Abstract
The newly identified type III interferon (IFN-λ) has antiviral activity against a broad spectrum of viruses. We thus examined whether IFN-λ has the ability to inhibit human immunodeficiency virus type 1 (HIV-1) infection of blood monocyte-derived macrophages that expressed IFN-λ receptors. Both IFN-λ1 and IFN-λ2, when added to macrophage cultures, inhibited HIV-1 infection and replication. This IFN-λ-mediated anti-HIV-1 activity is broad, as IFN-λ could inhibit infection by both laboratory-adapted and clinical strains of HIV-1. Investigations of the mechanism(s) responsible for the IFN-λ action showed that although IFN-λ had little effect on HIV-1 entry coreceptor CCR5 expression, IFN-λ induced the expression of CC chemokines, the ligands for CCR5. In addition, IFN-λ upregulated intracellular expression of type I IFNs and APOBEC3G/3F, the newly identified anti-HIV-1 cellular factors. These data provide direct and compelling evidence that IFN-λ, through both extracellular and intracellular antiviral mechanisms, inhibits HIV-1 replication in macrophages. These findings indicate that IFN-λ may have therapeutic value in the treatment of HIV-1 infection.
Innate immunity is the first line of defense against viral infections. Interferons (IFNs) are important players in host innate immunity, as they possess innate antiviral activity against a variety of viruses, including human immunodeficiency virus type 1 (HIV-1). While both type I IFNs (IFN-α, -β, -ω, -κ, -ɛ, -τ, -δ, and -ν subtypes) and type II IFN (IFN-γ) have been known for decades as the classical antiviral cytokines, a novel class of cytokines was recently discovered and named type III IFN (also called IFN-λ or interleukin-28/29 [IL-28/29]) (15, 34). IFN-λ is structurally and genetically close to the members of IL-10 family of cytokines but displays type I IFN-like antiviral activity and induction of typical IFN-inducible genes (2, 39). In humans, there are three genes encoding the three members of the type III IFN family, i.e., IFN-λ1, IFN-λ2, and IFN-λ3. IFN-λ shares a number of common biological functions with IFN-α/β, even though IFN-λ exerts its action through a receptor complex distinct from that for the type I IFNs (15, 34). Although type I and type III IFN receptors are unrelated, they trigger strikingly similar responses, mostly through the activation of signal transducer and activator of transcription 1 (STAT-1) and STAT-2, and to a lesser extent, that of STAT-3 (4, 8, 15, 16, 45).
IFN-λ expression depends on the same triggers (viral infection or Toll-like receptor ligands) and signal transduction pathways (23, 24, 43) that induce type I IFN expression. IFN-λ can be induced by viral infections and has potent antiviral activity against viral infections in vivo (8). Several reports have now demonstrated that IFN-λ has the ability to inhibit the replication of a number of viruses, including hepatitis C virus and hepatitis B virus (29), cytomegalovirus (4), herpes simplex virus type 2 (2), and vesicular stomatitis virus (4). However, it is still unclear whether IFN-λ has the ability to inhibit HIV-1 infection. Recently, one study reported that pretreatment of peripheral blood mononuclear cells with IFN-λ2 increased the expression of the CD4, CXCR4, and CCR5 genes, which was associated with enhanced HIV-1 binding and replication (32). In the present study, we investigated the effect of IFN-λ on HIV-1 infection of macrophages, a target of and long-lived reservoir for HIV-1. We also examined the mechanisms involved in IFN-λ action on HIV-1.
MATERIALS AND METHODS
Cells and viruses.
Peripheral blood samples were obtained from healthy donors and identified as HIV-1 antibody negative. The Institutional Review Board of the Children's Hospital of Philadelphia approved this research. Informed consent was obtained from the subjects. Monocytes were isolated from peripheral blood mononuclear cells as previously described (11). Briefly, mononuclear cells were separated by centrifugation (1,500 × g) for 45 min over lymphocyte separation medium (Organon Teknika Corp, Durham, NC). The mononuclear cell layer was collected and incubated with Dulbecco's modified Eagle medium (DMEM) in gelatin-coated flasks for 45 min at 37°C, and then nonadherent cells were washed off with DMEM. After detachment with EDTA, monocytes were resuspended in DMEM supplemented with 10% fetal bovine serum, glutamine (2 mmol/ml), penicillin (100 U/ml), streptomycin (100 μg/ml), and nonessential amino acids and plated in 96-well culture plates at a density of 105 cells/well. Following the initial purification, at least 98.5% cells were monocytes as determined by nonspecific esterase staining and fluorescence-activated cell sorting analysis with a monoclonal antibody against CD14 (Leu-M3) and low-density lipoprotein specific for monocytes and macrophages (10). The monocyte-derived macrophages were 7-day-cultured macrophages. MRC-5 cells (human diploid fibroblasts cells) were obtained from the Medical Research Council, London. BB19 cells (human brain capillary endothelial cells) were kindly provided by Dennis L. Kolson (Department of Neurology, University of Pennsylvania School of Medicine).
The macrophage-tropic R5 strains (Bal and JRFL) were obtained from the AIDS Research and Reference Reagent Program, National Institutes of Health (Bethesda, MD). Jago, another macrophage-tropic R5 strain, was obtained from the Center for AIDS Research at the School of Medicine, University of Pennsylvania. The HIV-1 Bal strain was initially isolated from human infant lung tissue. The HIV-1 Jago strain was isolated from cell-free cerebrospinal fluid from a patient with confirmed HIV-associated dementia (5). The HIV-1 JRFL strain was from the brain of a patient with AIDS dementia complex (33). HIV-1 p24 protein levels are as follows: Bal, 1,300 ng/ml; JRFL, 530 ng/ml; and Jago, 200 ng/ml.
