Abstract
Hepatitis B e antigen (HBeAg) is a secreted version of hepatitis B virus (HBV) core protein that promotes immune tolerance and persistent infection. It is derived from a translation product of the precore/core gene by two proteolytic cleavage events: removal of the amino-terminal signal peptide and removal of the carboxyl-terminal arginine-rich sequence. Four RXXR motifs are present at the carboxyl terminus of the HBeAg precursor, with the first two fused as 151RRGRSPR157. Genotype A possesses two extra amino acids at the first motif (151RRDRGRSPR159), which weakens the first motif and separates it from the second one. Western blot analysis of patient sera revealed a single HBeAg form for genotypes B to D but two additional forms of larger sizes for genotype A. Site-directed mutagenesis and transfection experiments with human hepatoma cell lines indicated that HBeAg of genotype B is derived from cleavage at the first (151RRGR154) motif. The major HBeAg form of genotype A corresponds to cleavage at the second (156RSPR159) motif, and the other two forms are cleavage products of the first (151RRDR154) and third (166RRRR169) motifs, respectively. Only the cleavage product of the third motif of genotype A was observed in furin-deficient LoVo cells, and an inhibitor of furin-like proprotein convertases blocked cleavage of the first and second motifs in human hepatoma cells. In conclusion, our study reveals genotypic differences in HBeAg processing and implicates furin as the major enzyme involved in the cleavage of the first and second RXXR motifs.
Approximately 2 billion people worldwide have been exposed to hepatitis B virus (HBV), and 350 million of them remain chronically infected. The long-term consequences of HBV infection include liver cirrhosis and hepatocellular carcinoma (21). The risk of developing chronic HBV infection correlates inversely with the age of transmission. In areas where HBV infection is endemic, such as China where over 10% of people are chronic carriers of the virus, HBV is transmitted both horizontally during early childhood and vertically from hepatitis B e antigen (HBeAg)-positive mothers during birth (68). In countries of intermediate HBV incidence such as South Africa, infection is acquired primarily horizontally during early childhood (28). In Western countries, HBV is transmitted mostly by drug abuse or unprotected sex during adulthood, and the majority of carriers will recover from the acute infection. In this regard, HBeAg is implicated as a major viral factor contributing to chronic infection following neonatal transmission (41).
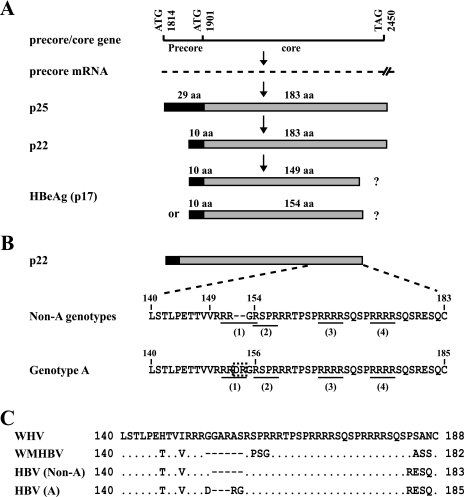
HBeAg was discovered by Magnius and Espmark over 30 years ago as a new serological marker for HBV infection (38), supplementing the then available assays for HBcAg (HBV core antigen or core protein) and HBsAg (HBV surface antigen or envelope protein). Soon after its discovery, the presence of HBeAg was found to correlate with liver diseases, whereas the loss of HBeAg followed by the rise of corresponding antibody (anti-HBe) was associated with normalization of liver function (22, 65). Later studies further revealed a correlation of HBeAg with high viremia titer and anti-HBe with reduced viral load. Since denaturation of HBcAg exposes HBe antigenicity (59), the two antigens were considered related. Subsequent transfection experiments established HBeAg as an alternative translation product of the core gene rather than a core protein derivative (39, 47, 49). The core gene of 183 codons (for most genotypes) is preceded by an in-frame precore AUG codon that extends the protein by 29 hydrophobic amino acid (aa) residues (Fig. 1A). Such a precore/core protein of 212 residues (p25) is translated from a 3.5-kb mRNA called precore RNA and converted to HBeAg by two proteolytic cleavage events in the secretory pathway. First, the N-terminal 19 residues encoded by the precore region serve as the signal peptide for translocation of the precore/core protein into the endoplasmic reticulum lumen, where the peptide is clipped away by a signal peptidase (Fig. 1A). The remaining polypeptide, p22, has 10 extra residues at its N terminus in comparison to that of the core protein (57). Next, about 30 residues are removed from the C terminus of p22 in a post-endoplasmic reticulum compartment (most likely the Golgi apparatus) to generate a mature HBeAg of 16 to 17 kb (60, 66).
FIG. 1.
Carboxyl-terminal sequences of e antigen precursors and sequential steps involved in HBeAg biosynthesis. (A) Schematic representation of the molecular events of HBeAg production. The number of residues in the core protein (183 aa) is based on non-A genotypes. (B) Comparison of the C-terminal sequences of p22 between genotype A and non-A genotypes. The four RXXR motifs are underlined, and the DR insertion in genotype A is boxed. (C) C-terminal sequence comparison of the e antigen precursors among WHV, WMHBV, and different HBV genotypes.
Although the bulk of the amino acid sequence of HBeAg is identical to that of the core protein, the two proteins fold very differently and have nonoverlapping biological functions. The HBeAg is a secreted soluble protein, whereas the core protein assembles into the core particle to accommodate pregenomic RNA encapsidation and genome replication. Core particle formation is preceded by dimer formation via an intermolecular disulfide bond involving a cysteine (Cys61) of the core protein, while its interaction with negatively charged pregenomic RNA is dependent on its arginine-rich C terminus (4, 45). For HBeAg, dimer formation is abrogated by the precore-derived cysteine (Cys-7 relative to the methionine of the core protein) which forms an intramolecular disulfide bond with Cys61 (46, 67). Furthermore, a hydrophobic sequence derived from the precore region (TrpLewTrp) prevents particle formation (67). Finally, HBeAg is unable to bind nucleic acid due to the lack of the arginine-rich sequence at its C terminus (4, 45). Despite their differences in higher structure, the major conformational epitope of the core particle coincides with one of the two HBeAg epitopes around position 80 of the core protein (50). Therefore, polyclonal antibodies raised against denatured core protein recognize HBeAg in immunoprecipitation (IP)-Western blot analysis and were used for the present study.
HBV-like hepatotropic DNA viruses (hepadnaviruses) have been found in woodchucks, ground squirrels, and ducks, all of which maintain the ability to express the e antigen (9, 18, 53). The e antigens of duck HBV (DHBV) and woodchuck hepatitis virus (WHV) are different from their HBV analogue in undergoing N-linked glycosylation (9, 52, 53). Surprisingly, experiments with the DHBV revealed that abolition of its e antigen expression neither impaired genome replication in transfected cells nor prevented viral infection of ducklings, although only a small number of ducks were followed and for just a short period of time (11, 53). Similarly, HBeAg expression was not required for HBV replication in cell culture (5, 63). However, the single chimpanzee injected with the HBeAg-negative mutant developed only transient infection (62). Subsequent studies with WHV, a much closer relative of HBV than DHBV, demonstrated that ablation of its e antigen expression markedly reduced the chronicity rate of neonatal infection (12, 14). This finding provided the most compelling evidence for the biological significance of e antigen expression, at least for mammalian hepadnaviruses. Immunological studies suggest that HBeAg serves as an immune tolerogen and can modulate the immune response to core protein. Whereas core protein induces Th1-type cytokines conducive to the clearance of infection, HBeAg induces a Th2-like response and can switch the immune response to core proteins from Th1 to Th2 type (13, 42). Epidemiological studies indicate that vertical transmission from HBeAg-negative mothers with low viremia was dependent on viral strains capable of HBeAg expression (8); HBV genotype G, which is incapable of HBeAg expression, usually coinfects with another HBV genotype (27, 34). Indeed, infection with HBV variants incapable of HBeAg expression has been associated with fulminant hepatitis, most likely due to the strong host immune response against the core protein (17, 35, 51, 58).
