Abstract
Minute virus of canines (MVC) is a member of the genus Bocavirus in the family Parvoviridae. We have molecularly cloned and sequenced the 5′- and 3′-terminal palindromes of MVC. The MVC genome, 5,404 nucleotides (nt) in length, shared an identity of 52.6% and 52.1% with that of human bocavirus and bovine parvovirus, respectively. It had distinct palindromic hairpins of 183 nt and 198 nt at the left-end and right-end termini of the genome, respectively. The left-end terminus was also found in two alternative orientations (flip or flop). Both termini shared extensive similarities with those of bovine parvovirus. Four full-length molecular clones constructed with different orientations of the left-end terminus proved to be infectious in Walter Reed canine cell/3873D (WRD) canine cells. Both MVC infection and transfection of the infectious clone in WRD cells revealed an identical RNA transcription profile that was similar to that of bovine parvovirus. Mutagenesis of the infectious clone demonstrated that the middle open reading frame encodes the NP1 protein. This protein, unique to the genus Bocavirus, was essential for MVC DNA replication. Moreover, the phospholipase A2 motif in the VP1 unique region was also critical for MVC infection. Thus, our studies revealed important information about the genus Bocavirus that may eventually help us to clone the human bocavirus and study its pathogenesis.
Bocavirus is a newly established genus in the subfamily Parvovirinae of the Parvoviridae family (61). It consists of two formal members, bovine parvovirus (BPV) and minute virus of canines (MVC) (61), and one tentative member, the human bocavirus (HBoV) (4). MVC was first recovered from canine fecal samples in 1970 (8). Although it was initially thought not to cause disease, recent studies of experimental infection revealed pathogenicity for newborn pups and fetuses (10). It caused enteritis with severe diarrhea (9, 44) and respiratory disease with breathing difficulty (31, 52); however, these symptoms were often associated with coinfection of other viruses (44). Pathological lesions in fetuses in experimental infections were found in the lungs and small intestine (10). Pathology caused by MVC was seen most commonly in animals between the ages of 1 to 5 weeks (10, 29, 31, 52). Serological evidence indicated that MVC was widespread in the dog population, with a 50 to 70% seroprevalence rate worldwide (11).
A virus that is associated with lower respiratory tract infections in humans was identified in 2005 and named HBoV because of its high similarity to BPV and MVC in its genomic sequence (4). The HBoV genome was detected in 1.5% to 19% of respiratory specimens from symptomatic hospitalized children (3, 7, 39). Most children infected with HBoV were younger than 24 months (4, 15, 39, 47, 60). Detection of antibodies against HBoV capsid suggested that children were infected with HBoV as early as 3 months of age, and most infections occurred before 1 year of age (25, 33, 34, 38). Isolation and culture of the virus have not been reported so far.
Our current knowledge of the genus Bocavirus is obtained mostly from studies of BPV. The BPV genome was sequenced some 20 years ago and is composed of 5,515 nucleotides (nt) (12, 13, 54). Three capsid proteins, VP1 (80 kDa), VP2 (72 kDa), and VP3 (62 kDa), have been detected in purified virions as well as during viral infection (32, 35). Analysis of the genomic sequence (13) revealed three open reading frames (ORFs). The left-hand ORF was predicted to encode the nonstructural protein NS1. The middle ORF (mid-ORF) of BPV is thought to encode the abundant 24-kDa nonstructural protein NP1, whose function is largely unknown (36). The right-hand ORF contains the coding sequences for the overlapping capsid protein genes VP1 and VP2. Only one promoter at a map unit of 4 (P4) was identified that transcribes one pre-mRNA. This pre-mRNA is processed to generate at least eight transcripts through alternative splicing as well as alternative polyadenylation (54).
