Abstract
The Arabidopsis flowering locus T (FT) gene encodes the mobile florigen essential for floral induction. While movement of the FT protein has been shown to occur within plants, systemic spread of FT mRNA remains to be unequivocally demonstrated. Utilizing novel RNA mobility assay vectors based on two distinct movement-defective viruses, Potato virus X and Turnip crinkle virus, and an agroinfiltration assay, we demonstrate that nontranslatable FT mRNA, independent of the FT protein, moves throughout Nicotiana benthamiana and mutant Arabidopsis plants and promotes systemic trafficking of viral and green fluorescence protein RNAs. Viral ectopic expression of FT induced flowering in the short-day N. tabacum Maryland Mammoth tobacco under long-day conditions. Recombinant Potato virus X bearing FT RNA spread and established systemic infection more quickly than the parental virus. The cis-acting element essential for RNA movement was mapped to the nucleotides 1 to 102 of the FT mRNA coding sequence. These data demonstrate that a plant self-mobile RNA molecule can mediate long-distance trafficking of heterologous RNAs and raise the possibility that FT RNA, along with the FT protein, may be involved in the spread of the floral stimulus throughout the plant.
RNA trafficking plays an important role in systemic signaling that controls plant development and defense against pathogen infection (25). Hundreds of RNA transcripts have been recently identified in phloem, suggesting phloem-mobile RNAs may act as long-distance signaling molecules in plants (4, 8). Indeed, systemic movement of a homeobox fusion transcript and gibberellic acid-insensitive RNA regulates leaf architecture (13, 19), a non-cell-autonomous mobile RNA represents a long-distance signal that modulates potato tuber formation (3), and small interfering RNAs are components of intercellular and systemic mobile signals for innate RNA silencing defense (9, 12, 14). RNA trafficking is also critical for plant viruses and viroids to establish systemic infection. It has been demonstrated that an RNA motif directs long-distance trafficking of a small naked RNA viroid (33, 44, 46). Moreover, a short RNA sequence is found to be involved in cell-to-cell movement of a plant viral RNA (24), and replication-independent viral RNA can move over long distances in plants (11).
In floral induction, the mobile florigen is encoded by the Arabidopsis flowering locus T (FT) gene. FT transcribes mRNA in the leaf, but its encoded FT protein functions in the shoot apices where flowers develop (1, 2, 40). The Arabidopsis FT protein and its orthologues have been shown to be involved in long-distance signaling in floral induction (7, 18, 22, 23, 29, 30, 37). However, whether FT mRNA is also capable of systemic spread remains to be demonstrated. We describe novel approaches which show that not only does FT RNA move over long distances but, remarkably, also facilitates the systemic spread of heterologous green fluorescent protein (GFP) mRNA and different viral RNAs in plants. The FT RNA movement does not rely on the expression of the FT protein. The FT RNA mobility is determined by a cis-acting element localized within nucleotides 1 to 102 of the FT mRNA coding sequence.
MATERIALS AND METHODS
Construction of RMA vectors.
The wild-type and mutant Arabidopsis FT genes were reverse transcription-PCR (RT-PCR) amplified using Pfu DNA polymerase and the primers PP354/PP356 or PP356/PP355, digested with BspEI and SalI, and cloned in-frame fused to the GFP coding sequence in the BspEI/SalI sites of PVX and PVX/GFP (38) to produce PVX/FT, PVX/mFT, PVX/GFP-FT, and PVX/GFP-mFT, respectively. Plasmid DNA of PVX/GFP, PVX/GFP-FT, and PVX/GFP-mFT were then digested with SalI and XhoI to remove the coat protein (CP) gene subgenomic RNA promoter and the CP gene and self-ligated to produce PVX/GFPΔCP, PVX/GFP-FTΔCP, and PVX/GFP-mFTΔCP. Expression of the GFP gene from PVX/GFPΔCP and the wild-type and mutated GFP-FT fusion gene from PVX/GFP-FTΔCP and PVX/GFP-mFTΔCP were under the control of an engineered CP subgenomic RNA promoter. For construction of TCV-based RNA mobility assay (RMA) vectors TCV/mFTΔCP, TCV/GFP-FTΔCP, and TCV/GFP-mFTΔCP, the Arabidopsis FT gene was RT-PCR amplified using Pfu DNA polymerase and the primers PP406 or PP407 and PP408, digested with BclI and PmeI, and cloned into the BglII/PmeI sites of TCV/ΔCP (34) or TCV/GFPΔCP (47). Using a similar PCR and cloning strategy, a series of TCV/trFTΔCP-based RMA vectors carrying truncated (tr) FT for mapping the cis-acting element required for FT RNA trafficking were constructed. All RMA constructs were confirmed by nucleotide sequencing. The primers used for the construction of RMA vectors are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a | Origin and modification |
|---|---|---|
| PP82 | CAGTGTTGGCTTGCAAACTAG | PVX |
| PP228 | GGGTAAGTTTTCCGTATGTTG | GFP |
| PP267 | AGAAAAGTCATGAAGGTTCTGCTAGCCACGG | TCV |
| PP269 | CAAGGTGCGCGAGGTTTACCAATC | PVX |
| PP271 | CGGCTACCACATCCAAGGAAGG | 18S rRNA |
| PP272 | GAGCTGGAATTACCGCGGCTG | 18S rRNA |
| PP354 | TCAAGATCCGGAATGTCTATAAATATAAGAGACCC | FT, BspEI |
| PP355 | TCAAGATCCGGATAGTCTATAAATATAAGAGACCC | FT, BspEI |
| PP356 | AGGAAGAAGTCGACTAAAGTCTTCTTCCTCCGCAG | FT, SalI |
| PP371 | ATGAGTAAAGGAGAAGAACTTTTCACTGGAG | GFP |
| PP372 | TTTGTGTCCAAGAATGTTTCCATCTTC | GFP |
| PP373 | GTATGCTGTTTCCGTTGTGATCTCTGTGAG | PVX |
| PP407 | TTCAAGTGATCAGTAGTCTATAAATATAAGAGAC | FT, BclI |
| PP408 | AAGGAAGTTTAAACCTAAAGTCTTCTTCCTCCGCA | FT, PmeI |
| PP433 | ATGCCTCTCCTACATACACTCAACACAG | TCV |
| PP434 | GTCTACGACATGTCTCTCTTTGCAG | TCV |
| PP473a | TTGGCCATAAGTAACCTTTAG | FT |
| PP512 | TCAAGATGATCATAGTCTATAAATATAAGAGACCC | FT, BclI |
| PP513 | TTAGTCACGTTTAAACGGCCATAAGTAACCTTTAG | FT, PmeI |
| PP514 | GTTACTTGATCATAGAGAGAGGTGACTAATGGCTT | FT, BclI |
| PP515 | AGGAAGTTTAAACCTAAAGTCTTCTTCCTCCGCAG | FT, PmeI |
| PP516 | AGATCCGTTTAAACCAAAGTATAGAAGTTCCTGAG | FT, PmeI |
| PP517 | AACTTCTGATCATGAGTTATGGTGGATCCAGATG | FT, BclI |
| PP518 | ATCTCAGTTTAAACGGTTGTTCCAGTTGTAGCAGG | FT, PmeI |
| PP519 | ACAACTTGATCATAGTTTGGCAATGAGATTGTGTG | FT, BclI |
| PP520 | CACCCTGGTTTAAACACTGTTTGCCTGCCAAGCTG | FT, PmeI |
| PP521 | GGCAGGTGATCATGATATGCACCAGGGTGGCGCCA | FT, BclI |
Introduced restriction endonuclease sites are in boldface, and the start and mutated nonsense stop codons are underlined.
