Abstract
In recent years, baculovirus has emerged as a tool for high-efficiency gene transfer into mammalian cells. However, the level of gene expression is often limited by the strength of the mammalian promoter used. Here, we show that the baculovirus RING protein IE2 is a strong, promiscuous trans-activator in mammalian cells, dramatically upregulating the cytomegalovirus (CMV) promoter in both Vero E6 and U-2OS cells. Further study of the cellular mechanism for the activation led to the discovery of a novel IE2 nuclear body structure which contains a high concentration of G-actin and closely associates with RNA polymerase II, PML, and SUMO1. IE2 mutagenesis studies indicated that the RING and coiled-coil domains of IE2 were necessary for nuclear body formation, as well as for strong activation of the CMV promoter in mammalian cells. Overall, this study shows that the IE2 trans-activator could significantly advance the use of baculovirus in mammalian gene transfer and protein production.
Baculovirus has a long history of being a versatile foreign protein expression system in either insect cells or larvae. In a landmark paper in 1995, Autographa californica multicapsid nucleopolyhedrosis virus (AcMNPV), a type species of baculovirus, was shown to transduce hepatocytes with high efficiency (14). Since then, applications for utilizing this remarkable virus in mammalian systems have become the subject of intense study. Numerous papers have been published on baculovirus mammalian gene transfer, with a view that baculovirus could become a nonpathogenic, specific targeting vehicle for gene therapy in the future (2, 11, 16, 17). The advantages of baculovirus over conventional mammalian vectors include (i) its ability to accommodate the long insertion of foreign gene constructs into its large (∼130-kb), double-stranded circular DNA, (ii) its low cytotoxicity toward mammalian cells even at a high multiplicity of infection (MOI), and (iii) its high specificity for host species, making it one of the safest eukaryotic pathogens to work with.
In almost all reported cases, recombinant baculoviruses intended for mammalian gene transfer have relied on either cytomegalovirus (CMV) immediate-early or simian virus 40 (SV40) early promoters to drive their target protein expression (4, 35). The baculovirus genome remains mostly silent throughout the transduction, with the exception of a few early genes, which are detected only at very low levels and do not alter the cells' potential for transcription or differentiation (10, 20). We showed in our previous experiments that during the transduction of susceptible Vero E6 cells, both of the two major AcMNPV transcriptional activators, IE1 and IE2, were necessary to achieve partial activation of the baculovirus genome, and they did so only if they were properly expressed using the mammalian CMV promoter (24).
With the predominant usage of the CMV promoter for baculovirus foreign gene expression in mammalian systems, we decided to test the abilities of both the IE1 and the IE2 protein for activation of this promoter in mammalian cells. IE1- or IE2-expressing plasmids were cotransfected into Vero E6 cells with reporter plasmids expressing luciferase under the CMV promoter. Unexpectedly, we found that IE2 was an extremely strong trans-activator in mammalian cells, capable of boosting the expression level of the luciferase gene over 122-fold in the presence of a baculovirus cis element, the homologous region (13) sequence.
IE2 is a highly structured gene with two nuclear localization sequences at its N-terminal end (19). It has a C3HC4-type RING finger domain which has been shown to cause S-phase cell cycle arrest in Spodoptera frugiperda IPLB-Sf21 (Sf21) cells (31), and the Bombyx mori NPV (BmNPV) IE2 homolog exhibits ubiquitin ligase activity in vitro (18). At its C terminus, there is a coiled-coil domain which is important for oligomerization of the IE2 protein within the insect cell nucleus, forming distinct nucleus foci (19). So far, its mechanism in baculovirus gene activation is unclear. Unlike traditional transcriptional factors, there is a possibility that IE2 activation may not require direct binding to the gene promoter. IE2 of Orgyia pseudotsugata MNPV was found to trans-activate the opep2 gene without direct binding to the opep2 promoter (34). Here, we attempt to elucidate the IE2 function in mammalian cells, specifically, how such a high level of stimulation of the CMV promoter can be achieved. In particular, we have examined the importance of both the RING domain and the coiled-coil domain in its function as a trans-activator.
Microscopic evidence has shown that IE2 forms distinct nuclear foci in insect cells. Although the function of these foci is unclear, focus formation was reported to be regulated by both the RING and the coiled-coil oligomerization domain (19). In addition, transiently expressed IE2 in BHK21 cells was found to associate with PML nuclear bodies (NBs) (29). As many PML-associated proteins are SUMOylated, we searched for potential SUMOylation site within IE2 by using SUMOplot software and found only one putative site. To find out whether these domains are involved in CMV promoter activation, we designed mutations within the RING and coiled-coil domains, as well as the predicted SUMOylation site, to test whether the ability to form the distinctive nuclear focus macrostructure correlates with the promoter activation activity of IE2 in mammalian cells.
From this study, we have found the IE2 functional domains necessary for strong CMV promoter activation by mutagenesis. By using detailed confocal microscopic imaging, we have also discovered a unique IE2 nuclear macrostructure which contains a high concentration of nuclear globular monomeric actin (G-actin) and associates with the larger foci of the activated RNA polymerase II (Pol II). Both PML and SUMO1 molecules had been found to associate with IE2 in vivo in insect and mammalian cells (29), but their function had not been previously identified. Here, we confirmed their association in Vero E6 cells and determined their effects by using RNA interference studies. The functional studies showed them likely to have a negative impact on viral gene expression in mammalian cells. Finally, we were able to confirm the role of RING ubiquitin ligase activity in promoter activation by IE2, through the addition of a proteasome inhibitor, MG132, to an in vivo expression assay.
MATERIALS AND METHODS
Baculovirus production in insect cells.
Recombinant AcMNPV was generated and propagated in Sf21 cells according to the standard protocols described by O'Reilly et al. (30). Cells were cultured at 26°C in TC100 insect medium, supplemented with 10% heat-inactivated fetal bovine serum (FBS). Virus titers were determined by quantitative PCR (25).
Plasmid and recombinant baculovirus generation.
The ie1 and ie2 genes were amplified from AcMNPV genomic DNA using the following primers: IE1F-NcoI, IE1R-XhoI, IE2F-NcoI, and IE2R-XhoI (for all primer sequences and vector maps, see Fig. S1 in the supplemental material). The coding sequences of ie1 and ie2 were inserted into the pTriEx-3 plasmid (Novagen) without the original stop codons. The resulting plasmids, pAcIE1 and pAcIE2, expressed IE1 and IE2 proteins containing His tags at their C-terminal ends.
The luciferase coding region was obtained by PCR from pTRE-luc with primers LucF and LucR (see Fig. S1 in the supplemental material) and inserted into pTriEx-3 to generate pAcL. The baculovirus enhancer sequence hr1 (homologous region 1) was PCR amplified with primer hr1F and hr1R (see Fig. S1 in the supplemental material) and cloned into pAcL to form pAhcL.
The pAcIE2, pAcL, and pAhcL plasmids were cotransfected with vAcRP23.Laz (Pharmingen), a linearized viral DNA of AcMNPV, into Sf21 cells by using Cellfectin (Life Technologies). The resulting recombinant baculoviruses, vAcIE2, vAcL, and vAhcL, were isolated through end point dilutions as described by O'Reilly et al. (30).
Six CMV promoter clones of various lengths (see Fig. 2B) were amplified from pTriEx-3 (Novagen) using primers CMV1∼6F-XhoI and CMVR-HindIII (see Fig. S1 in the supplemental material) and cloned into pGL-3 (Promega). The sequence of each clone was checked by DNA sequencing.