MVC was isolated and grown in the Walter Reed canine cell/3873D (WRD) cell line (8). An incomplete sequence of the genome of MVC that lacked critical sequences of the palindromic repeats (approximately 150 nt at both ends) was previously obtained (GenBank accession no. NC_004442) (58). This sequence showed 43% identity to that of BPV, while the NS1, VP1, and NP1 proteins were 33.6%, 41.4%, and 39% identical to those of BPV (58). The sequence of another MVC isolate (HM-6; GenBank accession no. AB158475) that also lacked both the left and right termini showed 96.3% identity to that of the prototype MVC sequence (48). An infectious clone for mutational studies is necessary to fully understand the molecular aspects of the genus Bocavirus and especially viral DNA replication. Although an infectious clone of BPV was reported (59), it was no longer available. Unfortunately, the palindromic repeats of HBoV are currently unknown. This seems to be due to difficulties in obtaining large quantities of virus, either by growing the virus in tissue culture or from clinical specimens that contain large quantities of virus, which are necessary to clone them. Because of similarities between HBoV and MVC both in the organization and sequence of the genomes and in the symptoms of diseases they cause, we decided to clone the MVC palindromic termini and thereafter construct the infectious clones described in this report. Characterizing a selected infectious clone provided us invaluable information about DNA replication and the functions of NP1 and VP1 of bocaviruses in general.
MATERIALS AND METHODS
Virus and cells.
The MVC virus used in this study was the original strain (GA3) isolated from lung tissue (10). It was grown in the WRD cell line (8) through two passages (MVC GA3 P2). Both the virus and the cell line were gifts from Colin Parrish at the James A. Baker Institute of Cornell University. The BPV used in this study was the original strain (HADEN) of BPV-1 isolated by F. R. Abinanti (1). The stock (Y3302) we used contained 2 × 104 FFU/ml titrated on bovine embryonic trachea (EBTr) cells (41). All cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum in 5% CO2 at 37°C.
Virus purification and quantification.
MVC was used to infect WRD cells in 20 flasks (175 cm2) at a multiplicity of infection of approximately 103 genomic copies/cell. The infected cells were harvested after the appearance of a clear cytopathic effect and suspended in 10 ml of phosphate-buffered saline. After freezing and thawing three times, released viruses were collected as supernatant with high-speed centrifugation (10,000 rpm for 1 h in a Sorvall RC-6 centrifuge). Virus-containing supernatant was adjusted to a density of approximately 1.40 g/ml by adding solid CsCl and was spun at 36,000 rpm (Sorvall TH641) for 36 h at 20°C. Fractions were collected in 0.5-ml increments and dialyzed against phosphate-buffered saline with subsequent quantification of MVC genomic copies by a real-time PCR assay. The MVC real-time PCR assay for absolute MVC genomic copies was developed following a previously described method (53) with a TaqMan probe of 5′-6-carboxyfluorescein-AACACACAAAGCGCGGCTACTCGG-6-carboxytetramethylrhodamine-3′ (nt 4376 to 4397 and nt 4401 to 4424), a forward primer of 5′-AGGACCATCGCTTGGATACATT-3′ (nt 4376 to 4397), and a reverse primer of 5′-TACTGGTCCGAGGGCTTGTT-3′ (nt 4445 to 4426). The fraction with a density of ∼1.39 g/ml contained the highest genomic copies of MVC at 4 × 108 per μl and was used to anneal the single-strand DNA (ssDNA) genome of MVC. All nucleotide numbers in this study refer to the full-length MVC clone (GenBank accession no. FJ214110) unless otherwise noted.
BPV was grown in EBTr cells and purified as described previously (41). The final purified fraction contained a high genomic copy of BPV (2.5 × 109 per μl), which was quantified by a real-time PCR method (53) using a TaqMan probe of 5′-6-carboxyfluorescein-TGCTAACAGTGAGTCTACAGTA-TAMRA-3′ (nt 2327 to 2348), a forward primer of 5′-GTTGCTGGCGATAGCGATACT-3′ (nt 2304 to 2324), and a reverse primer of 5′-CATGTCCTGAGTCGTTACTGCTAAACG-3′ (nt 2376 to 2350). Nucleotide numbers of BPV refer to accession no. DQ335247 in GenBank.
Cloning and sequencing the palindromic termini. (i) Sequencing the palindromic termini of MVC.
A total of 100 μl of MVC at 108 genomic copies/μl was heated at 95°C for 5 min and cooled slowly to form the double-stranded DNA (dsDNA) of the MVC replication form. In light of the previously published sequence of the MVC NSCap gene (accession no. NC_004442) (58), there are two BspEI sites on MVC genome, one site at nt 210 after the left-end terminus and another one at nt 4469 before the right-end terminus. Thus, annealed dsDNA was blunted and digested with BspEI. BspEI-digested DNA fragments were ligated into the SmaI/BspEI-digested vector of pBB1.4A. Positive clones were collected as MVCright clones or MVCleft clones based on sequence results using two sequencing primers that flanked the inserted terminus in pBB1.4A. Preliminary sequencing results revealed that there was a BglI site on the left-end terminus and an AseI site on the right-end terminus as shown in Fig. 1. Next, BglI-digested MVCleft clones were sequenced with the two sequencing primers from both directions, which gave rise to the whole sequence of the left-end terminus. Similarly, AseI-digested MVCright clones were sequenced for the right-end terminus.
