Abstract
Human T-cell leukemia virus type 1 (HTLV-1) infection causes adult T-cell leukemia and is associated with a variety of lymphocyte-mediated disorders. It has been hypothesized that a highly regulated pattern of HTLV-1 gene expression is critical for virus survival and disease pathogenesis. In this study, real-time reverse transcriptase PCR was used to determine the kinetics of viral gene expression in cells transiently transfected with an HTLV-1 proviral plasmid, in newly infected human peripheral blood mononuclear cells (PBMCs), and in PBMCs from newly infected rabbits. The HTLV-1 gene expression profiles in transiently transfected and infected cells were similar; over time, all transcripts increased and then maintained stable levels. gag/pol, tax/rex, and env mRNA were detected first and at the highest levels, whereas the expression levels of the accessory genes, including the antisense Hbz, were significantly lower than the tax/rex levels (ranging from 1 to 4 logs depending on the specific mRNA). In infected rabbits, tax/rex and gag/pol mRNA levels peaked early after inoculation and progressively decreased, which correlated inversely with the proviral load and host antibody response against viral proteins. Interestingly, Hbz mRNA was detectable at 1 week postinfection and increased and stabilized. The expression levels of all other HTLV-1 genes in infected rabbit PBMCs were at or below our limit of detection. This analysis provides insight into viral gene expression under various in vitro and in vivo experimental conditions. Our in vivo data indicate that in infected rabbits, Hbz mRNA expression over time directly correlates with the proviral load, which provides the first evidence linking Hbz expression to proviral load and the survival of the virus-infected cell in the host.
Human T-cell leukemia virus type 1 (HTLV-1) is a complex oncogenic retrovirus that causes adult T-cell leukemia/lymphoma (ATL) after a long clinically latent period (>30 years). HTLV-1 has the capacity to infect and transform primary human T lymphocytes both in cell culture and in infected individuals (18, 50). However, the relationship of specific viral gene expression and infected-cell survival, ultimately resulting in oncogenic transformation of T lymphocytes, is not completely understood.
HTLV-1 utilizes both strands of its proviral genome to express multiple gene products from unspliced mRNA and a complex array of alternative spliced mRNA. In addition to the gag, pol, and env genes that encode the structural and enzymatic proteins of all replication-competent retroviruses, HTLV-1 encodes the regulatory tax and rex genes and open reading frame I and II accessory genes from the positive genome strand. The negative-sense strand of the genome encodes the HTLV-1 B-Zip (Hbz) accessory gene. Tax increases the rate of viral gene transcription from the promoter located in the viral long terminal repeat (LTR) (9, 16, 21); furthermore, Tax modulates the transcription or activity of numerous cellular genes involved in cell growth and differentiation, cell cycle control, and DNA repair (8, 30, 36, 37, 41). Compelling evidence implicates Tax as the key viral protein required for cellular transformation and oncogenesis (17, 38, 39). The second regulatory protein, Rex, acts posttranscriptionally by preferentially binding, stabilizing, and selectively exporting the unspliced and incompletely spliced viral mRNA from the nucleus to the cytoplasm, essentially regulating production of the virion components (52).
To date, the accessory proteins (p12, p30, and p13) have not been detected in HTLV-infected or transformed cells, but functional activities have been revealed based on overexpression studies (33). p12 activates cells by regulating calcium signaling (15). p12 also enhances LFA-1 T-cell adhesion (24) and associates with cellular proteins, including the 16-kDa subunit of the vacuolar ATPase, interleukin-2 (IL-2) receptor β and γ chains, and the major histocompatibility complex class I heavy chain (22, 26, 34). p30 differentially modulates viral and/or cellular gene expression at the transcriptional level through association with p300/CBP and TIP60 and posttranscriptionally via binding and retaining tax/rex mRNA in the nucleus (4, 54). p13, localized to the mitochondria, has suppressive effects on cell growth in cell culture (42). Unlike p12, p30, and p13, the antisense HBZ protein is detected in most HTLV-1-transformed cell lines, HTLV-1 proviral-plasmid-transfected cells, and samples from ATL patients (2, 10, 31, 40). HBZ protein can interact with CREB and Jun family members, altering transcription factor binding and transactivation of both viral and cellular promoters (29). Hbz gene expression has also been linked to cellular proliferation (3, 40, 51). Interestingly, all of the accessory proteins are dispensable for viral replication and cellular transformation under standard cell culture conditions (2, 11, 14). However, in vivo studies using a rabbit model of infection have revealed that p12, p30, p13, and HBZ are important for the enhancement of infectivity and the establishment of persistent infection (2, 13, 20, 43), indicating that the activities of these proteins likely play key roles in viral infection, spread, and escape from the immune system.
Characterization of the viral gene expression profile throughout the processes of infection and immortalization/transformation is likely to provide key functional information on viral gene products and their potential contribution to HTLV pathogenesis. To date, such extensive analysis has not been undertaken due to a challenging assay system and the difficulty in detecting viral mRNA present in low abundance. The ultimate fate of HTLV-1-infected T-cells in vivo likely depends on their ability to balance proliferation, cell cycle control, and antiapoptotic signals that are mediated by viral and cellular proteins against the ability to evade the host immune response. Therefore, how the virus responds to environmental signals and regulates viral and cellular gene expression are critical to its long-term survival and persistence in infected individuals.
Real-time reverse transcriptase PCR (RT-PCR) is a highly sensitive method allowing the quantitation of low concentrations of mRNA transcripts and the discrimination of small changes in gene expression. We previously developed a series of oligonucleotide primer pairs and probes to quantitate all HTLV-1 mRNA species using TaqMan real-time RT-PCR (27). In the present study, we utilize this approach to measure the kinetics of viral gene expression in cells transiently transfected with an HTLV-1 proviral plasmid, in human peripheral blood mononuclear cells (PBMCs) newly infected with HTLV-1 in culture, and in PBMCs harvested from newly HTLV-1-inoculated rabbits. The HTLV-1 gene expression profiles in transiently transfected and infected cells were similar, with a general increase in all transcripts over time. gag/pol, tax/rex, and env mRNA were expressed first and at the highest levels. In infected rabbits, tax/rex and gag/pol mRNA peaked early after inoculation and decreased progressively, in contrast to an increasing proviral load and the host antibody response against viral proteins. The Hbz mRNA level was low but detectable 1 week postinfection and increased over time. Although our in vitro analysis revealed that tax/rex and gag/pol mRNA are expressed first, followed by the expression of all other transcripts (all increasing and then stabilizing over time), we detected no apparent regulatory control of specific transcripts. However, our in vivo analysis showed that tax/rex and gag/pol mRNA were expressed at the highest levels immediately after infection and then progressively declined over time, eventually stabilizing at low levels. Conversely, Hbz was expressed at a low level early after infection but continued to increase before reaching a plateau, in direct correlation with proviral load levels in infected rabbit PBMCs. Therefore, our results revealed an inverse correlation between tax/rex and Hbz mRNA expression over time, providing the first evidence linking Hbz expression to proviral load and the survival of the virus-infected cell in an infected host.
MATERIALS AND METHODS
Cells and plasmids.
293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and antibiotics (2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin). 729 is a human B-lymphoblast line (19). 729ACHneo (referred to as 729HTLV-1) is a 729-derived stable ACHneo transfectant cell line that produces HTLV-1 with the capacity to infect and transform human peripheral blood T lymphocytes (50). Both 729 and 729HTLV-1 cells were maintained in Iscove's medium supplemented with 10% FBS and antibiotics. Human and rabbit PBMCs were isolated by using Ficoll Hypaque (Amersham, Piscataway, NJ) and Percoll (Amersham, Piscataway, NJ), respectively, and cultured in RPMI 1640 medium supplemented with 20% FBS, antibiotics, and 10 U/ml recombinant human IL-2 (Roche Applied Biosciences, Indianapolis, IN).