Understanding the molecular mechanism of HBeAg expression and identification of host factors involved in HBeAg maturation may provide novel targets for therapeutic intervention. In this regard, a gap in our knowledge of the HBeAg biosynthetic pathway is the C-terminal cleavage site and the host enzyme involved. A study 25 years ago assigned the C terminus of serum HBeAg to V149 (60). However, this was based on the identification of free threonine and valine following carboxypeptidase A digestion of HBeAg coupled with the presence of the 146TTVV149 coding sequence in the core gene. It did not demonstrate unequivocally that valine was the most C-terminal residue of HBeAg. A later study found that the authentic HBeAg migrated more slowly than an artificial construct with the C terminus truncated at V149 (54). More recently, Messageot and colleagues combined site-directed mutagenesis with transfection experiments with a monkey kidney cell line to demonstrate that HBeAg is derived from cleavage at the 151RRGR154 sequence (40). This sequence represents the first (the most proximal) of four furin recognition sites (RXXR motifs) at the C terminus of p22 (Fig. 1B). The second motif, 154RSPR157, overlaps the first motif and thus represents an alternative cleavage site (151RRGRSPR157, where the first motif is underlined, and the second motif is in boldface type). In this regard, genotype A of HBV is unique in that it possesses six additional nucleotides (nt) in the core gene that introduces aspartate-arginine (DR) dipeptide into the HBeAg precursor. Interestingly, the insertion lies in the middle of the 151RRGR154 sequence, converting it to 151RRDR154 and releasing the second motif as a separate entity (151RRDRGRSPR159, where the insertion is in italics) (Fig. 1B). The consequence of this insertion for HBeAg processing and the role of furin as the candidate enzyme in the cleavage were investigated.
MATERIALS AND METHODS
Serum samples.
Serum samples were obtained for Western blot analysis from 12 HBeAg-positive patients infected with HBV genotype A, B, or D. Ten separate serum samples were used to generate HBeAg expression constructs of genotypes A to D. The protocol for serum sample collection was approved by the institutional review board of the University of Michigan, and written consent was obtained from the patients.
HBeAg expression constructs and site-directed mutants.
An HBV DNA fragment spanning the entire precore/core gene (nt 1785 to 2488 in the case of genotype A) was amplified from DNA extracted from serum samples using sense primer preCs (5′-CCGAACTCGAGGCATAAATTGGTCTGCGCACCAGCACCATGCAACTTTTTCACCTCTGCC-3′) and antisense primer preCas (5′-AAGCGAATTCAAGTTTCCCACCTTATGAGTCC-3′). The PCR products were cloned into the XhoI and EcoRI sites of pcDNA3.1 zeo(−) vector (Invitrogen, Carlsbad, CA), permitting protein expression under the cytomegalovirus (CMV) early promoter. Substitutions, insertions, and deletions were introduced by overlap-extension PCR using the Expand High Fidelity Plus amplification system (Roche) as described previously (1, 3, 48). All the DNA constructs were sequenced to confirm the lack of unwanted mutations. They were purified by the high-speed plasmid midi kit (Qiagen, Valencia, CA).
Furin expression constructs.
The human furin expression construct was obtained from OriGene (Rockville, MD). This clone lacked 6 nt (GTACAG) encoding V93 and Q94, which were reintroduced by site-directed mutagenesis. Chicken furin cDNA was a kind gift of A. Feldmann (16). The cDNA was cloned into pAdTrack-CMV vector, followed by recombination in Escherichia coli cells with the adenovirus backbone pAdEasy-1, as detailed elsewhere (20, 32). The recombinant DNA was transfected to human embryonic kidney (HEK) 293 cells to produce recombinant adenovirus.
Transient transfection and impacts of furin and proprotein convertase (PC) inhibitor.
Human liver cell lines Huh7 and HepG2 were maintained in Dulbecco's modified Eagle medium. The HEK 293 cell line (19) was maintained in minimum essential medium. Human colon cancer cell line LoVo was maintained in F-12K medium. All the media were supplemented with 10% fetal calf serum. Transient transfection was performed on cells seeded in six-well plates with 40 to 70% confluence at the time of transfection, using LT1 (Mirus) as an agent for transfection (1, 3, 48). Each well of six-well plates received 2 μg of HBeAg expression construct and 5 ng of cDNA encoding secreted alkaline phosphatase. To study the impact of exogenous furin on HBeAg processing, 1 μg of HBV construct was cotransfected with 1 μg of human furin cDNA or 1 μg of pcDNA vector to serve as controls. Alternatively, 1 day after transfection, adenovirus encoding chicken furin or empty adenovirus was added at a multiplicity of infection of 50. Culture medium was harvested at day 5 posttransfection. To determine the impact of the PC inhibitor on HBeAg processing, decanoyl-Arg-Val-Lys-Arg-chloromethylketone (dec-RVKR-cmk; Bachem, King of Prussia, PA) was dissolved in dimethyl sulfoxide at a concentration of 20 mM and added to cells at a concentration of 20 μM during medium change prior to transfection. It was added to the culture the day after transfection and again 2 days later.
HBeAg quantification and IP-Western blot analysis.
HBeAg was quantified by an enzyme-linked immunoassay (ETI-EBK Plus kit; DiaSorin, Stillwater, MN) using diluted samples to prevent signal saturation. For HBeAg IP, 1.4 ml of precleared culture supernatant was incubated overnight at 4°C with 1.4 μl of rabbit anti-core antibody (Dako, Carpinteria, CA), followed by the addition of 10 μl bed volume of protein G agarose beads and a further incubation of 4 h. The immobilized antigen-antibody complex was collected by low-speed centrifugation, washed once with phosphate-buffered saline, and subjected to sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis. Following transfer, the blots were blocked at room temperature for 1 h with 3% bovine serum albumin (BSA) dissolved in Tris-based saline-0.05% Tween 20 (TBST) buffer and incubated overnight at 4°C with the anti-core antibody diluted 1:2,000 in 3% BSA-TBST buffer. Subsequent steps were carried out at room temperature. The blots were washed for 40 min with TBST buffer and incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit antibodies (Amersham, Piscataway, NJ) diluted 1:50,000 to 1:80,000 in 3% BSA-TBST buffer. After another wash for 40 min, signals were revealed by enhanced chemiluminescence (ECL) using Western Lightning Plus-ECL reagents (Perkin-Elmer).
RESULTS
Genotype A is unique in harboring the DR insertion in the first putative furin recognition site.