FIG. 2.
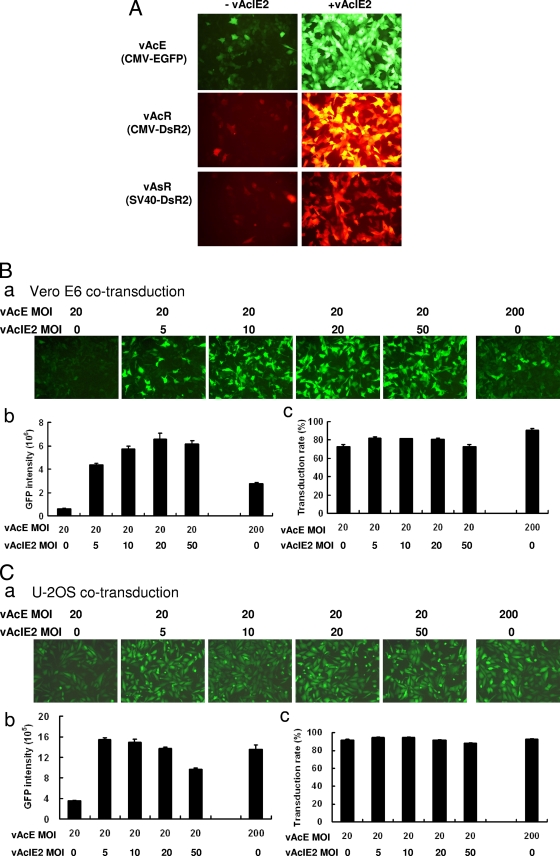
vAcIE2 can activate both CMV and SV40, with an effective dosage as low as an MOI of 5. (A) Fluorescence microscopy showed that IE2 strongly activated both CMV and SV40 promoters for gene expression. All viruses were added at an MOI of 150, and photos were taken at 43 h p.t. −, minus; +, plus. (B and C) vAcE at an MOI of 20 was added with vAcIE2 at an MOI of 0, 5, 10, 20 or 50. A vAcE control at an MOI of 200 was also included as a comparison. Significant upregulation of the CMV promoter was seen with an MOI as low as 5 for both Vero E6 (B, panel a) and U-2OS (C, panel a) cells by fluorescence microscopy. Flow cytometry analysis was used to calculate the fluorescence intensity (B and C, panels b) and transduction efficiency (B and C, panels c) of cells from each sample.
Luciferase assay.
Luciferase assays were conducted as described previously (38, 39), with modifications. Briefly, at 48 h posttransduction (p.t.), cells were washed with Dulbecco's phosphate-buffered saline (DPBS; Invitrogen) and lysed with 100 μl of cell culture lysis reagent (39). Cell lysates were centrifuged at 3,000 rpm for 30 min at 4°C before 20 μl of the supernatant was placed into the wells of a black 96-well microplate with 180 μl of luciferase assay reagent (39). Luciferase activity was measured with a luminometer (Lumat LB 9501; Berthold) by injecting 50 μl of 0.2 mM luciferin (Promega) into each well. The protein concentration of each sample was measured using the standard protocol for Coomassie protein assay reagent (Pierce). Relative luciferase units (RLU)/μg protein were obtained by dividing the measured luciferase units by the protein concentration of each sample. The RLU percentage was obtained by setting the highest RLU/μg protein value in a particular study to 100% and calculating the rest accordingly.
Transfection and transduction of mammalian cells.
Vero E6 cells were grown in minimal essential medium containing 10% FBS at 37°C with 5% CO2. U-2OS cells were grown in McCoy's medium containing 10% FBS at 37°C with 5% CO2. Transfection was performed using Lipofectamine 2000, following the protocol from the manufacturer (Invitrogen), at a total of 100 ng of DNA per 104 (Vero E6) or 8 × 103 (U-2OS) cells in 96-well plates. For transduction, cells were grown to about 50% confluence, and recombinant baculoviruses were added directly into the cell medium at various MOIs. After gentle mixing, the transduced cells were incubated at 37°C with 5% CO2 for 16 to 24 h if used for immunofluorescence (IF) experiments or for 48 h if used for the luciferase assay.
For PML and UBC9 small interfering RNA (siRNA) experiments, all siRNAs were synthesized with a T7 FastAmplify transcription kit (Epicentre) according to the manufacturer's instructions. The siRNAs used in this study were siUBC9 (5′-GGCCAGCCAUCACAAUCAAUU-3′ [sense] and 5′-UUGAUUGUGAUGGCUGGCCUC-3′ [antisense]), siPML (5′-CGTCTTTTTCGAGAGTCTGUU-3′ [sense] and 5′-CAGACTCTCGAAAAAGACGUU-3′ [antisense]), siGFP (5′-AUGAACUUCAGGGUCAGCUUG-3′ [sense] and 5′-CGGCAAGCUGACCCUGAAGUU-3′ [antisense]), and siLUC (5′-GUGCGUUGCUAGUACCAAC-3′ [sense] and 5′-GUUGGUACUAGCAACGCAC-3′ [antisense]).
Vero E6 cells (104) in 96-well plates were transfected with 50 nM of the siRNA using Lipofectamine 2000, by following the protocol from the manufacturer (Invitrogen). Baculovirus transduction (MOI = 50) was carried out 4 h after siRNA transfection.
Flow cytometry.
VeroE6 and U-2OS cells (105 per well) were seeded in 12-well culture plates and incubated for 12 h. Culture medium was removed before virus transduction and replaced with 1,000 μl of fresh medium with recombinant baculovirus vAcE or vAcIE2 at different MOIs. Cells were examined for enhanced green fluorescent protein (EGFP) expression by fluorescence microscopy at 24 h p.t. For flow cytometry, transduced cells were harvested with trypsin, washed with DPBS twice, and resuspended in 500 μl of DPBS. Cells were collected and analyzed by using an LSR II flow cytometer (Becton-Dickinson, San Jose, CA). The EGFP proteins within the cells were excited at 488 nm and detected at 525 nm. About 104 cells with three repeats were collected per specimen. The cells without virus transduction were the negative control. The transduction efficiency was obtained as a percentage: the number of EGFP-positive cells over the number of total cells collected. The level of EGFP expression was measured as the mean fluorescence intensity multiplied by the total cell number for each sample.
Vero E6 cell fractionation and Western blotting.
Vero E6 cells (106) in a T25 flask were transduced with wild-type (wt) or vAcIE2 baculovirus at an MOI of 50. After 48 h, cellular fractions were harvested according to the standard manufacturer's protocol by using a ProteoExtract subcellular proteome extraction kit (Calbiochem). The four fractions (cytosol, membrane/organelle, nucleus, and cytoskeleton) obtained were then subjected to Western blotting with an anti-His tag antibody (1:3,000; Serotec) used for IE2 detection and a mouse monoclonal anti-β-actin antibody (1:5,000; Abcam) used for β-actin detection.
IF and G-actin staining.