Reagents.
Recombinant human IFN-λ1 and IFN-λ2 were purchased from PeproTech Inc. (Rocky Hill, NJ). Recombinant human IFN-α 2a and IFN-β 1a were obtained from PBL Biomedical Laboratories (Piscataway, NJ). The primary antibody used for flow cytometric analysis was goat anti-human IFN-λ receptor (IL-10Rβ) (R&D Systems, Minneapolis, MN) at 1 μg per 2 × 105 cells. The secondary antibody used for flow cytometric analysis was Alexa 488-labeled chicken anti-goat immunoglobulin G (IgG) (Molecular Probes, Eugene, OR) at 2 μg per 2 × 105 cells. Normal goat serum (Sigma Chemical Co., St. Louis, MO) was used as a negative control. Goat anti-IFN-α/β receptor 1 (anti-IFN-α/βR1) was purchased from Sigma-Aldrich Inc. (St. Louis, MO). Rabbit anti-human APBEC3G/3F polyclonal antibodies were obtained from Immuno Diagnostics, Inc. (Woburn, MA). Monoclonal antibody to macrophage inflammatory protein 1α (MIP-1α), monoclonal antibody to MIP-1β, and monoclonal antibody to RANTES (regulated upon activation, normal T-cell expressed and secreted) were purchased from R&D Systems Inc. (Minneapolis, MN). Enzyme-linked immunosorbent assay (ELISA) kits for analysis of MIP-1α and MIP-1β proteins were purchased from Pierce Inc. (Rockford, IL). The secondary antibodies (horseradish peroxidase-conjugated goat anti-rabbit IgG and donkey anti-goat IgG) used for Western blotting were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA).
IFN-λ treatment and HIV-1 infection.
Macrophages maintained in culture for 7 days (105 cells/well in 96-well plates) were incubated either with or without IFN-λ1/λ2 (10 to 1,000 ng/ml) for 24 h before infection with different HIV-1 R5 strains (Bal, Jago, and JRFL). In order to minimize the variation of HIV-1 infectivity, the same batch of the virus stock was used for the infection experiments. The cells were infected with equal quantities of cell-free HIV-1 based on p24 protein content (30 ng/106 cells) for 2 h at 37°C in the presence or absence of IFN-λ. In order to minimize the variation of HIV-1 infectivity, we used the virus stock for each set of experiments. The cells were then washed three times with DMEM to remove unabsorbed virus, and fresh medium was added to cell cultures. The final wash was tested for viral reverse transcription (RT) activity and shown to be free of residual inoculum. Untreated cells served as controls. The cell cultures were replaced with fresh medium without IFN-λ every 4 days after HIV-1 infection. Supernatants were collected for HIV-1 RT activity assay 8 days after infection.
HIV-1 RT assay and p24 ELISA.
HIV-1 RT activity was determined based on the technique of Willey et al. (42) with modifications (11). Briefly, 10 μl of supernatants collected from HIV-infected macrophage cultures was added to 50 μl of a cocktail containing poly(A), oligo(dT), MgCl2, Nonidet P-40, and [32P]dTTP and incubated for 20 h at 37°C. Thirty microliters of the reaction mixture was then spotted on DE 81 paper and air dried. The filters were then washed in 2× standard saline citrate (SSC) (0.3 mol/liter NaCl, 0.03 mol/liter sodium citrate, pH 7) and 100% ethanol, dried, cut, and placed in a liquid scintillation counter (United Technologies Packard Inc., IL) for measurement of radioactivity. For HIV p24 assay, the culture supernatants were collected at the indicated time points after HIV-1 infection and analyzed by ELISA as described in the protocol provided by the manufacturer (Chiron Corp, Emeryville, CA).
RNA extraction and real-time RT-PCR.
Total RNA from macrophages was extracted with Tri-Reagent-BD (Molecular Research Center, Cincinnati, OH). Briefly, the total RNA was extracted by a single-step guanidium thiocyanate-phenol-chloroform extraction. Tri-Reagent (0.5 ml) was added to macrophage cultures (5 × 105 cells/well), and the cell lysates from two wells of the cultures were then pooled. After centrifugation at 13,000 × g for 15 min at 4°C, the RNA-containing aqueous phase was precipitated in isopropanol. RNA precipitates were then washed once in 75% ethanol and resuspended in 20 μl of RNase-free water. Total RNA (1 μg) was subjected to RT using the RT system (Promega, Madison, WI) with random primers for 1 h at 42°C. The reaction was terminated by incubating the reaction mixture at 99°C for 5 min, and the mixture was then kept at 4°C. The resulting cDNA was then used as a template for real-time PCR quantification. Real-time PCR was performed with 1/10 of the cDNA derived from 1 μg of RNA extracted from macrophages using the MyiQ single-Color real-time PCR detection system (Bio-Rad, Hercules, CA). The cDNA was amplified by PCR using the primers shown in Table 1, and the products were measured using SYBR green I (Bio-Rad Laboratories, Inc., Hercules, CA). The data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and presented as the change in induction relative to that of untreated control cells.
TABLE 1.