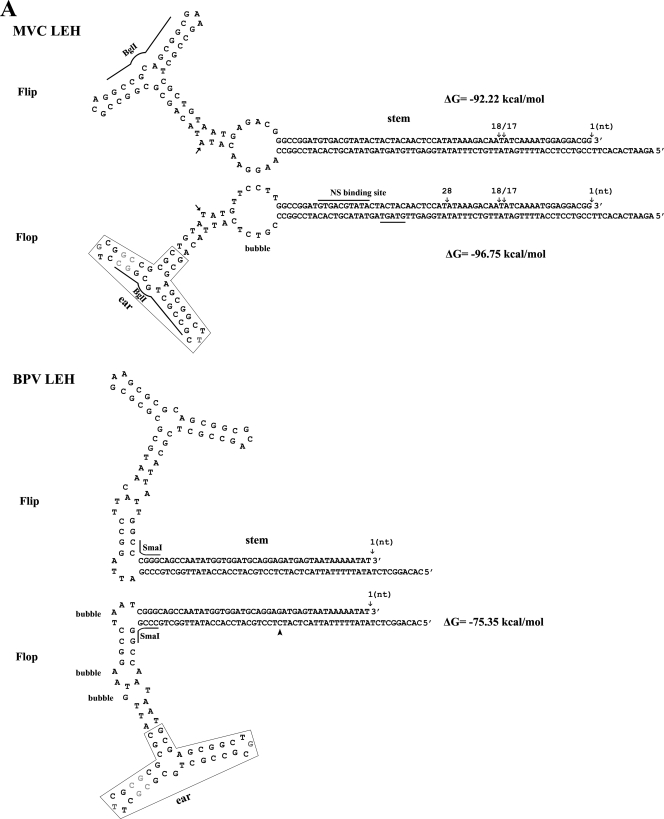
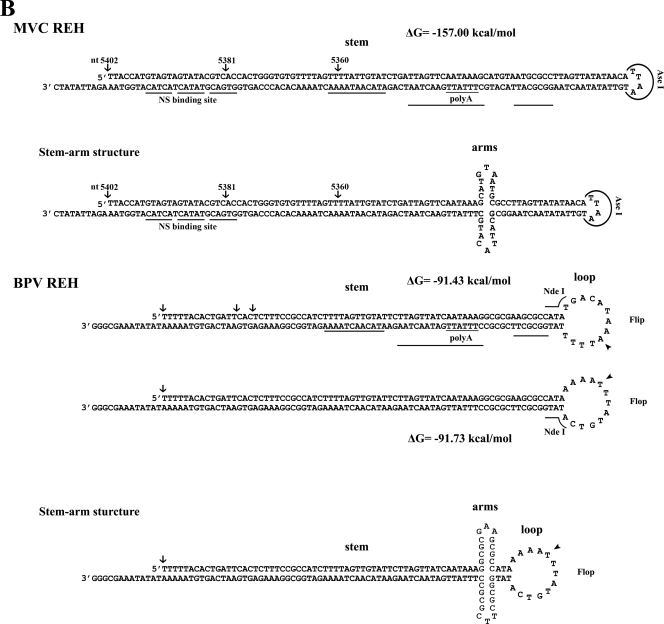
FIG. 1.
Sequences and structures of MVC palindromic repeats. (A) The sequence of the MVC and BPV left-end hairpin (LEH) is shown in flip and flop orientations. The left-end terminus of MVC in flip or flop and the left-end terminus of BPV in flop were terminated at various ends as shown by arrows with an indication of their ended nucleotide numbers. The left-end terminus of BPV shown in flip refers to the previously published sequence (12, 54). The corrected nucleotide in BPV left-end terminus is indicated by the arrowhead. The putative NS1 binding site of MVC is indicated. The rabbit-ear structure that shares a great identity between MVC and BPV is shown. BglI and SmaI that were used to digest the left-end end hairpin for sequencing are shown. (B) Sequences of the right-end hairpin (REH) of MVC and BPV are shown in a stem or stem-arm structure. The right-end terminus of MVC is shown in a stem or stem-arm structure. The right-end terminus of BPV is shown in flip or flop orientation. The stem-arm structure of BPV is shown only in the flop orientation. The right-end terminus was terminated at various ends as shown by arrows with an indication of their respective nucleotide numbers. AseI and NdeI that were used to digest the right-end hairpin for sequencing are shown. The corrected nucleotide in BPV right-end terminus was indicated by the arrowhead. The DNAMAN program (Lynnon, Co., Quebec, Canada) was used to predict the secondary structures of both termini of MVC and BPV.