Eight-well Lab-Tek II chamber slides (Nunc) were seeded with Vero E6 cells (4 × 104/well), and the cells were transduced with recombinant baculoviruses (MOI = 50). At 16 to 24 h p.t., the cells were fixed with 4% paraformaldehyde, washed three times with DPBS buffer (Invitrogen), and permeabilized by incubation with −20°C 100% acetone. After three 5-min washes in DPBS, the cells on the slides were blocked with blocking buffer (10% FBS in DPBS) for 1 h and then incubated with the appropriate primary antibodies for 1 h. Antibodies used included mouse anti-His-tagged antiserum (Serotec), rabbit anti-PML antiserum (Santa Cruz), rabbit anti-SUMO1 antiserum (Santa Cruz), and rabbit anti-Pol II antiserum C21, which recognizes mainly phosphorylated Pol II and was a kind gift from C.-K. Shen (Academia Sinica). All primary antibodies were diluted 1:100 in blocking buffer. Cells were then washed three times with washing buffer (0.1% Tween 20 in DPBS [DPBST]) and incubated with 1:100 dilutions of Alexa Fluor 405 goat anti-mouse immunoglobulin G (IgG) or Alexa Fluor 555 goat anti-rabbit IgG (Invitrogen). At the same time, G-actin was specifically stained with Alexa Fluor 488-conjugated DNase I (1:500; Invitrogen). After 1 h, cells were washed three times in washing buffer and sealed with aqueous mounting medium (HIS002B; Serotec). Fluorescent images were visualized with a Zeiss laser confocal microscope (LSM510).
RNA FISH.
The gene sequence of ie2 was cloned into pGEM-T Easy vector (Promega), linearized by restriction digests at NcoI or PstI sites, and transcribed in vitro using either SP6 or T7 RNA Pol. In this way, we generated both antisense and sense strains of ie2 transcripts. The transcripts were labeled with Alexa Fluor 594 dye, purified, and used for RNA hybridization as described in the fluorescence in situ hybridization (FISH) Tag RNA kit (Invitrogen) with slight modifications. Briefly, RNA FISH was performed with IF slides after the IF and G-actin staining. After the last washing step, slides were refixed with 4% paraformaldehyde and washed three times with DPBS. Hybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 100 μg/ml fragmented salmon DNA, 50 μg/ml heparin, and 0.1% Tween 20) was added to the slide, which was heated at 55°C for 5 min. Probes were diluted in hybridization buffer (∼1 ng/μl) and denatured by heating at 80°C for 2 min before cooling on ice. Denatured probes were added to the slide and incubated overnight at 37°C. The next day, the probes were removed, and samples were then washed three times with hybridization buffer at 42°C. The washing steps were followed by two 5-min washes with 50% DPBST/50% hybridization buffer at room temperature. Samples were then washed three times with DPBS before being sealed with SlowFade Gold antifade reagent (Invitrogen). Images were obtained with a Zeiss laser confocal microscope (LSM510).
IE2 mutagenesis study.
PCR site-directed point mutagenesis was performed to obtain both the IE2C230S (cysteine 230-to-serine) RING domain mutant and the IE2SUMO (KKSE-to-RRSE) SUMO site mutant. For IE2C230S, two DNA fragments were PCR amplified from the ie2 gene, using primer pairs IE2F-NcoI/CS-R and CS-F/IE2R-XhoI. For IE2SUMO, primer pairs IE2F-NcoI/SUMO-R and SUMO-F/IE2R-XhoI were used (for all primer sequences, see Fig. S1 in the supplemental material). The two fragments for each IE2 mutant clone were annealed through bridging PCR. The resulting templates were reamplified with IE2F-NcoI and IE2R-XhoI. The IE2 mutant fragments were then cloned into TriEx-3 vector between the NcoI and XhoI sites. For the coiled-coil domain deletion clone, PCR fragments were amplified using IE2F-NcoI and CdelR-XhoI (see Fig. S1 in the supplemental material) and cloned into the TriEx-3 vector. All three mutants contained His tags at their C termini for identification.
MG132 drug assay.
Ninety-six-well plates were seeded with Vero E6 cells (104), and the cells were transduced with vAcL and either wt or vAcIE2 baculovirus at an MOI of 50. After overnight incubation, fresh medium containing 5 μm of MG132 (Sigma) was used to replace the virus-containing medium. The cells were incubated for a further 48 h before the luciferase assay.
RESULTS
IE2 is a strong activator of the CMV promoter in mammalian cells.
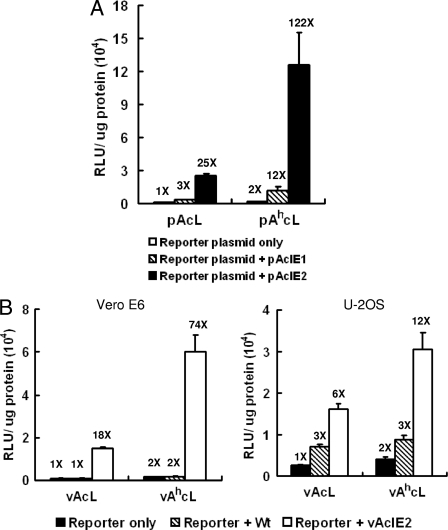
We have previously found that IE1 and IE2 can activate some early baculovirus promoters within Vero E6 cells (24). To test whether they also activate the mammalian CMV promoter, we cotransfected IE1- or IE2-expressing plasmids with the reporter plasmid pAcL (where CMV drives the luciferase gene) or pAhcL (where CMV drives luciferase with an additional hr1 baculovirus enhancer) (26). We found that IE2 dramatically improved the CMV promoter-driven luciferase activity, which increased 122-fold in the presence of the hr1 sequence and over 25-fold without the hr1 sequence (Fig. 1A). The IE1-expressing plasmid, on the other hand, improved luciferase activity only 3-fold without the hr1 sequence and 12-fold with the hr1 sequence (Fig. 1A). The cotransfection assays suggested that IE1 or IE2 by itself was the only viral component needed for CMV promoter activation in Vero E6 cells.
FIG. 1.
IE2 is a strong activator of the CMV promoter in mammalian cells. (A) Plasmids expressing IE1 (pAcIE1) or IE2 (pAcIE2) (70 ng/103 cells) were cotransfected with reporter plasmids pAcL or pAhcL (30 ng/103 cells). Luciferase assays performed at 48 h p.t. showed a dramatic CMV promoter activation by IE2. Luciferase activity was activated to 25 times (X) the basal level with IE2 alone or 122 times the basal level when hr enhancer was also present. (B) Luciferase reporter viruses vAcL and vAhcL (MOI = 20) were cotransduced with IE2-expressing recombinant baculovirus vAcIE2 (MOI of 20 for VeroE6; MOI of 10 for U-2OS). A luciferase assay was performed at 48 h p.t., and the CMV promoters were activated up to 74 times (Vero E6) or 12 times (U-2OS) the basal level by vAcIE2.
We further tested the ability of IE2 to activate the CMV promoter in the form of the recombinant baculovirus vAcIE2, where a copy of the ie2 gene was placed under the CMV promoter. Cotransduction of the vAcIE2 virus with two reporter viruses, vAcL and vAhcL, showed that IE2 was a very strong activator of the CMV promoter in Vero E6 cells, capable of upregulating CMV promoter activity 74-fold with (or 18-fold without) the hr1 sequence (Fig. 1B). In U-2OS cells, baculovirus generally has a better transduction rate, resulting in higher basal reporter expression, but IE2 was still able to enhance the CMV promoter-driven luciferase expression 12-fold with (or 6-fold without) the hr1 sequence (Fig. 1B). We also noticed that the addition of wt baculovirus also improved luciferase activity in U-2OS cells. It is possible that the wt baculovirus was able to express a small amount of IE2 from its original promoter in U-2OS cells, and this finding might explain the higher recombinant protein expression level observed for these cells during baculovirus gene transfer.