RMAs.
RNA transcripts from each movement-deficient recombinant PVX and TCV vector were produced by in vitro transcription as previously described (34), pretreated with RNase-free DNase (Promega), and then mechanically inoculated onto N. benthamiana or A. thaliana ft-10 mutant plants at 5 to 6 or 15 to 16 leaves, respectively, in repeated experiments. N. benthamiana plants aged at only 24 days after sowing seeds and were too young to initiate flowering. For PVX-based RMA, total RNAs (50 ng) extracted from inoculated and newly growing young leaves separately collected at 7 days postinoculation (dpi) were pretreated with RNase-free DNase (Promega) and used for RT-PCR detection (30 cycles) (34) with the primers PP354 and PP356 for FT, PP371 and PP372 for GFP, PP269 and PP373 for PVX, and PP271 and PP272 for 18S rRNA. Epidermal cells with GFP expression and the movement of GFP-tagged viruses were examined with a Zeiss Axiophot microscope with filters for GFP (excitation, 450 to 490 nm; long-pass emission, 520 nm) or under long-wavelength UV light (Upland UVP model B 100AP) through a yellow filter (Kodak no. 58) and photographed with a Nikon CoolPix 995 digital camera (38).
For TCV-based RMAs, inoculated, newly growing young leaves and shoot apices were carefully collected at 7 dpi from mock- and virus-inoculated N. benthamiana and mutant ft-10 A. thaliana plants and used for total RNA extraction. Total RNAs were treated with RNase-free DNase (Promega), and RT-PCR (30 cycles) detection was performed with the primers PP354 and PP356 for FT, PP371 and PP372 for GFP, PP433 and PP434 for TCV RNA, PP267 and PP473a for TCV-mFT RNA, PP267 and PP228 for TCV-GFP RNA, and PP271 and PP272 for 18S rRNA. The resultant specific TCV-mFT RT-PCR products were purified and verified by direct sequencing.
In two separate mapping experiments, plants were inoculated with recombinant viral RNAs that were produced by in vitro transcription from each TCV/trFTΔCP-based RMA vectors and pretreated with RNase-free DNase (Promega). Total RNAs were extracted at 7 dpi from inoculated and uninoculated young leaves, pretreated with RNase-free DNase (Promega), and used for RT-PCR (30 cycles) assays with primer sets listed in Table 2.
TABLE 2.
Mapping cis-acting element for FT RNA movement
| RMA vectora | FT RNA 5′-3′ coordinates | Primer pair used for:
|
Predicted size (bp) of RT-PCR productb | |
|---|---|---|---|---|
| PCR and cloning | RT-PCR detection | |||
| TCV/FTn102ΔCP* | 1-102 | PP512/PP513 | PP267/PP513 | 375 |
| TCV/FTn201ΔCP* | 1-201 | PP512/PP516 | PP267/PP516 | 474 |
| TCV/FTn300ΔCP* | 1-300 | PP512/PP518 | PP267/PP518 | 573 |
| TCV/FTn399ΔCP* | 1-399 | PP512/PP520 | PP267/PP520 | 672 |
| TCV/FT103cΔCP† | 103-528 | PP514/PP515 | PP267/PP515 | 699 |
| TCV/FT202cΔCP† | 202-528 | PP517/PP515 | PP267/PP515 | 600 |
| TCV/FT301cΔCP† | 301-528 | PP519/PP515 | PP267/PP515 | 500 |
| TCV/FT400cΔCP† | 400-528 | PP521/PP515 | PP267/PP515 | 402 |
*, A stop codon was introduced to replace the native FT start codon; †, a stop codon was placed immediately upstream of the truncated FT RNA. Thus, all truncated FT RNAs produced from the eight RMA vectors are nontranslatable.
The predicted size of each RT-PCR product includes TCV and cloning sequences (273 bp) plus the corresponding truncated FT sequence.
Transient agroinfiltration assay of RNA mobility.
The GFP-FT and GFP-mFT fusion genes were isolated from PVX/GFP-FT and PVX/GFP-mFT and inserted into between the Cauliflower mosaic virus 35S promoter and terminator-poly(A) signal sequence of a binary vector in Agrobacterium tumefaciens LBA4404 (34) to produce 35S-GFP-FT-poly(A) and 35S-GFP-mFT-poly(A). In two experiments, leaves of 24-day-old N. benthamiana plants were syringe infiltrated with Agrobacterium cultures carrying 35S-GFP-poly(A) (34), 35S-GFP-FT-poly(A) and 35S-GFP-mFT-poly(A). At 4 days postinfiltration, agroinfiltrated leaf tissues and noninfiltrated newly developed young leaves were collected for protein and RNA analysis.
Induction of flowering by viral transient expression of the Arabidopsis FT protein.
In three experiments, young short-day (SD) N. tabacum Maryland Mammoth plants were mock inoculated or infected with PVX/FT, PVX/mFT, PVX/GFP, or PVX/GFP-FT and maintained in an insect-free containment glasshouse at 25°C with a long-day (LD; 16-h) photoperiod. Local and systemic symptoms, including chlorotic lesions on inoculated leaves and mild chlorosis on young leaves, developed after 7 to 14 dpi. All PVX/FT-infected plants started to shoot at ∼20 dpi and flowered at ∼35 dpi; this was photographically recorded with a Nikon digital CoolPix 995 camera. Total RNAs (50 ng) extracted from systemically infected young leaves collected at 42 dpi were treated with RNase-free DNase (Promega) and used for RT-PCR detection (30 cycles) (34) with the primers PP82 and PP356 for virus-carried FT RNA. The resultant specific RT-PCR products were purified and verified by direct sequencing. The viral transient FT protein was analyzed by Western blotting detection.
Western blotting.
To investigate GFP, FT, GFP-FT fusion protein, and PVX CP expression, total proteins were extracted from leaf tissues as described previously (15). Western blot analyses of protein aliquots (10 μg) were performed with polyclonal antibodies specifically raised against GFP, the Arabidopsis FT and PVX CP, and detected using a goat anti-rabbit immunoglobulin G conjugated with alkaline phosphatase (Sigma) and BCIP/NBT substrates (Roche) as described previously (39).