Furin, the candidate enzyme responsible for the conversion of p22 into HBeAg, preferentially cleaves after the R-X-K/R-R (P4-P3-P2-P1) consensus sequence but can tolerate a nonbasic residue at the P2 position (40, 43, 44). The C terminus of p25 contains four such R-X-X-R motifs, 151RRGR154, 154RSPR157, 164RRRR167, and 172RRRR175 (Fig. 1B). Inspection of 981 full-length HBV sequences deposited in GenBank revealed a DR insertion within the first RXXR motif (151RRGR154) of the majority of genotype A clones (171/175) but none of the 180, 328, 171, 71, 29, 13, and 14 clones belonging to genotypes B to H, respectively. The remaining four genotype A clones turned out to be recombinants with a genotype D-specific sequence in the core gene.
Only genotype A produced multiple-size forms of HBeAg.
We used a polyclonal anti-core antibody (Dako) for the IP and Western blot analysis of HBeAg. Similarly, others have used polyclonal anti-core antibody for the detection of the WHV e antigen (9). Although the Dako antibody detects both the core protein and HBeAg, the precore-core construct does not produce core protein due to the optimal sequence context surrounding the precore AUG codon (1). Moreover, the core protein is bigger than all the size forms of HBeAg produced from genotype A and thus would not complicate data interpretation. The CMV-driven core protein construct produced only the full-length core protein without any degradation product (data not shown).
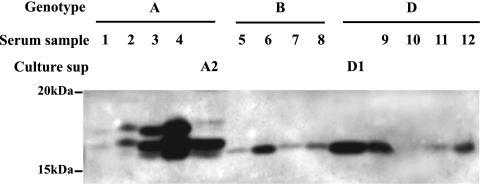
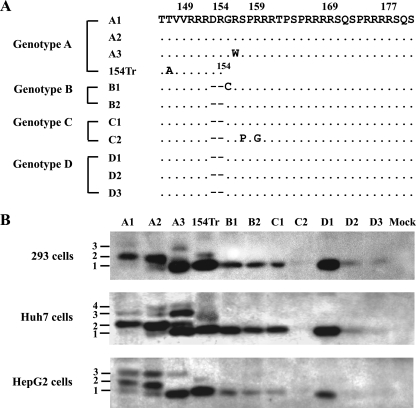
IP-Western blot analysis revealed a single HBeAg form from all the serum samples of genotype B or D infection. In contrast, two to three bands were visible for the four genotype A samples, with the fast-migrating species having the same mobility as HBeAg produced from other genotypes (Fig. 2). The variability in signal intensity among samples probably reflects different HBeAg titers or is caused by amino acid substitutions within the major core protein epitope. We found that Dako polyclonal antibody was inefficient at detecting the core protein and HBeAg of a naturally occurring HBV isolate due to a single E77Q substitution in the core gene (Kim et al., unpublished data). In order to further characterize the different HBeAg forms by genetic manipulation, we amplified the precore/core gene from 10 additional serum samples to generate HBeAg expression constructs under the CMV promoter. Three of the 10 clones harbored various substitutions in the C terminus of p22 (Fig. 3A). The constructs were transfected to two human liver cell lines (HepG2 and Huh7) and one human kidney cell line (HEK 293). Consistent with results from blood samples, constructs of genotypes B to D produced a single HBeAg species (Fig. 3B, lanes B1 to D3). The three genotype A clones produced multiple bands instead, with the fastest migrating band (band 1) being indistinguishable from the single HBeAg species produced by other genotypes (Fig. 3B, lanes A1 to A3). The relative intensities of the three bands showed clonal and cell type variations. Band 2 was the dominant species for A1 and A2 constructs in HEK 293 and Huh7 cells, but in HepG2 cells the relative intensity of band 3 increased. Interestingly, the A3 clone lost band 2 in HEK 293 and HepG2 cells, which was accompanied by an increase in band 1; in Huh7 cells, band 2 was diminished, with a concomitant increase in bands 1 and 3. Since A3 harbored an R156W mutation at the p4 position of the second RXXR motif (156RSPR159), this finding suggests that band 2 is generated by cleavage at the RSPR motif.
FIG. 2.
IP-Western blot analysis of HBeAg present in patient serum samples. HBeAg secreted from Huh7 cells transfected with genotype A (A2) and D (D1) constructs served as controls. Culture sup, culture supernatant.
FIG. 3.
Molecular forms of HBeAg secreted from human liver and kidney cell lines following transfection with precore/core constructs under the CMV promoter. (A) C-terminal sequences of P22 from 10 expression constructs, with numbering based on genotype A. 154Tr is an artificial construct with protein translation terminated after R154. (B) IP-Western blot analysis of HBeAg secreted from transfected HEK 293, Huh7, and HepG2 cells, respectively. The different size forms of HBeAg are labeled 1 to 4, from small to large. Mock, mock transfected.
The single HBeAg species of genotype B is generated by cleavage at the 151RRGR154 motif.
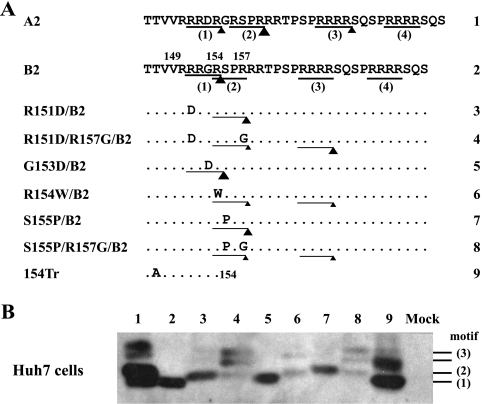
HBeAg produced from genotypes B to D comigrated with the major band produced by an artificial construct terminating after R154 in the core gene (Fig. 3B, compare lane 154Tr with B1). In this regard, Messageot et al. (40) identified R154 as the HBeAg cleavage site for genotype D in a monkey kidney cell line (COS). To further clarify this issue, we introduced mutations into the 151RRGR154 and 154RSPR157 motifs of clone B2 and determined their impacts on HBeAg production from Huh7 cells. Either an R151D mutation at the P4 position or an S155P mutation at the P1′ position of the 151RRGR154 motif caused replacement of the original HBeAg species (band 1) with a slower migrating band (band 2) (Fig. 4B, compare lanes 3 and 7 with lane 2). This is consistent with an inhibitory effect of a hydrophobic residue at the P1′ position on furin-like PCs (31). Combining the R151D or S155P mutation with an R157G mutation, which targets the second motif (154RSPR157), reduced the intensity of band 2 and generated two bands of even higher molecular weight (double band 3) (Fig. 4, lanes 4 and 8). The R154W mutation, which targets the first motif via the P4 position and the second motif via the P1 position, generated weak band 2 and another band of higher molecular weight (Fig. 4, lane 6). Together these observations establish the 151RRGR154 motif, the first of four RXXR motifs, as the cleavage site for wild-type genotype B. However, mutation of this motif does not abolish HBeAg processing but rather shifts the cleavage site to a distant RXXR motif.
FIG. 4.
Impact of mutating the first and second RXXR motifs on HBeAg production by a genotype B clone. (A) Amino acid sequences of clone B2 and its site-directed mutants, with the four RXXR motifs numbered. Cleavage efficiency in transfected Huh7 cells is indicated by the size of the triangle. (B) IP-Western blot analysis of HBeAg secreted from Huh7 cells. The numbers shown to the right correspond to the RXXR motifs shown in panel A. Mock, mock transfected.