To test whether IE2 activation of a heterologous viral promoter was limited to the CMV promoter, we cotransduced vAcIE2 with three different reporter viruses, vAcR (for which CMV drives DsR2), vAsR (for which SV40 drives DsR2), and vAcE (for which CMV drives EGFP) (for schematics of the constructs, see Fig. S1 in the supplemental material). The addition of vAcIE2 virus led to much-higher intensities of green or red fluorescence from the expressed EGFP and DsR2 proteins (Fig. 2A). We also found that an MOI as low as 5 was sufficient for significant CMV promoter activation by vAcIE2 (Fig. 2B and C) in Vero E6 and U-2OS cells. EGFP fluorescence intensities of cells cotransduced with vAcE at an MOI of 20 plus vAcIE2 at an MOI of 5 were better than (Vero E6) or comparable to (U-2OS) those of cells transduced with vAcE only at an MOI of 200 (Fig. 2B, panels a and b, and Fig. 2C, panels a and b). This result was due not to improved transduction efficiencies, as the transduction rates of EGFP-expressing cells did not vary much between each sample (Fig. 2B and C, panels c) but, rather, to improved EGFP expression levels in the vAcE/vAcIE2-transduced cells.
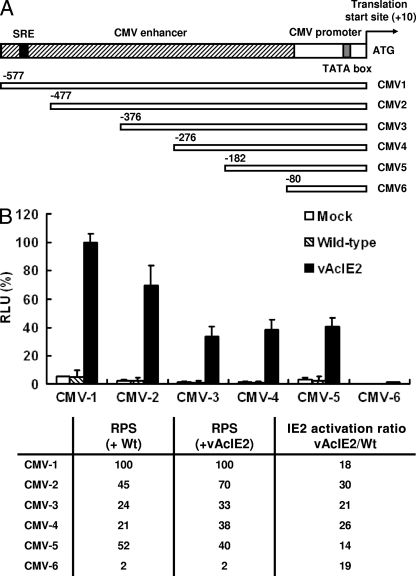
It is clear that not only the CMV promoter but also the SV40 promoter showed a remarkable degree of upregulation by IE2. We theorized that IE2 could either recognize sequences in both CMV and SV40 promoters or act through mechanisms which do not require promoter binding via a specific DNA sequence. To determine whether such a specific sequence exists on the CMV promoter, upstream progressive deletions of the CMV promoter linked to the luciferase gene were coexpressed with IE2 (Fig. 3A). The percent promoter strength of individual deleted CMV (CMV-2 to CMV-6) promoters relative to that of the full-length CMV-1 promoter was calculated as the relative promoter strength (RPS). We found two regions (from −182 to −80 bp and from −577 to −376 bp) of the CMV promoter which are important for CMV promoter strength, as represented by the drops in RPS when these two regions were deleted (Fig. 3B). However, activation ratios between vAcIE2- and wt-transduced cells remained similar throughout all six CMV-Luc clones. For example, the expression levels of the shortest promoter, CMV-6, were very weak in both cells transduced by vAcIE2 and wt viruses, but the activation of vAcIE2 versus that of the wt was still 19-fold (Fig. 3B). This was very similar to that by IE2 under the CMV-1 full-length promoter (18-fold). These data suggested that IE2 either acts on the basal −80 promoter region, which was the common denominator between all six CMV-Luc clones, or functions in a posttranscriptional manner. As IE2 also can activate the SV40 promoter, it most likely acts in a non-sequence-specific way, through interactions with yet-unknown host mechanisms.
FIG. 3.
Interaction of IE2 with the CMV promoter. (A) A series of deletion fragments of the CMV promoter were cloned into the pGL-3 luciferase reporter vector. SRE, serum response element. (B) The resulting luciferase expressing vectors were transfected into Vero E6 cells, followed by mock, wt, or vAcIE2 transduction at an MOI of 50. The graph shows the relative luciferase activity from each sample, with CMV-1 set as 100% activity. We calculated the RPS of each promoter for each deletion clone and the ability of IE2 to activate each clone as IE2 activation ratios, calculated as the luciferase level with vAcIE2 transduction/luciferase level with wt transduction.
IE2 associates with monomeric G-actin within the nucleus.
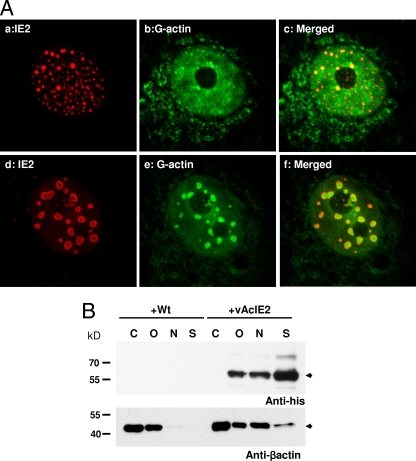
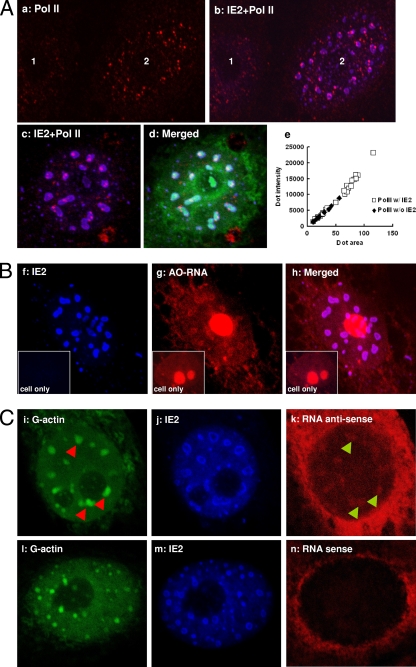
A recent paper has demonstrated a role for nuclear actin in transcription control (37). Nuclear actin forms part of preinitiation complexes (PICs) and is necessary for RNA Pol II-mediated transcription (15). Here, we examined actin distribution within the IE2-expressing cells. IF staining combined with confocal microscopy showed that IE2 nuclear foci appeared as numerous tiny dots during early transduction. These nuclear foci decreased in number but increased in volume, eventually developing into large NBs with a hollow center (Fig. 4A). When stained with DNase I conjugate, which binds monomeric G-actin with an affinity of about 5 × 10−8 M, we discovered that G-actin associated very strongly with IE2 nuclear structures and, more surprisingly, appeared to be trapped within these novel hollow-centered NBs formed by IE2 at later time points (Fig. 4A, panel f). The sizes of the IE2 NBs seemed to correspond to the amounts of IE2 protein expressed. A higher MOI produces larger NBs. We also stained vAcIE2-transduced cells with phalloidin; however, no significant change in F-actin distribution was observed (data not shown).
FIG. 4.
IE2 foci interact with G-actin within Vero E6 cell nuclei. (A) Panels a to c show numerous smaller IE2 foci as stained by IF at 16 h p.t. (IE2, red; G-actin, green). Panels d to f show vAcIE2-transduced cells at 20 h p.t. The larger IE2 NBs appeared to contain G-actin (green). (B) Actin redistribution in vAcIE2-transduced cells at 48 h p.t. Cells were fractionated into cytosolic (C), organellar (O), nuclear (N), and cytoskeletal (S) fractions. Western blots detected His-tagged IE2 in noncytosolic fractions and a significant β-actin shift into the nuclear fractions in vAcIE2-transduced cells.
