Abstract
Although the major component of the prion is believed to be the oligomer of PrPSc, little information is available concerning regions on the PrPSc molecule that affect prion infectivity. During the analysis of PrPSc molecules from various prion strains, we found that PrPSc of the Chandler strain showed a unique property in the conformational-stability assay, and this property appeared to be useful for studying the relationship between regions of the PrPSc molecule and prion infectivity. Thus, we analyzed PrPSc of the Chandler strain in detail and analyzed the infectivities of the N-terminally denatured and truncated forms of proteinase K-resistant PrP. The N-terminal region of PrPSc of the Chandler strain showed region-dependent resistance to guanidine hydrochloride (GdnHCl) treatment. The region approximately between amino acids (aa) 81 and 137 began to be denatured by treatment with 1.5 M GdnHCl. Within this stretch, the region comprising approximately aa 81 to 90 was denatured almost completely by 2 M GdnHCl. Furthermore, the region approximately between aa 90 and 137 was denatured completely by 3 M GdnHCl. However, the C-terminal region thereafter was extremely resistant to the GdnHCl treatment. This property was not observed in PrPSc molecules of other prion strains. Denaturation of the region between aa 81 and 137 by 3 M GdnHCl significantly prolonged the incubation periods in mice compared to that for the untreated control. More strikingly, the denaturation and removal of this region nearly abolished the infectivity. This finding suggests that the conformation of the region between aa 81 and 137 of the Chandler strain PrPSc molecule is directly associated with prion infectivity.
Prion diseases, such as scrapie, bovine spongiform encephalopathy (BSE), and Creutzfeldt-Jakob disease, are fatal neurodegenerative disorders characterized by the accumulation of a disease-specific, abnormal isoform of the prion protein, PrPSc, in the central nervous system, astrogliosis, neuronal vacuolation, and neuronal cell death. PrPSc is believed to be generated from a cellular form of prion protein, PrPC, by a posttranslational modification including conformational transformation. Although the prion entity, the causative agent of prion diseases, remains to be elucidated, PrPSc is believed to be a major component of the prion.
Direct interaction between PrPC and preexisting PrPSc precedes the transformation of PrPC into newly generated PrPSc. Data on the regions of PrPC that are indispensable for PrPSc formation and prion propagation have been accumulated using neuroblastoma cells persistently infected with prions and transgenic (Tg) mice expressing mutant PrPs. Although the extreme N-terminal region, amino acids (aa) 23 to 32, modulates prion propagation (8, 9, 34), the region between aa 32 and approximately aa 90 is not essential for the production of PrPSc and the propagation of the prion (9, 18, 22, 39). The region of residues 114 to 121, the most amyloidgenic region of PrP, is essential for the conversion of PrPC into PrPSc (11, 23). A deletion mutant lacking residues 23 to 88 and 141 to 176 can convert into PrPSc and support prion propagation in Tg mice, suggesting that the region of residues 141 to 176 is not essential for prion propagation (22, 34). The cysteine residue at 178 that forms an intramolecular disulfide bond with another cysteine residue at 213 is essential for PrPSc formation (22). Additionally, amino acid substitutions at 167 and 218 prevent PrPSc formation and show a dominant-negative effect on prion propagation (15, 28). Due to the difficulty of direct manipulation of PrPSc, the regions of PrPSc that are important for prion infectivity have not been elucidated. It is well accepted that not the removal of the protease-sensitive N-terminal domain (aa 23 to around aa 90) from PrPSc but the denaturation of the remaining C-terminal domain diminishes prion infectivity. However, the relationship between prion infectivity and the regions of PrPSc is largely unclear.
From the analysis of biochemical properties of PrPSc molecules of various prion strains, we found that PrPSc of the Chandler strain has region-dependent resistance to denaturation by guanidine hydrochloride (GdnHCl). This property allows for the denaturation and removal of specific regions of PrPSc. In this study, we describe the unique conformational stability of PrPSc of the Chandler strain and demonstrate that the region approximately between aa 81 and 137 of PrPSc is important for the infectivity of the Chandler prion strain.
MATERIALS AND METHODS
Mouse and prion strains.
Mouse-adapted prion strains 22L (7), Chandler (17), Fukuoka-1 (35), G1 (unpublished data), and Obihiro (31) were used in this study. These mouse-adapted strains were propagated in female Jcl:ICR mice (CLEA Japan) except where otherwise specified. In some cases, C56BL/6J mice (CLEA Japan) and RIII/J and I/LnJ mice (Jackson Laboratories) were used for prion propagation. In addition, mouse-adapted BSE prion strains, designated KUS-m and TE-m, which were derived from samples obtained in Japanese BSE cases KUS and TE by a third serial passage in RIII/J and C57BL/6J mice, respectively, were used. All procedures for animal experiments were carried out according to protocols approved by the Institutional Committee for Animal Experiments.
Antibodies.
Anti-PrP monoclonal antibodies (MAbs) 110, 118, 147, 31C6, 43C5, and 44B1 (16) were used. In addition, B103 rabbit polyclonal antibodies (pAb) raised against the bovine PrP synthetic peptide comprising aa 103 to 121, which corresponds to aa 90 to 109 of mouse PrP, were used (13).
Conformational-stability assay.
Conformational-stability assays were carried out as described by Legname et al. (19, 20) with some modifications. The brains of mice infected with prions were homogenized in phosphate-buffered saline (PBS) to make 10% homogenates. Aliquots of the homogenates were stored at −30°C until use. The 10% brain homogenates (50 μl) were mixed with equal volumes of various concentrations of GdnHCl (0 to 8 M) and incubated at 37°C for 1 h. Samples were then diluted by the addition of 850 μl of NTS buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% Triton X-100, and 0.5% sodium deoxycholate). To adjust the final GdnHCl concentration to 0.4 M, 50 μl of various concentrations of GdnHCl (0 to 8 M) were added to the samples. The samples were then digested with proteinase K (PK; Roche) at 20 μg/ml for 30 min at 37°C. After the termination of PK activity by adding Pefabloc (Roche) to obtain a final concentration of 2 mM, 500 μl of a 5:1 mixture of 2-butanol and methanol was added and the samples were mixed well and kept for 10 min at ambient temperature. PrPSc was pelleted by centrifugation at 20,000 × g for 10 min at 20°C. The resulting pellet was dissolved in 1× sodium dodecyl sulfate (SDS) sample buffer (62.5 mM Tris-HCl [pH 6.8], 5% glycerol, 3 mM EDTA, 4% β-mercaptoethanol, 0.04% bromophenol blue, 5% SDS, 4 M urea) by being boiled for 5 min. SDS-polyacrylamide gel electrophoresis and immunoblotting were carried out as described elsewhere (38). The chemiluminescence intensities of bands of PrPSc were measured with a LAS-3000 chemiluminescence image analyzer (Fujifilm). Quantitative analyses of the blots were carried out with Image Reader LAS-3000 software, version 1.11 (Fujifilm). The sigmoidal patterns of denaturation curves were plotted using a nonlinear least-squares fit. The concentrations of GdnHCl required to denature 50% of PrPSc ([GdnHCl]1/2 values) were estimated from the denaturation curves, and statistical analysis was carried out by a one-way analysis of variance followed by a Newman-Keuls test.