We noticed that the truncation mutant 154Tr produced a slower migrating band (Fig. 3 to 7), which was much weaker than the major band in most experiments except for Fig. 4B. The nature of this form of increased size remains unclear, but according to the NetOGlyc 3.1 prediction program T142 and T146 are putative sites of O-linked glycosylation (25). If the larger form of 154Tr is indeed glycosylated, PC cleavage at R154 probably prevents glycosylation at these sites, because HBeAg of non-A genotypes contains a single band comigrating with the lower band of 154Tr.
FIG. 7.
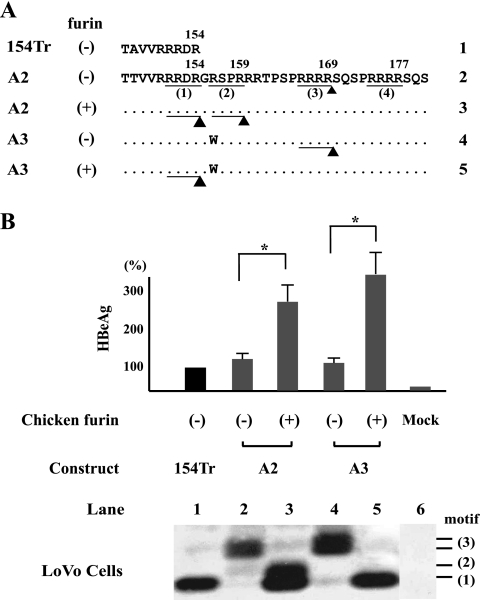
Furin confers cleavage at the first and second motifs of genotype A. The furin-deficient LoVo cells were transfected with the A2 or A3 construct and subsequently infected with the adenovirus vector or recombinant adenovirus expressing chicken furin. 154Tr served as a size marker. (A) Schematic representation of cleavage in the absence or presence of chicken furin. (B) Quantification of secreted HBeAg by a commercial kit and IP-Western blot analysis of the various HBeAg forms. Mock, mock transfected.
Neither a suboptimal first motif nor a separate second motif alone is sufficient to generate multiple HBeAg forms.
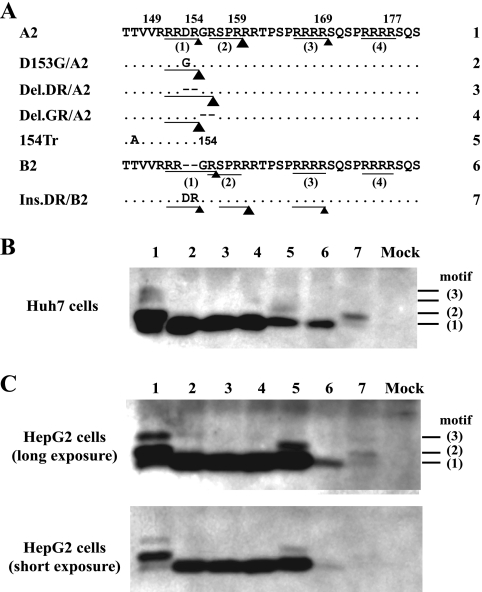
In the non-A genotypes, the first RXXR motif (151RRGR154) is fused to the second one (154RSPR157), thus rendering the cleavage at these two sites mutually exclusive (151RRGRSPR157). The DR insertion in genotype A has two consequences. First, it converts the 151RRGR154 motif into the less-favorable 151RRDR154 (acidic residues suppress cleavage by furin). Second, it generates a 1-aa spacer between the first and second motifs (151RRDRGRSPR159). As expected, deleting the unique DR sequence from clone A2 of genotype A converted multiple HBeAg species into a single one that comigrated with HBeAg produced from genotype B (Fig. 5B and C, compare lanes 1, 3, and 6). Interestingly, the same effect could be achieved by deletion of the GR dipeptide (Fig. 5, lane 4) or introduction of a D153G mutation (Fig. 5, lane 2). Since the GR deletion fuses the first and second motifs without altering the suboptimal first motif (151RRDRSPR157), while the D153G mutation improves the first motif without fusing the two motifs (151RRGRGRSPR159), multiple HBeAgs produced by genotype A are due to a suboptimal first motif in conjunction with accessibility of the second motif. Either condition alone is insufficient. Certainly, for the D153G mutant we cannot exclude the possibility that some molecules were cut at both the 151RRGR154 site and the 156RSPR159 site, but the HBeAg size is determined by the proximal cleavage event if more than one site is cleaved. In agreement with results for genotype A, a G153D mutation in genotype B had no effect on the HBeAg cleavage pattern (Fig. 4B, lane 5), while insertion of the DR sequence increased the size of its HBeAg (Fig. 5B and C, lane 7).
FIG. 5.
Mutations capable of converting the multiple HBeAg forms of genotype A into a single-size form. (A) Amino acid sequences of three site-directed mutants of clone A2 and one mutant of clone B2. The triangle size indicates the efficiency of cleavage in Huh7 cells. (B and C) HBeAg expression from Huh7 and HepG2 cells, respectively. Numbers shown to the right indicate the motifs recognized. Mock, mock transfected.
Multiple HBeAg bands of genotype A correspond to cleavage at the first, second, and third motifs.
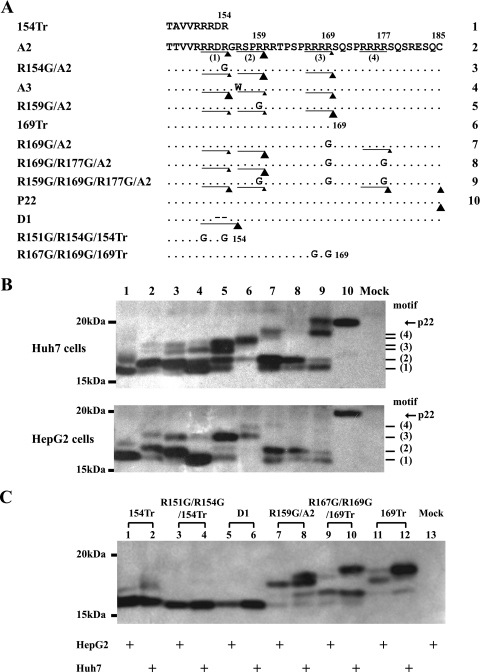
Next, we used a panel of site-directed mutants of clone A2 to map the cleavage sites for various forms of HBeAg produced by genotype A. Each of the four RXXR motifs was mutated to RXXG, either alone or in various combinations (Fig. 6A). For HepG2 cells, clone A2 produced three HBeAg bands numbered from small to large (Fig. 6B, bottom, lane 2). An R154G mutation of the first motif (151RRDR154) markedly increased band 2 at the expense of band 1 (Fig. 6B, lane 3), suggesting that band 1 is a cleavage product of the first motif. An R159G mutation targeting the second motif (156RSPR159) abolished band 2 and enhanced band 3 (Fig. 6B, lane 5), thus linking band 2 with cleavage of the second motif. The ability of the R169G mutation to abolish the third band confirmed it as a cleavage product of the third motif (166RRRR169) (Fig. 6B, lane 7).
FIG. 6.