To confirm the observation that high concentrations of G-actin were located within the IE2 NBs, Vero E6 cells transduced by either wt AcMNPV or vAcIE2 were harvested at 48 h p.t. and fractionated into cytosolic, organellar, nuclear, and cytoskeleton matrix fractions. Each fraction was then probed for the presence of His-tagged IE2 and β-actin (which includes both G-actin and F-actin forms) by Western blotting (Fig. 4B). Significantly, a strong β-actin band was present in the nuclear fraction of vAcIE2-transduced cells, while only a weak band was detected in the same fraction from wt-transduced cells. This result correlated well with the confocal study and showed that the expression of IE2 resulted in β-actin migration into the cell nucleus and subsequent association with the IE2 nuclear structures. The presence of IE2 in the cytoskeleton matrix fraction could indicate either that a high concentration of detergent was necessary to dissolve the compact IE2 nuclear macrostructure or that there was a strong association between IE2 and the intermediate filaments of cells.
IE2 NBs are sites of active transcription.
The cytoplasmic terminal domain of Pol II is phosphorylated during active transcription (22). We used antibody against this specific area in IF microscopy to determine whether these IE2 NBs are active transcription sites. Indeed, activated Pol II was strongly associated with, or more likely embedded into, both the smaller IE2 foci and the larger IE2 NBs (Fig. 5A, panels b and c). Interestingly, Pol II foci associated with IE2 appeared to be significantly larger than ones not associated with IE2, and the majority of the larger Pol II dots appeared to be in contact with the IE2 NBs (Fig. 5A, panels b and c). When Fig. 5A, panel a, was analyzed with MetaMorph software (Molecular Devices), the result showed that all the larger Pol II dots were in contact with IE2 NBs (Fig. 5A, panel e).
FIG. 5.
IE2 NBs are active transcription centers. (A) Panels a and b, untransduced (1) or vAcIE2-transduced (2) Vero E6 cells were examined by IF at 16 h p.t. Only nuclei are visible in these photos. Transduced cells showed a greater number of enlarged activated Pol II dots (red), which were associated with IE2 NBs. Panels c and d, an enlarged nucleus showing that activated Pol II, like G-actin (green), locates within the IE2 NBs (blue). IF performed at 20 h p.t. Panel e, a MetaMorph analysis showed that all the larger Pol II dots associate with (w/) IE2, while the Pol II without (w/o) IE2 dots tend to be smaller. (B) vAcIE2-transduced cell nucleus showing that the transcribed RNA, which emits red fluorescence with acridine orange staining (AO-RNA), colocalized with the IE2 NBs. An untransduced cell is shown in each inset as a control. (C) RNA FISH with antisense (i to k) and sense (l to n) probes (red) against the CMV promoter-driven ie2 gene. The antisense probe hybridized with abundant RNA transcripts within IE2 NBs (blue) and colocalized with G-actin (green). The sense probe acted as the negative control. Arrowheads were used to show the relative positions of IE2 NBs and RNA clusters.
Acridine orange is a nucleic acid cationic dye which emits red fluorescence when associated with RNA. IE2-expressing cells fixed and stained with this compound showed RNA associated with the inner ring of the IE2 NBs (Fig. 5B). An RNA FISH probe specific for the IE2 transcript (IE2 antisense probe), which was driven by the CMV promoter, also detected RNA transcripts associated with the IE2 NBs (Fig. 5C, panel k), while there was no such association for the sense control probe (Fig. 5C, panel n). The fluorescence of the RNA probes at the RNA-rich nuclear rim was probably due to nonspecific binding of the RNA probes with cellular RNAs.
Together, these results showed that the IE2 NBs were sites of viral gene transcription; moreover, nascent viral RNAs were likely produced within these IE2 NBs, juxtaposed to the recruited monomeric G-actin and Pol II.
Both the RING domain and the coiled-coil domain are essential for IE2 activation of the CMV promoter.
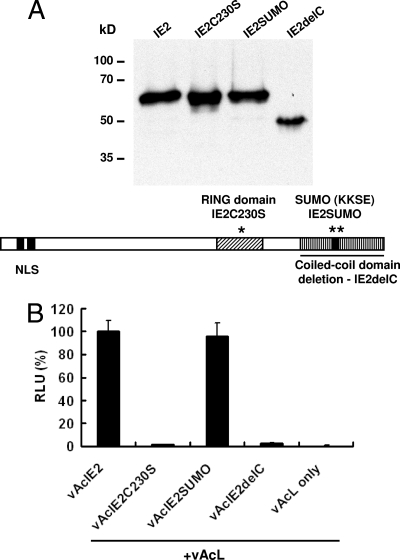
To study if the structure of IE2 NB has some functional significance regarding viral promoter activation in mammalian cells, we constructed three mutants of IE2 by PCR mutagenesis (Fig. 6A). Recombinant baculovirus vAcIE2C230S expressed an IE2 mutant (IE2C230S) with the zinc-chelating cysteine 230 replaced by serine. The vAcIE2delC virus expressed a mutant (IE2delC) with the coiled-coil domain deletion, resulting in a mutant IE2 without its oligomerization domain. Last, the mutant virus vAcIE2SUMO expressed an IE2SUMO mutant which contained arginine substitutions of lysine residues at position 360 and 361, thus removing the predicted SUMOylation site. We aimed to test whether the ability to form the distinctive NB structure and associate with G-actin was connected to the promoter activation function of IE2 in mammalian cells.
FIG. 6.
IE2 mutagenesis study. (A) Three mutant IE2 proteins, IE2C230S, IE2SUMO and IE2delC, with mutations (IE2C230S and IE2SUMO) or a deletion (IE2delC) within the RING, SUMO, and coiled-coil domains, respectively, were expressed by recombinant baculoviruses. The Western blot shows that all three mutant proteins were produced at similar levels and expected sizes. KKSE, lys-lys-ser-glu. (B) IE2 mutant viruses were cotransduced with vAcL and tested by luciferase assays. vAcIE2SUMO showed CMV promoter activation levels similar to those of vAcIE2, while both vAcIE2C230S and vAcIE2delC lost their activity.
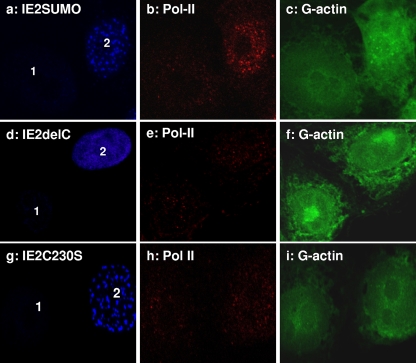
Luciferase assays showed that both the RING and the coiled-coil domain IE2 mutants lost their CMV promoter activation ability, while the IE2SUMO mutant remained as effective as wild-type IE2 (Fig. 6B). Interestingly, IF microscopy showed that the IE2SUMO mutant retained its NB structure and was capable of elevating the amount of activated Pol II (Fig. 7, panel b) and associating with G-actin foci in the transduced cell (Fig. 7, panel c, cell 2).
FIG. 7.
The distribution of IE2 mutants, Pol II, and G-actin in the nucleus. The behavior of IE2SUMO (a to c) appears to be very similar to that of wt IE2, forming NBs which associate with Pol II and G-actin. IE2delC is expressed as a diffuse pattern within the nucleus (d to f). IE2C230S formed irregular nuclear foci (g to i), which were weakly associated with G-actin. There was no obvious difference in activated Pol II levels between untransduced (1) and transduced (2) cell nuclei for IE2delC and IE2C230S mutants.