Deglycosylation.
The 10% brain homogenates (250 μl) were mixed with equal volumes of the NTS buffer and digested with PK at 20 μg/ml for 1 h at 37°C. Proteolysis was terminated by the addition of Pefabloc to a final concentration of 4 mM. Samples were then mixed with 1/5 volume of 5× denaturation buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA, 5% SDS, 10% β-mercaptoethanol) and 5 U of N-glycosidase F (Roche) and incubated for 16 h at 37°C. Proteins were precipitated by the addition of 1/2 volume of a 5:1 mixture of 2-butanol and methanol followed by centrifugation at 20,000 × g for 10 min at 20°C.
Preparation of cell lysates.
A Neuro2a mouse neuroblastoma subclone persistently infected with the Chandler strain (ScN2a-5) (38) was used. ScN2a-5 cells grown in 10-cm dishes were collected by using a cell scraper and pelleted by centrifugation at 300 × g for 5 min. The cells were washed once with PBS and pelleted again by centrifugation. The resulting pellets were lysed with 1 ml of lysis buffer (10 mM Tris-HCl [pH 7.5], 0.5% Triton X-100, 0.5% sodium deoxycholate, 150 mM NaCl, 5 mM EDTA) for 30 min on ice. Nuclei and cell debris were removed by low-speed centrifugation at 300 × g, and supernatants were further centrifuged at 100,000 × g for 30 min at 4°C. The resulting pellets were suspended in 50 μl of PBS and used for conformational-stability assays as the PrPSc-enriched fraction.
Bioassay.
The 10% brain homogenates (540 μl) were mixed with equal volumes of various concentrations (0 to 6 M) of GdnHCl solution and then incubated at 37°C for 1 h. Samples were then diluted by the addition of 9.18 ml of NTS buffer, and 540 μl of various concentrations of GdnHCl solution were added to adjust the final concentration of GdnHCl to 0.4 M. The mixtures were ultracentrifuged at 197,000 × g for 2.5 h at 4°C, and the resulting pellet was resuspended in 540 μl of PBS and used for the bioassay. Small aliquots of the samples were digested with PK and analyzed by immunoblotting to confirm the existence of PrPSc. To prepare the PK-treated inoculums for the bioassay, 540-μl aliquots of 10% brain homogenates were treated with GdnHCl as described above. After the GdnCHl treatment, samples were digested with 10 μg/ml of PK for 1 h at 37°C, and digestion was stopped by adding Pefabloc to a final concentration of 2 mM. Samples were ultracentrifuged, and the resulting pellets were resuspended in PBS as described above. Samples (20 μl) were intracerebrally inoculated into 4-week-old female Jcl:ICR mice.
RESULTS
Conformational stabilities of PrPSc molecules of the mouse-adapted prion strains.
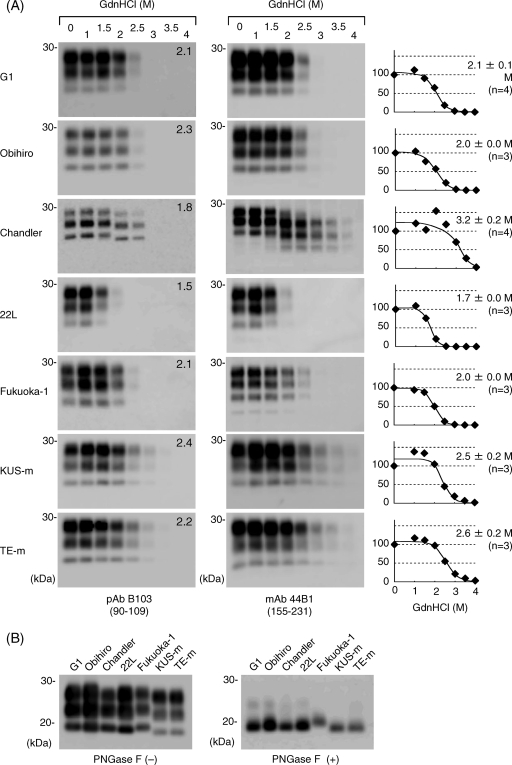
To examine the biochemical differences of PrPSc molecules from various mouse-adapted prion strains, conformational-stability assays were carried out to assess the resistance of PrPSc to denaturation by GdnHCl (Fig. 1A). When immunoblots were probed with pAb B103 and MAb 44B1, which recognize aa 90 to 109 and aa 155 to 231 of mouse PrP, respectively, the amounts of PrPSc molecules of the G1, Obihiro, and Fukuoka-1 strains were found to be nearly unchanged by treatment with up to 2 M GdnHCl. The treatment with 2.5 M GdnHCl led to the first decrease in the amount of PrPSc, and only a trace amount of PrPSc was detected after treatment with 3 M GdnHCl. The [GdnHCl]1/2 was estimated from the denaturation curve for each prion strain (Fig. 1A). The [GdnHCl]1/2 values for the G1, Obihiro, and Fukuoka-1 strains based on the results obtained with MAb 44B1 ranged from 2.0 to 2.1 M, and there was no significant difference among them (Table 1). This finding indicates that these strains have similar levels of resistance to GdnHCl treatment. In contrast, the [GdnHCl]1/2 for the 22L strain was significantly lower than those for the G1, Obihiro, and Fukuoka-1 strains, indicating that PrPSc of the 22L strain is less stable than those of other strains. Moreover, the [GdnHCl]1/2 values for BSE strains KUS-m and TE-m were higher than those for other mouse-adapted prion strains except the Chandler strain. The incubation period for each prion strain and the [GdnHCl]1/2 values are summarized in Table 1. Although the [GdnHCl]1/2 values for the G1, Obihiro, and Fukuoka-1 strains are comparable, the G1 strain had an extremely long incubation period.
FIG. 1.