The pBB1.4A cloning vector was constructed by inserting a DNA fragment that was composed of a XhoI-EcoRV-NotI-SmaI-linked 1.4-kb DNA fragment amplified from the adenovirus 5 E1A gene (nt 1013 to 2400; accession no. AC_000008) with a right-hand linker of BspEI-BsrGI-SphI-KpnI-HindIII-XhoI into SalI-digested pM20, the B19 infectious clone (68). The forward sequencing primer that was located approximately 176 nt before the insertion described above was 5′-CTGCGCGTAACCACCACA-3′; the reverse sequencing primer that was about 134 nt after the insertion described above was 5′-CATGGGGTCAGGTGGGAC-3′.
All clones containing terminal repeats were sequenced by MCLAB (www.mclab.com) using the selected method for the GC-rich/hairpin structure.
(ii) Sequencing the palindromic termini of BPV.
A total of 100 μl of purified BPV at 108 genomic copies/μl was used to prepare dsDNA of BPV as described above. Based on the previously published sequence of the BPV genome (accession no. DQ335247), annealed dsDNA was blunted and digested with EcoRI. There are two EcoRI sites on BPV, one site at nt 978 after the left-end terminus and another one at nt 5067 before the right-end terminus. EcoRI-digested DNA fragments were ligated into the SmaI/EcoRI-digested vector of pBB1.4B. Positive clones were collected as BPVright clones or BPVleft clones based on sequence results using the two sequencing primers. Preliminary sequencing results showed a SmaI site on the left-end terminus and an NdeI site on the right-end terminus as shown in Fig. 1. Finally, the whole sequence of the left-end hairpin was obtained by sequencing SmaI-digested fragments of BPVleft clones, and the right-end hairpin sequence was obtained by sequencing NdeI-digested fragments of BPVright clones.
The pBB1.4B cloning vector was constructed by inserting a DNA fragment that was composed of a SalI-NotI-SmaI-linked 1.4-kb DNA fragment amplified from the adenovirus 5 E1A gene (nt 1013 to 2400; accession no. AC_000008) with a right-hand linker of EcoRI-NheI-SphI-KpnI-HindIII-XhoI into SalI-digested pM20.
All sequence alignments described in this study were performed by Vector NTI (Invitrogen).
Plasmid constructions and infectious clones of MVC.
MVCleft clones that contained various left-end termini were digested with BspEI and SphI and were ligated to BspEI-SphI-digested MVC CapNS DNA (nt 4617 to 1014), which resulted in clones of MVCleftSph. Next, MVCright clones were digested with SphI and EcoRV and were ligated to SphI/(blunted) HindIII-digested MVCleftSph clones, which resulted in full-length clones of MVC with various right-end and left-end termini in the flip or flop orientation. MVC CapNS DNA was amplified with primers based on the published MVC NSCap sequence (accession no. NC_004442) (58).
A representative sequence of the full MVC genome with the left-end terminus in flip orientation was deposited in GenBank (accession no. FJ214110). The clone containing the whole MVC genome of nt 1 to 5402 (Flip1 to Flip5402) was selected and used as an infectious clone of MVC, pIMVC. Sites of EcoRV and NotI before the MVC genome were deleted in pIMVC.
Mutants based on the infectious clone of pIMVC.
All mutants based on pIMVC and the detailed mutations therein are diagramed (see Fig. 4 and 5, respectively).