Primers used for quantitative RT-PCR
| Target gene | Primera | Nucleotide sequence |
|---|---|---|
| GAPDH | F | 5′-GGTGGTCTCCTCTGACTTCAACA-3′ |
| R | 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ | |
| IL-10R | F | 5′-GGCTGAATTTGCAGATGAGCA-3′ |
| R | 5′-GAAGACCGAGGCCATGAGG-3′ | |
| IL-28R | F | 5′-ACCTATTTTGTGGCCTATCAGAGCT-3′ |
| R | 5′-CGGCTCCACTTCAAAAAGGTAAT-3′ | |
| IFN-α | F | 5′-TTTCTCCTGCCTGAAGAACAG-3′ |
| R | 5′-GCTCATGATTTCTGCTCTGACA-3′ | |
| IFN-β | F | 5′-AAAGAAGCAGCAATTTTCAGC-3′ |
| R | 5′-CCTTGGCCTTCAGGTAATGCA-3′ | |
| APOBEC3G | F | 5′-TCAGAGGACGGCATGAGACTTAC-3′ |
| R | 5′-AGCAGGACCCAGGTGTCATTG-3′ | |
| APOBEC3F | F | 5′-TTC GAG GCC AGG TGT ATT CC-3′ |
| R | 5′-GGC AGC TGG TTG CCA CAG A-3′ |
F, forward; R, reverse.
Flow cytometric analysis.
Macrophages (2 × 105) were incubated with antibody (goat anti-human) to IFN-λ receptor (IL-10Rβ) for 20 min at 4°C, followed by incubation with secondary antibodies (chicken anti-goat) under the same conditions. Isotype-matched antibody was used as a negative control. Stained cells were analyzed on an Epics-Elite flow cytometer (Beckman Coulter Electronics, Hialeah, FL).
ELISA for CC chemokines.
ELISA for analysis of MIP-1α and MIP-1β proteins was performed as described in the protocol provided by the manufacturer (R&D Systems Inc., Minneapolis, MN). Briefly, 50 μl of supernatants was added to antibody-coated wells and incubated for 1 h at room temperature. The plates were washed with the provided buffer solution and incubated with 100 μl of biotinylated antibody reagent for 1 h at room temperature. The plate was washed again, treated with 100 μl of prepared streptavidin-horseradish peroxidase solution, and incubated for 30 min at room temperature. After an additional wash, 100 μl of 3,3′,5,5′-tetramethyl benzidine dihydrochloride substrate solution was added to each well, and color was allowed to develop at room temperature for 30 min. The reaction was stopped by the addition of 100 μl of stop solution to each well. The plate was read on a microplate reader (ELX800; Bio-Tek Instruments, Inc., Winooski, VT).
Statistical analysis.
Where appropriate, data were expressed as means ± standard deviations (SD). For comparison of the means of two groups, statistical significance was assessed by analysis of variance with post hoc testing performed with the Bonferroni/Mann-Whitney test. Calculations were performed with the use of State Statistical Software (Stata Corp., College Station, TX). Statistical significance was defined as a P value of <0.05.
RESULTS AND DISCUSSION
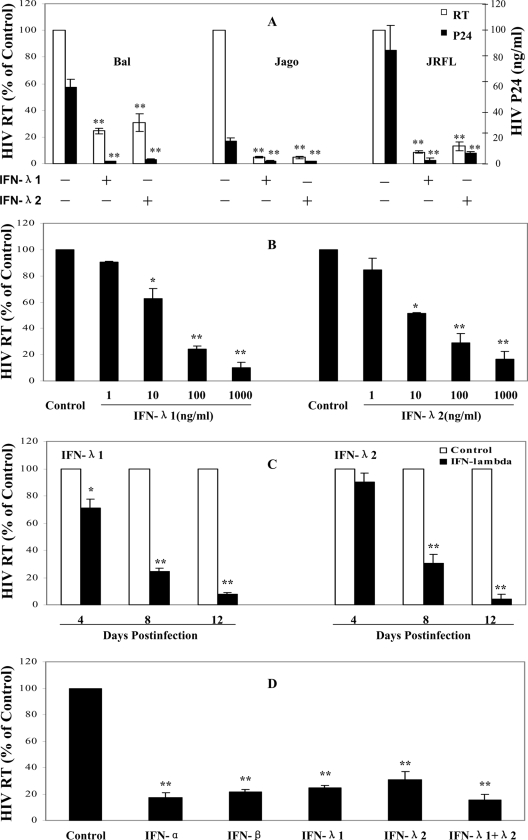
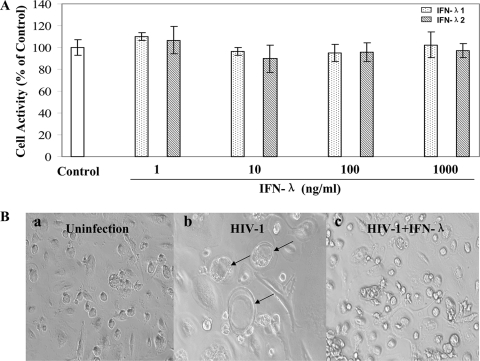
Monocytes isolated from the peripheral blood of different donors were cultured in vitro for 7 days, when the majority of cells differentiated into macrophages. To determine whether virus particles were released from macrophages infected with the HIV-1 strains (Bal, Jago, and JRFL), the supernatants were collected at 8 days postinfection and tested for the presence of p24 viral antigen, using a commercially available ELISA (Chiron Corp, Emeryville, CA). Levels of RT activity present in HIV-1-infected macrophage culture supernatants were determined by the methods of Willey et al. (42) with modifications. Significant RT activity and p24 antigen expression were observed at 8 days postinfection with HIV-1 R5 strains (Fig. 1). Treatment of 7-day-cultured macrophages with IFN-λ1 or IFN-λ2 significantly inhibited infection of different HIV-1 R5 strains (Bal, Jago, and JRFL) as evidenced by p24 protein and RT activity (Fig. 1A). This inhibitory effect on HIV-1 by IFN-λ was dose and time dependent (Fig. 1B and C). IFN-λ1 or IFN-λ2 at a concentration of 100 ng/ml was shown to have anti-HIV activity similar to that of IFN-α or IFN-β at a concentration of 1,000 IU/ml (Fig. 1D), although it is unclear whether the concentrations used for these IFNs are comparable with regard to anti-HIV efficiency. It is apparent that the combination of IFN-λ1 and IFN-λ2 inhibited HIV replication to a greater degree than IFN-λ1 or IFN-λ2 alone. However, this combined effect of IFN-λ1 and IFN-λ2 on HIV is not statistically significant compared with either IFN-λ1 (P = 0.13) or IFN-λ2 (P = 0.21) alone. This inhibitory effect of IFN-λ on HIV-1 was not due to cytotoxicity, since IFN-λ at concentrations of 1,000 ng/ml or lower had no cytotoxic effect on macrophages (Fig. 2A). Morphologically, HIV-1 Bal-infected macrophage cultures without IFN-λ treatment demonstrated characteristic giant syncytium formation (Fig. 2B, panel b), whereas IFN-λ-treated macrophages failed to develop HIV-1-induced giant syncytia (Fig. 2B, panel c).