RESULTS AND DISCUSSION
Arabidopsis FT RNA facilitates the movement of heterologous GFP and viral RNAs in N. benthamiana.
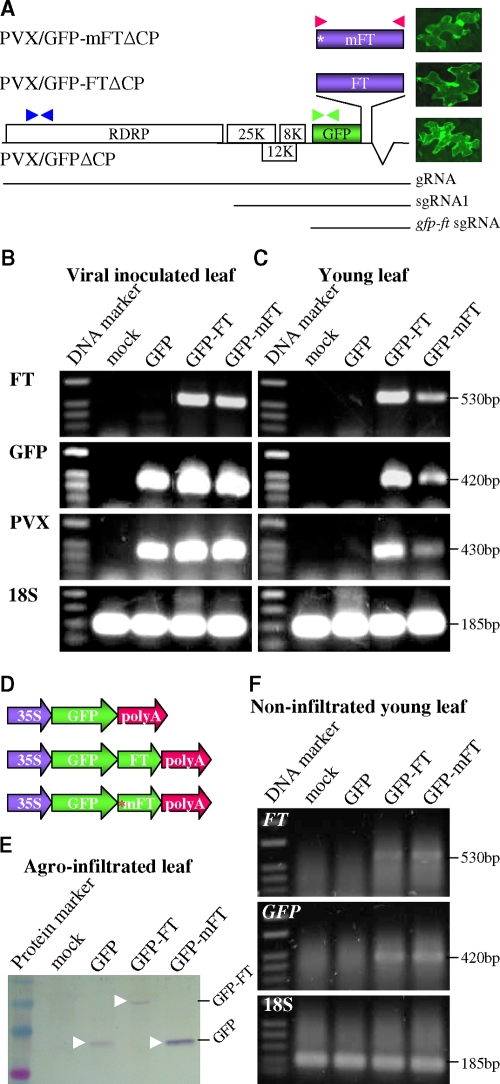
To investigate plant RNA trafficking, we developed a GFP-tagged movement-defective Potato virus X (PVX/GFPΔCP)-based RMA (Fig. 1A). Deletion of viral CP prevented PVX/GFPΔCP movement and restricted viral RNAs to single N. benthamiana epidermal cells. Thus, unlike the wild-type virus, PVX/GFPΔCP was incapable of accessing the phloem to move systemically (10). It should be noted that free GFP, once loaded from the mesophyll into the sieve tube, can move over long distances in the phloem (16, 17, 31, 32). PVX/GFP-FTΔCP and PVX/GFP-mFTΔCP had, respectively, the translatable or nontranslatable Arabidopsis FT that was fused in-frame to the GFP coding sequence (Fig. 1A). Individual epidermal cells expressing transient GFP or GFP-FT fusion protein from PVX/GFPΔCP, PVX/GFP-mFTΔCP, or PVX/GFP-FTΔCP showed green fluorescence (Fig. 1A). However, cell-to-cell movement of PVX/GFP-FTΔCP or PVX/GFP-mFTΔCP or intercellular spread of GFP or GFP-FT among epidermal cells was not detected by epifluorescence microscopy (Fig. 1A). We also examined the upper leaves for systemic movement of GFP or GFP-FT protein and these recombinant viruses but failed to observe any GFP in any type of cells.
FIG. 1.
(A) PVX-based RMA vectors. Viral 7.0-kb genomic gRNA, 2.6-kb subgenomic sgRNA1, and 1.4-kb GFP-FT sgRNA are indicated. The CP gene was deleted. The positions of a stop codon (*) replacing FT start codon in PVX/GFP-mFTΔCP, and three sets of RT-PCR primers for the detection of FT (▸◂ [red], PP354/PP356), GFP (▸◂ [green], PP371/PP372), or PVX (▸◂ [blue], PP269/PP373) RNAs are indicated. Individual epidermal cells expressing free GFP or GFP-FT fusion protein showed green fluorescence. (B and C) RT-PCR analysis of FT, GFP, and PVX RNA and 18S rRNA in inoculated and newly growing young leaves, including shoot apices of N. benthamiana. RNAs were extracted from leaves of plants mock inoculated (mock) or inoculated with PVX/GFPΔCP (GFP), PVX/GFP-FTΔCP (GFP-FT), or PVX/GFP-mFTΔCP (GFP-mFT). The 1-kb DNA ladder and sizes of RT-PCR products are indicated. (D) Construction of 35S promoter-controlled FT gene expression cassettes for a transient agroinfiltration assay of RNA mobility. The position of an introduced stop codon (*) replacing the FT gene start codon in 35S-GFP-mFT-poly(A) is indicated. (E) Expression of free GFP or GFP-FT fusion protein. Total proteins were extracted from N. benthamiana leaves infiltrated with agrobacteria carrying the 35S-controlled gene expression cassette and were analyzed by Western blotting with a GFP-specific antibody. The positions of free GFP and GFP-FT fusion protein are indicated. (F) FT and GFP mRNAs in nonagroinfiltrated newly developed systemic young leaves were analyzed by RT-PCR. Samples are from plants with mock infiltration (mock) or infiltrated with agrobacteria carrying 35S-GFP-poly(A) (GFP), 35S-GFP-FT-poly(A) (GFP-FT), or 35S-GFP-mFT-poly(A) (GFP-mFT). The sizes of RT-PCR products, free and GFP-FT fusion proteins, and the prestained protein markers or 1-kb DNA ladder are indicated.
We then used a more sensitive RT-PCR assay to test for RNA movement. Accumulation of recombinant viral RNAs from each vector was readily detected in inoculated leaves by RT-PCR using FT-, GFP-, and PVX-specific primers (Fig. 1A and B). However, no spread of genomic RNA (gRNA) and subgenomic RNA (sgRNA) of PVX/GFPΔCP to newly formed young leaves occurred (Fig. 1C). In striking contrast, inclusion of FT RNA in PVX/GFP-FTΔCP enabled GFP-FT sgRNA and PVX/GFP-FTΔCP gRNA to move systemically from inoculated leaves to young leaves (Fig. 1C). Furthermore, PVX/GFP-mFTΔCP gRNA and sgRNAs that contained nonsense mutations precluding FT protein synthesis also spread systemically (Fig. 1C), demonstrating that nontranslatable mFT RNA was able to move and promote long-distance trafficking of heterologous GFP and PVX RNAs. FT and mFT possessed similar abilities to facilitate the spread of heterologous RNAs, although the levels of mobile RNAs detected in the systemic leaves varied between plants for the FT and mFT constructs. The absence of GFP signal in neighboring epidermal cells and upper leaves after FT RNA-mediated movement from lower parts into younger tissues suggests that CP may be required for vascular exit, which could be a novel role for CP in viral movement, although we cannot exclude the possibility that it is due to the limited sensitivity of fluorescence detection of free GFP and GFP-FT fusion proteins.