The HTLV-1 proviral plasmid ACHneo has been described previously (1). Plasmids containing specific HTLV-1 cDNA gene sequences and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA gene sequences have been described previously (27) and were used to generate standard curves for the TaqMan real-time PCR assays. Plasmid ML789 is pCRScript derived (Stratagene, La Jolla, CA) and contains a rabbit GAPDH gene fragment that was amplified from rabbit PBMC genomic DNA.
Transient transfection, p19 ELISA and Western blotting.
293T cells (8 × 105) were transfected with 8 μg of the ACHneo plasmid by using the Lipofectamine method (Invitrogen, Carlsbad, CA). Cells and culture supernatants were harvested at 4, 12, 20, 28, 44, and 60 h posttransfection. p19 enzyme-linked immunosorbent assay (ELISA) (Zeptometrix Corporation, Buffalo, NY) was performed to quantitate HTLV-1 Gag protein production in the culture supernatants. Cell pellets were lysed for Western blot analysis to detect HTLV-1 p24, Tax, and Rex protein expression. Western blots were performed as described previously (53). The following dilutions were used for individual antibodies against different viral proteins: HTLV-1 rabbit anti-Rex1 polyclonal antisera, 1:1,000; HTLV-1 mouse anti-Tax1 monoclonal antibody (NIH catalog no. 1318), 1:1,000; and HTLV-1 mouse anti-p24 (clone 46/3.24.4; ZeptoMetrix Corp, Buffalo, NY), 1:1,000. Horseradish peroxidase-conjugated anti-immunoglobulin G (IgG; mouse or rabbit as appropriate) was used as the secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and proteins were visualized by using an electrochemiluminescence Western blot analysis system (Amersham Biosciences, Piscataway, NJ).
Long-term immortalization assay.
Irradiated 729HTLV-1 producer cells (1 × 106) were cocultured with 2 × 106 freshly isolated PBMCs in the presence of IL-2 (10 U/ml) in 24-well culture plates. An immortalization growth curve was generated by enumerating viable cells per well (an average of three wells per time point) by trypan blue exclusion at weekly intervals. HTLV-1 expression was confirmed by detection of p19 Gag protein in culture supernatants by ELISA at weekly intervals. Immortalized cells were defined as cells with continuous proliferation >8 weeks postcoculture.
Rabbit inoculation procedures.
Twelve-week-old specific-pathogen-free New Zealand White rabbits (Hazelton, Kalamazoo, MI) were inoculated via the lateral ear vein with 107 gamma-irradiated (7,500 rad) 729HTLV-1 (nine rabbits) or uninfected 729 control cells (two rabbits). At 0, 1, 2, 4, 6, and 8 weeks postinoculation, 15 ml of blood was drawn from the central ear artery of each animal for testing. This protocol was approved by University Laboratory Animal Resources (ULAR) of The Ohio State University. Plasma serum reactivity to specific viral antigenic determinants was detected by using a commercial HTLV-1 Western blot assay (GeneLabs Diagnostics, Singapore) with a modification in which goat anti-human IgG conjugated with alkaline phosphatase was replaced with goat anti-rabbit IgG conjugated with alkaline phosphatase (Chemicon, Temecula, CA). Serum (dilution of 1:200) showing reactivity to Gag (p24 or p19) and Env (gp21 or gp46) antigens was classified as positive.
DNA/RNA preparation and cDNA synthesis.
Genomic DNA was extracted from 107 rabbit PBMCs by using an Easy-DNA kit (Invitrogen Corp., Carlsbad, CA). Total RNA was extracted from transfected 293T cells, HTLV-1-immortalized human T lymphocytes, or rabbit PBMCs by using an RNeasy kit and subjected to DNase-treated columns. Poly(A)+ mRNA was isolated from total mRNA of transfected cells or immortalized cell lines by using an Oligotex kit (Qiagen, Inc., Valencia, CA). First-strand cDNAs were prepared with a SuperScript first-strand synthesis system for RT-PCR (Invitrogen Corp., Carlsbad, CA) using random hexamers and 200 ng (total mRNA) or 80 ng [poly(A)+ mRNA] of input RNA per reaction mixture according to the manufacturer's instructions.
TaqMan real-time PCR and real-time RT-PCR.
TaqMan real-time PCR (Applied Biosystems, Foster City, CA) was performed to quantitate the proviral copy number per cell in infected rabbit PBMCs. An amount of 500 ng of rabbit genomic DNA was put into each reaction mixture as template for TaqMan real-time PCR using HTLV-1-specific primers #19 and #20 and probe TMP-3 as previously described (27). Absolute copy number was determined by extrapolation against a standard curve generated from log10 dilutions of plasmid DNA containing known copies of the gag/pol sequence (27). Each reaction was performed in duplicate, and the results were averaged. The copy number-per-cell value for a sample was generated based on the estimation that 1 μg rabbit PBMC DNA is equivalent to 134,600 cells. The specific primers, probes, and protocol for TaqMan real-time RT-PCR to quantitate all HTLV-1 mRNA and human GAPDH mRNA were previously described (27). Although several Hbz transcripts have been described, i.e., major spliced (SP-1), minor spliced (SP-2), and unspliced (10, 46), this study focused on the detection of the major spliced transcript since it has been shown to be expressed at the highest concentration in ATL (46) and HTLV-1 T-cell lines (27), its expression correlated directly with protein expression in HTLV-1-infected cells (3), and it was shown to have growth-promoting effects in T cells (3, 51). Rabbit GAPDH mRNA was quantified by using primers rGAPDH-S (5′-GATGCTGGTGCCGAGTACGTGG-3′) and rGAPDH-AS (5′-GTGGTGCAGGATGCGTTGCTGA-3′) and probe BY-1 (5′FAM-ACCACCATGGAGAAGGCCGGG-TAMRA3′). The total mRNA copy number was determined by using a standard curve generated from log10 dilutions of a plasmid containing the corresponding cDNA sequence for each mRNA and normalized to 1 million copies of human or rabbit GAPDH mRNA.
RESULTS
Kinetic analysis of HTLV-1 gene expression in transiently transfected 293T cells.
Multiple gene products from unspliced mRNA and a complex array of alternative spliced mRNA are produced by HTLV-1 (Fig. 1). Although HTLV-1 gene expression appears to be highly regulated, a kinetic expression profile of all the viral genes has been difficult to assess experimentally. We previously generated and characterized a series of oligonucleotide primer pairs and probes to specifically quantitate all HTLV-1 mRNA species by using TaqMan real-time RT-PCR (27). We initially determined the HTLV-1 gene expression profile at defined time points following the transient transfection of 293T cells with the HTLV-1 proviral plasmid, ACHneo. Cells were transfected and harvested at 0, 4, 12, 20, 28, and 44 h posttransfection, and poly(A)+ mRNA was isolated. We confirmed that the poly(A)+ mRNA samples from each time point were free of DNA contamination as determined by the failure of gag/pol-specific primers to amplify a fragment by standard RT-PCR in the absence of RT (data not shown). cDNA was synthesized from poly(A)+ mRNA by using RT and random hexamers and further subjected to real-time PCR with specific primers and probes to quantitate the cellular GAPDH mRNA (to be used as a normalization control) and each specific viral mRNA. As early as 4 h posttransfection, we were able to detect unspliced gag/pol mRNA and tax/rex doubly spliced mRNA (Table 1). At 4 h, tax/rex mRNA was the most abundant transcript, at levels approximately two- to threefold higher than the level of full-length gag/pol mRNA (5.5 × 102 versus 1.93 × 102 per 106 copies of GAPDH mRNA). However, over time (12 to 44 h), tax/rex and gag/pol mRNA increased by several logs, with the full-length gag/pol mRNA consistently expressed at the highest concentration. Expression of env and the accessory genes was first detected at 12 h and increased slightly over the time course; however, their levels ranged from 1 to 4 logs lower than the levels of tax/rex depending on the specific mRNA. p30 gene expression remained below our detection limit throughout the time course (<25 copies). Failure to detect p30 expression is consistent with the results of our previous study, in which p30 mRNA expression levels in HTLV-1-transformed cell lines were 1 to 2 logs lower than other accessory gene expression levels (27).