FIG. 1.
Effect of IFN-λ on HIV-1 infection of macrophages. (A) Three HIV-1 R5 strains (Bal, Jago, and JRFL) were used to infect 7-day-cultured macrophages with or without IFN-λ pretreatment (100 ng/ml) for 24 h. HIV-1 RT activity and p24 production were determined at day 8 postinfection. (B) Dose-dependent effect of IFN-λ on HIV-1. Macrophages were preincubated with IFN-λ1/λ2 at the indicated concentrations for 24 h prior to HIV-1 infection (Bal strain). HIV-1 RT activity was determined at day 8 postinfection. (C) Time-dependent effect of IFN-λ on HIV-1. Macrophages were pretreated with IFN-λ (100 ng/ml) for 24 h and then infected with the HIV-1 Bal strain. HIV-1 RT activity was measured at the indicated time points postinfection. (D) Effect of IFNs on HIV-1 infection of macrophages. Macrophages were treated with or without IFN-α (1,000 IU/ml), IFN-β (1,000 IU/ml), IFN-λ1 (100 ng/ml), IFN-λ2 (100 ng/ml), and IFN-λ1/λ2 (100 ng/ml each) for 24 h and then infected with the HIV-1 Bal strain. Culture supernatants were collected for determination of HIV-1 RT activity at day 8 postinfection. The data shown are expressed as percent HIV RT activity in IFN-treated cultures compared to control cultures (without IFN treatment, which is defined as 100%; RT activity range, 8,000 to 90,000 cpm). The results shown are means ± SD for triplicate cultures and are representative of experiments using cells from three different donors (IFN-treated macrophages versus untreated control: **, P < 0.01. *, P < 0.05).
FIG. 2.
Effect of IFN-λ on cell viability and HIV-induced syncytium formation in macrophages. (A) Cytotoxic effect of IFN-λ on macrophages. Macrophages were cultured in the presence or absence of IFN-λ1 or IFN-λ2 for 72 h. The cytotoxicity of macrophages was measured by the CellTiter 96 AQueous assay. The data are expressed as percent cell viability compared to the control (without IFN-λ treatment, which is defined as 100%). The results shown are means ± SD for triplicate cultures. (B) Effect of IFN-λ on HIV-induced syncytium formation in macrophages. The morphology of untreated and uninfected (a), untreated and HIV-1-infected (Bal strain) (b), and IFN-λ1/λ2-treated (100 ng/ml each) and infected macrophages (c) was observed and photographed under a light microscope (magnification, ×200) at 8 days postinfection. The arrows show giant syncytium formation.
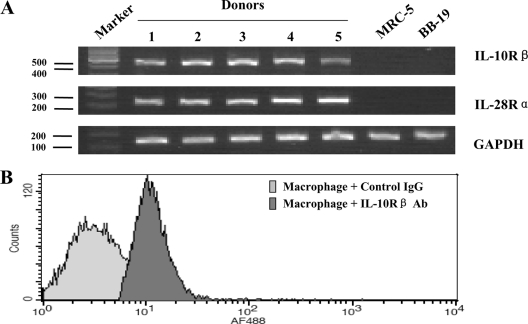
Biological functions of IFN-λ are mediated through its receptors, which are composed of two chains, IL-28Rα and IL-10Rβ. IL-28Rα is IFN-λ specific, and IL-10Rβ is shared among IL-10, IL-22, and IL-26. Both IL-28Rα and IL-10Rβ are necessary to form a functional IFN-λ receptor. However, while IL-10Rβ is ubiquitously expressed, IL-28Rα expression is not detected in some cell types, mainly fibroblastic and endothelial cells, which are unresponsive to IFN-λ (39). Monocytes freshly isolated from human blood do not express IL-28Rα but become positive when the cells differentiate into dendritic cells upon IL-4 and granulocyte-macrophage colony-stimulating factor treatment (8). To determine if the IFN-λ receptor complex consisting of IL-10Rβ and IL-28Rα is expressed in macrophages, we examined 7-day-cultured macrophages by RT-PCR and flow cytometry assay. As shown in Fig. 3A, the negative control cells, human diploid fibroblasts (MRC-5) and human brain capillary endothelial cells (BB19), had no detectable IL-28Rα and IL-10Rβ transcripts. In contrast, macrophages derived from monocytes isolated from five different donors expressed mRNA transcripts for both IL-28Rα and IL-10Rβ (Fig. 3A). The expression of these IFN-λ receptors was further confirmed by the observation that macrophages expressed IFN-λ receptor (IL-10Rβ) at the protein level (Fig. 3B). The identification of IFN-λ receptor expression by macrophages is essential for supporting the specific antiviral action of IFN-λ on HIV-1.
