Gao, and B. Chandran. 2005. Envelope glycoprotein gB of Kaposi's sarcoma-associated herpesvirus is essential for egress from infected cells. J. Virol. 7910952-10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laquerre, S., R. Argnani, D. B. Anderson, S. Zucchini, R. Manservigi, and J. C. Glorioso. 1998. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J. Virol. 726119-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, S. K., and R. Longnecker. 1997. The Epstein-Barr virus glycoprotein 110 carboxy-terminal tail domain is essential for lytic virus replication. J. Virol. 714092-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, W. C., and A. O. Fuller. 1993. Herpes simplex virus type 1 and pseudorabies virus bind to a common saturable receptor on Vero cells that is not heparan sulfate. J. Virol. 675088-5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopper, M., and T. Compton. 2004. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J. Virol. 788333-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manservigi, R., P. G. Spear, and A. Buchan. 1977. Cell fusion induced by herpes simplex virus is promoted and suppressed by different viral glycoproteins. Proc. Natl. Acad. Sci. USA 743913-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106167-180. [DOI] [PubMed] [Google Scholar]
- 44.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 45.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 812017-2027. [DOI] [PubMed] [Google Scholar]
- 46.Navarro, D., P. Paz, S. Tugizov, K. Topp, J. La Vail, and L. Pereira. 1993. Glycoprotein B of human cytomegalovirus promotes virion penetration into cells, transmission of infection from cell to cell, and fusion of infected cells. Virology 197143-158. [DOI] [PubMed] [Google Scholar]
- 47.Nowak, B., C. Sullivan, P. Sarnow, R. Thomas, F. Bricout, J. C. Nicolas, B. Fleckenstein, and A. J. Levine. 1984. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology 132325-338. [DOI] [PubMed] [Google Scholar]
- 48.Ohizumi, Y., H. Suzuki, Y. Matsumoto, Y. Masuho, and Y. Numazaki. 1992. Neutralizing mechanisms of two human monoclonal antibodies against human cytomegalovirus glycoprotein 130/55. J. Gen. Virol. 732705-2707. [DOI] [PubMed] [Google Scholar]
- 49.Peeters, B., N. de Wind, M. Hooisma, F. Wagenaar, A. Gielkens, and R. Moormann. 1992. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J. Virol. 66894-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pertel, P. E. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 764390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rauh, I., and T. C. Mettenleiter. 1991. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J. Virol. 655348-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rauh, I., F. Weiland, F. Fehler, G. M. Keil, and T. C. Mettenleiter. 1991. Pseudorabies virus mutants lacking the essential glycoprotein gII can be complemented by glycoprotein gI of bovine herpesvirus 1. J. Virol. 65621-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarmiento, M., M. Haffey, and P. G. Spear. 1979. Membrane proteins specified by herpes simplex viruses. III. Role of glycoprotein VP7(B2) in virion infectivity. J. Virol. 291149-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaffer, P. A., G. M. Aron, N. Biswal, and M. Benyesh-Melnick. 1973. Temperature-sensitive mutants of herpes simplex virus type 1: isolation, complementation and partial characterization. Virology 5257-71. [DOI] [PubMed] [Google Scholar]
- 55.Schaffer, P. A., V. C. Carter, and M. C. Timbury. 1978. Collaborative complementation study of temperature-sensitive mutants of herpes simplex virus types 1 and 2. J. Virol. 27490-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoppel, K., E. Hassfurther, W. Britt, M. Ohlin, C. A. Borrebaeck, and M. Mach. 1996. Antibodies specific for the antigenic domain 1 of glycoprotein B (gpUL55) of human cytomegalovirus bind to different substructures. Virology 216133-145. [DOI] [PubMed] [Google Scholar]
- 57.Schumacher, D., B. K. Tischer, W. Fuchs, and N. Osterrieder. 2000. Reconstitution of Marek's disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 7411088-11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tugizov, S., D. Navarro, P. Paz, Y. Wang, I. Qadri, and L. Pereira. 1994. Function of human cytomegalovirus glycoprotein B: syncytium formation in cells constitutively expressing gB is blocked by virus-neutralizing antibodies. Virology 201263-276. [DOI] [PubMed] [Google Scholar]
- 59.Tugizov, S., Y. Wang, I. Qadri, D. Navarro, E. Maidji, and L. Pereira. 1995. Mutated forms of human cytomegalovirus glycoprotein B are impaired in inducing syncytium formation. Virology 209580-591. [DOI] [PubMed] [Google Scholar]
- 60.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 7810960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, X., D. Y. Huang, S. M. Huong, and E. S. Huang. 2005. Integrin αvβ3 is a coreceptor for human cytomegalovirus. Nat. Med. 11515-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, X., S. M. Huong, M. L. Chiu, N. Raab-Traub, and E. S. Huang. 2003. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 424456-461. [DOI] [PubMed] [Google Scholar]
- 64.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 10012396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 762316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]