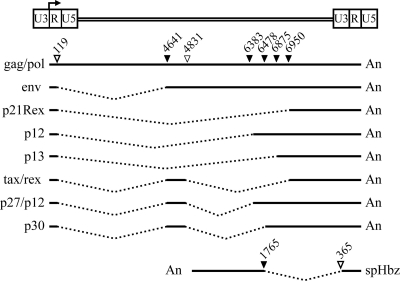
FIG. 1.
Provirus genome of HTLV-1 and its unspliced, singly spliced, and doubly spliced mRNA. At least eight positive-sense transcripts and one negative-sense transcript are expressed by HTLV-1. The genomic unspliced mRNA encodes the Gag, Pol, and Pro proteins. Four singly spliced mRNA species are the result of splicing of exon 1 (nucleotides 1 to 119) to major splice acceptors at positions 4641 (Env), 6383 or 6478 (p12), 6950 (p21rex), and 6875 (p13). The three doubly spliced mRNA include exon 1, exon 2 (4641 to 4831), and a third exon that starts at position 6950 (Tax/Rex), 6478 (p30), or 6383 (p27/p12). The Hbz singly spliced major antisense transcript initiates at multiple sites in the 3′ LTR and utilizes a splice donor site at position 365 and a splice acceptor site at nucleotide position 1765 (Hbz unspliced and minor spliced transcripts not shown). Nucleotide numbering starts at the beginning of the R region for the positive-sense transcripts and the last nucleotide of U5 in the 3′ LTR for the antisense transcript. Black lines designate exons, and dotted lines designate introns. Major utilized splice donor (open triangles) and splice acceptor (closed triangles) sites are indicated above the unspliced mRNA. An, poly(A) adenylation; spHbz, spliced Hbz.
TABLE 1.
Kinetic analysis of HTLV-1 transcript expression in transfected 293T cellsa
| mRNA transcript | Copy no. at indicated time (h) posttransfection
|
|||||
|---|---|---|---|---|---|---|
| 0 | 4 | 12 | 20 | 28 | 44 | |
| gag/pol | 0.00E+00 | 1.93E+02 | 9.33E+04 | 2.55E+05 | 2.09E+05 | 1.22E+05 |
| tax/rex | 0.00E+00 | 5.25E+02 | 4.11E+04 | 7.77E+04 | 1.26E+05 | 1.08E+05 |
| env | 0.00E+00 | 0.00E+00 | 2.07E+03 | 7.57E+03 | 7.83E+03 | 5.27E+03 |
| p21rex | 0.00E+00 | 0.00E+00 | 1.82E+01 | 1.08E+02 | 3.37E+02 | 7.44E+02 |
| p12 | 0.00E+00 | 0.00E+00 | 7.66E+02 | 1.88E+03 | 1.91E+03 | 3.28E+03 |
| p27/p12 | 0.00E+00 | 0.00E+00 | 2.08E+02 | 8.22E+02 | 9.71E+02 | 3.22E+02 |
| p13 | 0.00E+00 | 0.00E+00 | 4.23E+01 | 1.44E+02 | 2.43E+02 | 2.14E+03 |
| p30 | 0.00E+00 | 0.87E+00 | 1.06E+00 | 1.78E+00 | 2.10E+00 | 3.65E+00 |
| Hbz | 0.00E+00 | 0.00E+00 | 4.71E+01 | 1.33E+02 | 6.92E+01 | 2.30E+02 |
293T cells were transfected with the HTLV-1 proviral clone ACHneo, and RNA was isolated at indicated time points posttransfection. Copy numbers shown are the averages from two transfections and are normalized to 1 million copies of GAPDH. Detection limit cutoff is 25 copies.
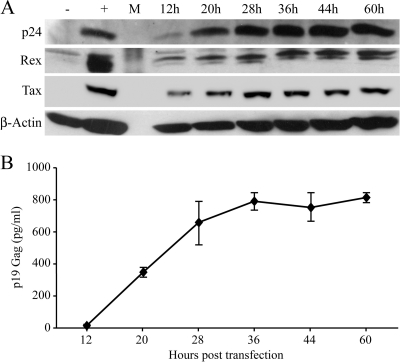
To determine if viral protein production kinetics correlated with mRNA expression kinetics, Western blotting was performed on transfected cell lysates to detect HTLV-1 p24 Gag, Tax, and Rex proteins. Viral protein expression was initially detected in cell lysates at 12 h posttransfection. In direct correlation with mRNA expression, protein levels increased and then plateaued at later time points (Fig. 2A). Quantitation of p19 Gag in the supernatants of transfected cells, which is a measure of new virion production, showed a similar pattern (Fig. 2B). Taken together, our results revealed that, following transient transfection of cells, all HTLV-1 gene transcripts increased over time, eventually reaching a plateau at 28 to 44 h. gag/pol, tax/rex, and env mRNA were expressed first and at the highest levels, whereas expression levels of the accessory genes, including the antisense Hbz, were significantly lower than those of tax/rex.
FIG. 2.
Time course of HTLV-1 protein expression following transient transfection. 293T cells (8 × 105) were transfected with 8 μg HTLV-1 ACHneo plasmid by using Lipofectamine in 100-mm plates. Culture supernatants and cell lysates were harvested at 12, 20, 28, 36, 44, and 60 h posttransfection. (A) Detection of HTLV-1 p24, Rex, and Tax protein expression in transfected-cell lysates by Western blot analysis. β-Actin levels were assessed as a loading control. +, present; −, absent; M, marker. (B) p19 Gag ELISA was used to quantify HTLV-1 viral particle production in culture supernatant. p19 was first detected in the supernatant at 12 h posttransfection. Error bars indicate standard deviations of the results of two independent transfections performed in triplicate.
Kinetic analysis of HTLV-1 gene expression in PBMCs infected with HTLV-1 in cell culture.