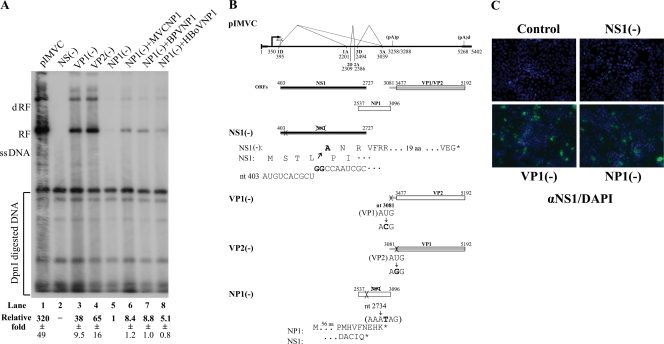
FIG. 4.
NP1 was essential for MVC replication. (A) Southern blot analysis. WRD cells were transfected with the constructs as indicated, which are diagramed in panel B. Hirt DNA was prepared and digested with DpnI at 2 days posttransfection. The blot was probed with the MVC NSCap probe (Fig. 3B). Detected bands are indicated with respective designations to the left. Quantifications of the abundance of RF DNA were standardized relatively to DpnI-digested plasmid DNA. The relative abundances (fold) that appeared in comparison with RF DNA generated from NP1(−) (lane 5) are shown as the average ± the standard deviation. −, RF DNA negative (lane 2). At least three separate experiments were performed. One microgram of NP1-expressed constructs was cotransfected where needed. (B) Diagrams of mutants on pIMVC. The MVC genome on pIMVC is shown to scale with the indication of three main ORFs, NS1, NP1, and VP1/VP2. The sites of mutations in individual mutants are indicated by nucleotide numbers, the resulting mutations of amino acids are shown with nucleotide(s) in bold, and shifted encoding frames are shown for mutants of NS1(−) and NP1(−), respectively. (C) Immunofluorescence assay. Mutants of NS(−), VP1(−), and NP1(−) were transfected into WRD cells in chamber slides. NS1 immunofluorescence was developed as described in the legend to Fig. 2C. Images were taken by an Eclipse SE TE2000-S UV microscope (Nikon) at a ×20 magnification.
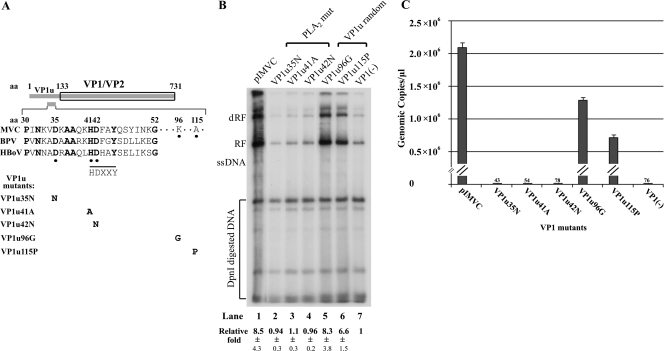
FIG. 5.
The PLA2 motif on VP1u was critical for MVC infection. (A) Diagrams of mutations on pIMVC. The ORF of VP1/VP2 is shown to scale. The HDXXY motif and the surrounding region of the MVC, BPV, and HBoV VP1u are diagramed with indications of amino acids that were mutated. Conserved amino acids among bocaviruses are bold, and three conserved amino acids (DXXXXXHD) among bocaviruses and sPLA2s are underscored with dots. Mutated amino acids in bold are listed within individual mutants corresponding to the position in the VP1u region under the diagram. (B) Southern blot analysis. WRD cells were transfected with the mutants as indicated and are diagramed in panel A. Hirt DNA treatment, Southern blot analysis, and quantification were followed as described in the legend to Fig. 4A. The relative amounts of RF DNA (fold) that resulted in comparison with RF DNA generated from the VP1(−) mutant (lane 7) are shown as the average ± the standard deviation. (C) Quantification of MVC progeny infectious virions from transfection of VP1u mutants. VP1u mutants and pIMVC were transfected to WRD cells in 60-mm dishes. At 3 days posttransfection, cell lysates were prepared by three cycles of freezing and thawing. An amount of 1 ml was used to infect WRD cells in 60-mm dishes. At 3 days postinfection, cell lysates were prepared and clarified by brief centrifugation. A total of 100 μl of each supernatant was treated with benzonase (5 units/μl; EMD Sciences) to digest free DNA overnight at 37°C. Viral DNA extracted from treated samples was subjected to quantitative PCR.
P5 RNA probe construct.
The P5 probe clone was constructed by cloning MVC nt 2344 to 2548 into BamHI/HindIII-digested vector pGEM4Z (Promega, Madison, WI).
Expression plasmids.