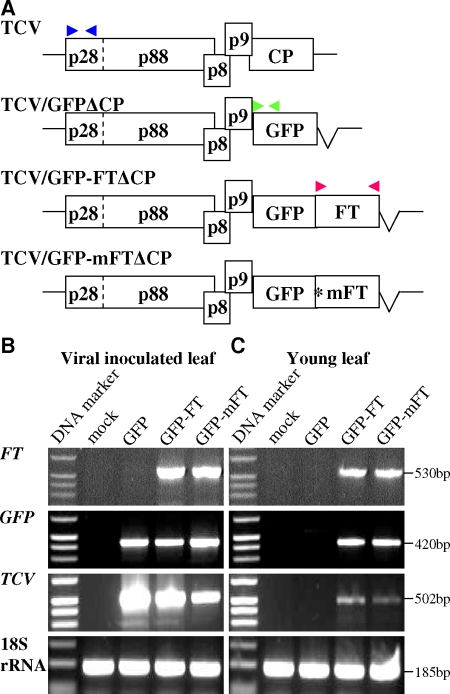
FT and mFT RNA movement and FT-mediated heterologous RNA trafficking were also demonstrated in a distinct Turnip crinkle virus (TCV)-based RMA (Fig. 2). In a similar experimental design, we used a GFP-tagged movement-deficient TCVΔCP-based RMA vector (Fig. 2A) and also showed that the Arabidopsis FT RNA promoted systemic movement of TCV RNA. RT-PCR detection of FT, GFP, and TCV RNA and 18S rRNA (18S) in inoculated leaves (Fig. 2B) and particularly in the newly formed young leaves and shoot apices (Fig. 2C) of N. benthamiana plants infected with TCV/GFPΔCP, TCV/GFP-FTΔCP, or TCV/GFP-mFTΔCP mirrored the results obtained with the PVX-based RMA.
FIG. 2.
The Arabidopsis FT RNA supports TCV RNA movement. (A) GFP-tagged movement-deficient TCV-based RMA vectors. The position of an introduced stop codon (*) replacing the FT gene start codon in TCV/GFP-mFTΔCP and three sets of RT-PCR primers for detections of FT (▸◂ [red], PP354/PP356), GFP (▸◂ [green], PP371/PP372) or TCV (▸◂ [blue], PP433/PP434) are indicated. (B and C) RT-PCR analysis of FT, GFP and TCV RNA and 18S rRNA (18S) in inoculated (B) and newly formed young leaves and shoot apices (C) of N. benthamiana plants. Total RNAs used as RT-PCR templates were extracted from inoculated or young leaves of plants with mock inoculation (mock) or inoculated with RNA transcripts from TCV/GFPΔCP (GFP), TCV/GFP-FTΔCP (GFP-FT), or TCV/GFP-mFTΔCP (GFP-mFT). The 1-kb DNA ladder and the sizes of RT-PCR products are indicated.
Mobility of Arabidopsis FT RNA is independent of viral RNA sequences.
The self-mobility of FT RNA and the associated heterologous RNAs was further confirmed by an agroinfiltration assay. N. benthamiana leaves were infiltrated with Agrobacterium tumefaciens carrying the 35S promoter-controlled GFP or GFP-tagged FT gene expression cassettes, respectively (Fig. 1D). Western blot analysis showed that expression of free GFP occurred in leaves infiltrated with agrobacterium carrying 35S-GFP-poly(A) and 35S-GFP-mFT-poly(A), while the larger GFP-FT fusion protein was expressed from 35S-GFP-FT-poly(A) (Fig. 1E). These data clearly demonstrated that the replacement of the initiation codon with a stop codon in the FT open reading frame blocked the translation of FT protein from the 35S-GFP-mFT construct. We also examined the Arabidopsis FT sequence and found no internal in-frame start codons; thus, it is unlikely that mFT RNA would be translated from internal initiation. Consistent with the results obtained from the viral RMAs, following agroinfiltration, both FT (mFT) and FT (mFT)-GFP RNAs spread systemically to young leaves that had developed on the plant after the agroinfiltration (Fig. 1F). It should be pointed out that systemic movement of FT (mFT) and FT (mFT)-GFP RNA seems to be stronger in virus-based RMAs than in the agroinflitration assays. This may be due to differences in the site (nucleus versus cytoplasm) and the means of production of the mobile RNA molecules (35S promoter-controlled transcription versus recombinant viral RNA replication and transcription) in agroinfiltrated cells compared to viral inoculated cells. Nevertheless, our data indicate that no viral sequences and proteins are necessary for systemic spread of FT RNA.
Viral transient expression of Arabidopsis FT protein promotes floral induction.
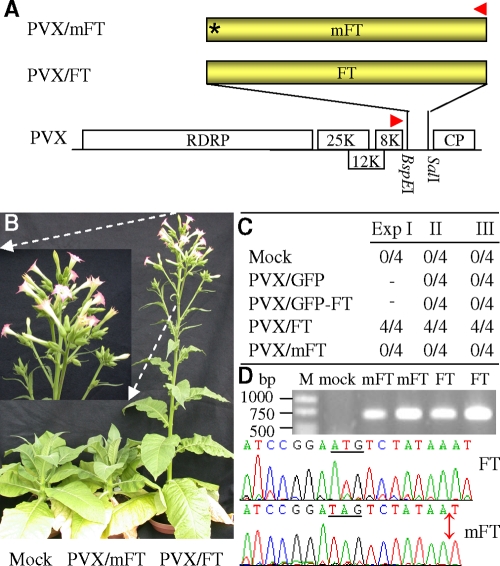
Arabidopsis FT is required to induce flowering. However, it did not stimulate early flowering in day neutral N. benthamiana expressing GFP-FT, probably due to interference by the GFP fusion. To test this, we infected SD N. tabacum Maryland Mammoth tobacco (21) with PVX/FT (Fig. 3A) and PVX/GFP-FT (see Fig. 6A). Viral ectopic expression of the GFP-FT fusion protein was unable to induce flowering under noninducing LD conditions (Fig. 3B and C); however, all Maryland Mammoth plants infected with PVX/FT that had the capacity to produce free FT protein flowered in LD, while control plants infected with either PVX/mFT carrying a mutated nontranslatable FT mRNA or PVX/GFP remained vegetative (Fig. 3B and C). Viral delivery of wild-type and mutated FT RNA was readily detected in systemically infected leaves by RT-PCR and further confirmed by direct sequencing of the specific RT-PCR products (Fig. 3D). Free FT protein expressed from PVX/FT but not from PVX/mFT was also detected by Western blotting with an antiserum specifically raised against FT (Fig. 4D). The fact that PVX/FT could induce flowering but PVX/mFT could not shows that the Arabidopsis FT RNA alone is not sufficient to initiate flowering but that its protein product expressed from PVX/FT is necessary for floral induction.
FIG. 3.