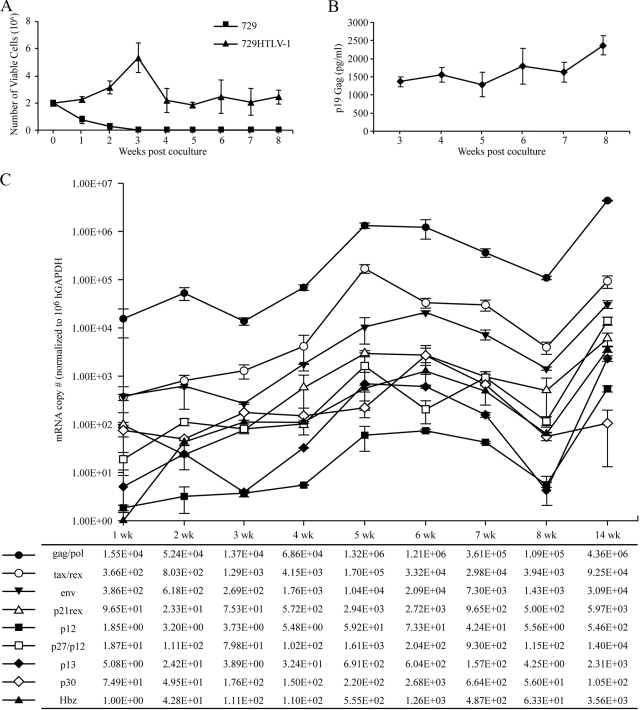
We next characterized HTLV-1 gene expression in newly infected cells and throughout the immortalization process in vitro. Freshly isolated human PBMCs were cocultivated with lethally irradiated 729HTLV-1 producer cells in the presence of a low concentration of recombinant IL-2 (10 U/ml). Cell number and viability were monitored at weekly intervals to follow the immortalization process and the characteristic expansion of T-cells from the PBMC mixed-cell population. The growth curve of a representative assay indicates a progressive loss of viable cells over time in cocultures containing irradiated uninfected 729 cells and PBMCs (Fig. 3A). In contrast, immortalization is clearly apparent for the PBMC/729HTLV-1 coculture, in which the cell number increased early, followed by a cell number decrease, as the newly infected cells underwent a proliferative burst and subsequent crisis stage. After the crisis stage, the cell number remained fairly constant as cell growth was offset by cell death. Surviving cells were proliferating [as shown by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assays], harbored provirus, and were considered to be immortalized by HTLV-1 (49, 50). A measurable increase in cell number is typically not observed until after 10 to 12 weeks in culture (3, 48). We detected p19 Gag in the culture supernatants, confirming HTLV-1 infection, replication, and virion production (Fig. 3B).
FIG. 3.
HTLV-1 T-lymphocyte immortalization assay. Freshly isolated human PBMCs (2 × 106) were cultured with 106 γ-irradiated 729HTLV-1 producer cells in 24-well plates. (A) Representative growth curve is presented to show cell viability as determined by trypan blue exclusion at weekly intervals. Numbers shown at each time point are the average of the results from three random wells, with error bars denoting standard deviations. (B) p19 Gag ELISA was used to quantify HTLV-1 virion production by detecting Gag protein in the culture supernatants. Numbers shown at each time point are the averages of the results from three random wells, with error bars denoting standard deviations. First time point begins at 3 weeks when there is no remaining signal in the supernatant from irradiated producer cells (49). (C) Real-time RT-PCR was performed on cells from triplicate wells at weekly intervals (1 to 8) and at a final 14-week time point to quantify the HTLV-1 transcript levels throughout the immortalization process. The data are presented graphically, with error bars denoting standard deviations, and with the average numbers displayed below. Human GAPDH mRNA was used as the internal control, and numbers presented are normalized to 1 × 106 copies of human GAPDH mRNA.
PBMCs from random cocultured wells were harvested each week for poly(A)+ mRNA extraction. Poly(A)+ mRNA was subjected to real-time RT-PCR for quantitation of each specific HTLV-1 mRNA. HTLV-1 mRNA expression following infection of PBMCs was consistent with the cell growth and immortalization observations (Fig. 3C): HTLV-1 mRNA levels increased from weeks 1 to 5, significantly decreased during the typical cell growth crisis stage (weeks 6, 7, and 8), and slowly recovered to relatively high steady-state levels as the surviving newly immortalized cells expanded. The general gene expression profile in newly infected PBMCs was similar to that in cells transiently transfected with proviral plasmid: full-length gag/pol mRNA (approximately 106 copies per 106 copies of GAPDH mRNA) > doubly spliced tax/rex mRNA (105 copies) > singly spliced env mRNA (104 copies). The levels of transcripts encoding the accessory proteins (p21, p12, p30, and p13), including the major antisense spliced mRNA encoding HBZ, were significantly lower than tax/rex mRNA (ranging from 1 to 4 logs depending on the specific mRNA). We might expect viral gene expression to be dynamic and regulated at both the transcriptional (primarily Tax mediated) and posttranscriptional (Rex mediated) level as viral protein products are produced. For example, as Rex increases and functions to facilitate export of the incompletely spliced mRNA, viral mRNA concentrations will be redistributed, with more gag/pol and env mRNA at the expense of tax/rex and other completely spliced mRNA. This in turn would reduce Tax and alter viral transcription. However, other than gag/pol and tax/rex transcripts being expressed first and consistently at the highest levels throughout the time course, there did not appear to be a highly regulated viral gene expression pattern following transfection or infection of cells in vitro.
Kinetic analysis of HTLV-1 gene expression in PBMCs from HTLV-1-inoculated rabbits.
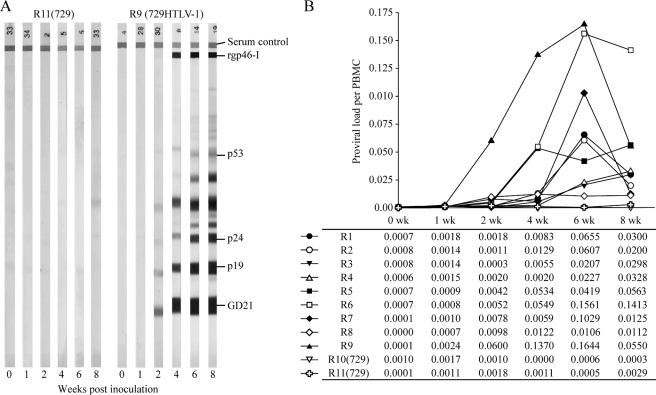
To evaluate the HTLV-1 gene expression profile in vivo, we utilized our established rabbit animal model. HTLV-1 consistently infects rabbits and results in a persistent infection in T lymphocytes. In addition, primary rabbit T lymphocytes can be immortalized/transformed in culture as has been reported for human cells (12). Twelve-week-old New Zealand White rabbits were inoculated with 1 × 107 γ-irradiated 729HTLV-1 producer cells (nine rabbits) or uninfected 729 cells as a control (two rabbits). Rabbit blood was drawn at weeks 0, 1, 2, 4, 6, and 8 after inoculation, and plasma and PBMCs were isolated for further analysis. To determine the serologic response of rabbits to HTLV-1 infection, we measured anti-HTLV-1 antibody responses in rabbits by Western blot analysis. A representative seroconversion pattern from each of the inoculated groups is shown in Fig. 4A (rabbit plasma was diluted 1:200). Seroconversion was detected in the 729HTLV-1-inoculated rabbits starting at week 2, and antibody titers rose over the time course of the experiment. As expected, we were unable to detect any antibody response in rabbits inoculated with control 729 cells. To further confirm infection status and quantitate the HTLV-1 proviral load in inoculated rabbits over time, DNA was extracted from isolated PBMCs and subjected to real-time PCR (Fig. 4B). Integrated HTLV-1 proviral DNA was detected as early as 2 weeks postinoculation, and although the loads varied for individual rabbits, they increased over time. By week 8, the HTLV-1 proviral loads in the nine infected rabbits varied from 0.02 to 0.14 copies per cell, with a mean proviral load value of 0.043 (43 infected cells per 103 rabbit PBMCs).
FIG. 4.