MVCNP1 was constructed by inserting MVC NP1 ORF (nt 2517 to 3096) plus a C-terminal flag into pcDNA3, while HBoVNP1 was constructed by inserting the HBoV NP1 ORF (nt 2410 to 3067; accession no. DQ000496) plus a C-terminal HA into pcDNA3. The BPVNP1 construct was described previously (54). pGEXNS1c112 was constructed by inserting the C-terminal sequence of NS1 (nt 2389 to 2724) into the pGEX-4T-3 expression vector (GE Healthcare Bio-Sciences Corp., Piscataway, NJ).
MVC DNA and RNA analysis.
WRD cells were cultured to approximately 50% confluence in 60-mm dishes and infected with MVC at a multiplicity of infection of 100 (genomic copies/cell) or transfected with 2 μg of plasmid DNA using Lipofectamine and Plus reagent (Invitrogen) (28). Total RNA was isolated from either infected cells or transfected cells 3 days later using the TRIzol reagent (Invitrogen) (28). For Northern blot analyses, mRNA was purified with the FastTrack MAG Micro mRNA isolation kit (Invitrogen).
Northern blot analyses were done exactly as previously described (50), using mRNA samples and 32P-labeled DNA probes as indicated. All Northern probes were created by PCR amplification of the region specified and diagramed (see Fig. 3). RNase protection assays were performed as previously described (46, 57). Low-molecular-weight DNA (Hirt DNA) was extracted from transfected WRD cells as described previously (28), and Southern blots were performed as described previously (50) with the MVC NSCap probe diagramed in Fig. 3.
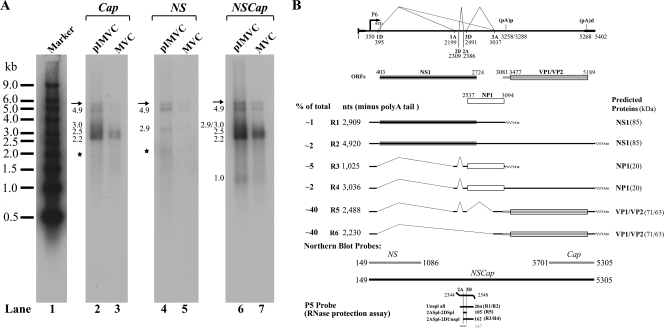
FIG. 3.
Transcription profile of MVC. (A) Northern blot analysis of MVC mRNAs. (A) mRNA purified from 20 μg of total RNA was used for Northern blotting. The blot was hybridized to three DNA probes, NS, Cap, and NSCap, respectively, which spanned various regions of the MVC genome, as indicated. Probes are shown at the bottom of panel B. RNA bands detected by each probe are indicated by their respective sizes in kilobases to the left of each lane. Smear RNA bands in the regions, indicated by asterisks in lanes 2 and 4, are likely degraded RNAs. Bands indicated by arrows are likely the ssDNA of MVC that was copurified with oligo(dT)-coated magnetic beads. The RNA marker ladder is shown in lane 1 (54). (B) Genetic map of MVC. The MVC genome is shown to scale with transcription landmarks identified by RT-PCR and confirmed by RNase protection assay and Northern blot analysis, including the P6 promoter, the splice donors (D) and acceptors (A), (pA)p, and (pA)d. Six RNA species detected significantly in Northern blots are diagramed to display their identities and respective sizes. The putative ORFs that each can encode are also diagramed together with the predicted sizes of translated proteins. The approximate percentages of individual RNA species generated during infection or transfection, as quantified by Northern blot analyses, are shown to the right.
Signals of RNase protection assays, Northern blots, and Southern blots were quantified with the Typhoon 941 phosphor imager and ImageQuant TL software v2005 (GE Healthcare).
Production of antibody against MVC NS1 and immunofluorescence assay.