Assignment of the multiple HBeAg species from wild-type genotype A to three RXXR motifs. (A) Amino acid sequences of clones A2 and A3, site-directed mutants of A2, as well as truncation mutants 154Tr and 169Tr, which serve as size markers. The two variant forms of these deletion mutants, R151G/R154G/154Tr and R167G/R169G/169Tr, are resistant to C-terminal trimming by basic carboxypeptidases. The size of the triangle indicates the abundance of the corresponding product in Huh7 cells. (B) HBeAg expression in Huh7 cells (top) and HepG2 cells (bottom). The numbers shown to the side correspond to motifs shown in panel A under A2. (C) Parallel analysis of HBeAg forms produced in HepG2 and Huh7 cells and impact of mutations that prevent C-terminal trimming of the Tr154 and Tr169 mutants. Mock, mock transfected.
For Huh7 cells, cleavage at the second motif was not completely abolished by an R156W or R159G mutation alone (Fig. 6B, top, lanes 4 and 5). Moreover, the R169G mutant displayed a double band of even slower mobility (band 4) (Fig. 6B, lane 7), which was abolished when the R169G mutation was combined with an additional R177G mutation targeting the fourth motif (Fig. 6B, lane 8). Thus, band 4 most likely represents cleavage at the fourth (174RRRR177) motif. However, the band reappeared with the R159G/R169G/R177G triple mutant (lane 9). This mutant produced an even larger species that comigrated with unprocessed p22 derived from cell lysate (Fig. 6B, compare lanes 9 and 10). This is consistent with the earlier report that p22 can be secreted, albeit at low efficiency (57). Finally, cleavage at the third motif generated a doublet in Huh7 cells (25), with the fast-migrating species corresponding to the single band produced by HepG2 cells (Fig. 6C, lanes 7 and 8). Since the third motif contains tetrameric arginines, the fast-migrating species could represent a further processed form where the newly exposed arginines are trimmed away by a basic carboxypeptidase. Alternatively, the slow-migrating species may represent a glycosylated form because residues T142, T146, T147, and T162 (and three serine residues downstream of the third RXXR motif) are potential O-linked glycosylation sites (25). Parallel analysis of HBeAg produced in HepG2 and Huh7 cells revealed comigration of the single band produced in HepG2 cells with the lower band of Huh7 cells (Fig. 6C, lanes 7 and 8). The R167G/R169G mutant of 169Tr, which could not be attacked by a basic carboxypeptidase, was inconclusive as to whether the size difference could be attributed to a difference in the C termini. On the other hand, this mutant showed increased processing of its internal 156RSPR159 motif in comparison to that of the original 169Tr mutant (Fig. 6C, lane 10 versus lane 12), suggesting that blocking carboxypeptidase cleavage facilitated endopeptidase cleavage at a nearby site.
Furin can cleave the RRDR, RRGR, and RSPR motifs present in the HBeAg precursor.
The human colon cancer cell line LoVo is deficient in furin expression (61) and produced a very different pattern of HBeAg than HepG2, Huh7, and HEK 293 cells. Transfection of clone A2 of genotype A into this cell line generated a doublet of HBeAg consistent with cleavage at the third motif (166RRRR169) (Fig. 7B, lane 2). Introduction of chicken furin by adenovirus vector not only markedly increased HBeAg titer based on an enzyme-linked immunoassay but also led to the replacement of the original forms with two smaller ones corresponding to cleavage at the first (151RRDR154) and second (156RSPR159) motifs (Fig. 7B, lane 3). For clone A3, which contains an R156W mutation that prevents cleavage of the second motif, chicken furin generated a cleavage product of the first (151RRDR154) motif only (Fig. 7B, lane 5). Similar experiments with a genotype D clone also revealed the ability of chicken furin to recognize the RRGR motif (data not shown).
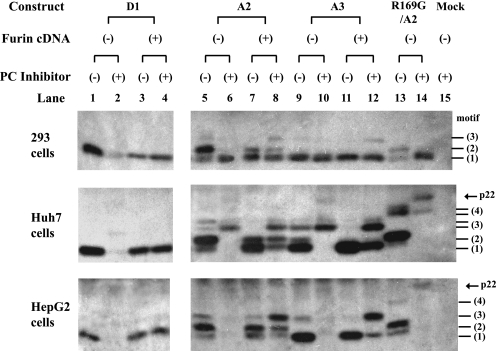
In a complementary approach, we determined whether HBeAg processing in HepG2, Huh7, and HEK 293 cells could be blocked by dec-RVKR-cmk, a substrate inhibitor of furin and other PCs such as PC6, PC7, and PACE4. The chemical abolished HBeAg expression from genotype D in all the three cell lines (Fig. 8, compare lanes 1 and 2), which could be overcome by cotransfection of human furin cDNA (Fig. 8, lane 4). The results for genotype A were cell type dependent. Whereas dec-RVKR-cmk blocked all the HBeAg forms in HepG2 cells (Fig. 8, bottom, lanes 6, 10, and 14), it failed to abolish cleavage at the third motif for Huh7 cells (Fig. 8, middle) or the first motif for HEK 293 cells (Fig. 8, top). One possible explanation is that in addition to furin (or a furin-like PC), HEK 293 cells contain a dec-RVKR-cmk-resistant PC that can cleave the RRDR but not the RRGR motif. Cotransfection of human furin cDNA in the absence of a furin inhibitor increased the smallest form of HBeAg corresponding to cleavage at the first RXXR motif (Fig. 8, top and middle, lane 7), while furin overexpression in the presence of dec-RVKR-cmk increased larger HBeAg forms (Fig. 8, lane 8). These results are most consistent with a competition between p22 and the inhibitor and possibly also among the four RXXR motifs of p22 for cleavage.
FIG. 8.
Effects of a PC inhibitor and human furin overexpression on HBeAg processing in HEK 293, Huh7, and HepG2 cells. Cells were transfected in duplicate with HBeAg expression constructs together with an empty plasmid or a plasmid encoding human furin. One set of samples was treated with dec-RVKR-cmk. HBeAg was detected by IP-Western blot analysis. Mock, mock transfected.
DISCUSSION
The HBV genome ranges from 3,182 to 3,248 bp in a genotype-specific manner (29). Thus, genotypes D and E/G have, respectively, a 33-nt and 3-nt deletion in the preS1 region. Genotype G possesses a 36-nt insertion at the 5′ end of the core gene, while genotype A has a 6-nt insertion near the 3′ end of the core gene. The origins of these insertions/deletions and their biological consequences remain largely enigmatic. We recently found that removing the 36 nt from genotype G markedly reduced core protein expression and genome replication, thus validating its essential role for genotype G survival (34). In the present study, we focused on the impact of the genotype A-specific 6-nt insertion on HBeAg expression.
Most of the previous studies revealed HBeAg as a single molecular species, and a recent work mapped the single HBeAg form from genotype D as a cleavage product of R154 in the first of four RXXR motifs (40). However, studies as early as 1988 reported multiple-size forms of HBeAg (50, 57), although the structural basis has never been experimentally tested. We found that all of the 171 nonrecombinant genotype A sequences deposited in GenBank possess the 6-nt insertion that adds the DR dipeptide to the core protein and HBeAg precursor. However, alignment of the mammalian hepadnaviral genomes revealed this position to be the site of deletional (or insertional) events. Compared with the core genes of the WHV and ground squirrel hepatitis virus, the core genes of the wooly monkey HBV (WMHBV) and HBV of non-A genotypes harbor deletions of 6 and 5 aa, respectively (Fig. 1C). Genotype A HBV harbors a shorter deletion of 3 aa. In addition, WMHBV and genotype A HBV display amino acid substitutions adjacent to the site of deletion. These deletions and substitutions alter the RXXR motifs and are most likely to affect e antigen processing. For most HBV genotypes, the first and second RXXR motifs are fused (151RRGRSPR157), thus preventing simultaneous cleavage at both motifs. Preferred cleavage at R154 rather than R157 is apparently due to the better sequence context of RRGR (three arginines) than RSPR (two arginines). The extra DR sequence found in genotype A (151RRDRGRSPR159) not only weakens the first motif through a negatively charged residue but also physically separates the two motifs. We showed that combination of these two events, rather than each condition alone, shifted the major cleavage site to R159 in the second RXXR motif (Fig. 5). Since RSPR is not a strong motif, additional forms of HBeAg are produced by alternative cleavage at R154 of the first motif or R169 at the third motif.