Assessment of HTLV-1 infection in rabbits. Twelve-week-old New Zealand White rabbits were inoculated with 1 × 107 γ-irradiated 729HTLV-1 producer cells (nine total rabbits) or uninfected 729 cells as a control (two total rabbits). Following inoculation, 15 ml of blood was drawn from each rabbit at week 0, 1, 2, 4, 6, and 8 for collection of sera and rabbit PBMCs. (A) Sera from inoculated rabbits were tested for reactivity to specific HTLV-1 proteins by Western blotting. Results for a representative rabbit from each group, as indicated, are shown with reactive viral proteins labeled on the right. rgp46, HTLV-1-specific recombinant envelope surface protein; p53, Gag precursor; p24, capsid; p19, matrix; GD21, recombinant envelope transmembrane. Results labeled “Serum control” indicate comparable concentrations of serum Ig levels among the samples. (B) Genomic DNA was isolated from rabbit PBMCs and subjected to TaqMan real-time PCR using HTLV-1-specific primers #19 and #20 and probe TMP-3; the standard curve was generated by serial 10-fold dilutions of plasmid DNA (27). The proviral load is plotted against weeks postinfection, with the actual copy number per cell displayed below. The copy number per cell value for a sample was generated based on the estimation that 1 μg of PBMC DNA is equivalent to 134,600 cells. R1 to R11, rabbits 1 to 11.
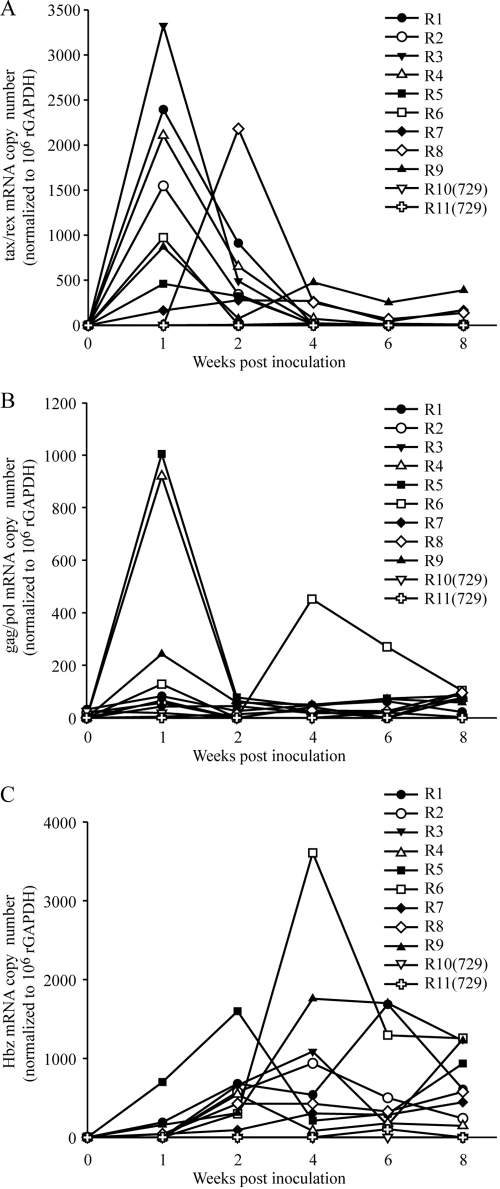
To evaluate the kinetics of HTLV-1 gene expression in inoculated rabbits, total mRNA was extracted from rabbit PBMCs harvested at week 0, 1, 2, 4, 6, and 8 postinoculation and subjected to real-time RT-PCR. tax/rex mRNA was expressed very early after infection and at the highest levels of all viral mRNA detected (3,000 copies per 106 GAPDH mRNA copies). After 1 to 2 weeks, tax/rex mRNA progressively decreased and stabilized at relatively low levels (Fig. 5A). gag/pol mRNA expression mirrored that of tax/rex mRNA expression but with an average magnitude that was approximately fourfold lower at its peak (Fig. 5B). With the exception of Hbz mRNA, all other HTLV-1 mRNA in rabbit PBMCs, including those encoding Env and the accessory proteins, were below our limit of detection (25 copies; data not shown). Hbz mRNA was detectable at low levels early after inoculation and slowly increased and stabilized (Fig. 5C). Interestingly, at 8 weeks postinfection, the Hbz mRNA was expressed at the highest concentration of all mRNA measured (an average of ninefold more than tax/rex mRNA). We have shown that the level of the major spliced Hbz mRNA transcript in HTLV-1-transformed human and rabbit T-cell lines and newly immortalized human T lymphocytes directly correlates with HBZ protein production (3; data not shown). However, we have been unable to detect HBZ protein in PBMCs isolated from infected rabbits, which is likely a reflection of the limited number of infected cells in PBMCs of inoculated rabbits.
FIG. 5.
Kinetics and profile of HTLV-1 mRNA expression in infected rabbits. RNA extracted from rabbit PBMCs (harvested from rabbits at week 0, 1, 2, 4, 6, and 8 postinoculation as described in Fig. 4 legend) were subject to real-time RT-PCR to quantitate tax/rex mRNA expression (A), gag/pol and full-length genomic mRNA expression (B), and Hbz antisense mRNA expression (C). Numbers shown are values normalized to 1 million copies of rabbit GAPDH mRNA. R1 to R11, rabbits 1 to 11.
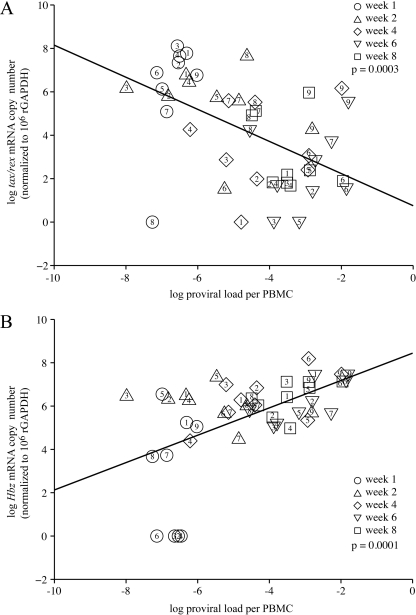
We next compared tax/rex and Hbz gene expression directly to proviral load in an effort to understand the relationship between specific gene expression and viral spread and persistence. Typical of HTLV-1 infection, the proviral load in PBMCs was low early after infection and increased over time until a stable proviral load (“set point”) unique to each infected rabbit was reached (Fig. 4B). We noted that the rabbits that expressed the highest levels of tax/rex mRNA at the early time points had lower proviral loads at later weeks; this is likely reflective of a more robust immune response and the elimination of infected cells (compare Fig. 4B with Fig. 5A). Our data also indicated that tax/rex mRNA expression was highest early after infection and then decreased, which correlated inversely with the proviral load (Fig. 6A). In contrast, Hbz mRNA expression started out low and then slowly increased and stabilized in direct correlation with the proviral load (Fig. 6B). We did note that rabbit 8 was a bit of an outlier for tax/rex expression kinetics and rabbit 6 was an outlier for both gag/pol and Hbz expression kinetics (Fig. 5). Elimination of the results for these two rabbits in our analysis whose results are presented in Fig. 6 did not significantly alter the curve or P values and thus did not alter the interpretation (data not shown). Interestingly, the timing of expression of Hbz is consistent with its reported cellular proliferative function and its maintenance in ATL cells in which the Tax oncoprotein is rarely expressed.
FIG. 6.