Unlike IE2SUMO, the IE2delC protein failed to activate the CMV promoter and form distinct IE2 NBs, confirming the essential role of IE2 oligomerization in both situations (Fig. 6B; Fig. 7, panels d to f). But the most interesting observations came from experiments with the IE2C230S RING domain mutant. In a previous study, a similar mutant from BmNPV was shown to form abnormally large foci compared to those seen with wt IE2 (19). Here, we also observed focus formation for the AcMNPV IE2C230S mutant, but we observed no NB formation (Fig. 7, panel g; Fig. 8B, panels i and m). Although the IE2C2305 nuclear focus still weakly associated with G-actin, it no longer formed a shell-like structure around it, indicating that a functional RING domain was essential for the observed structure of G-actin-centered IE2 NBs within the nucleus. More importantly, no significant addition of enlarged activated Pol II foci was observed with these defective IE2 structures in the transduced cells (Fig. 7, panel h, cell 2) versus observations for the untransduced cell (Fig. 7, panel h, cell 1). This correlates well with the result from Fig. 6B and showed that the IE2C230S mutant lost its ability to activate the CMV promoter.
FIG. 8.
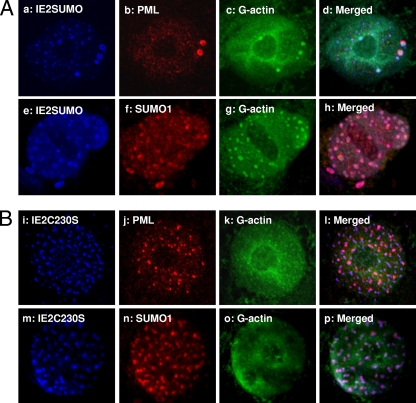
IE2C230S and IE2SUMO both associate with PML and SUMO1. (A) PML (a to d) and SUMO1 (e to h) associate with the IE2SUMO mutant. (B) The IE2C230S mutant lacking well-defined NB structures also colocalizes with PML and SUMO1, even though it has lost its activation activity.
Together, these results suggested that the formation of IE2 NBs requires both the oligomerization and RING domains of IE2. It is possible that the ability to form G-actin-containing NBs is linked to the function of IE2 as a trans-activator, as both IE2C230S and IE2Cdel mutants failed to form NBs and also lost their promoter-activating function.
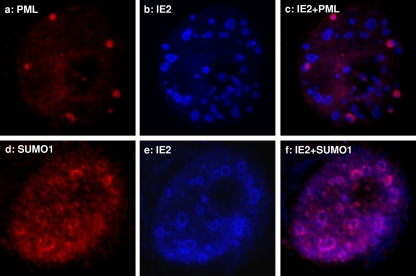
IE2 NBs associate with PML and SUMO1.
Upon infection, the DNA replication centers of mammalian DNA viruses, such as human herpesvirus 1 (HSV-1), SV40, and adenovirus, are frequently observed to lie adjacent to the PML NBs (28). Some of these viruses also express viral proteins which disrupt PML NBs, likely to avoid interferon-induced antiviral cellular defenses (6). A previous study using insect TN-368 cells showed a close association between IE2 foci, PML, and SUMO1 (27). Here, using mammalian cells, we also observed that IE2 NBs were in close contact with SUMO1 and PML NBs (Fig. 9, panels c and f). Both appeared to partially cover, or associate with, the outer edges of the IE2 NBs. It is possible that the SUMO1 observed was actually SUMOylated PML, but we could not rule out the possibility that other SUMOylated factors may bind to the IE2 NBs or that IE2 itself may be SUMOylated.
FIG. 9.
Association of PML and SUMO1 with IE2. IF studies with PML (a to c) and SUMO1 (d to f) antibodies. The condensed, larger PML NBs appear to associate with the outer boundary of IE2 NBs, either fully or partially covering them. SUMO1 appears as discontinuous beads around the IE2 NBs.
From the mutagenesis study, we found that all mutant IE2s capable of forming nuclear foci (IE2SUMO and IE2C230S) appeared to associate closely with SUMO1 and PML NBs (Fig. 8), regardless of whether they retained their functions as trans-activators of the CMV promoter. The IE2delC mutant didn't appear to induce PML or SUMO1 focus formation, as it was expressed in a diffuse pattern inside the nucleus (see Fig. S2 in the supplemental material). PML has been associated with interferon-mediated antiviral activity (8) and is viewed as part of the cellular defense machinery against viral infection. It is also a repressor of translation initiation through the binding of its RING domain to eIF4E, resulting in the inhibition of nucleocytoplasmic mRNA transport (5). Upon baculovirus transduction, the size of PML NBs significantly increased in HepG2 cells (23). However, no cytopathic effect has been observed, even when very high virus titers, up to an MOI of 1,000, were used in the mammalian cell transductions (14). There is as yet no defined function for PML NBs regarding baculovirus transduction.
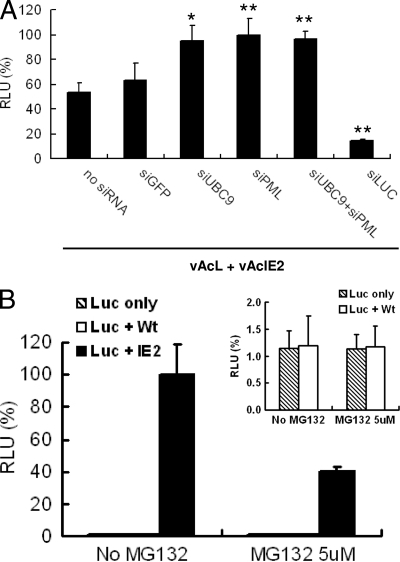
PML contains three SUMOylation sites and one noncovalent binding motif, which are essential for the formation of PML NBs (33). To answer the question concerning roles of PML- and SUMO1-containing factors in IE2 activation of the CMV promoters, we added siRNAs specific for PML and UBC9 (the E2 enzyme involved in SUMOylation) to the luciferase functional assay. Figure 10A shows that both siPML and siUBC9 improved the luciferase activity of the reporter virus vAcL. Also, there was no additive effect from adding both siRNAs, suggesting that it is mainly PML NBs which direct the host antiviral defense against viral promoter activation, as the more general inhibition of cellular SUMOylation mechanism caused by siUBC9 had no additional effect. The effect was specific for IE2 activation of the CMV promoter, as both siRNAs had no effect on CMV promoter activity in the absence of vAcIE2 (see Fig. S3 in the supplemental material). Thus, we show here PML NB most likely plays a negative role in baculovirus-mediated gene transcription in mammalian cells.
FIG. 10.
IE2 activation of the CMV promoter is enhanced by siPML and siUBC9 but inhibited by MG132. (A) siRNAs against pml and sumo1 genes were transfected into Vero E6 cells, followed by vAcIE2+vAcL cotransduction. A luciferase assay detected enhanced effects from both siRNAs on IE2 trans-activation of the CMV promoter. siGFP (against egfp) was used as a negative control, while siLUC (against luciferase) was a positive control for siRNA efficiency. Data are presented as percentages of RLU in the absence of siRNA (n = 4). * and **, P < 0.05 and P < 0.001, respectively, compared with the no-siRNA sample (t test). (B) MG132, which blocks the proteasome degradation pathway, had a negative effect on IE2 trans-activation. This result suggested that the ubiquitin ligase activity of IE2 RING domain plays an active role in CMV promoter activation. MG132 did not affect luciferase expression in the control groups (inset).
The ubiquitin-proteasome pathway is involved in IE2 trans-activator activity.