The cleavage product at R169 of the third RXXR motif was not abundant for genotype A, and the cleavage product at the fourth motif (R177) was not observed at all for the wild-type construct. For non-A genotypes, there is no evidence for cleavage of either the third or the fourth motif. Nevertheless, the R159G mutation in clone A2 markedly increased cleavage at R169 of genotype A, while an R169G mutation generated a larger HBeAg species consistent with cleavage at R177 of the fourth RXXR motif (Fig. 6). Similarly, mutating the first and second RXXR motifs in genotype B produced larger HBeAg species (Fig. 4). The rare detection of large-sized HBeAg for the wild-type clones could be explained by competition among the four RXXR motifs for PC binding and cleavage or cleavage of a single p22 molecule at multiple sites. In the latter case, only the most proximal cleavage event can be visualized by Western blot analysis. Messageot and colleagues observed a 20-kDa intermediate inside transfected cells but not in culture supernatant (40), which is consistent with sequential cleavage from the C terminus of p22. Nevertheless, cleavage at the third and fourth motifs is not required for cleavage at the second motif, as evidenced by the R169G/R177G/A2 mutant (Fig. 6B, lane 8).
What are the enzymes responsible for HBeAg processing? Among the seven PCs capable of cleaving precursor proteins via basic amino acids in the substrates, furin, PC5/PC6-B, and PC7/LPC are associated with the constitutive secretory pathway and are candidates for HBeAg maturation (55). Observation of the cleavage product of the 166RRRR169 motif in LoVo cells (Fig. 7) and also in Huh7 cells treated with dec-RVKR-cmk (Fig. 8) suggests that the RRRR motif can be cleaved by a PC other than furin. On the other hand, cleavage of the RRGR, RRDR, and RSPR motifs does require furin or a related PC, as evidenced by the lack of cleavage products in LoVo cells which could be overcome by chicken furin (Fig. 7). A combination of PC inhibitor with human furin cDNA in human liver and kidney cell lines also supports this conclusion, although HEK 293 cells might possess another PC capable of processing the 151RRDR154 motif and being resistant to dec-VKR-cmk (Fig. 8). In this regard, although HEK 293 cells were derived from human embryonic kidney cells, they display characteristics of neuronal cells as well (7, 19, 56). Considering that transfection with PC5 or PC7 cDNA had no effect on dec-RVKR-cmk-treated Huh7 cells (K. Ito, unpublished observation), furin appears to be the major enzyme involved in HBeAg processing for both genotype A and non-A genotypes. This point requires further validation using small interfering RNA technology.
The cleavage product at the 151RRDR154 motif was more abundant in HepG2 cells than in Huh7 cells. This could be attributed to higher furin expression levels in HepG2 cells, an issue that remains to be validated by comparative Western blot analysis. We found that furin overexpression led to increased cleavage product of the first motif in LoVo, HepG2, and Huh7 cells (Fig. 7B and 8, middle and bottom). HepG2 cells also differed from Huh7 cells in that the cleavage product of the third motif (R169) migrated as a single band rather than as a doublet. Either the fast-migrating species in Huh7 cells (and the single species found in HepG2 cells) corresponds to a mature form for which the C-terminal four arginines have been removed by a basic carboxypeptidase or the slow-migrating form represents a modified version with O-linked glycosylation. This is possible considering that the e antigens of both WHV and DHBV are modified by N-linked glycosylation (9, 52, 53).
HBeAg becomes an immune target when the host mounts an anti-HBe immune response. Since HBeAg expression is not directly involved in the viral life cycle, HBV can shut off HBeAg expression by nonsense or frameshift mutations in the precore region (6, 10, 64). However, the precore region is also part of an RNA secondary structure in the pregenomic RNA called the ɛ signal, which is essential for HBV genome replication (26). That explains why a G1896A nonsense mutation, which for most genotypes improves rather than disrupts a base pair in the ɛ signal, is the most common HBeAg-abolishing mutation. However, genotype A and some isolates of genotypes C and F rarely circulate as a G1896A mutant because of the disruption of that particular base pair (2, 33, 37). Here we demonstrate that genotype A also differs from other HBV genotypes in the molecular structure of HBeAg. Whether the multiple-size forms of HBeAg produced by genotype A alter HBeAg function is unknown. Native Alaskans with chronic HBV infection are infected with genotypes A, B, C, D, and F. In this patient population, where the host genetic background and age at the time of infection are similar, genotype A was associated with faster HBeAg clearance than other HBV genotypes (36). Recent clinical trials showed that genotype A was also associated with a higher rate of HBeAg loss than other genotypes (B, C, and D) after interferon but not lamivudine therapy (15, 23, 24, 30). It remains to be established whether such differences are related to the different molecular forms of HBeAg produced from genotype A, the difference in HBeAg titer, or the reduced ability of genotype A to switch off HBeAg expression.
Acknowledgments
We thank A. Feldmann for chicken furin cDNA, T. He and B. Vogelstein for recombinant adenovirus vectors, J. Li for the preparation of adenovirus construct expressing chicken furin, and M. Hussain for technical assistance.
This work was supported by the American Cancer Society award RSG 06-059-01-MBC and the National Institutes of Health grant R21CA133976 to S.T. and by a National Institutes of Health Cooperative grant UO1 DK57577 to A.S.-F.L.