Conformational stabilities of PrPSc molecules of various prion strains. (A) Immunoblots for the conformational-stability assay. Brain homogenates from prion-infected mice (prion strains are indicated to the left) were treated with 0 to 4 M GdnHCl (as indicated at the top) and subjected to PK digestion. PrPSc was detected by either pAb B103 (left column) or MAb 44B1 (right column). Epitopes for antibodies are indicated in parentheses. Independent assays of each strain with MAb 44B1 were carried out at least three times (the number of assays is indicated in parentheses to the right of the graphs), and based on the quantitative results for the blots probed with MAb 44B1, the denaturation curves were plotted using a nonlinear least-squares fit. [GdnHCl]1/2 values (means ± SD) are indicated for each graph. Numbers in the top right corners of the blots probed with pAb B103 are the [GdnHCl]1/2 values (in molars). (B) Molecular masses of PrPSc molecules. Brain homogenates from prion-infected mice (prion strains are indicated at the top) were treated with PK, and the immunoblot was probed with pAb B103. To compare the molecular masses of the PK-resistant cores of PrPSc molecules more precisely, PK-treated samples were further treated with peptide-N-glycosidase F (PNGase F) (right). −, absent; +, present.
TABLE 1.
Conformational stabilities and incubation periods of prion strains
| Prion straina | Mouse strain for propagation | No. of serial passagesb | [GdnHCl]1/2(M) determined with:
|
Mean incubation period (days) ± SD | No. of micee | |
|---|---|---|---|---|---|---|
| pAb B103 | MAb 44B1 or 31C6d | |||||
| G1 | slc:ICR | 4 | 2.1 | 2.1 ± 0.1 | 326 ± 53 | 5 |
| Obihiro | Jcl:ICR | >5 | 2.3 | 2.0 ± 0.0 | 153 ± 7 | 24 |
| Chandlerc | Jcl:ICR | >5 | 1.8 | 3.2 ± 0.2* | 150 ± 2 | 15 |
| I/LnJ | 2 | 2.2 | >3.5† | 227 ± 7 | 4 | |
| C57BL/6J | 3 | 2.3 | 3.5† | 153 ± 6 | 6 | |
| 22L | Jcl:ICR | 2 | 1.5 | 1.7 ± 0.0** | 144 ± 3 | 5 |
| Fukuoka-1 | Jcl:ICR | 2 | 2.1 | 2.0 ± 0.0 | 146 ± 8 | 8 |
| KUS-m | RIII/J | 3 | 2.4 | 2.5 ± 0.2* | 165 ± 11 | 6 |
| TE-m | C57BL/6J | 3 | 2.2 | 2.6 ± 0.2* | 168 ± 4 | 6 |
Strain G1 was obtained from sheep with experimental scrapie; the 22L and Fukuoka-1 strains were derived from prions passaged in mice carrying Prnpa/a but different from Jcl:ICR mice; and KUS-m and TE-m were obtained from BSE field cases KUS and TE.
History (number) of serial passages in the mouse strain listed to the left.
Chandler strain prions passaged in Jcl:ICR mice were then passaged in I/LnJ or C57BL/6J mice.
The [GdnHCl]1/2 values were estimated from the denaturation curves plotted by using blots probed with MAb 44B1 (values shown are means ± SD of results from at least three independent assays) and MAb 31C6 (values determined with this MAb are indicated by †). *, higher than value for G1 (P < 0.05); **, lower than value for G1 (P < 0.05).
Number of mice used for the calculation of the incubation period.
Among the PrPSc molecules of the prion strains used in this study, PrPSc of the Chandler strain showed a unique alteration in molecular mass with the increase of the GdnHCl concentration. When the blots were probed with pAb B103, the PrPSc bands detected in the samples treated with 2.0 and 2.5 M GdnHCl were approximately 1 to 2 kDa smaller than those in samples treated with lower concentrations, and PrPSc was almost undetectable after the 3 M GdnHCl treatment. When the blots were probed with MAb 44B1, the PrPSc bands detected in samples treated with more than 2.0 M GdnHCl were approximately 6 to 7 kDa smaller than those in samples treated with lower concentrations, and these bands were still detected even after the treatment with 3.5 M GdnHCl. The [GdnHCl]1/2 for PrPSc of the Chandler strain was estimated to be 3.2 M from the results obtained with MAb 44B1.
Further characterization of the GdnHCl resistance of PrPSc of the Chandler strain.
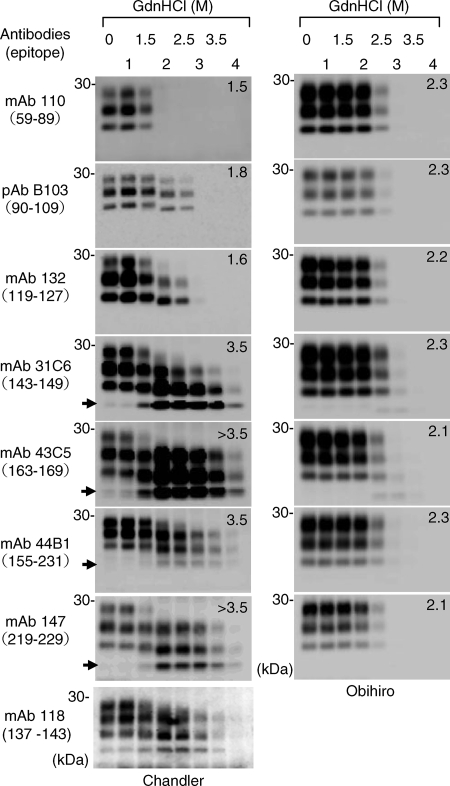
The results of the conformational-stability assays suggested that the N- and C-terminal regions of PK-resistant PrPSc of the Chandler strain have different levels of resistance to GdnHCl treatment. Thus, we analyzed PrPSc of the Chandler strain more precisely with six additional MAbs (Fig. 2). By using MAb 110, recognizing repetitive amino acid sequences at positions 59 to 65 and 83 to 89, PrPSc was undetected after treatments with more than 2 M GdnHCl. The major N terminus of the PK-resistant core of PrPSc molecules (designated PrP27-30) of the ME7 and Obihiro strains is reported to be Gly at position 81 (10, 12). Moreover, the molecular mass of deglycosylated Chandler PrPSc is identical to that of the Obihiro strain PrPSc (Fig. 1B). Taken together, these observations indicate the major N terminus of the PK-resistant core of the Chandler PrPSc to be at position 81. We assumed, therefore, that the 1- to 2-kDa-smaller PrPSc bands detected with pAb B103 after 2.0 and 2.5 M GdnHCl treatments resulted from the denaturation and removal of the region from aa 81 to a residue around aa 90 (herein referred to as aa 90) of mouse PrPSc. The PrPSc patterns detected by MAb 132 appeared to be almost identical to those detected by pAb B103, indicating that the region between aa 90 and the epitope for MAb 132 (aa 119 to 127) was almost denatured by treatment with more than 3 M GdnHCl. After treatments with more than 2 M GdnHCl, the presence of the approximately 6- to 7-kDa-smaller PrPSc bands was evident on the blots probed with MAb 31C6 (recognizing aa 143 to 149) and MAbs recognizing the C-terminal region thereafter (MAbs 43C5, 44B1, and 147). With 2.0 and 2.5 M GdnHCl treatments, the 6- to 7-kDa-smaller PrPSc bands are thought to overlap with the 1- to 2-kDa-smaller PrPSc bands that were detected with pAb B103 and MAb 132. Therefore, the presence of the 6- to 7-kDa-smaller PrPSc was more obvious after treatment with more than 3 M GdnHCl, at which the N-terminal region of the PK-resistant core of PrPSc between aa 81 and the epitope for MAb 132 was denatured and undetectable after PK digestion. MAb 118, which recognizes aa 137 to 143 of mouse PrP, also reacted with the 6- to 7-kDa-smaller PrPSc bands (Fig. 2). This result suggests that the truncated PK-resistant PrPSc lacks the N-terminal region up to around aa 127 to 137, although the exact N terminus (hereinafter referred to as aa 137) remains to be determined. Taken together, these results indicate that the PK-resistant core of PrPSc (aa 81 to 231) of the Chandler strain has region-dependent conformational stability under conditions of GdnHCl treatment. The region of aa 81 to 90 of PrPSc is the most sensitive to GdnHCl and is denatured almost completely by 2 M GdnHCl. The region between aa 90 and 137 is denatured almost completely by more than 3 M GdnHCl, while the remaining C-terminal region of PrPSc is highly resistant to GdnHCl. The N-terminally truncated nonglycosylated PrPSc was detectable after 1.5 M GdnHCl treatment (Fig. 2, blots probed with MAbs 31C6, 43C5, 44B1, and 147), suggesting that the region between aa 81 and 137 begins to be denatured with 1.5 M GdnHCl treatment. In contrast to PrPSc of the Chandler strain, PrPSc of the Obihiro strain was nearly undetectable after 3 M GdnHCl treatment, regardless of the antibodies used, and the [GdnHCl]1/2 values estimated from the different blots were comparable (Fig. 2).