Ectopic expression of FT induces flowering. (A) The translatable and mutated (*) nontranslatable Arabidopsis FT coding sequences were cloned into wild-type PVX vector to produce PVX/FT and PVX/mFT, respectively. (B and C) Floral induction caused by viral expression of FT protein. Young SD N. tabacum Maryland Mammoth plants were mock inoculated or infected with PVX/FT or PVX/mFT and grown under a noninducing LD photoperiod. Twelve plants infected by PVX/FT in three separate experiments started bolting at ∼20 dpi, flowered at ∼35 dpi, and were photographed at 42 dpi (B and inset image). Tobacco mock inoculated or infected with PVX/mFT, PVX/GFP, or PVX/GFP-FT did not flower (B and C). (D) Detection of viral transient FT RNA. Viral transient FT RNA was detected by RT-PCR using primers PP82 (▸) and PP356 (◂) in systemic young leaves from two separate plants infected with PVX/mFT (mFT) or PVX/FT (FT) but not in a mock-infected plant (mock). The position and the sizes of 1-kb DNA ladder (lane M) are indicated. Direct sequencing of RT-PCR products (648 bp) verified the presence of virally expressed wild-type and mutant FT RNA in flowering and nonflowering plants, respectively. The native FT ATG (underlined) in PVX/FT and its TAG replacement (underlined) together with a nucleotide deletion (double-arrow) in PVX/mFT are indicated.
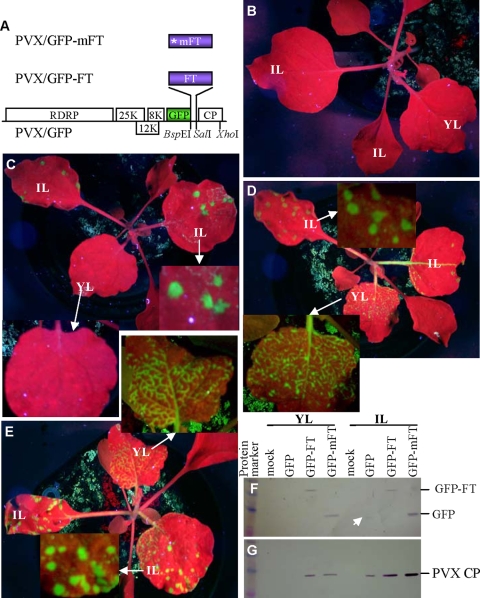
FIG. 6.
Arabidopsis FT RNA enhances the systemic spread of PVX. PVX-based FT expression cassettes are schematically represented (A). The translatable and nontranslatable Arabidopsis FT coding sequences were fused in-frame with the GFP coding sequence of PVX/GFP by using the BspEI and SalI sites. The unique XhoI site used to produce CP-defective RMA vectors (Fig. 1A) is indicated. Twenty-four-day-old N. benthamiana plants were mock inoculated (B) or infected with PVX/GFP (C), PVX/GFP-FT (D), or PVX/GFP-mFT (E). (C to E) Local infection of inoculated leaves (IL) induced GFP-expressing lesions. However, only plants challenged with PVX/GFP-FT or PVX/GFP-mFT quickly established systemic infection at 7 dpi, showing GFP green fluorescence in newly developed young leaves (YL) (D and E). Moreover, protein were extracted from inoculated and systemic young leaves of plants with mock (mock) or viral infection, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and detected by Western blotting with antiserum raised against GFP (F) and PVX CP (G). The positions of free GFP, GFP-FT fusion protein, and the PVX CP are indicated. The production of viral CP, free GFP, and GFP-FT fusion proteins associated with PVX/GFP (GFP), PVX/GFP-FT (GFP-FT) or PVX/GFP-mFT (GFP-mFT) was consistent with symptom development. These data indicate that the Arabidopsis FT RNA with or without its translatable capacity enhances systemic spread of PVX.
FIG. 4.
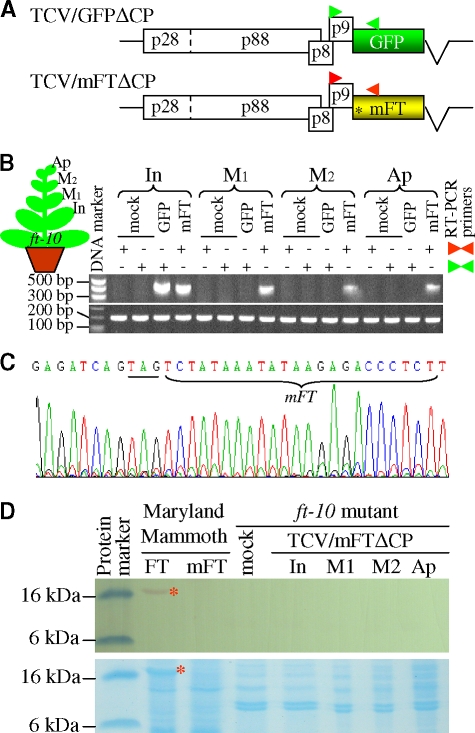
(A) Movement-deficient TCV-based RMA vectors. The parental TCV vector and long-distance movement-deficient TCV/GFPΔCP were previously constructed (34). The position of the introduced stop codon (*) replacing ATG in the FT gene in TCV/mFTΔCP and two sets of RT-PCR primers are indicated. (B and C) RT-PCR analysis of FT-TCV and GFP-TCV RNA (top panel) using primers PP267/PP473a (▸◂ [green]) or PP267/PP228 (▸◂ [red]) and 18S rRNA (bottom panel) in inoculated (In) and newly formed young leaves (M1 and M2) and shoot apices (Ap) of ft-10 A. thaliana mutant plants. RNAs were extracted from tissues of plants mock-inoculated (mock) or inoculated with TCV/GFPΔCP (GFP) or TCV/mFTΔCP (mFT). The positions and sizes of the 1-kb DNA ladder are indicated. (C) Direct sequencing of mFT RT-PCR products (372 bp) generated from total RNA extracted from young leaves of Arabidopsis mutant plants inoculated with TCV/mFTΔCP. (D) Western blot analysis indicated no endogenous or viral transient FT protein was expressed in the mock-inoculated leaves (mock) or in different leaves (In, M1, and M2) and shoot apices (AP) of ft-10 plants inoculated with TCV/mFTΔCP. Viral ectopic expression of FT protein (red asterisk) was detected in N. tabacum Maryland Mammoth plants infected with PVX/FT (FT) but not with PVX/mFT (mFT), a finding consistent with that the FT protein is required for floral induction (Fig. 3B). Coomassie blue-stained gel indicates the equal loading of protein samples. The positions and sizes of the protein markers are indicated.
Systemic movement of FT RNA does not require FT protein.