Glutathione S-transferase (GST)-fused NS1 C-terminal 112 amino acids (GST-NS1c112aa) was expressed in Escherichia coli BL21 cells transformed with pGEXNS1c112 and was purified following instructions from the GST fusion system handbook (GE Healthcare). A GSTrap FF 1-ml column was used for purification of the GST fusion protein by the Biologic LP system (Bio-Rad). The purified GST fusion NS1 protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and appeared as a single band at a size of approximately 47 kDa. Purified GST-NS1c112aa (500 μg/ml) was emulsified with TiterMax Gold adjuvant (Norcross, GA) at a ratio of 1:1 and was injected into three Sprague-Dawley rats subcutaneously at a dose of 400 μg/rat. Six weeks later, blood was taken from the tail vein. Sera were tested for MVC NS1-specific antibody by enzyme-linked immunosorbent assay coated with purified GST-NS1c112aa and immunofluorescence assay with MVCNS1-transfected or MVC-infected WRD cells in chamber slides (Lab-Tek II; Nalge Nunc). Animals with the highest titers were chosen to boost antibody production by injecting 50 μg purified protein through the tail vein. After 2 weeks, boosted animals were euthanized and the terminal blood was collected and the sera harvested. All the animal experiments were approved by the KUMC IACUC.
Immunofluorescence assays were performed essentially as previously described (28).
RESULTS
Sequencing of the palindromic termini of MVC and BPV revealed conserved structures and motifs in the genus Bocavirus.
BPV virions are packaged in a way such that 90% of them contain negative-sense DNA and 10% contain positive-sense DNA (12, 13). Therefore, assuming there is a similar maturation mechanism for MVC, we used the strategy of annealing negative- and positive-sense DNA to clone the hairpin structure of MVC. A total of seven clones of the left-end terminus were isolated. Sequencing demonstrated that three were in the flop orientation and four in the flip orientation (Fig. 1A), and they had different end points. Our cloned left-end termini ended at two major sites, nt 1 and nt 18/17, respectively, while one terminus ended at nt 29. The longest left-end terminus had a palindromic structure with 183 nt; similar to BPV, the shortest left-end terminus had only 155 nt (Fig. 1A). The secondary structures of the left-end termini were predicted based on the minimal free energy (ΔG) of the structure. The stem of the MVC left-end terminus contained a “bubble” region and an asymmetric T, which were similar to the features on the left-end hairpin of the minute virus of mice (MVM) (20). In contrast, the BPV left-end stem had three bubble structures, and no unpaired T residues were found (Fig. 1A). The bubble and asymmetric T residue are critical structures in MVM replication of the left-end hairpin (20). Surprisingly, the “rabbit ear” sequences on the left-end terminus of both MVC and BPV were almost identical; only 2 nt at the turn-around region and a pair of G-C dinucleotides were obviously inverted, indicating that sequences of the left-end terminus might share a high degree of conservation in the genus Bocavirus. In other parvoviruses, such as the MVM-like parvoviruses, almost identical rabbit ear structures were seen (5). Conservation of the rabbit ear structures in these viruses suggests both common functions provided by the structures as well as essential functions provided by these regions of the termini. In adeno-associated virus (AAV), the large Rep protein binds to the turn-around region of the ear structures (66). Whether NS1 of the bocaviruses binds to the rabbit ear structures is unknown. However, it is likely that these structures confer to the bocaviruses a similar strategy of DNA replication. Three repeats of the GT motif (GTGAC/GTATA/GTAGT) were found on the “stem” before the bubble (Fig. 1A), which may be an MVC NS1 binding site. Similar repeats (GTAGT/GTATA/GTCAC) were also present on the right-end terminus (Fig. 1B). Conserved NS1 binding sites of BPV were not obvious on both ends of the BPV genome.
Only one orientation of the MVC right-end terminus was obtained and it was 198 bases in length (Fig. 1B). Three clones with various ends were obtained. The right-end terminus of MVC also shared a high similarity with that of BPV, including the distal polyadenylation site [(pA)d] and a stretch of conserved sequences (TTTTA/TTGTAT/TTAGTT/AATAAA/GCGCC), which could represent a general feature of the bocaviruses. These conserved sequences were located presumably after the NS1 nicking site, and thus the stable structure (ΔG = −157 kcal/mol) formed as a long stem is probably more important than these sequences. However, they may be signals for correct polyadenylation at the AAUAAA poly(A) site. It should be emphasized that, among parvoviruses, only the bocaviruses have the distal poly(A) signal on the right-end palindromic terminus.