FIG. 2.
Region-dependent conformational stability of PrPSc of the Chandler strain. Brain homogenates from mice infected with the Chandler (left) and Obihiro (right) strains were subjected to the conformational-stability assay, and immunoblots were probed with the various anti-PrP antibodies indicated to the left. Epitopes for antibodies are indicated in parentheses. Due to relatively weak reactivity, five times the tissue equivalents of those for the blots for the other MAbs were loaded for MAb 118. Numbers in the top right corners of the blots are the [GdnHCl]1/2 values (in molars). Arrows, the N-terminally truncated nonglycosylated PrPSc.
The 6- to 7-kDa-smaller unglycosylated PrPSc was occasionally detected by MAbs recognizing the C-terminal region of PrP without GdnHCl pretreatment, but usually at a very low level. On the other hand, this band was not detected by antibodies recognizing the N-terminal region of PrP (MAbs 110 and 132 and pAb B103). These findings suggest that processing of the region up to aa 137 of the Chandler PrPSc occurs in the brain tissues, albeit at a very low level. Alternatively, the processing may occur during the sample preparation or autolysis.
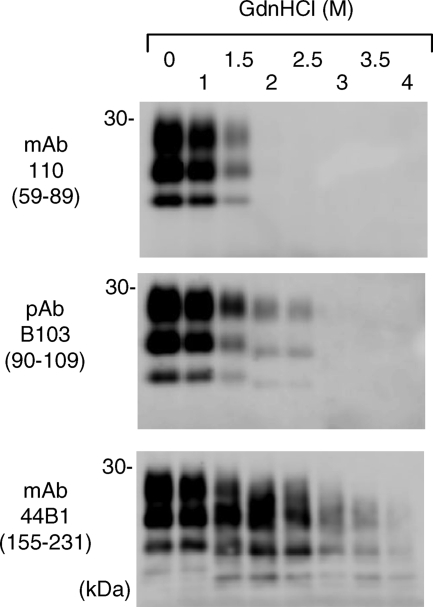
Conformational stability of PrPSc in cells infected with the Chandler strain.
Next, we examined whether PrPSc in cells persistently infected with the Chandler strain shows the region-dependent conformational stability. PrPSc-enriched fractions obtained from ScN2a-5 cell lysates were subjected to conformational-stability assays (Fig. 3). MAb 110 detected the PK-resistant PrPSc bands with up to 1.5 M GdnHCl treatment, and the 1- to 2-kDa-smaller PrPSc bands were detected by pAb B103 with 2 and 2.5 M GdnHCl treatments. Furthermore, the 6- to 7-kDa-smaller N-terminally truncated PrPSc bands were detected by MAb 44B1 even after 3 and 3.5 M GdnHCl treatments. These results were consistent with those for PrPSc obtained from the brains of mice infected with the Chandler strain, indicating that the unique conformational stability was maintained in cultured cells.
FIG. 3.
Region-dependent conformational stability of PrPSc in cells persistently infected with the Chandler strain. PrPSc-enriched fractions obtained from ScN2a-5 cells were subjected to the conformational-stability assay. The antibodies used are listed to the left, and epitopes for antibodies are indicated in parentheses.
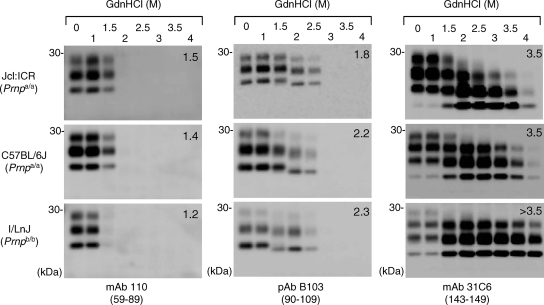
Conformational stability of the Chandler PrPSc in mice with different PrP genotypes.
To examine whether the region-dependent conformational stability was maintained in mice with different genotypes, assays were carried out using brains of C57BL/6J (Prnpa/a) and I/LnJ (Prnpb/b) mice infected with the Chandler strain (Fig. 4). The patterns of PrPSc from C57BL/6J mice were almost identical to those of PrPSc from Jcl:ICR mice. In contrast, the N-terminal region of PrPSc from I/LnJ mice was less resistant to GdnHCl than those of PrPSc molecules from Jcl:ICR and C57BL/6J mice; the [GdnHCl]1/2 value for PrPSc from I/LnJ mice (1.2 M) was lower than those for PrPSc molecules from Jcl:ICR and C57BL/6J mice (1.5 and 1.4 M, respectively), and PrPSc from I/LnJ mice was undetected by MAb 110 after the 1.5 M GdnHCl treatment. In addition, the C terminus of PrPSc from I/LnJ mice appeared to be more stable than those of PrPSc molecules from Jcl:ICR and C57BL/6J mice. Although a slight difference in the sensitivity to GdnHCl was observed, it should be emphasized that the sequential shift in molecular mass with an increase of GdnHCl concentration was reproduced for the Chandler PrPSc propagated in mice with the Prnpb/b genotype; the 1- to 2-kDa-smaller PrPSc bands were detected with pAB B103 after 1.5 and 2 M GdnHCl treatments, and the intensity of the 6- to 7-kDa-smaller unglycosylated PrPSc bands detected with MAb 31C6 increased remarkably after treatment with GdnHCl at 1.5 M or higher. These results suggested that the region-dependent conformational stability of the PrPSc from the Chandler strain was maintained in mice with different PrP genotypes.