Relationship between HTLV-1 proviral load and tax/rex or Hbz mRNA levels in infected rabbits. We used log transformation of both proviral load (data used are those in Fig. 4) and tax/rex or Hbz mRNA (data used are those in Fig. 5) to stabilize the variance. Linear mixed modeling was employed to take account of correlated multiple observations in a time series from the same rabbit. (A) The data show a negative correlation between log tax/rex mRNA expression and log proviral load in HTLV-1-infected rabbits (P = 0.0003). (B) The data show a direct correlation between log Hbz mRNA and log proviral load in HTLV-1-infected rabbits (P = 0.0001). Each weekly time point is designated by a distinct symbol, as indicated, and the result for each individual infected rabbit per weekly time point is denoted (1 to 9) inside the symbol.
DISCUSSION
Although compelling evidence indicates that the HTLV-1 Tax protein is critical for viral oncogenesis, the viral gene expression profile throughout the immortalization process and the relationships of individual gene expression to infected cell survival and cellular transformation have not been assessed. Alternative splicing of mRNA is important for specific gene regulation (28), and HTLV-1 produces a set of closely related mRNA by alternative splicing. We and others have hypothesized that a precise blueprint of the viral gene expression profile might provide useful information for dissecting the functional role of specific viral genes in the process of viral infection and cellular transformation (17, 23, 33). In this study, real-time RT-PCR was used to determine the kinetics of viral gene expression using three systems: 293T cells transiently transfected with HTLV-1 proviral plasmids, human PBMCs infected with HTLV-1 in vitro, and PBMCs harvested from HTLV-1-inoculated rabbits. Our results provide a composite picture of HTLV-1 gene expression under multiple experimental systems and, most notably, provide the first kinetics analysis of mRNA expression in a newly infected host. More importantly, we provide the first evidence linking Hbz expression to proviral load and the survival of the virus-infected cell in vivo.
Transient transfection of 293T cells with the HTLV-1 plasmid revealed tax/rex completely spliced mRNA as early as 4 h posttransfection at approximately twofold greater levels than unspliced gag/pol/genome mRNA. Subsequently (12 to 44 h), transcription of these two mRNA increased dramatically and then stabilized with the gag/pol mRNA expressed at the highest concentration. Doubly or completely spliced mRNA (encoding Tax and Rex) was expressed first, followed by a significant increase in unspliced and incompletely spliced transcripts. These results fit a model where, initially, Tax enhances viral transcription, but as Rex increases in concentration, it functions to inhibit splicing and facilitate export of the unspliced and singly spliced mRNA (5, 23, 52). Expression of env and the other accessory genes was detected at 12 h, increased slightly over time, and plateaued at levels significantly (1 to 4 logs) lower than tax/rex or gag/pol. Thus, it is clear from our results that the incompletely spliced and doubly spliced transcripts are not generated at the same rate from full-length transcripts.
Kinetic analysis of HTLV-1 mRNA expression in newly infected human PBMCs showed a specific-gene expression pattern for HTLV-1 mRNA that was similar to the results from proviral-plasmid-transfected 293T cells, where transcript levels were consistent with the cell growth and the immortalization process. All HTLV-1 mRNA levels increased between weeks 1 and 5, with gag/pol, tax/rex, and env expressed at the highest concentrations. However, during the typical cell growth crisis stage between weeks 6 and 8, all transcript levels declined significantly (mirroring cell growth) but slowly recovered to steady-state and relatively high levels as the surviving newly immortalized cells expanded. The levels of pX transcripts (encoding Tax/Rex and accessory gene products) in newly immortalized human cells were consistent with previous studies of HTLV-1 gene expression in established transformed cell lines: tax/rex > p21rex ≥ p12, p27/p12, p13, and p30 (27, 35). However, our data provide the first kinetic expression analysis of the antisense Hbz major spliced transcript, which we observed to mirror the expression pattern and levels of the other accessory gene transcripts, with gradual increase over time and stabilization approximately two logs below tax/rex mRNA levels. Although functional overexpression studies revealed that p30 and HBZ can modulate viral gene expression by distinct mechanisms at the transcriptional level (p30 and HBZ) and the posttranscriptional level (p30) (2, 4, 32, 53, 54), there was no obvious alteration in overall or specific gene expression that could be directly correlated with their mRNA expression patterns. One study showed that in the context of a proviral clone, the repressive effects of HBZ on Tax transcription were not apparent following transient transfection (2). It is possible that these proteins may not achieve a threshold level or be expressed within the proper microenvironment to function in vitro, results consistent with both p30 and HBZ being dispensable in cell culture but required for efficient infectivity and persistence in infected rabbits (2, 7, 14).
Rabbits inoculated with HTLV-1 induce a significant antibody response to viral antigens and become persistently infected. Proviral loads were variable among individual rabbits but in general increased over time before stabilizing or reaching a set point. Our kinetics analysis of HTLV-1 mRNA expression in newly infected rabbits indicated that tax/rex mRNA was the most abundant mRNA detected and had expression levels that correlated inversely with proviral loads. tax/rex mRNA peaked very early and after 1 to 2 weeks postinfection progressively decreased and stabilized at relatively low levels. gag/pol mRNA expression mirrored that of tax/rex mRNA expression but with an average magnitude approximately fourfold lower at its peak. DNA methylation of retroviruses is one host defense mechanism for inactivating retrovirus gene expression (47) which may ultimately allow virus-infected cells to escape from the host immune system and establish a latent state. Consistent with the mRNA expression pattern we observed in infected rabbits over time, a previous study reported that the gag/pol and env regions of the HTLV-1 proviruses in infected individuals at the time of seroconversion are methylated and that methylation continues to progress into the 5′ LTR (45). Moreover, DNA methylation of the HTLV-1 5′ LTR has been shown to silence viral gene transcription in leukemia cells (25, 44). The finding that the 3′ LTR is unmethylated in all ATL cases and carriers (45) is consistent with the expression of Hbz in ATL cells and its constant and stable expression in HTLV-1-infected rabbits. The relative expression of tax/rex mRNA was significantly lower in vivo (approximately 1 to 2 logs) than in cell culture. This difference can be attributed to a lower ratio of infected-to-uninfected cells within PBMCs but also likely reflects immune regulation and clearance of infected cells. Tax has the remarkable ability to promote the proliferation of infected cells. However, it is also a major target of cytotoxic T lymphocytes in vivo (6). Interestingly, rabbits that display a higher tax/rex mRNA level at earlier weeks tend to have a lower proviral load at later weeks, clearly suggesting negative pressure on tax/rex mRNA-expressing cells.
With the exception of Hbz mRNA, all other HTLV-1 mRNA in rabbit PBMCs, including those encoding Env and the accessory proteins, were below our limit of detection; this was consistent with the low percentage of infected cells in vivo and the low level of expression of these specific mRNA in cultured or transfected cells. In contrast to tax/rex and gag/pol mRNA, Hbz mRNA expression appeared to start low and slowly increase and stabilize over time, showing a direct correlation with proviral load and thus supporting the conclusion that Hbz mRNA expression remains relatively constant in the infected cell and increases with infected-cell numbers. Hbz is expressed in almost all ATL cells, whereas tax/rex is expressed at low levels or rarely (10, 31, 40). A snapshot of Hbz and tax/rex mRNA expression levels in primary ATL cells revealed high Hbz and low tax/rex mRNA expression levels (46). Consistent with our results, these authors also showed that Hbz mRNA expression levels correlated significantly with infected cell numbers. Taken together, the data strongly implicate a key role for Hbz expression in infected-cell survival and disease. Indeed, suppression of Hbz gene expression by small interfering RNA inhibited the proliferation of primary HTLV-1-immortalized T lymphocytes and transformed cell lines derived from ATL patients (3, 40). Moreover, the knockdown of Hbz expression significantly decreased tumor formation and tissue infiltration in a mouse tumorgenicity transplant model (3).