The virus-based RMAs, together with the agroinfiltration assay, demonstrate that Arabidopsis FT RNA not only is self-mobile but also can mediate systemic trafficking of heterologous RNAs in N. benthamiana. To eliminate the possibility that endogenous plant-derived FT protein might be involved in facilitation of the FT RNA movement, we used the TCV-based RMA vector TCV/mFTΔCP containing a nontranslatable FT gene (Fig. 4A). Deletion of the CP gene in TCVΔCP prevents its spreading from infected cells to distal parts of plants (6). However, in contrast to the PVX-based RMA, TCV is able to infect A. thaliana (36) and TCVΔCP can establish local infection in this host (unpublished data). We inoculated the Arabidopsis ft-10 T-DNA insertion mutant (43) with RNA transcripts of TCV/mFTΔCP or TCV/GFPΔCP. In inoculated leaves, accumulation of recombinant viral RNAs from both RMA vectors was readily detected by RT-PCR, while no specific recombinant viral RNA was detectable in mock-inoculated control plants (Fig. 4B). Not surprisingly, no systemic spread of TCV/GFPΔCP RNA to noninoculated leaves occurred. However, the nontranslatable mutant mFT RNA in TCV/mFTΔCP moved and facilitated long-distance movement of viral RNAs to young leaves and the shoot apices (Fig. 4B). The nonsense mutation prohibiting FT protein synthesis from TCV/mFTΔCP was maintained in the Arabidopsis mutant (Fig. 4C). Moreover, no viral transiently expressed or endogenous FT protein was detected in different leaf tissues of mock-inoculated and virus-treated ft-10 mutant plants, a finding consistent with the mutation in ft-10 (Fig. 4D). Long-distance trafficking of virus-derived mFT RNA from inoculated leaves to noninoculated leaves and the shoot apices was also detected in ft-1 mutant plants (X. Liu, C. Li, S. Jackson, and Y. Hong, unpublished data). On the other hand, when fusing GFP downstream of FT, the nonsense mutation also eliminates GFP expression (K. Zhang and Y. Hong, unpublished data). Thus, we conclude that the systemic FT RNA trafficking does not require the FT protein.
Mapping of the cis-acting element within Arabidopsis FT RNA.
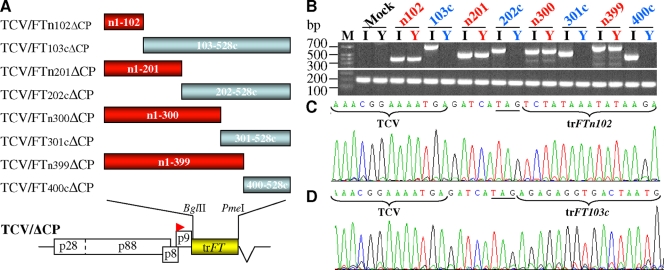
To elucidate what controls long-distance spread of FT RNA, we constructed a series of TCV/ΔCP-based RMA vectors carrying truncated nontranslatable FT fragments (Fig. 5A). We found that viral-derived FT RNAs from TCV/FTn102ΔCP, TCV/FTn201ΔCP TCV/FTn300ΔCP and TCV/FTn399ΔCP were capable of movement and facilitating systemic movement of TCV RNA. This was evident by positive RT-PCR detections of TCV and virus-derived FT RNAs in the noninoculated, newly developing and growing young leaves (Fig. 5B). All of the truncated FT RNAs possess nucleotides 1 to 102 of FT RNA (Fig. 5A). In striking contrast, FT RNAs generated from TCV/FT103cΔCP, TCV/FT202cΔCP, TCV/FT301cΔCP, and TCV/FT400cΔCP that lack the 5′-terminal 102 nucleotides were completely immobile. Consequently, RT-PCRs failed to detect any virus-derived FT-related RNA in the young leaves of virus-treated plants although recombinant viral RNAs accumulated in the inoculated leaves (Fig. 5B). Direct sequencing of RT-PCR products confirmed the identities of the mobile and immobile TCV and virus-derived FT RNA molecules (Fig. 5C and D). Thus, the element required for systemic FT RNA movement was unequivocally mapped to the 102-nucleotide sequence at the 5′ terminus of FT mRNA.
FIG. 5.
Functional mapping of the cis-acting element that controls the FT RNA movement. (A) TCV/trFTΔCP-based RMA vectors carrying truncated (tr) nontranslatable Arabidopsis FTs. (B) Detection of virus-derived FT RNA (top panel) by RT-PCR in systemic young leaves (Y) from plants inoculated with TCV/FTn102ΔCP (n102), TCV/FTn201ΔCP (n201), TCV/FTn300ΔCP (n300), or TCV/FTn399ΔCP (n399); but not from plants inoculated with TCV/FT103cΔCP (103c), TCV/FT202cΔCP (202c), TCV/FT301cΔCP (301c) or TCV/FT400cΔCP (400c). Recombinant viral RNA of each TCV/trFTΔCP was readily detectible in inoculated leaves (I). No specific virus-derived FT RNA was detected in mock-inoculated plants. RT-PCR analysis of 18S rRNA (bottom panel) is included as an RNA control. (C and D) Direct sequencing of RT-PCR products verified the presence of virus-derived truncated FT RNA. Example sequence panels are shown for truncated FT RNA expressed in the young leaves of plants inoculated by TCV/FTn102ΔCP (C) or in the TCV/FT103cΔCP-inoculated leaves (D). The TAG stop codon is underlined and TCV and virus-derived FT sequences are indicated.
It should be noted that TCV without CP moves readily in dcl2/dcl4 mutated Arabidopsis that is defective in RNA silencing. Thus, the TCV CP is mainly providing a silencing-suppression function which allows virus movement, although it may also be offering some movement support (28). This is consistent with the idea that the CP silencing-suppressor possesses a differential role in viral intercellular spread (35). However, the Arabidopsis FT RNA and indeed FT protein have no effect on RNA silencing (K. Zhang, C. Li, and Y. Hong, unpublished data). Therefore, FT RNA is only providing movement function and the FT RNA-mediated systemic spread of TCV seems to operate with a distinct mechanism.
The Arabidopsis FT RNA enhances systemic spread of PVX.
The translational and nontranslational Arabidopsis FT coding sequences were fused in-frame with the GFP coding sequence of PVX/GFP to produce PVX/GFP-FT and PVX/GFP-mFT (Fig. 6A), respectively. In repeated experiments, 24-day-old Nicotiana benthamiana plants were mock inoculated (Fig. 6B) or infected with RNA transcripts produced by in vitro transcription from PVX/GFP (Fig. 6C), PVX/GFP-FT (Fig. 6D), or PVX/GFP-mFT (Fig. 6E). Local infection of the inoculated leaves induced GFP-expressing green lesions (ca. 10 lesions per inoculated leaf) 3 to 5 dpi. However, only plants challenged with PVX/GFP-FT or PVX/GFP-mFT quickly established systemic infection at 7 dpi, showing GFP green fluorescence in newly developing young leaves. Development of systemic symptoms in plants infected by PVX/GFP took 2 to 3 extra days. Furthermore, the accumulations of free GFP and GFP-FT fusion proteins (Fig. 6F) and viral CP (Fig. 6G) in the inoculated and systemic young leaves after mock inoculation (mock) or infection of PVX/GFP (GFP), PVX/GFP-FT (GFP-FT), or PVX/GFP-mFT (GFP-mFT) were consistent with the development of viral symptoms. Western blot assays of total proteins extracted from inoculated and systemic young leaves using GFP- and PVX CP-antiserum indicated that only a trace amount of free GFP and PVX CP was detected in the inoculated leaves but not in the systemic young leaves of PVX/GFP-infected plants. However, viral CP as well as the GFP-FT fusion protein or free GFP expressed from PVX/GFP-FT or PVX/GFP-mFT, respectively, could be readily detected in both inoculated and systemic leaves.