FIG. 4.
Region-dependent conformational stability of the Chandler PrPSc in mice with different genetic backgrounds. Brain homogenates from Jcl:ICR (Prnpa/a), C57BL/6J (Prnpa/a), and I/LnJ (Prnpb/b) mice infected with the Chandler strain were subjected to the conformational-stability assay. The antibodies used and their epitopes (in parentheses) are indicated. Numbers in the top right corners of the blots are the [GdnHCl]1/2 values (in molars).
Effect of denaturation and removal of the N-terminal region of PrPSc on prion infectivity.
To examine whether the denaturation of specific regions of PrPSc affects the prion infectivity, brain homogenates from mice infected with the Chandler strain were treated with GdnHCl and subjected to bioassays. Small aliquots were analyzed by immunoblotting to confirm the region-specific denaturation of PrPSc in the inoculums (Fig. 5A). Survival times of mice inoculated with samples treated with 1 and 1.5 M GdnHCl were equivalent to those of mice inoculated with the non-GdnHCl-treated control (Table 2). Compared to the survival time of mice receiving the control (0 M; mean survival time ± standard deviation [SD], 159 ± 14 days), the survival time of mice receiving samples treated with 2 M GdnHCl seemed to be prolonged (176 ± 12 days); however, the difference was not statistically significant (P > 0.05). In contrast, significant prolongation of the survival time (206 ± 25 days; P < 0.01) after the 3 M GdnHCl treatment was observed. These results suggest that the denaturation of aa 81 to 137 of PrPSc greatly influences the prion infectivity. To confirm the involvement of aa 81 to 137 in the prion infectivity more precisely, this region was removed by treatment with 3 M GdnHCl followed by PK digestion. The expected size shift of PrPSc in the inoculums was confirmed prior to the bioassay. Furthermore, the intensities of the PrPSc bands in samples treated with 0 and 3 M GdnHCl were relatively equivalent, indicating that equal molar amounts of PK-resistant PrPSc existed in the inoculums (Fig. 5B). These samples were intracerebrally inoculated into mice to examine the prion infectivity (Table 2). In contrast to mice receiving the non-GdnHCl-treated control (survival time, 170 ± 11 days), mice inoculated with the sample treated with 3 M GdnHCl exhibited an attack rate of 40% and a mean survival time of 235 days (n = 2). Furthermore, two of five mice were still alive at 365 days postinoculation (dpi) (Table 2). These results suggest that the infectivity of the N-terminally truncated PK-resistant PrPSc lacking aa 81 to 137 was extremely low.
FIG. 5.
Region-specific denaturation or removal of PrPSc in inoculums for the bioassay. (A) Confirmation of region-specific denaturation. Brains of mice infected with the Chandler strain were treated with various concentrations of GdnHCl (without PK treatment), and the fraction containing PrPSc was recovered by ultracentrigation. Small aliquots of the inoculums were treated with PK and analyzed by immunoblotting with MAb 44B1. (B) Confirmation of the removal of aa 81 to 137. Brain homogenates from mice infected with the Chandler and Obihiro strains were treated with 0 or 3 M GdnHCl and subjected to PK digestion. After the termination of proteolysis, samples were ultracentrifuged to collect the fraction containing PrPSc. Small aliquots of the inoculums were analyzed by immunobloting with MAb 44B1. Equal amounts of brain tissues were loaded into the lanes.
TABLE 2.
Effects of GdnHCl treatment and PK digestion on prion infectivity
| Strain | GdnHCl concn (M) | PK digestiona | No. of infected mice/no. of mice in groupb | Mean survival time (dpi) ± SD |
|---|---|---|---|---|
| Chandler | 0 | − | 4/4 | 159 ± 14 |
| 1 | − | 5/5 | 150 ± 9 | |
| 1.5 | − | 7/7 | 165 ± 12 | |
| 2 | − | 4/4 | 176 ± 12 | |
| 3 | − | 5/5 | 207 ± 25 | |
| 0 | + | 6/6 | 170 ± 11 | |
| 3 | + | 2/5c | 234, 236, >365 | |
| Obihiro | 0 | + | 5/5 | 152 ± 7 |
| 3 | + | 5/5 | 186 ± 11 |
+, performed; −, not performed.
Number of mice which showed typical clinical manifestations of scrapie and/or were positive for PrPSc by immunoblotting/number of mice used in the bioassay.
Two mice showed typical clinical manifestations and were positive for PrPSc (at 234 and 236 dpi), and one mouse was found dead without having shown any symptoms at 336 dpi and was negative for PrPSc. The remaining two mice were still alive without any symptoms >365 dpi.
The immunoreactivity of PK-resistant PrPSc of the Obihiro strain, in contrast to that of PrPSc of the Chandler strain, decreased less than 1% from that in the original samples when the samples were treated with 3 M GdnHCl and subjected to PK digestion (Fig. 5B). Consistent with the decrease in the amount of PrPSc, the survival time was prolonged for 34 days by treatment with 3 M GdnHCl (Table 2). From the dose-survival time standard curve for the Obihiro strain in ICR mice, the 34-day prolongation was estimated to represent more than a 2-log reduction in infectivity.
DISCUSSION
Prion strains have been distinguished by their biological properties, including incubation periods and neuropathological lesion profiles in mice experimentally inoculated with test samples (3, 4, 6, 7). However, these types of experiments are time-consuming, and the results are difficult to standardize among laboratories. Biochemical properties of PrPSc, such as molecular mass, glycoforms, PK resistance, and sensitivity to denaturants, often differ among prion strains (2, 5, 14, 25-27, 29), although relationships between the biochemical and biological properties are unclear. Elucidating the strain-specific biochemical properties as well as direct relationships between biochemical and biological properties will facilitate the distinction of prion strains without time-consuming bioassays and an understanding of the mechanisms involved in prion strains. From our analyses of the stabilities of PrPSc molecules to the treatment of GdnHCl with a panel of anti-PrP antibodies, we found that PrPSc of the Chandler strain possesses unique region-dependent conformational stability. The region of aa 81 to 137 of PrPSc begins to be denatured by 1.5 M GdnHCl and is almost completely denatured and becomes PK sensitive with 3 M GdnHCl treatment. In contrast, the C-terminal region (after aa 137) is extremely resistant to denaturation (Fig. 6).