In summary, this study provides a quantitative analysis of HTLV-1 gene expression using various experimental systems. Our in vivo data indicate that in immune-competent HTLV-1-infected rabbits, tax/rex expression peaks early after infection and progressively decreases over time, which correlates inversely with the proviral load. Uniquely, Hbz mRNA expression started out low and then slowly increased and stabilized in direct correlation with the proviral load, providing the first evidence linking Hbz expression to proviral load and cell survival. These results are consistent with the maintenance of expression of Hbz in most ATL cells and support an important role for Hbz in leukemogenesis.
Acknowledgments
We thank Li Xie for technical assistance, Kate Hayes-Ozello for editorial comments on the manuscript, and Tim Vojt for figure preparation.
This work was supported by grants from the National Institutes of Health (CA100730 and CA077556).
Footnotes
Published ahead of print on 4 January 2009.
REFERENCES
- 1.Anderson, M. D., J. Ye, L. Xie, and P. L. Green. 2004. Transformation studies with a human T-cell leukemia virus type 1 molecular clone. J. Virol. Methods 116195-202. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, J., B. Yamamoto, M. Li, A. J. Phipps, I. Younis, M. D. Lairmore, and P. L. Green. 2006. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood 1073976-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, J., B. Zimmerman, M. Li, M. D. Lairmore, and P. L. Green. 2008. Human T-cell leukemia virus type 1 antisense encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood 1123788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awasthi, S., A. Sharma, K. Wong, J. Zhang, E. F. Matlock, L. Rogers, P. Motloch, S. Takemoto, H. Taguchi, M. D. Cole, B. Luscher, O. Dittrich, H. Tagami, Y. Nakatani, M. McGee, A. M. Girard, L. Gaughan, C. N. Robson, R. J. Monnat, Jr., and R. Harrod. 2005. A human T-cell lymphotropic virus type 1 enhancer of Myc-transforming potential stabilizes Myc-TIP60 transcriptional interactions. Mol. Cell. Biol. 256178-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakker, A. L. X., C. T. Ruland, D. W. Stephens, A. C. Black, and J. D. Rosenblatt. 1996. Human T-cell leukemia virus type 2 Rex inhibits pre-mRNA splicing in vitro at an early stage of spliceosome formation. J. Virol. 705511-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bangham, C. R., and M. Osame. 2005. Cellular immune response to HTLV-1. Oncogene 246035-6046. [DOI] [PubMed] [Google Scholar]
- 7.Bartoe, J. T., B. Albrecht, N. D. Collins, M. D. Robek, L. Ratner, P. L. Green, and M. D. Lairmore. 2000. Functional role of pX open reading frame II of human T-lymphotropic virus type 1 in maintenance of viral loads in vivo. J. Virol. 741094-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boxus, M., J. C. Twizere, S. Legros, J. F. Dewulf, R. Kettmann, and L. Willems. 2008. The HTLV-1 Tax interactome. Retrovirology 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cann, A. J., J. D. Rosenblatt, W. Wachsman, N. P. Shah, and I. S. Y. Chen. 1985. Identification of the gene responsible for human T-cell leukemia virus transcriptional regulation. Nature 318571-574. [DOI] [PubMed] [Google Scholar]
- 10.Cavanagh, M.-H., S. Landry, B. Audet, C. Arpin-Andre, P. Hivin, M.-E. Pare, J. Thete, E. Wattel, S. Marriott, J.-M. Mesnard, and B. Barbeau. 2006. HTLV-I antisense transcripts initiating in the 3′ LTR are alternatively spliced and polyadenylated. Retrovirology 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, N. D., C. D'Souza, B. Albrecht, M. D. Robek, L. Ratner, W. Ding, P. L. Green, and M. Lairmore. 1999. Proliferation response to interleukin-2 and Jak/Stat activation of T cells immortalized by human T-cell lymphotropic virus type 1 is independent of open reading frame I expression. J. Virol. 739642-9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, N. D., G. C. Newbound, L. Ratner, and M. D. Lairmore. 1996. In vitro CD4+ lymphocyte transformation and infection in a rabbit model with a molecular clone of human T-cell lymphotropic virus type 1. J. Virol. 707241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins, N. D., G. C. Newbound, B. Albrecht, J. Beard, L. Ratner, and M. D. Lairmore. 1998. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood 914701-4707. [PubMed] [Google Scholar]
- 14.Derse, D., J. Mikovits, and F. Ruscetti. 1997. X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology 237123-128. [DOI] [PubMed] [Google Scholar]
- 15.Ding, W., B. Albrecht, R. Luo, W. Zhang, J. R. L. Stanley, G. C. Newbound, and M. D. Lairmore. 2001. Endoplasmic reticulum and cis-Golgi localization of human T-lymphotropic virus type 1 p12I: association with calreticulin and calnexin. J. Virol. 757672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felber, B. K., H. Paskalis, C. Kleinman-Ewing, F. Wong-Staal, and G. N. Pavlakis. 1985. The pX protein of HTLV-I is a transcriptional activator of its long terminal repeats. Science 229675-679. [DOI] [PubMed] [Google Scholar]
- 17.Feuer, G., and P. L. Green. 2005. Comparative biology of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2. Oncogene 245996-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green, P. L., and I. S. Y. Chen. 1994. Molecular features of the human T-cell leukemia virus: mechanisms of transformation and leukemogenicity, p. 227-311. In J. A. Levy (ed.), The Retroviridae, vol. 3. Plenum Press, New York, NY. [Google Scholar]
- 19.Green, P. L., Y. Xie, and I. S. Y. Chen. 1990. The internal methionine codons of the human T-cell leukemia virus type II rex gene are not required for p24Rex production or virus replication and transformation. J. Virol. 644914-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiraragi, H., S.-J. Kim, A. J. Phipps, M. Silic-Benussi, V. Ciminale, L. Ratner, P. L. Green, and M. D. Lairmore. 2006. Human T-lymphotropic virus type 1 mitochondrion-localizing protein p13II is required for viral infectivity in vivo. J. Virol. 803469-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue, J. I., M. Yoshida, and M. Seiki. 1987. Transcriptional (p40x) and post-transcriptional (p27xIII) regulators are required for the expression and replication of human T-cell leukemia virus type I genes. Proc. Natl. Acad. Sci. USA 843653-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, J. M., J. C. Mulloy, V. Ciminale, J. Fullen, C. Nicot, and G. Franchini. 2000. The MHC class I heavy chain is a common target of the small proteins encoded by the 3′ end of HTLV type 1 and HTLV type 2. AIDS Res. Hum. Retrovir. 161777-1781. [DOI] [PubMed] [Google Scholar]
- 23.Kashanchi, F., and J. N. Brady. 2005. Transcriptional and post-transcriptional gene regulation of HTLV-1. Oncogene 245938-5951. [DOI] [PubMed] [Google Scholar]
- 24.Kim, S. J., A. M. Nair, S. Fernandez, L. Mathes, and M. D. Lairmore. 2006. Enhancement of LFA-1-mediated T cell adhesion by human T lymphotropic virus type 1 p12I. J. Immunol. 1765463-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koiwa, T., A. Hamano-Usami, T. Ishida, A. Okayama, K. Yamaguchi, S. Kamihira, and T. Watanabe. 2002. 5′-Long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 769389-9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koralnik, I. J., J. C. Mulloy, T. Andresson, J. Fullen, and G. Franchini. 1995. Mapping of the intermolecular association of human T cell leukaemia/lymphotropic virus type I p12I and the vacuolar H+-ATPase 16 kDa subunit protein. J. Gen. Virol. 761909-1916. [DOI] [PubMed] [Google Scholar]