The BmNPV IE2 RING domain has been shown in an in vitro study to have ubiquitin ligase activity (18). It is likely that the AcMNPV IE2 RING protein also has the same function. Here, we asked whether the ubiquitin-proteasome pathway plays a role in IE2 promoter activation. The vAcIE2-transduced cells were incubated with medium containing 5 μM of the proteasome inhibitor MG132. As shown in Fig. 10B, the addition of MG132 had a detrimental effect on IE2 activation of the CMV promoter, prompting a 60% drop in reporter luciferase activity. Conversely, MG132 had no effect on the luciferase virus alone or the luciferase virus plus WT virus control groups (Fig. 10B, top right panel). These results suggested that the proteasome degradation pathway plays a positive role in CMV promoter activation by IE2 and that the ubiquitin ligase activity of IE2 RING domain is likely to be functional in mammalian cells. Moreover, the mechanism of CMV promoter activation by IE2 probably involved the proteasome degradation pathway through its RING domain ubiquitin ligase activity.
DISCUSSION
In this study, we have discovered a baculovirus RING protein, IE2, which forms NBs and acts as an extremely strong transcriptional activator in mammalian cells. Although the formation of IE2 nuclear structures has been observed before in both insect and mammalian systems (23, 29), no clear function has been assigned to these structures. IE2 protein is thought to enhance DNA replication during the baculovirus infection cycle (12), and IE2 nuclear structures have been observed to associate with either viral DNA replication sites or PML/SUMO1 in insect TN-368 cells (27). We found that in mammalian Vero E6 cells, overexpressed IE2 also forms distinct NBs, and detailed confocal studies enabled us to identify a unique hollow-centered structure which contains G-actin and recruits active RNA Pol II for the strong activation of viral gene promoters.
We have shown that the gene activation function of IE2 is closely correlated with the function of its RING domain. A single point mutation in this domain abolished its ability to activate the CMV promoter, as well as the formation of the IE2 NB structure. Although the IE2C230S mutant still associates with G-actin weakly, the lack of proper IE2 NB structure due to a defective RING domain may very well contribute to its loss of function. Many mammalian viruses also have early genes which contain a RING domain-like structure and form NBs. For example, the ICP0 of HSV-1 is a RING finger ubiquitin ligase which also activates the expression of a variety of HSV-1 genes (7). Both IE2 and the HSV-1 ICP0 are coded by immediate-early genes (12, 36). ICP0 is most famous for its ability to disrupt SUMO1-modified PML NBs in vivo, and its RING domain is essential for this activity (9). Interestingly, we also observed a close association, but not a colocalization, between PML NBs and IE2 NBs, and PML NBs did not seem to distinguish between functional IE2 and the nonfunctional IE2C230S mutant. From this observation as well as the positive effect of PML siRNA on IE2 activity, we suggest that PML NBs may play an antiviral defense role during baculovirus transduction and would attach to foreign viral structures regardless of its functionality. This view correlates well with a recent paper which showed that baculovirus transduction causes an increase in PML NB size in HepG2 cells (23). It is a puzzle why such an increase in PML NB size, which usually results from cellular stress and the virus-induced interferon response, did not lead to an observable cytopathic effect. One possible explanation is that the ubiquitin ligase activity of IE2 may in fact be functional and target PML in the nucleus, causing the breakdown of PML through the ubiquitin degradation pathway. This mechanism may moderate the cellular defense effect of PML NBs and result in the low cytopathic rate observed in baculovirus transduction studies. The MG132 experiment seemed to support this view, as it showed that the protein degradation pathway played a role in the function of IE2. Further research will be necessary to determine if PML is indeed the target of IE2 or if other cellular antiviral factors are also involved.
In our studies, we found that the IE2 NB structure acted as a transcriptionally active compartment within the nucleus, containing the PIC component G-actin, activated Pol II, and nascent RNA transcripts. The high concentration of nuclear G-actin within these NBs could also explain the previous observation that baculovirus transduction alters host cell chromatin distribution (23). The role of nuclear actin in transcription, mRNA processing, and chromatin remodeling is a field of intense debate and recent study (1, 3). Although it is known that nuclear actin forms part of the PIC with Pol II (15), it was unclear whether the polymeric or the monomeric actin was involved in the transcription process. Also, although there is immunostaining evidence that monomeric actin was associated with the chromatin-remodeling complex (32), no direct in vivo evidence has been found to prove that monomeric actin contributes to chromatin remodeling. Here, we showed a connection between IE2 protein-induced transcription activation and monomeric G-actin redistribution, and this connection could suggest a role for monomeric actin in cellular transcription machinery. Also, we would like to suggest that the observed baculovirus transduction-induced chromatin reorganization by Laakkonen et al. (23) could have been a direct result of G-actin deprivation from the nucleus due to IE2 NB formation, as IE2 translation was detected in HepG2 cells in their study.
The formation of IE2 NB could also have functioned as a defense against the antiviral function of PML NBs and other SUMOylated transcriptional repressors. This idea is supported by both the RNA interference and the MG132 experiments, which showed a negative role of PML and a positive effect of IE2 RING domain ubiquitin ligase activity, respectively. MG132 blocking of the proteasome degradation pathway may have prevented IE2-mediated degradation of host antiviral factors, with PML NB proteins as possible targets.
In conclusion, we have described a novel G-actin-associated nuclear domain, the hollow-centered IE2 NB, which dramatically improves the desired protein yield of baculovirus-mediated gene transfer in mammalian cells. We have elucidated the functional domains of IE2 necessary for this activation, namely the RING and the coiled-coil domains. A similar dependency on RING domain-assembled supramolecules for biochemical activity has been reported for many RING proteins, e.g., PML, Z, and BRCA1/BARD1 (21). However, we believe this is the first report that a RING transcriptional factor actively associates with a high concentration of nuclear G-actin within its assembled structure, and this compartmentalization of nuclear space may represent a novel mechanism for strong transcriptional activation. As many mammalian viral proteins are known to form nuclear substructures, it would be a major advance in our understanding of gene expression if a link between these NB-forming transcriptional factors, nuclear G-actin, and transcriptional regulation could be established.
Supplementary Material
Acknowledgments
We thank James Shen and Hsiu-Ming Shih for valuable discussions and suggestions; Andrew Tiffany, Miranda Loney, and Harry Wilson for revisions; and the Academia Sinica IMB Imaging Core for help with the confocal imaging.
This research was funded by grants NSC95-2313-B-005-063-MY3 from the National Science Council and 94S-1303 from the Academia Sinica.