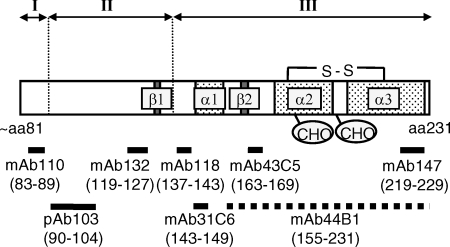
FIG. 6.
Schematic representation of region-specific denaturation of the Chandler PrPSc. The PK-resistant core of the Chandler PrPSc (from aa ∼81 to 231) is depicted, with the locations of two β-strands (β1 and β2), three α-helices (α1 to α3), two N glycosylation sites (CHO), and an intramolecular disulfide bond (S-S). The locations of epitopes are indicated by thick lines labeled with amino acid positions (in parentheses). The epitope for MAb 44B1, which recognizes a discontinuous epitope, is indicated by a dashed line, while those for other antibodies that recognize linear epitopes are indicated by solid lines. Region I (aa 81 to 90), indicated above, was denatured almost completely by treatment with up to 2 M GdnHCl, and the removal of this region generates the 1- to 2-kDa-smaller PK-resistant PrPSc. Region II (aa 90 to 137) was denatured almost completely by treatment with up to 3 M GdnHCl, and the removal of regions I and II consequently generates the 6- to 7-kDa-smaller PK-resistant PrPSc (region III, aa 137 to the C terminus) that is highly resistant to denaturation but lacks prion infectivity.
When the blots in Fig. 2 were carefully examined, in the Chandler PrPSc sample treated with 2 and 2.5 M GdnHCl, the 1- to 2-kDa-smaller diglycosylated PrPSc was detected with MAbs 31C6 and 44B1 while the corresponding bands were unclear with MAbs 147 and 43C5. This result suggests that the C-terminal region is also truncated in certain fractions of PrPSc. However, we think that the C-terminal truncation is not a major effect for the following reasons. First, the affinity of the MAbs and the amount of the 1- to 2-kDa-smaller PrPSc influenced the result. The affinity of MAb 147 is lower than that of MAbs 31C6 and 44B1 (K. Sakata and M. Horiuchi, unpublished results); therefore, it is possible that MAb 147 could not visualize the relatively small amount of the 1- to 2-kDa-smaller PrPSc in the samples treated with 2 and 2.5 M GdnHCl. Second, the conformation of the particular region of PrP on the blot may influence the interpretation of the results. The immunoreactivity of the 6- to 7-kDa-smaller PrPSc increased when MAbs recognizing the middle part of PrP (MAbs 31C6 and 43C5) were used; this tendency was especially obvious with MAb 43C5 (Fig. 2). We cannot explain the exact reason for this effect at the moment. However, the results suggest that the epitope of MAb 43C5 on the 6- to 7-kDa-smaller PrPSc on the blot may be more easily accessible than that on the regular and the 1- to 2-kDa-smaller PrPSc molecules. If these two types of molecules exist in the limited area of the blot, the reaction of the MAb to the easily accessible epitope will be pronounced. Although we do not exclude the possibility of the C-terminal truncation, further fine-detail experiments will be required to address the C-terminal truncation.
The sequential size shift of PK-resistant PrPSc according to the denaturation profile was not observed in our study of other mouse-adapted prion strains, natural and experimental sheep scrapie and Japanese BSE cases (data not shown). Additionally, this property was maintained in mice with different Prnp genotypes and in cells persistently infected with the Chandler strain. Therefore, these results suggest that the region-dependent conformational stability is specific to PrPSc of the Chandler strain. In contrast, the conformational-stability assay of the RML prion, which is thought to be synonymous with or very close to the Chandler strain, showed no region-dependent conformational stability (19, 36). One possibility that explains this discrepancy is the use of different antibodies for PrPSc detection; Legname et al. (19) and Thackray et al. (36) used the Fab fragment HuM-D18, which recognizes aa 132 to 156, and MAb 683, which recognizes aa 168 to 172, respectively. Both antibodies recognize the C-terminal region after the epitope for MAb 132 and thus should detect the molecular size changes in PrPSc molecules that possess region-dependent conformational stability, as found in the Chandler strain. As these molecular size changes were not detected in those studies, it is unlikely that the difference in antibodies accounts for the discrepancy. Alternatively, genetic backgrounds of mice used for prion propagation may cause the difference in the conformational stability. It has been reported previously that the biochemical properties of PrPSc vary depending on the cell and tissue types for prion propagation without changing biological properties (1). Indeed, the mice used for the propagation of the RML prion in the previous study (CD-1 Swiss mice) were different from those used in this study (Jcl:ICR and C57BL/6J). Thus, further analysis of the Chandler strain propagated in various mouse strains, as well as analyses of other mouse-adapted prion strains, especially those of the lineage of the Chandler strain, such as 139A (6), will be required to conclude that the region-dependent conformational stability is specific to the Chandler strain.
Legname et al. (20) reported linear correlation between the [GdnHCl]1/2 values and incubation periods. In contrast, no linear correlation was observed in our results (n = 9; r = 0.007). We think that the sample size in our study was too small to make any conclusion. In particular, few data are available for strains showing longer incubation periods or higher [GdnHCl]1/2 values at present. Therefore, further accumulation of data will be required to assess the correlation between incubation periods and conformational stabilities of PrPSc.
PrPSc includes PK-sensitive and PK-resistant molecules (2, 29, 30, 37). Both types of PrPSc are infectious, and PK digestion alone decreases prion infectivity to some extent (2, 32). However, it is well known that the PK-resistant core of PrPSc, PrP27-30, which is produced by the removal of the PK-sensitive N-terminal region of PrPSc (from aa 23 to around aa 90), possesses prion infectivity. Prions propagated in Tg mice expressing PrP that lacks aa 23 to 88 can propagate in mice expressing wild-type PrP (18). These previous results indicate that this N-terminal region of PrPSc is not essential for the infectivity of the prion. However, analyzing the relationship between other regions of PrPSc and infectivity by making deletions or mutations has been difficult. In this study, we utilized the region-dependent conformational stability of the Chandler PrPSc and truncated the PrPSc directly at the N-terminal region up to around aa 137 to produce the N-terminally truncated PK-resistant PrPSc; this approach allowed us to then analyze the influence of this region on prion infectivity. Compared to the regular PK-resistant core of PrPSc that is produced by PK digestion without GdnHCl treatment, the N-terminally truncated PK-resistant PrPSc had extremely low infectivity despite the existence of the C-terminal region as PK-resistant fragments (Table 2). Since we have not produced a dose-incubation period standard curve for the Chandler strain in Jcl:ICR mice, we cannot estimate the exact reduction in infectivity. However, the attack rate and the survival time suggested that the infectivity decreased to nearly the detection limit in the bioassay. This result provides direct evidence that the region of aa 81 to 137 of PK-resistant PrPSc is critical for prion infectivity, although evidence for other prion strains remains to be elucidated. However, PK treatment alone reduced the infectivity of the Chandler strain (mean survival times, 159 and 170 days for mice receiving samples without and with PK treatment, respectively) (Table 2), indicating that the PK-sensitive PrPSc fraction possessing prion infectivity was present in the brain homogenates of the Chandler strain-infected mice. Our results clearly showed that the region of aa 81 to 137 of the PK-resistant core of the Chandler PrPSc is important for infectivity; however, it remains unclear whether the same conclusion is applicable to the infectivity of the PK-sensitive PrPSc fraction.