We provide here compelling evidence that Arabidopsis FT RNA is capable of systemic movement and that FT RNA can also act as a cis transportation carrier for heterologous RNAs. The FT RNA mobile function is independent of the FT protein. Consistent with this idea, an engineered plant RNA virus carrying Arabidopsis FT RNA spread more quickly than its parental virus to establish infection of young tissues. It should be noted that the CP genes of PVX and TCV participate in virus-plant interactions and contribute to symptom development (5, 10, 20, 45); thus, it was not surprising that the CP-deleted recombinant viruses that were transported through the plant via FT RNA in cis could still not start a proper infection. Moreover, we demonstrate that the systemic mobility of FT RNA is determined by a cis-acting element of 102 nucleotides at the 5′ terminus of FT RNA. The positive identification of cis-acting element for FT RNA trafficking provides a unique opportunity to dissect the molecular mechanism that governs cellular RNA signaling in plants. It is possible that the cis-RNA sequence may bind host proteins to form an RNA-protein complex for FT RNA spread. Indeed, some plant and viral proteins can bind RNAs and facilitate their intercellular and long-distance trafficking (26, 27, 41, 42). The discovery that a host RNA molecule can mediate systemic trafficking of heterologous RNAs is also significant. In particular, facilitation of viral RNA movement by the Arabidopsis FT RNA raises the possibility that viruses and plants might have evolved similar mechanisms for systemic RNA trafficking.
There is now collective evidence that the Arabidopsis FT protein and its tomato SFT and rice Hd3a orthologs may act as a non-cell-autonomous flower-inducing signal that can move from the end of the vasculature into the meristematic tissue in the shoot apical meristem (7, 18, 22, 23, 29, 30, 37). Elegant experiments wherein the intercellular movement of FT protein is prevented by either a large C-terminal fusion and/or a nuclear localization signal have demonstrated that FT protein needs to move, and is sufficient on its own, to induce flowering (18, 29). FT- and Hd3a-GFP fusion proteins were found to cross a graft union from a donor scion and reach the apex of the recipient plant (7, 37), and FT-derived peptides have been identified in phloem exudates from Cucurbita (23), suggesting that the protein is transported through the phloem. Once the FT protein enters the apical meristem it interacts with the bZIP transcription factor FD to activate floral identity genes such as APETELA 1 (1, 40). However, these studies do not definitely rule out a role for FT mRNA as part of the mobile florigenic signal. In this context, it is worthwhile noting that mRNA of the rice FT ortholog Hd3a has been detected at extremely low levels in shoot apices of rice even though the Hd3a promoter is not active in the shoot apical meristem (37). The evidence presented here that the Arabidopsis FT RNA is able to move systemically is in contrast to these recent reports where FT RNA movement could not be detected (7, 18, 22, 23, 29, 30), possibly due to the different host plants and experimental systems used. For example, high levels of viral FT expression, even from immobilized viruses in single cells, could increase the levels of systemic FT RNA over the detection limit. Our data raise the possibility that systemic movement of the FT RNA may also contribute to the long-distance florigen signaling, although the FT protein is still required to trigger flowering, as shown by the nonflowering of PVX/mFT-inoculated plants.
Acknowledgments
We thank B. Thomas, T. M. A. Wilson, and S. Bright for critical reading of the manuscript and D. Baulcombe for providing the original PVX vector.
C.L. is supported by the Warwick University Postgraduate Studentship. This work was supported in part by the Warwick HRI-UK Biotechnology and Biological Sciences Research Council core funding, a Warwick Venture grant, and a Warwick Research Development Fund grant to Y.H.
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Abe, M., Y. Kobayashi, S. Yamamoto, Y. Daimon, A. Yamaguchi, Y. Ikeda, H. Ichinoki, M. Notaguchi, K. Goto, and T. Araki. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 3091052-1056. [DOI] [PubMed] [Google Scholar]
- 2.An, H., C. Roussot, P. Suárez-López, L. Corbesier, C. Vincent, M. Piñeiro, S. Hepworth, A. Mouradov, S. Justin, C. Turnbull, and G. Coupland. 2004. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 1313615-3626. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, A. K., M. Chatterjee, Y. Yu, S.-G. Suh, W. A. Miller, and D. J. Hannapel. 2006. Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 183443-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buhtz, A., F. Springer, L. Cappell, D. C. Baulcombe, and J. Kehr. 2008. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53739-749. [DOI] [PubMed] [Google Scholar]
- 5.Chapman, S., G. Hills, J. Watts, and D. Baulcombe. 1992. Mutational analysis of the coat protein gene of potato virus X: effects on virion morphology and viral pathogenicity. Virology 191223-230. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, Y., A. Gisel, and P. C. Zambryski. 2000. Cell-to-cell and systemic movement of recombinant green fluorescent protein-tagged Turnip crinkle viruses. Virology 273258-266. [DOI] [PubMed] [Google Scholar]
- 7.Corbesier, L., C. Vincent, S. Jang, F. Fornara, Q. Fan, I. Searle, A. Giakountis, S. Farrona, L. Gissot, C. Turnbull, and G. Coupland. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 3161030-1033. [DOI] [PubMed] [Google Scholar]
- 8.Deeken, R., P. Ache, I. Kajahn, J. Klinkenberg, G. Bringmann, and R. Hedrich. 2008. Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 55746-759. [DOI] [PubMed] [Google Scholar]
- 9.Dunoyer, P., C. Himber, and O. Voinnet. 2005. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nature Genetics 371356-1360. [DOI] [PubMed] [Google Scholar]
- 10.Fedorkin, O., A. Solovyev, N. Yelina, A. Zamyatnin, Jr., R. Zinovkin, K. Mäkinen, J. Schiemann, and S. Y. Morozov. 2001. Cell-to-cell movement of potato virus X involves distinct functions of the coat protein. J. Gen. Virol. 82449-458. [DOI] [PubMed] [Google Scholar]
- 11.Gopinath, K., and C. C. Kao. 2007. Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell 191179-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton, A. J., O. Voinnet, L. Chappell, and D. C. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 214671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haywood, V., T.-S. Yu, N.-C. Huang, and W. J. Lucas. 2005. Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 4249-68. [DOI] [PubMed] [Google Scholar]