Footnotes
Published ahead of print on 4 February 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bettinger, B. T., D. M. Gilbert, and D. C. Amberg. 2004. Actin up in the nucleus. Nat. Rev. Mol. Cell Biol. 5410-415. [DOI] [PubMed] [Google Scholar]
- 2.Boyce, F. M., and N. L. Bucher. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA 932348-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, M., and X. Shen. 2007. Nuclear actin and actin-related proteins in chromatin dynamics. Curr. Opin. Cell Biol. 19326-330. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, T., C. Y. Xu, Y. B. Wang, M. Chen, T. Wu, J. Zhang, and N. S. Xia. 2004. A rapid and efficient method to express target genes in mammalian cells by baculovirus. World J. Gastroenterol. 101612-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, N., M. Sharma, A. Kentsis, J. M. Perez, S. Strudwick, and K. L. Borden. 2001. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 204547-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 207266-7273. [DOI] [PubMed] [Google Scholar]
- 7.Everett, R. D. 1987. The regulation of transcription of viral and cellular genes by herpesvirus immediate-early gene products (review). Anticancer Res. 7589-604. [PubMed] [Google Scholar]
- 8.Everett, R. D., and M. K. Chelbi-Alix. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89819-830. [DOI] [PubMed] [Google Scholar]
- 9.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 135062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, R., T. Matsuyama, J. Yamagishi, K. Sahara, S. Asano, and H. Bando. 2006. Expression of Autographa californica multiple nucleopolyhedrovirus genes in mammalian cells and upregulation of the host β-actin gene. J. Virol. 802390-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, H., Y. Wang, N. Li, W.-P. Peng, Y. Sun, G.-Z. Tong, and H.-J. Qiu. 2007. Efficient gene delivery into mammalian cells mediated by a recombinant baculovirus containing a whispovirus ie1 promoter, a novel shuttle promoter between insect cells and mammalian cells. J. Biotechnol. 131138-143. [DOI] [PubMed] [Google Scholar]
- 12.Gomi, S., C. E. Zhou, W. Yih, K. Majima, and S. Maeda. 1997. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology 23035-47. [DOI] [PubMed] [Google Scholar]
- 13.Guarino, L. A., M. A. Gonzalez, and M. D. Summers. 1986. Complete sequence and enhancer function of the homologous DNA regions of Autographa californica nuclear polyhedrosis virus. J. Virol. 60224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, C., V. Sandig, G. Jennings, M. Rudolph, P. Schlag, and M. Strauss. 1995. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc. Natl. Acad. Sci. USA 9210099-10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann, W. A., L. Stojiljkovic, B. Fuchsova, G. M. Vargas, E. Mavrommatis, V. Philimonenko, K. Kysela, J. A. Goodrich, J. L. Lessard, T. J. Hope, P. Hozak, and P. de Lanerolle. 2004. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 61094-1101. [DOI] [PubMed] [Google Scholar]
- 16.Hu, Y. C. 2006. Baculovirus vectors for gene therapy. Adv. Virus Res. 68287-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huser, A., and C. Hofmann. 2003. Baculovirus vectors: novel mammalian cell gene-delivery vehicles and their applications. Am. J. Pharmacogenomics 353-63. [PubMed] [Google Scholar]
- 18.Imai, N., N. Matsuda, K. Tanaka, A. Nakano, S. Matsumoto, and W. Kang. 2003. Ubiquitin ligase activities of Bombyx mori nucleopolyhedrovirus RING finger proteins. J. Virol. 77923-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai, N., S. Matsumoto, and W. Kang. 2005. Formation of Bombyx mori nucleopolyhedrovirus IE2 nuclear foci is regulated by the functional domains for oligomerization and ubiquitin ligase activity. J. Gen. Virol. 86637-644. [DOI] [PubMed] [Google Scholar]
- 20.Kenoutis, C., R. C. Efrose, L. Swevers, A. A. Lavdas, M. Gaitanou, R. Matsas, and K. Iatrou. 2006. Baculovirus-mediated gene delivery into mammalian cells does not alter their transcriptional and differentiating potential but is accompanied by early viral gene expression. J. Virol. 804135-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kentsis, A., R. E. Gordon, and K. L. Borden. 2002. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl. Acad. Sci. USA 9915404-15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 142452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laakkonen, J. P., M. U. Kaikkonen, P. H. Ronkainen, T. O. Ihalainen, E. A. Niskanen, M. Hakkinen, M. Salminen, M. S. Kulomaa, S. Yla-Herttuala, K. J. Airenne, and M. Vihinen-Ranta. 2008. Baculovirus-mediated immediate-early gene expression and nuclear reorganization in human cells. Cell. Microbiol. 10667-681. [DOI] [PubMed] [Google Scholar]
- 24.Liu, C. Y., C. H. Wang, J. C. Wang, and Y. C. Chao. 2007. Stimulation of baculovirus transcriptome expression in mammalian cells by baculoviral transcriptional activators. J. Gen. Virol. 882176-2184. [DOI] [PubMed] [Google Scholar]
- 25.Lo, H. R., and Y. C. Chao. 2004. Rapid titer determination of baculovirus by quantitative real-time polymerase chain reaction. Biotechnol. Prog. 20354-360. [DOI] [PubMed] [Google Scholar]
- 26.Lo, H. R., C. C. Chou, T. Y. Wu, J. P. Yuen, and Y. C. Chao. 2002. Novel baculovirus DNA elements strongly stimulate activities of exogenous and endogenous promoters. J. Biol. Chem. 2775256-5264. [DOI] [PubMed] [Google Scholar]
- 27.Mainz, D., I. Quadt, and D. Knebel-Morsdorf. 2002. Nuclear IE2 structures are related to viral DNA replication sites during baculovirus infection. J. Virol. 765198-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20660-667. [DOI] [PubMed] [Google Scholar]
- 29.Murges, D., I. Quadt, J. Schroer, and D. Knebel-Morsdorf. 2001. Dynamic nuclear localization of the baculovirus proteins IE2 and PE38 during the infection cycle: the promyelocytic leukemia protein colocalizes with IE2. Exp. Cell Res. 264219-232. [DOI] [PubMed] [Google Scholar]
- 30.O'Reilly, D. R., L. K. Miller, and Luckow, V. A. 1994. Baculovirus expression vectors: a laboratory manual. Oxford University Press, New York, NY.
- 31.Prikhod'ko, E. A., and L. K. Miller. 1998. Role of baculovirus IE2 and its RING finger in cell cycle arrest. J. Virol. 72684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoenenberger, C. A., S. Buchmeier, M. Boerries, R. Sutterlin, U. Aebi, and B. M. Jockusch. 2005. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J. Struct. Biol. 152157-168. [DOI] [PubMed] [Google Scholar]
- 33.Shen, T. H., H. K. Lin, P. P. Scaglioni, T. M. Yung, and P. P. Pandolfi. 2006. The mechanisms of PML-nuclear body formation. Mol. Cell 24331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shippam-Brett, C. E., L. G. Willis, and D. A. Theilmann. 2001. Analysis of sequences involved in IE2 transactivation of a baculovirus immediate-early gene promoter and identification of a new regulatory motif. Virus Res. 7513-28. [DOI] [PubMed] [Google Scholar]
- 35.Spenger, A., W. Ernst, J. P. Condreay, T. A. Kost, and R. Grabherr. 2004. Influence of promoter choice and trichostatin A treatment on expression of baculovirus delivered genes in mammalian cells. Protein Expr. Purif. 3817-23. [DOI] [PubMed] [Google Scholar]
- 36.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 672571-2585. [DOI] [PubMed] [Google Scholar]
- 37.Vartiainen, M. K., S. Guettler, B. Larijani, and R. Treisman. 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 3161749-1752. [DOI] [PubMed] [Google Scholar]
- 38.Wu, C. P., J.-Y. Wang, T.-Y. Huang, H.-R. Lo, and Y.-C. Chao. 2008. Identification of baculoviral factors required for the activation of enhancer-like polyhedrin upstream (pu) sequence. Virus Res. 1387-16. [DOI] [PubMed] [Google Scholar]
- 39.Wu, Y.-L., C. Y. Y. Liu, C. P. Wu, C.-H. Wang, S.-T. Lee, and Y.-C. Chao. 2008. Cooperation of ie1 and p35 genes in the activation of baculovirus AcMNPV and HzNV-1 promoters. Virus Res. 135247-254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.