FIG. 3.
Expression of IFN-λ receptor in macrophages. (A) RT-PCR analysis of IFN-λ receptor gene transcripts in macrophages. Total cellular RNA was extracted from negative control cells (MRC-5 and BB19) and macrophages derived from monocytes from five different donors. Sizes are estimated from a DNA ladder (100-bp fragments) subjected to coelectrophoresis as markers. RT-PCR-amplified products as indicated were visualized on a 1.5% agarose gel. (B) Flow cytometry analysis of IFN-λ receptor (IL-10Rβ) expression in macrophages. IL-10Rβ expression was detected by incubating macrophages with antibody (Ab) directed against IL-10Rβ. Nonspecific goat IgG served as a negative control.
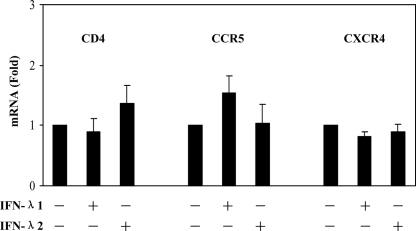
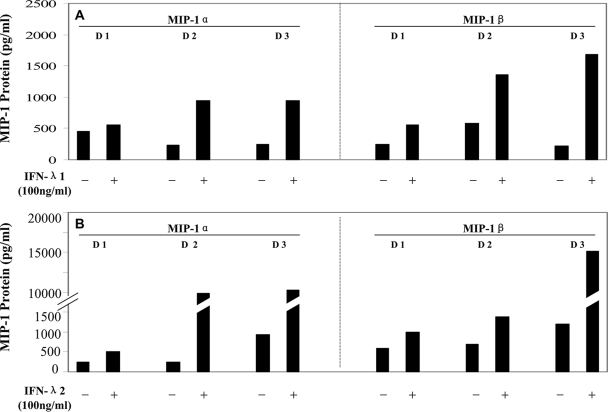
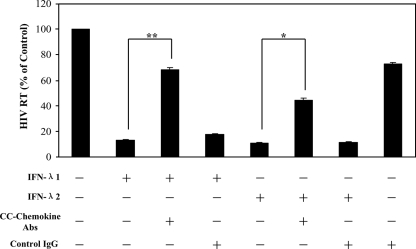
We next examined the mechanisms involved in IFN-λ-mediated anti-HIV-1 activity in macrophages. We were particularly interested in the role of IFN-λ in the modulation of the CCR5 receptor, the coreceptor for HIV-1 R5 strain entry into macrophages, and of CC chemokines (MIP-1α, MIP-1β, and RANTES), the ligands for CCR5 receptor on macrophages (1). CC chemokines are important inhibitors of R5 strains of HIV-1 in cells of macrophage lineage (1, 6, 14). Although IFN-λ had little effect on the expression of HIV-1 receptors (CD4, CCR5, and CXCR4) (Fig. 4), both IFN-λ1 and IFN-λ2 effectively induced the expression of MIP-1α and MIP-1β protein in macrophages (Fig. 5). The role of CC chemokines in the IFN-λ-mediated anti-HIV-1 effect was confirmed in the experiments showing that antibodies to CC chemokines (MIP-1α/β and RANTES) partially blocked IFN-λ-mediated HIV-1 inhibition in macrophages (Fig. 6).
FIG. 4.
Effect of IFN-λ on the expression of CD4, CCR5, and CXCR4 in macrophages. Macrophages were cultured in the presence or absence of IFN-λ (100 ng/ml) for 24 h. Total cellular RNA extracted from cell cultures was subjected to the real-time PCR for CD4, CCR5, CXCR4, and GAPDH RNA quantification. The data are expressed as CD4, CCR5, or CXCR4 mRNA levels relative (fold) to the control (without IFN-λ treatment, which is defined as 1). The results shown are means ± SD for triplicate cultures and are representative of three experiments with cells from three different donors.
FIG. 5.
Effect of IFN-λ on MIP-1α/β protein expression in macrophages. Macrophages derived from monocytes from three different donors (D) were incubated with or without IFN-λ1 (A) or IFN-λ2 (B) for 24 h, and the culture supernatants were collected for the analysis of MIP-1α/β production.
FIG. 6.
Effect of antibodies to CC chemokines on IFN-λ-mediated anti-HIV-1 activity. Macrophages were incubated with or without IFN-λ1 or IFN-λ2 (100 ng/ml) and/or antibodies (Abs) (25 μg/ml each) to human CC chemokines (MIP-1α, MIP-1β, and RANTES) for 24 h prior to HIV-1 infection (Bal strain). IgG antibody was used as the control. HIV-1 RT activity in culture supernatants was measured at day 8 postinfection. The results shown are means ± SD for triplicate cultures and are representative of experiments using cells from three different donors (**, P < 0.01; *, P < 0.05).