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Ahn, S. H., A. Kramvis, S. Kawai, H. C. Spangenberg, J. Li, G. Kimbi, M. Kew, J. Wands, and S. Tong. 2003. Sequence variation upstream of precore translation initiation codon reduces hepatitis B virus e antigen production. Gastroenterology 1251370-1378. [DOI] [PubMed] [Google Scholar]
- 2.Arauz-Ruiz, P., H. Norder, K. A. Visona, and L. O. Magnius. 1997. Genotype F prevails in HBV infected patients of Hispanic origin in Central America and may carry the precore stop mutant. J. Med. Virol. 51305-312. [PubMed] [Google Scholar]
- 3.Bang, G., K. H. Kim, M. Guarnieri, F. Zoulim, S. Kawai, J. Li, J. Wands, and S. Tong. 2005. Effect of mutating the two cysteines required for HBe antigenicity on hepatitis B virus DNA replication and virion secretion. Virology 332216-224. [DOI] [PubMed] [Google Scholar]
- 4.Birnbaum, F., and M. Nassal. 1990. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J. Virol. 643319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blum, H. E., T. J. Liang, E. Galun, and J. R. Wands. 1991. Persistence of hepatitis B viral DNA after serological recovery from hepatitis B virus infection. Hepatology 1456-63. [DOI] [PubMed] [Google Scholar]
- 6.Brunetto, M. R., M. Stemler, F. Bonino, F. Schodel, F. Oliveri, M. Rizzetto, G. Verme, and H. Will. 1990. A new hepatitis B virus strain in patients with severe anti-HBe positive chronic hepatitis B. J. Hepatol. 10258-261. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, S. A., J. Lin, E. Y. Dobrikova, and M. Gromeier. 2005. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J. Virol. 796281-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candotti, D., K. Danso, and J. P. Allain. 2007. Maternofetal transmission of hepatitis B virus genotype E in Ghana, west Africa. J. Gen. Virol. 882686-2695. [DOI] [PubMed] [Google Scholar]
- 9.Carlier, D., O. Jean-Jean, and J. M. Rossignol. 1994. Characterization and biosynthesis of the woodchuck hepatitis virus e antigen. J. Gen. Virol. 75171-175. [DOI] [PubMed] [Google Scholar]
- 10.Carman, W. F., M. R. Jacyna, S. Hadziyannis, P. Karayiannis, M. J. McGarvey, A. Makris, and H. C. Thomas. 1989. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet ii588-591. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C., G. Enders, R. Sprengel, N. Peters, H. E. Varmus, and D. Ganem. 1987. Expression of the precore region of an avian hepatitis B virus is not required for viral replication. J. Virol. 613322-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, H. S., M. C. Kew, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1992. The precore gene of the woodchuck hepatitis virus genome is not essential for viral replication in the natural host. J. Virol. 665682-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, M. T., J. N. Billaud, M. Sallberg, L. G. Guidotti, F. V. Chisari, J. Jones, J. Hughes, and D. R. Milich. 2004. A function of the hepatitis B virus precore protein is to regulate the immune response to the core antigen. Proc. Natl. Acad. Sci. USA 10114913-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cote, P. J., B. E. Korba, R. H. Miller, J. R. Jacob, B. H. Baldwin, W. E. Hornbuckle, R. H. Purcell, B. C. Tennant, and J. L. Gerin. 2000. Effects of age and viral determinants on chronicity as an outcome of experimental woodchuck hepatitis virus infection. Hepatology 31190-200. [DOI] [PubMed] [Google Scholar]
- 15.Erhardt, A., D. Blondin, K. Hauck, A. Sagir, T. Kohnle, T. Heintges, and D. Haussinger. 2005. Response to interferon alfa is hepatitis B virus genotype dependent: genotype A is more sensitive to interferon than genotype D. Gut 541009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldmann, A., M. K. Schafer, W. Garten, and H. D. Klenk. 2000. Targeted infection of endothelial cells by avian influenza virus A/FPV/Rostock/34 (H7N1) in chicken embryos. J. Virol. 748018-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedt, M., P. Gerner, E. Lausch, H. Trubel, B. Zabel, and S. Wirth. 1999. Mutations in the basic core promotor and the precore region of hepatitis B virus and their selection in children with fulminant and chronic hepatitis B. Hepatology 291252-1258. [DOI] [PubMed] [Google Scholar]
- 18.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 19.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 3659-74. [DOI] [PubMed] [Google Scholar]
- 20.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 952509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollinger, F. B., and T. J. Liang. 2001. Hepatitis B virus, p. 2971-3035. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 22.Hoofnagle, J. H., G. M. Dusheiko, L. B. Seeff, E. A. Jones, J. G. Waggoner, and Z. B. Bales. 1981. Seroconversion from hepatitis B e antigen to antibody in chronic type B hepatitis. Ann. Intern. Med. 94744-748. [DOI] [PubMed] [Google Scholar]
- 23.Hou, J., R. Schilling, H. L. Janssen, B. E. Hansen, R. Heijtink, E. Sablon, R. Williams, G. K. Lau, S. W. Schalm, and N. V. Naoumov. 2007. Genetic characteristics of hepatitis B virus genotypes as a factor for interferon-induced HBeAg clearance. J. Med. Virol. 791055-1063. [DOI] [PubMed] [Google Scholar]
- 24.Janssen, H. L., M. van Zonneveld, H. Senturk, S. Zeuzem, U. S. Akarca, Y. Cakaloglu, C. Simon, T. M. So, G. Gerken, R. A. de Man, H. G. Niesters, P. Zondervan, B. Hansen, and S. W. Schalm. 2005. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet 365123-129. [DOI] [PubMed] [Google Scholar]
- 25.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15153-164. [DOI] [PubMed] [Google Scholar]
- 26.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 93389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato, H., E. Orito, R. G. Gish, N. Bzowej, M. Newsom, F. Sugauchi, S. Suzuki, R. Ueda, Y. Miyakawa, and M. Mizokami. 2002. Hepatitis B e antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology 35922-929. [DOI] [PubMed] [Google Scholar]
- 28.Kiire, C. F. 1996. The epidemiology and prophylaxis of hepatitis B in sub-Saharan Africa: a view from tropical and subtropical Africa. Gut 38(Suppl. 2)S5-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramvis, A., M. Kew, and G. Francois. 2005. Hepatitis B virus genotypes. Vaccine 232409-2423. [DOI] [PubMed] [Google Scholar]
- 30.Lau, G. K., T. Piratvisuth, K. X. Luo, P. Marcellin, S. Thongsawat, G. Cooksley, E. Gane, M. W. Fried, W. C. Chow, S. W. Paik, W. Y. Chang, T. Berg, R. Flisiak, P. McCloud, and N. Pluck. 2005. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 3522682-2695. [DOI] [PubMed] [Google Scholar]
- 31.Lazure, C., D. Gauthier, F. Jean, A. Boudreault, N. G. Seidah, H. P. Bennett, and G. N. Hendy. 1998. In vitro cleavage of internally quenched fluorogenic human proparathyroid hormone and proparathyroid-related peptide substrates by furin. Generation of a potent inhibitor. J. Biol. Chem. 2738572-8580. [DOI] [PubMed] [Google Scholar]
- 32.Li, J., S. Tong, H. B. Lee, A. L. Perdigoto, H. C. Spangenberg, and J. R. Wands. 2004. Glycine decarboxylase mediates a postbinding step in duck hepatitis B virus infection. J. Virol. 781873-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, J.-S., S.-P. Tong, Y.-M. Wen, L. Vitvitski, Q. Zhang, and C. Trépo. 1993. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J. Virol. 675402-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, K., F. Zoulim, C. Pichoud, K. Kwei, S. Villet, J. Wands, J. Li, and S. Tong. 2007. Critical role of the 36-nucleotide insertion in hepatitis B virus genotype G in core protein expression, genome replication, and virion secretion. J. Virol. 819202-9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, T. J., K. Hasegawa, N. Rimon, J. R. Wands, and E. Ben-Porath. 1991. A hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. N. Engl. J. Med. 3241705-1709. [DOI] [PubMed] [Google Scholar]