The denaturation of this region by 3 M GdnHCl treatment appeared to be less effective than the removal of this region in reducing prion infectivity. However, considering the effect of GdnHCl on PrPSc aggregates, the denaturation itself appears to result in a substantial loss of infectivity (Table 2). The GdnHCl treatment has two expected effects: the dissociation of large PrPSc aggregates into small aggregates and the denaturation of the PrPSc molecules. Hence, without PK digestion, small aggregates consisting of PrPSc with incompeletely denatured aa 81 to 137 may remain and infectivity may be observed. Such small PrPSc aggregates should be PK sensitive, and therefore, the infectivity should be diminished after PK digestion (32). Alternatively, this region may have been somewhat refolded after the GdnHCl treatment, which would lead to infectivity.
Several distinct domains of PrPC are reported to be involved in the direct interaction with PrPSc (21, 33), whereas domains on PrPSc that are involved in binding to PrPC remain undetermined. The N-terminally truncated PrPSc may be useful for the analysis of the PrPC binding domain on the PrPSc molecule. Here, we showed an example of a possible biochemical approach to PrPSc manipulation, in which we directly produced the N-terminally truncated PrPSc from native PrPSc. It has been reported previously that some conditions (e.g., pH) in protease digestion affect the N-terminal truncation of the PK-resistant core of PrPSc (24). Thus, further investigation of region-specific denaturation and proteolysis may be useful not only for the analysis of prion strains but also for the manipulation of PrPSc.
Acknowledgments
We thank Katsumi Doh-ura (Tohoku University) and Noriyuki Nishida (Nagasaki University) for providing Fukuoka-1 and 22L strains, respectively.
This work was supported by a grant from the global COE Program (F-001) and a grant-in-aid for science research (A; grant no. 18208026) and a grant-in-aid for exploratory research (grant no. 20658070) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. This work was also supported by a grant from the Ministry of Health, Labor and Welfare of Japan (grant no. 20330701). This work was also partly supported by a grant-in-aid from the BSE Control Project of the Ministry of Agriculture, Forestry and Fisheries of Japan and a grant for Strategic Cooperation to Control Emerging and Re-emerging Infections and the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 28 January 2009.
REFERENCES
- 1.Arima, K., N. Nishida, S. Sakaguchi, K. Shigematsu, R. Atarashi, N. Yamaguchi, D. Yoshikawa, J. Yoon, K. Watanabe, N. Kobayashi, S. Mouillet-Richard, S. Lehmann, and S. Katamine. 1999. Biological and biochemical characteristics of prion strains conserved in persistently infected cell cultures. J. Virol. 797104-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessen, R. A., and R. F. Marsh. 1994. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 687859-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce, M., A. Chree, I. McConnell, J. Foster, G. Pearson, and H. Fraser. 1994. Transmission of bovine spongiform encephalopathy and scrapie to mice: strain variation and the species barrier. Philos. Trans. R. Soc. Lond. B 343405-411. [DOI] [PubMed] [Google Scholar]
- 4.Bruce, M. E. 1993. Scrapie strain variation and mutation. Br. Med. Bull. 49822-838. [DOI] [PubMed] [Google Scholar]
- 5.Collinge, J., K. C. Sidle, J. Meads, J. Ironside, and A. F. Hill. 1996. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 383685-690. [DOI] [PubMed] [Google Scholar]
- 6.DeArmond, S. J., S. L. Yang, A. Lee, R. Bowler, A. Taraboulos, D. Groth, and S. B. Prusiner. 1993. Three scrapie prion isolates exhibit different accumulation patterns of the prion protein scrapie isoform. Proc. Natl. Acad. Sci. USA 906449-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickinson, A. G. 1976. Scrapie in sheep and goats. Front. Biol. 44209-241. [PubMed] [Google Scholar]
- 8.Fischer, M., T. Rülicke, A. Raeber, A. Sailer, M. Mose, B. Oesch, S. Brandner, A. Aguzzi, and C. Weissmann. 1996. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 151255-1264. [PMC free article] [PubMed] [Google Scholar]
- 9.Flechsig, E., D. Shmerling, I. Hegyi, A. J. Raeber, M. Fischer, A. Cozzio, C. von Mering, A. Aguzzi, and C. Weissmann. 2000. Prion protein devoid of the octapeptide repeat region restores susceptibility to scrapie in PrP knockout mice. Neuron 27399-408. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi, H., T. Yokoyama, M. Takata, Y. Iwamaru, M. Imamura, Y. Ushiki, and M. Shinagawa. 2005. The N-terminal cleavage site of PrPSc from BSE differs from that of PrPSc from scrapie. Biochem. Biophys. Res. Commun. 3281024-1027. [DOI] [PubMed] [Google Scholar]
- 11.Hölscher, C., H. Delius, and A. Bürkle. 1998. Overexpression of nonconvertible PrPc Δ114-121 in scrapie-infected mouse neuroblastoma cells leads to trans-dominant inhibition of wild-type PrPSc accumulation. J. Virol. 721153-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hope, J., G. Multhaup, L. J. Reekie, R. H. Kimberlin, and K. Beyreuther. 1988. Molecular pathology of scrapie-associated fibril protein (PrP) in mouse brain affected by the ME7 strain of scrapie. Eur. J. Biochem. 172271-277. [DOI] [PubMed] [Google Scholar]
- 13.Horiuchi, M., N. Yamazaki, T. Ikeda, N. Ishiguro, and M. Shinagawa. 1995. A cellular form of prion protein (PrPC) exists in many non-neuronal tissues of sheep. J. Gen. Virol. 762583-2587. [DOI] [PubMed] [Google Scholar]
- 14.Horiuchi, M., T., Nemoto, N. Ishiguro, H. Furuoka, S. Mohri, and M. Shinagawa. 2002. Biological and biochemical characterization of sheep scrapie in Japan. J. Clin. Microbiol. 403421-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko, K., L. Zulianello, M. Scott, C. M. Cooper, A. C. Wallace, T. L. James, F. E. Cohen, and S. B. Prusiner. 1997. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. USA 9410069-10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, C.-L., A. Umetani, T. Matsui, N. Ishiguro, M. Shinagawa, and M. Horiuchi. 2004. Antigenic characterization of an abnormal isoform of prion protein using a new diverse panel of monoclonal antibodies. Virology 32040-51. [DOI] [PubMed] [Google Scholar]