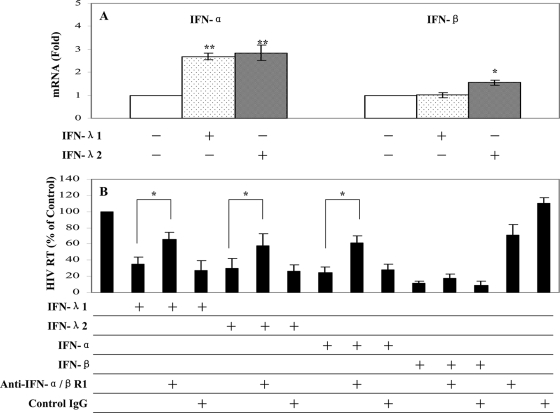
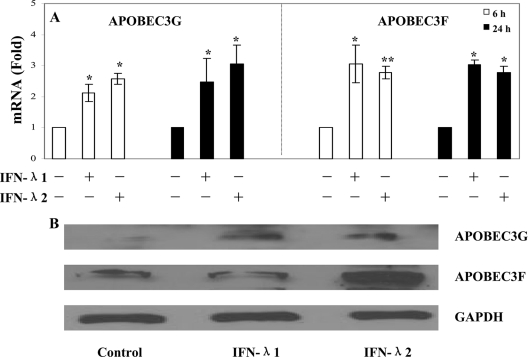
Our further studies showed that IFN-λ had the ability to induce the intracellular expression of type I IFNs (Fig. 7A). Preincubation of macrophages with antibody to IFN-α/βR1 prior to HIV-1 infection could significantly reverse IFN-λ-mediated anti-HIV-1 activity (Fig. 7B). Type I IFNs (IFN-α/β) are well known for their ability to inhibit a wide range of viruses (37), including HIV-1 (3, 7, 19, 27, 36, 38). IFN-α inhibits HIV-1 replication in macrophages at different stages by suppressing RT (35) and preventing transcription of integrated provirus (26). Similarly, IFN-β also inhibits HIV-1 replication in macrophages (9). The importance of type I IFNs in the immunopathogenesis of HIV-1 infection is further highlighted by the in vivo observation (13) that type I IFN production is profoundly and transiently impaired in primary HIV-1 infection. Thus, the induction of intracellular IFN-α/β expression by IFN-λ provides an additional mechanism for the anti-HIV-1 action of IFN-λ in macrophages. Although the anti-HIV-1 mechanism(s) of type I IFNs remains to be determined, several cellular factors, such as microRNAs, BST2, PKR, OAS1, and ISG15, have recently been identified as type I IFN-inducible anti-HIV-1 elements in HIV-1-infected target cells (12, 20-22, 25, 28, 30, 31, 40, 41). Among these factors, APOBEC3G and APOBEC3F are particularly important, as they have been recently shown to have the ability to restrict HIV-1 replication in both CD4+ T cells and macrophages. APOBEC3G can either edit the newly synthesized viral DNA or have an inhibitory effect at another site(s) of the HIV-1 life cycle (17, 18, 44). Therefore, it is important to determine whether IFN-λ has the ability to enhance the expression of these newly identified innate anti-HIV-1 factors in macrophages. Our data that IFN-λ enhanced APOBEC3G and APOBEC3F expression at both the mRNA and protein levels (Fig. 8) support the notion that elevated levels of APOBEC3G/3F may contribute to IFN-λ-mediated anti-HIV-1 activity in macrophages.
FIG. 7.
Effect of IFN-λ on the expression of IFN-α/β in macrophages. (A) IFN-α/β mRNA expression. Macrophages were cultured in the presence or absence of IFN-λ (100 ng/ml) for 6 h. Total cellular RNA extracted from cell cultures was subjected to real-time PCR for IFN-α, IFN-β, and GAPDH RNA quantification. The data are expressed as IFN-α or IFN-β mRNA levels relative (fold) to the control (without IFN-λ treatment, which is defined as 1). The results shown are means ± SD for triplicate cultures and are representative of three experiments with cells from three different donors (IFN-treated macrophages versus untreated control: *, P < 0.05; **, P < 0.01). (B) Antibody to IFN-α/βR1 blocks IFN-λ-mediated anti-HIV activity in macrophages. Macrophages were incubated with or without goat anti-IFN-α/βR1 (10 μg/ml) for 1 h prior to treatment with IFN-λ. IFN-α (1,000 IU/ml) or IFN-β (1,000 IU/ml) treatment of macrophages was used as controls. Goat IgG was used as a control IgG to determine the specificity of the antibody to anti-IFN-α/βR1. Macrophages treated with IFN-λ (100 ng/ml) for 24 h were infected with the HIV-1 Bal strain. Culture supernatants were collected for HIV-1 RT activity at day 8 postinfection. The data shown are expressed as percent HIV RT activity in IFN-treated cultures compared to control cultures (without IFN treatment, which is defined as 100%). The results shown are means ± SD for triplicate cultures and are representative of experiments using cells from three different donors (**, P < 0.01; *, P < 0.05).
FIG. 8.
Effect of IFN-λ on the expression of APOBEC3G/3F in macrophages. (A) APOBEC3G/3F mRNA expression. Macrophages were cultured in the presence or absence of IFN-λ (100 ng/ml) for 6 h and 24 h. Total cellular RNA extracted from cell cultures was subjected to real-time PCR for APOBEC3G, APOBEC3F, and GAPDH RNA quantification. The data are expressed as APOBEC3G or APOBEC3F mRNA levels relative (fold) to the control (without IFN-λ treatment, which is defined as 1). The results shown are means ± SD for triplicate cultures and are representative of three experiments with cells from three different donors (IFN-treated macrophages versus untreated control: *, P < 0.05; **, P < 0.01). (B) APOBEC3G/3F protein expression. Total proteins extracted from macrophages cultured in the presence or absence of IFN-λ (100 ng/ml) for 24 h were subjected to Western blot assay using antibodies against APOBEC3G, APOBEC3F, and GAPDH. The results shown are representative of two independent experiments.