- 36.Livingston, S. E., J. P. Simonetti, L. R. Bulkow, C. E. Homan, M. M. Snowball, H. H. Cagle, S. E. Negus, and B. J. McMahon. 2007. Clearance of hepatitis B e antigen in patients with chronic hepatitis B and genotypes A, B, C, D, and F. Gastroenterology 1331452-1457. [DOI] [PubMed] [Google Scholar]
- 37.Lok, A. S., U. Akarca, and S. Greene. 1994. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc. Natl. Acad. Sci. USA 914077-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnius, L. O., and J. A. Espmark. 1972. New specificities in Australia antigen positive sera distinct from the Le Bouvier determinants. J. Immunol. 1091017-1021. [PubMed] [Google Scholar]
- 39.McLachlan, A., D. R. Milich, A. K. Raney, M. G. Riggs, J. L. Hughes, J. Sorge, and F. V. Chisari. 1987. Expression of hepatitis B virus surface and core antigens: influences of pre-S and precore sequences. J. Virol. 61683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messageot, F., S. Salhi, P. Eon, and J. M. Rossignol. 2003. Proteolytic processing of the hepatitis B virus e antigen precursor. Cleavage at two furin consensus sequences. J. Biol. Chem. 278891-895. [DOI] [PubMed] [Google Scholar]
- 41.Milich, D., and T. J. Liang. 2003. Exploring the biological basis of hepatitis B e antigen in hepatitis B virus infection. Hepatology 381075-1086. [DOI] [PubMed] [Google Scholar]
- 42.Milich, D. R., M. K. Chen, J. L. Hughes, and J. E. Jones. 1998. The secreted hepatitis B precore antigen can modulate the immune response to the nucleocapsid: a mechanism for persistence. J. Immunol. 1602013-2021. [PubMed] [Google Scholar]
- 43.Molloy, S. S., P. A. Bresnahan, S. H. Leppla, K. R. Klimpel, and G. Thomas. 1992. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 26716396-16402. [PubMed] [Google Scholar]
- 44.Nakayama, K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nassal, M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 664107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nassal, M., and A. Rieger. 1993. An intramolecular disulfide bridge between Cys-7 and Cys61 determines the structure of the secretory core gene product (e antigen) of hepatitis B virus. J. Virol. 674307-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ou, J. H., O. Laub, and W. J. Rutter. 1986. Hepatitis B virus gene function: the precore region targets the core antigen to cellular membranes and causes the secretion of the e antigen. Proc. Natl. Acad. Sci. USA 831578-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parekh, S., F. Zoulim, S. H. Ahn, A. Tsai, J. Li, S. Kawai, N. Khan, C. Trepo, J. Wands, and S. Tong. 2003. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J. Virol. 776601-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roossinck, M. J., S. Jameel, S. H. Loukin, and A. Siddiqui. 1986. Expression of hepatitis B viral core region in mammalian cells. Mol. Cell. Biol. 61393-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salfeld, J., E. Pfaff, M. Noah, and H. Schaller. 1989. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J. Virol. 63798-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato, S., K. Suzuki, Y. Akahane, K. Akamatsu, K. Akiyama, K. Yunomura, F. Tsuda, T. Tanaka, H. Okamoto, Y. Miyakawa, and M. Mayumi. 1995. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann. Intern. Med. 122241-248. [DOI] [PubMed] [Google Scholar]
- 52.Schlicht, H. J. 1991. Biosynthesis of the secretory core protein of duck hepatitis B virus: intracellular transport, proteolytic processing, and membrane expression of the precore protein. J. Virol. 653489-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlicht, H. J., J. Salfeld, and H. Schaller. 1987. The duck hepatitis B virus pre-C region encodes a signal sequence which is essential for synthesis and secretion of processed core proteins but not for virus formation. J. Virol. 613701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlicht, H. J., and G. Wasenauer. 1991. The quaternary structure, antigenicity, and aggregational behavior of the secretory core protein of human hepatitis B virus are determined by its signal sequence. J. Virol. 656817-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seidah, N. G., and A. Prat. 2002. Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 3879-94. [DOI] [PubMed] [Google Scholar]
- 56.Shaw, G., S. Morse, M. Ararat, and F. L. Graham. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16869-871. [DOI] [PubMed] [Google Scholar]
- 57.Standring, D. N., J. H. Ou, F. R. Masiarz, and W. J. Rutter. 1988. A signal peptide encoded within the precore region of hepatitis B virus directs the secretion of a heterogeneous population of e antigens in Xenopus oocytes. Proc. Natl. Acad. Sci. USA 858405-8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stuyver, L., S. De Gendt, J. F. Cadranel, C. Van Geyt, G. Van Reybroeck, R. Dorent, I. Gandjbachkh, M. Rosenheim, F. Charlotte, P. Opolon, J. M. Huraux, and F. Lunel. 1999. Three cases of severe subfulminant hepatitis in heart-transplanted patients after nosocomial transmission of a mutant hepatitis B virus. Hepatology 291876-1883. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi, K., Y. Akahane, T. Gotanda, T. Mishiro, M. Imai, Y. Miyakawa, and M. Mayumi. 1979. Demonstration of hepatitis B e antigen in the core of Dane particles. J. Immunol. 122275-279. [PubMed] [Google Scholar]
- 60.Takahashi, K., A. Machida, G. Funatsu, M. Nomura, S. Usuda, S. Aoyagi, K. Tachibana, H. Miyamoto, M. Imai, T. Nakamura, Y. Miyakawa, and M. Mayumi. 1983. Immunochemical structure of hepatitis B e antigen in the serum. J. Immunol. 1302903-2907. [PubMed] [Google Scholar]
- 61.Takahashi, S., K. Kasai, K. Hatsuzawa, N. Kitamura, Y. Misumi, Y. Ikehara, K. Murakami, and K. Nakayama. 1993. A mutation of furin causes the lack of precursor-processing activity in human colon carcinoma LoVo cells. Biochem. Biophys. Res. Commun. 1951019-1026. [DOI] [PubMed] [Google Scholar]
- 62.Tong, S. P., B. Brotman, J. S. Li, L. Vitvitski, D. Pascal, A. M. Prince, and C. Trepo. 1991. In vitro and in vivo replication capacity of the precore region defective hepatitis B virus variants. J. Hepatol 13(Suppl. 4)S68-S73. [DOI] [PubMed] [Google Scholar]
- 63.Tong, S. P., C. Diot, P. Gripon, J. Li, L. Vitvitski, C. Trepo, and C. Guguen-Guillouzo. 1991. In vitro replication competence of a cloned hepatitis B virus variant with a nonsense mutation in the distal pre-C region. Virology 181733-737. [DOI] [PubMed] [Google Scholar]
- 64.Tong, S. P., J. S. Li, L. Vitvitski, and C. Trepo. 1990. Active hepatitis B virus replication in the presence of anti-HBe is associated with viral variants containing an inactive pre-C region. Virology 176596-603. [DOI] [PubMed] [Google Scholar]
- 65.Trepo, C. G., L. O. Magnius, R. A. Schaefer, and A. M. Prince. 1976. Detection of e antigen and antibody: correlations with hepatitis B surface and hepatitis B core antigens, liver disease, and outcome in hepatitis B infections. Gastroenterology 71804-808. [PubMed] [Google Scholar]
- 66.Wang, J., A. S. Lee, and J. H. Ou. 1991. Proteolytic conversion of hepatitis B virus e antigen precursor to end product occurs in a postendoplasmic reticulum compartment. J. Virol. 655080-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wasenauer, G., J. Kock, and H. J. Schlicht. 1992. A cysteine and a hydrophobic sequence in the noncleaved portion of the pre-C leader peptide determine the biophysical properties of the secretory core protein (HBe protein) of human hepatitis B virus. J. Virol. 665338-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao, G. B. 1996. Importance of perinatal versus horizontal transmission of hepatitis B virus infection in China. Gut 38(Suppl. 2)S39-S42. [DOI] [PMC free article] [PubMed] [Google Scholar]