- 17.Kim, C.-L., A. Karino, N. Ishiguro, M. Shinagawa, M. Sato, and M. Horiuchi. 2004. Cell-surface retention of PrPC by anti-PrP antibody prevents protease-resistant PrP formation. J. Gen. Virol. 853473-3482. [DOI] [PubMed] [Google Scholar]
- 18.Legname, G., I. V. Baskakov, H. O. Nguyen, D. Riesner, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 2004. Synthetic mammalian prions. Science 305673-676. [DOI] [PubMed] [Google Scholar]
- 19.Legname, G., H. O. Nguyen, I. V. Baskakov, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 2005. Strain-specified characteristics of mouse synthetic prions. Proc. Natl. Acad. Sci. USA 1022168-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legname, G., H. O. Nguyen, D. Peretz, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 2006. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc. Natl. Acad. Sci. USA 10319105-19110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moroncini, G., N. Kanu, L. Solforosi, G. Abalos, G. C. Telling, M. Head, J. Ironside, J. P. Brockes, D. R. Burton, and R. A. Williamson. 2004. Motif-grafted antibodies containing the replicative interface of cellular PrP are specific for PrPSc. Proc. Natl. Acad. Sci. USA 10110404-10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muramoto, T., M. Scott, F. E. Cohen, and S. B. Prusiner. 1996. Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc. Natl. Acad. Sci. USA 9315457-15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norstrom, E. M., and J. A. Mastrianni. 2005. The AGAAAAGA palindrome in PrP is required to generate a productive PrPSc-PrPC complex that leads to prion propagation. J. Biol. Chem. 28027236-27243. [DOI] [PubMed] [Google Scholar]
- 24.Notari, S., S. Capellari, A. Giese, I. Westner, A. Baruzzi, B. Ghetti, P. Gambetti, H. A. Kretzschmar, and P. Parchi. 2004. Effects of different experimental conditions on the PrPSc core generated by protease digestion: implications for strain typing and molecular classification of CJD. J. Biol. Chem. 27916797-16804. [DOI] [PubMed] [Google Scholar]
- 25.Pan, T., P. Wong, B. Chang, C. Li, R. Li, S. C. Kang, T. Wisniewski, and M. S. Sy. 2005. Biochemical fingerprints of prion infection: accumulations of aberrant full-length and N-terminally truncated PrP species are common features in mouse prion disease. J. Virol. 79934-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parchi, P., A. Giese, S. Capellari, P. Brown, W. Schulz-Schaeffer, O. Windl, I. Zerr, H. Budka, N. Kopp, P. Piccardo, S. Poser, A. Rojiani, N. Streichemberger, J. Julien, C. Vital, B. Ghetti, P. Gambetti, and H. Kretzschmar. 1999. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol. 46224-233. [PubMed] [Google Scholar]
- 27.Peretz, D., M. R. Scott, D. Groth, R. A. Williamson, D. R. Burton, F. E. Cohen, and S. B. Prusiner. 2001. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 10854-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrier, V., K. Kaneko, J. Safar, J. Vergara, P. Tremblay, S. J. DeArmond, F. E. Cohen, S. B. Prusiner, and A. C. Wallace. 2002. Dominant-negative inhibition of prion replication in transgenic mice. Proc. Natl. Acad. Sci. USA 9913079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safar, J., H. Wille, V. Itri, D. Groth, H., Serban, M. Torchia, F. E. Cohen, and S. B. Prusiner. 1998. Eight prion strains have PrPSc molecules with different conformations. Nat. Med. 41157-1165. [DOI] [PubMed] [Google Scholar]
- 30.Safar, J. G., M. D. Geschwind, C. Deering, S. Didorenko, M. Sattavat, H. Sanchez, A. Serban, M. Vey, H. Baron, K. Giles, B. L. Miller, S. J. Dearmond, and S. B. Prusiner. 2005. Diagnosis of human prion disease. Proc. Natl. Acad. Sci. USA 1023501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinagawa, M., K. Takahashi, S. Sasaki, S. Doi, H. Goto, and G. Sato. 1985. Characterization of scrapie agent isolated from sheep in Japan. Microbiol. Immunol. 29543-551. [DOI] [PubMed] [Google Scholar]
- 32.Silveira, J. R., G. J. Raymond, A. G. Hughson, R. E. Race, V. L. Sim, S. F. Hayes, and B. Caughey. 2005. The most infectious prion protein particles. Nature 437257-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solforosi, L., A. Bellon, M. Schaller, J. T. Cruite, G. C. Abalos, and R. A. Williamson. 2007. Toward molecular dissection of PrPC-PrPSc interactions. J. Biol. Chem. 2827465-7471. [DOI] [PubMed] [Google Scholar]
- 34.Supattapone, S., P. Bosque, T. Muramoto, H. Wille, C. Aagaard, D. Peretz, H. O. Nguyen, C. Heinrich, M. Torchia, J. Safar, E. F. Cohen, S. J. DeArmond, S. B. Prusiner, and M. Scott. 1999. Prion protein of 106 residues creates an artificial transmission barrier for prion replication in transgenic mice. Cell 96869-878. [DOI] [PubMed] [Google Scholar]
- 35.Tateishi, J., M. Ohta, M. Koga, Y. Sato, and Y. Kuroiwa. 1979. Transmission of chronic spongiform encephalopathy with kuru plaques from humans to small rodents. Ann. Neurol. 5581-584. [DOI] [PubMed] [Google Scholar]
- 36.Thackray, A. M., L. Hopkins, M. A. Klein, and R. Bujdoso. 2007. Mouse-adapted ovine scrapie prion strains are characterized by different conformers of PrPSc. J. Virol. 8112119-12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzaban, S., G. Friedlander, O. Schonberger, L. Horonchik, Y. Yedidia, G. Shaked, R. Gabizon, and A. Taraboulos. 2002. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 4112868-12875. [DOI] [PubMed] [Google Scholar]
- 38.Uryu, M., A. Karino, Y. Kamihara, and M. Horiuchi. 2007. Characterization of prion susceptibility in Neuro2a mouse neuroblastoma cell subclones. Microbiol. Immunol. 51661-669. [DOI] [PubMed] [Google Scholar]
- 39.Zulianello, L., K. Kaneko, M. Scott, S. Erpel, D. Han, F. E. Cohen, and S. B. Prusiner. 2000. Dominant-negative inhibition of prion formation diminished by deletion mutagenesis of the prion protein. J. Virol. 744351-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]