Collectively, our data provide the first experimental evidence at the cellular and molecular levels that IFN-λ has the ability to suppress HIV-1 infection of macrophages. There are at least two distinct mechanisms involved in IFN-λ-mediated anti-HIV-1 activity: the induction of extracellular factors, e.g., CC chemokines, which block HIV-1 entry into macrophages, and the activation of intracellular innate immunity, e.g., the induction of type I IFNs and anti-HIV-1 cellular factors (APOBEC3G and APOBEC3F). These anti-HIV-1 mechanisms of IFN-λ action offer an attractive alternative for HIV-1 therapy, as it would be difficult for HIV-1 to develop resistance to the actions at both the extracellular and intracellular levels in the context of innate immune activation. However, future studies are needed to determine the impact of IFN-λ on HIV-1 infection and replication in the ex vivo and in vivo systems. Study of the antiviral properties of IFN-λ will be essential not only for our understanding of the fundamental and biological functions of IFN-λ but also for exploring the potential to develop IFN-λ-based treatment for HIV-1 infection.
Acknowledgments
This work was supported by National Institutes of Health grants DA 12815, DA 25477, and DA 22177 (to W.-Z. Ho) and by a Foerderer Murray Award from the Children's Hospital of Philadelphia (to W.-Z. Ho). Wei Hou is the recipient of a scholarship from the China Scholarship Council.
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 2721955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Ank, N., H. West, C. Bartholdy, K. Eriksson, A. R. Thomsen, and S. R. Paludan. 2006. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 804501-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baca-Regen, L., N. Heinzinger, M. Stevenson, and H. E. Gendelman. 1994. Alpha interferon-induced antiretroviral activities: restriction of viral nucleic acid synthesis and progeny virion production in human immunodeficiency virus type 1-infected monocytes. J. Virol. 687559-7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brand, S., F. Beigel, T. Olszak, K. Zitzmann, S. T. Eichhorst, J. M. Otte, J. Diebold, H. Diepolder, B. Adler, C. J. Auernhammer, B. Goke, and J. Dambacher. 2005. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am. J. Physiol. Gastrointest. Liver Physiol. 289G960-G968. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W., J. Sulcove, I. Frank, S. Jaffer, H. Ozdener, and D. L. Kolson. 2002. Development of a human neuronal cell model for human immunodeficiency virus (HIV)-infected macrophage-induced neurotoxicity: apoptosis induced by HIV type 1 primary isolates and evidence for involvement of the Bcl-2/Bcl-xL-sensitive intrinsic apoptosis pathway. J. Virol. 769407-9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocchi, F., A. L. DeVico, A. Garzino-Demo, S. K. Arya, R. C. Gallo, and P. Lusso. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 2701811-1815. [DOI] [PubMed] [Google Scholar]
- 7.Coccia, E. M., B. Krust, and A. G. Hovanessian. 1994. Specific inhibition of viral protein synthesis in HIV-infected cells in response to interferon treatment. J. Biol. Chem. 26923087-23094. [PubMed] [Google Scholar]
- 8.Dumoutier, L., A. Tounsi, T. Michiels, C. Sommereyns, S. V. Kotenko, and J. C. Renauld. 2004. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J. Biol. Chem. 27932269-32274. [DOI] [PubMed] [Google Scholar]
- 9.Gessani, S., P. Puddu, B. Varano, P. Borghi, L. Conti, L. Fantuzzi, G. Gherardi, and F. Belardelli. 1994. Role of endogenous interferon-beta in the restriction of HIV replication in human monocyte/macrophages. J. Leukoc. Biol. 56358-361. [DOI] [PubMed] [Google Scholar]
- 10.Hassan, N. F., D. E. Campbell, and S. D. Douglas. 1986. Purification of human monocytes on gelatin-coated surfaces. J. Immunol. Methods 95273-276. [DOI] [PubMed] [Google Scholar]
- 11.Ho, W. Z., J. Lioy, L. Song, J. R. Cutilli, R. A. Polin, and S. D. Douglas. 1992. Infection of cord blood monocyte-derived macrophages with human immunodeficiency virus type 1. J. Virol. 66573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, J., F. Wang, E. Argyris, K. Chen, Z. Liang, H. Tian, W. Huang, K. Squires, G. Verlinghieri, and H. Zhang. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 131241-1247. [DOI] [PubMed] [Google Scholar]
- 13.Kamga, I., S. Kahi, L. Develioglu, M. Lichtner, C. Maranon, C. Deveau, L. Meyer, C. Goujard, P. Lebon, M. Sinet, and A. Hosmalin. 2005. Type I interferon production is profoundly and transiently impaired in primary HIV-1 infection. J. Infect. Dis. 192303-310. [DOI] [PubMed] [Google Scholar]
- 14.Kelly, M. D., H. M. Naif, S. L. Adams, A. L. Cunningham, and A. R. Lloyd. 1998. Dichotomous effects of beta-chemokines on HIV replication in monocytes and monocyte-derived macrophages. J. Immunol. 1603091-3095. [PubMed] [Google Scholar]
- 15.Kotenko, S. V., G. Gallagher, V. V. Baurin, A. Lewis-Antes, M. Shen, N. K. Shah, J. A. Langer, F. Sheikh, H. Dickensheets, and R. P. Donnelly. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 469-77. [DOI] [PubMed] [Google Scholar]
- 16.Lasfar, A., A. Lewis-Antes, S. V. Smirnov, S. Anantha, W. Abushahba, B. Tian, K. Reuhl, H. Dickensheets, F. Sheikh, R. P. Donnelly, E. Raveche, and S. V. Kotenko. 2006. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 664468-4477. [DOI] [PubMed] [Google Scholar]