- 14.Himber, C., P. Dunoyer, G. Moissiard, C. Ritzenthaler, and O. Voinnet. 2003. Transitivity-dependent and independent cell-to-cell movement of RNA silencing. EMBO J. 224523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, Y., K. Saunders, M. R. Hartley, and J. Stanley. 1996. Resistance to geminivirus infection by virus-induced expression of a plant cytotoxin. Virology 220119-127. [DOI] [PubMed] [Google Scholar]
- 16.Imlau, A., E. Truernit, and N. Sauer. 1999. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11309-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itaya, A., G. Liang, Y.-M. Woo, R. S. Nelson, and B. Ding. 2000. Nonspecific intercellular protein trafficking probed by green-fluorescent protein in plants. Protoplasma 213165-175. [Google Scholar]
- 18.Jaeger, K. E., and P. A. Wigge. 2007. FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 171050-1054. [DOI] [PubMed] [Google Scholar]
- 19.Kim, M., W. Canio, S. Kessler, and N. Sinha. 2001. Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293287-289. [DOI] [PubMed] [Google Scholar]
- 20.Kong, Q., J. W. Oh, and A. E. Simon. 1995. Symptom attenuation by a normally virulent satellite RNA of turnip crinkle virus is associated with the coat protein open reading frame. Plant Cell 71625-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang, A., M. K. Chailakhyan, and I. A. Frolova. 1977. Promotion and inhibition of flower formation in a day neutral plant in grafts with a short-day plant and a long-day plant. Proc. Natl. Acad. Sci. USA 742412-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lifschitz, E., T. Eviatar, A. Rozman, A. Shalit, A. Goldshmidt, Z. Amsellem, J. P. Alvarez, and Y. Eshed. 2006. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 1036398-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, M. K., H. Belanger, Y. J. Lee, E. Varkonyi-Gasic, K. Taoka, E. Miura, B. Xoconostle-Cázares, K. Gendler, R. A. Jorgensen, B. Phinney, T. J. Lough, and W. J. Lucas. 2007. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 191488-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lough, T. J., R. H. Lee, S. J. Emerson, R. L. Forster, and W. J. Lucas. 2006. Functional analysis of the 5′ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology 351455-465. [DOI] [PubMed] [Google Scholar]
- 25.Lough, T. J., and W. J. Lucas. 2006. Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57203-232. [DOI] [PubMed] [Google Scholar]
- 26.Lucas, W. J. 2006. Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344169-184. [DOI] [PubMed] [Google Scholar]
- 27.Lucas, W. J., S. Bouché-Pillon, D. P. Jackson, L. Nguyen, L. Baker, B. Ding, and S. Hake. 1995. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 2701980-1983. [DOI] [PubMed] [Google Scholar]
- 28.Manfre, A. J., and A. E. Simon. 2008. Importance of coat protein and RNA silencing in satellite RNA/virus interactions. Virology 379161-167. [DOI] [PubMed] [Google Scholar]
- 29.Mathieu, J., N. Warthmann, F. Küttner, and M. Schmid. 2007. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 171055-1060. [DOI] [PubMed] [Google Scholar]
- 30.Notaguchi, M., M. Abe, T. Kimura, Y. Daimon, T. Kobayashi, A. Yamaguchi, Y. Tomita, K. Dohi, M. Mori, and T. Araki. 2008. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 491645-1658. [DOI] [PubMed] [Google Scholar]
- 31.Oparka, K. J., A. G. Roberts, P. Boevink, S. Santa Cruz, I. Roberts, K. S. Pradel, A. Imlau, G. Kotlizky, N. Sauer, and B. Epel. 1999. Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97743-754. [DOI] [PubMed] [Google Scholar]
- 32.Peleg, G., D. Malter, and S. Wolf. 2007. Viral infection enables phloem loading of GFP and long-distance trafficking of the protein. Plant J. 51165-172. [DOI] [PubMed] [Google Scholar]
- 33.Qi, Y., T. Pélissier, A. Itaya, E. Hunt, M. Wassenegger, and B. Ding. 2004. Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 161741-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryabov, E. V., R. Van Wezel, J. Walsh, and Y. Hong. 2004. Cell-to-cell, but not long-distance, spread of RNA silencing that is induced in individual epidermal cells. J. Virol. 783149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, Y., E. V. Ryabov, R. Van Wezel, C. Li, M. Jin, W. Wang, Z. Fan, and Y. Hong. 2009. Suppression of local RNA silencing is not sufficient to promote cell-to-cell movement of Turnip crinkle virus in Nicotiana benthamiana. Plant Signal. Behav. 415-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon, A. E. 1994. Interaction between Arabidopsis thaliana and virus, p. 685-704. In E. Meyerowitz and C. Sommerville (ed.), Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Tamaki, S., S. Matsuo, H. L. Wong, S. Yokoi, and K. Shimamoto. 2007. Hd3a protein is a mobile flowering signal in Rice. Science 3161033-1036. [DOI] [PubMed] [Google Scholar]
- 38.Van Wezel, R., X. Dong, H. Liu, P. Tien, J. Stanley, and Y. Hong. 2002. Mutations of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene silencing suppression. Mol. Plant-Microbe interact. 15203-208. [DOI] [PubMed] [Google Scholar]
- 39.Van Wezel, R., and Y. Hong. 2004. Virus survival of RNA silencing without deploying protein-mediated suppression in Nicotiana benthamiana. FEBS Lett. 56265-70. [DOI] [PubMed] [Google Scholar]
- 40.Wigge, P. A., M. C. Kim, K. E. Jaeger, W. Busch, M. Schmid, J. U. Lohmann, and D. Weigel. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 3091056-1059. [DOI] [PubMed] [Google Scholar]
- 41.Xoconostle-Cázares, B., Y. Xiang, R. Ruiz-Medrano, H. L. Wang, J. Monzer, B. C. Yoo, K. C. McFarland, V. R. Franceschi, and W. J. Lucas. 1999. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 28394-98. [DOI] [PubMed] [Google Scholar]
- 42.Yoo, B. C., F. Kragler, E. Varkonyi-Gasic, V. Haywood, S. Archer-Evans, Y. M. Lee, T. J. Lough, and W. J. Lucas. 2004. A systemic small RNA signaling system in plants. Plant Cell 161979-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo, S. K., K. S. Chung, J. Kim, J. H. Lee, S. M. Hong, S. J. Yoo, S. Y. Yoo, J. S. Lee, and J. H. Ahn. 2005. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139770-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, X., A. J. Archual, A. A. Amin, and B. Ding. 2008. A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 2035-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, F., and A. E. Simon. 2003. Enhanced viral pathogenesis associated with a virulent mutant virus or a virulent satellite RNA correlates with reduced virion accumulation and abundance of free coat protein. Virology 3128-13. [DOI] [PubMed] [Google Scholar]
- 46.Zhong, X., X. Tao, J. Stombaugh, N. Leontis, and B. Ding. 2007. Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J. 263836-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, Y., E. Ryabov, X. Zhang, and Y. Hong. 2008. Influence of viral genes on the cell-to-cell spread of RNA silencing. J. Exp. Bot. 592803-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]