- 27.Li, M., and P. L. Green. 2007. Detection and quantitation of HTLV-1 and HTLV-2 mRNA species by real-time RT-PCR. J. Virol. Methods 142159-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez, A. J. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32279-305. [DOI] [PubMed] [Google Scholar]
- 29.Mesnard, J. M., B. Barbeau, and C. Devaux. 2006. HBZ, a new important player in the mystery of adult T-cell leukemia. Blood 1083979-3982. [DOI] [PubMed] [Google Scholar]
- 30.Mulloy, J. C., T. Kislyakova, A. Cereseto, L. Casareto, A. LoMonico, J. Fullen, M. V. Lorenzi, A. Cara, C. Nicot, C.-Z. Giam, and G. Franchini. 1998. Human T-cell lymphotropic/leukemia virus type 1 Tax abrogates p53-induced cell cycle arrest and apoptosis through its CREB/ATF functional domain. J. Virol. 728852-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murata, K., T. Hayashibara, K. Sugahara, A. Uemura, T. Yamaguchi, H. Harasawa, H. Hasegawa, K. Tsuruda, T. Okazaki, T. Koji, T. Miyanishi, Y. Yamada, and S. Kamihira. 2006. A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J. Virol. 802495-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicot, C., J. M. Dundr, J. R. Johnson, J. R. Fullen, N. Alonzo, R. Fukumoto, G. L. Princler, D. Derse, T. Misteli, and G. Franchini. 2004. HTLV-1-encoded p30II is a post-transcriptional negative regulator of viral replication. Nat. Med. 10197-201. [DOI] [PubMed] [Google Scholar]
- 33.Nicot, C., R. L. Harrod, V. Ciminale, and G. Franchini. 2005. Human T-cell leukemia/lymphoma virus type 1 nonstructural genes and their functions. Oncogene 246026-6034. [DOI] [PubMed] [Google Scholar]
- 34.Nicot, C., J. C. Mulloy, M. G. Ferrari, J. M. Johnson, K. Fu, R. Fukumoto, R. Trovato, J. Fullen, W. J. Leonard, and G. Franchini. 2001. HTLV-1 p12(I) protein enhances STAT5 activation and decreases the interleukin-2 requirement for proliferation of primary human peripheral blood mononuclear cells. Blood 98823-829. [DOI] [PubMed] [Google Scholar]
- 35.Princler, G. L., J. G. Julias, S. H. Hughes, and D. Derse. 2003. Roles of viral and cellular proteins in the expression of alternatively spliced HTLV-1 pX mRNAs. Virology 317136-145. [DOI] [PubMed] [Google Scholar]
- 36.Ramadan, E., M. Ward, X. Guo, S. S. Durkin, A. Sawyer, M. Vilela, C. Osgood, A. Pothen, and O. J. Semmes. 2008. Physical and in silico approaches identify DNA-PK in a Tax DNA-damage response interactome. Retrovirology 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ressler, S., G. F. Morris, and S. J. Marriott. 1997. Human T-cell leukemia virus type 1 Tax transactivates the human proliferating cell nuclear antigen promoter. J. Virol. 711181-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robek, M. D., and L. Ratner. 1999. Immortalization of CD4+ and CD8+ T lymphocytes by human T-cell leukemia virus type 1 Tax mutants expressed in a functional molecular clone. J. Virol. 734856-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross, T. M., M. Narayan, Z. Y. Fang, A. C. Minella, and P. L. Green. 2000. Tax transactivation of both NFκB and CREB/ATF is essential for human T-cell leukemia virus type 2-mediated transformation of primary human T cells. J. Virol. 742655-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satou, Y., J. Yasunaga, M. Yoshida, and M. Matsuoka. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. USA 103720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitt, I., O. Rosin, P. Rohwer, M. Gossen, and R. Grassmann. 1998. Stimulation of cyclin-dependent kinase activity and G1- to S-phase transition in human lymphocytes by the human T-cell leukemia/lymphotropic virus type 1 Tax protein. J. Virol. 72633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silic-Benussi, M., I. Cavallari, T. Zorzan, E. Rossi, H. Hiraragi, A. Rosato, K. Horie, D. Saggioro, M. D. Lairmore, L. Willems, L. Chieco-Bianchi, D. M. D'Agostino, and V. Ciminale. 2004. Suppression of tumor growth and cell proliferation by p13II, a mitochondrial protein of human T cell leukemia virus type 1. Proc. Natl. Acad. Sci. USA 1016629-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silverman, L. R., A. J. Phipps, A. Montgomery, L. Ratner, and M. D. Lairmore. 2004. Human T-cell lymphotropic virus type 1 open reading frame II-encoded p30II is required for in vivo replication: evidence of in vivo reversion. J. Virol. 783837-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeda, S., M. Maeda, S. Morikawa, Y. Taniguchi, J. Yasunaga, K. Nosaka, Y. Tanaka, and M. Matsuoka. 2004. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int. J. Cancer 109559-567. [DOI] [PubMed] [Google Scholar]
- 45.Taniguchi, Y., K. Nosaka, J. Yasunaga, M. Maeda, N. Mueller, A. Okayama, and M. Matsuoka. 2005. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Usui, T., K. Yanagihara, K. Tsukasaki, K. Murata, H. Hasegawa, Y. Yamada, and S. Kamihira. 2008. Characteristic expression of HTLV-1 basic zipper factor (HBZ) transcripts in HTLV-1 provirus-positive cells. Retrovirology 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verma, M. 2003. Viral genes and methylation. Ann. N. Y. Acad. Sci. 983170-180. [DOI] [PubMed] [Google Scholar]
- 48.Xie, L., and P. L. Green. 2005. Envelope is a major viral determinant of the distinct in vitro cellular transformation tropism of human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2. J. Virol. 7914536-14545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie, L., B. Yamamoto, A. Haoudi, O. J. Semmes, and P. L. Green. 2006. PDZ binding motif of HTLV-1 Tax promotes virus-mediated T-cell proliferation in vitro and persistence in vivo. Blood 1071980-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye, J., L. Silverman, M. D. Lairmore, and P. L. Green. 2003. HTLV-1 Rex is required for viral spread and persistence in vivo but is dispensable for cellular immortalization in vitro. Blood 1023963-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida, M., Y. Satou, J. Yasunaga, J. Fujisawa, and M. Matsuoka. 2008. Transcriptional control of spliced and unspliced human T-cell leukemia virus type 1 bZIP factor (HBZ) gene. J. Virol. 829359-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Younis, I., and P. L. Green. 2005. The human T-cell leukemia virus Rex protein. Front. Biosci. 10431-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Younis, I., L. Khair, M. Dundr, M. D. Lairmore, G. Franchini, and P. L. Green. 2004. Repression of human T-cell leukemia virus type 1 and 2 replication by a viral mRNA-encoded posttranscriptional regulator. J. Virol. 7811077-11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, W., J. W. Nisbet, B. Albrecht, W. Ding, F. Kashanchi, J. T. Bartoe, and M. D. Lairmore. 2001. Human T-lymphotropic virus type 1 p30II regulates gene transcription by binding CREB binding protein/p300. J. Virol. 759885-9895. [DOI] [PMC free article] [PubMed] [Google Scholar]