- 17.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 42499-103. [DOI] [PubMed] [Google Scholar]
- 18.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 11421-31. [DOI] [PubMed] [Google Scholar]
- 19.Meylan, P. R., J. C. Guatelli, J. R. Munis, D. D. Richman, and R. S. Kornbluth. 1993. Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and -gamma in primary human macrophages. Virology 193138-148. [DOI] [PubMed] [Google Scholar]
- 20.Mosoian, A., A. Teixeira, C. S. Burns, G. Khitrov, W. Zhang, L. Gusella, P. Klotman, and M. Klotman. 2007. Influence of prothymosin-alpha on HIV-1 target cells. Ann. N. Y. Acad. Sci. 1112269-285. [DOI] [PubMed] [Google Scholar]
- 21.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451425-430. [DOI] [PubMed] [Google Scholar]
- 22.Okumura, A., G. Lu, I. Pitha-Rowe, and P. M. Pitha. 2006. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. USA 1031440-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onoguchi, K., M. Yoneyama, A. Takemura, S. Akira, T. Taniguchi, H. Namiki, and T. Fujita. 2007. Viral infections activate types I and III interferon genes through a common mechanism. J. Biol. Chem. 2827576-7581. [DOI] [PubMed] [Google Scholar]
- 24.Osterlund, P., V. Veckman, J. Siren, K. M. Klucher, J. Hiscott, S. Matikainen, and I. Julkunen. 2005. Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J. Virol. 799608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng, G., K. J. Lei, W. Jin, T. Greenwell-Wild, and S. M. Wahl. 2006. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 20341-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poli, G., P. Biswas, and A. S. Fauci. 1994. Interferons in the pathogenesis and treatment of human immunodeficiency virus infection. Antiviral Res. 24221-233. [DOI] [PubMed] [Google Scholar]
- 27.Poli, G., J. M. Orenstein, A. Kinter, T. M. Folks, and A. S. Fauci. 1989. Interferon-alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science 244575-577. [DOI] [PubMed] [Google Scholar]
- 28.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 891-47. [DOI] [PubMed] [Google Scholar]
- 29.Robek, M. D., B. S. Boyd, and F. V. Chisari. 2005. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 793851-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55255-281. [DOI] [PubMed] [Google Scholar]
- 32.Serra, C., A. Biolchini, A. Mei, S. Kotenko, and A. Dolei. 2008. Type III and I interferons increase HIV uptake and replication in human cells that overexpress CD4, CCR5, and CXCR4. AIDS Res. Hum. Retroviruses 24173-180. [DOI] [PubMed] [Google Scholar]
- 33.Sharpless, N. E., W. A. O'Brien, E. Verdin, C. V. Kufta, I. S. Chen, and M. Dubois-Dalcq. 1992. Human immunodeficiency virus type 1 tropism for brain microglial cells is determined by a region of the env glycoprotein that also controls macrophage tropism. J. Virol. 662588-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheppard, P., W. Kindsvogel, W. Xu, K. Henderson, S. Schlutsmeyer, T. E. Whitmore, R. Kuestner, U. Garrigues, C. Birks, J. Roraback, C. Ostrander, D. Dong, J. Shin, S. Presnell, B. Fox, B. Haldeman, E. Cooper, D. Taft, T. Gilbert, F. J. Grant, M. Tackett, W. Krivan, G. McKnight, C. Clegg, D. Foster, and K. M. Klucher. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 463-68. [DOI] [PubMed] [Google Scholar]
- 35.Shirazi, Y., and P. M. Pitha. 1992. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J. Virol. 661321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirazi, Y., and P. M. Pitha. 1993. Interferon alpha-mediated inhibition of human immunodeficiency virus type 1 provirus synthesis in T-cells. Virology 193303-312. [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi, T., and A. Takaoka. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14111-116. [DOI] [PubMed] [Google Scholar]
- 38.Tissot, C., and N. Mechti. 1995. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J. Biol. Chem. 27014891-14898. [DOI] [PubMed] [Google Scholar]
- 39.Uze, G., and D. Monneron. 2007. IL-28 and IL-29: newcomers to the interferon family. Biochimie 89729-734. [DOI] [PubMed] [Google Scholar]
- 40.Van Damme, N., D. Goff, C. Katsura, R. L. Jorgenson, R. Mitchell, M. C. Johnson, E. B. Stephens, and J. Guatelli. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, F. X., J. Huang, H. Zhang, X. Ma, and H. Zhang. 2008. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J. Gen. Virol. 89722-730. [DOI] [PubMed] [Google Scholar]
- 42.Willey, R. L., D. H. Smith, L. A. Lasky, T. S. Theodore, P. L. Earl, B. Moss, D. J. Capon, and M. A. Martin. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, K., A. Puel, S. Zhang, C. Eidenschenk, C. L. Ku, A. Casrouge, C. Picard, H. von Bernuth, B. Senechal, S. Plancoulaine, S. Al-Hajjar, A. Al-Ghonaium, L. Marodi, D. Davidson, D. Speert, C. Roifman, B. Z. Garty, A. Ozinsky, F. J. Barrat, R. L. Coffman, R. L. Miller, X. Li, P. Lebon, C. Rodriguez-Gallego, H. Chapel, F. Geissmann, E. Jouanguy, and J. L. Casanova. 2005. Human TLR-7-, -8-, and -9-mediated induction of IFN-alpha/beta and -lambda is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity 23465-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 42494-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, Z., O. J. Hamming, N. Ank, S. R. Paludan, A. L. Nielsen, and R. Hartmann. 2007. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 817749-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]