We obtained only one clone with the BPV left-end end hairpin in flop orientation (Fig. 1A). One nucleotide was corrected in the stem of the left-end hairpin, as indicated in Fig. 1A, compared to the sequence previously published (12). Four clones with the right-end terminus of BPV were obtained, which contained various ends (Fig. 1B). Compared with the previously published sequence of the right-end terminus (12), 1 nt was corrected in the “loop” of the right-end hairpin present in both the flip and flop orientation as indicated in Fig. 1B. The BPV right-end terminus could form a stem-arm structure composed of 10 G-C bonds between the poly(A) site and the loop; however, the potential arm structure on the right-end terminus of MVC was less likely to be formed thermodynamically, since it had only four G-C and four A-T bonds. A similar arm structure, which forms an asymmetric cruciform configuration of the right-end hairpin of MVM, had been implicated as being important for MVM replication (6, 18, 19), but whether this stem-arm rearrangement is required for DNA replication of BPV or MVC has not been experimentally demonstrated.
Construction of MVC infectious clones.
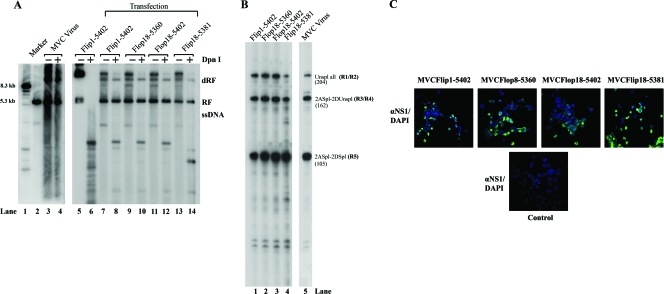
Four full-length clones of MVC were constructed. Of these, two had the flop left-end terminus structure and the other two had the flip orientation. Differing ends of both termini were included among the clones. We transfected these four clones into WRD cells, and MVC DNA replication was confirmed by Southern blot analysis with extracted Hirt DNA (Fig. 2A). Newly synthesized DpnI-resistant MVC DNA was detected in cells transfected with all four clones (Fig. 2A, lanes 8, 10, 12, and 14). Interestingly, the monomer ssDNA of MVC either in transfection of the infectious clones or in MVC infection was not clearly detectable by Southern blot analysis (Fig. 2A). Similar difficulties in demonstration of ssDNA of AAV5 have previously been encountered; however, ssDNA of BPV was apparently detectable in BPV-infected EBTr cells (J. Qiu, unpublished data). Next, we used cell lysates prepared from the transfected cells to infect WRD cells. At 3 days postinfection, significant cytopathic effects were observed in all four reinfections (data not shown). MVC infection was confirmed by production of MVC mRNAs (R1 to R5) using the RNase protection assay (Fig. 2B) and by expression of MVC NS1 using the immunofluorescence assay (Fig. 2C). These results demonstrated that all four clones were infectious in WRD cells. Although the presence of different ends of either the left-end or the right-end terminus or different orientations of the left-end terminus (flip or flop) did not make significant differences in the infections, we chose the longest clone (nt 1 to 5402) with the left-end terminus in flip orientation as the infectious clone, named pIMVC throughout this study.
FIG. 2.
Four MVC full-length clones were replication-competent and infectious in WRD cells. Four MVC infectious clones as indicated were transfected into WRD cells. (A) Southern blot analysis. At 2 days posttransfection, Hirt DNA samples were extracted from transfected cells and MVC-infected cells and were subjected to digestion by DpnI as indicated. Blots were hybridized with the MVC NSCap probe as diagramed in Fig. 3. Hirt DNA extracted from MVC-infected WRD cells served as a positive control of DpnI-resistant MVC RF DNA (lane 4). Plasmid DNA of Flip1-5402 was used as a negative control for DpnI digestion. Lane 1 and lane 2 are size markers of 8.3 kb and 5.3 kb, respectively. dRF, double replicative form of MVC DNA. It is likely that the Flip18-5381 clone contains mutations in the backbone (lane 14). (B) RNase protection assay. Four samples of cell lysate were prepared from transfected cells and were used to infect WRD cells. At 3 days postinfection, total RNA was isolated and detected for MVC-specific mRNAs by the RNase protection assay using the probe 5 that is diagramed in Fig. 3B. Total RNA isolated from MVC-infected WRD cells served as a control (lane 5). The sizes of protected bands in parentheses with their respective designations are shown to the right. (C) Immunofluorescence assay. Four samples of cell lysate from transfected cells were used to infect WRD cells in chamber slides. After fixation, slides were incubated with a rat polyclonal anti-NS1 antibody at a dilution of 1:100 and a fluorescein isothiocyanate-conjugated secondary antibody. The blue background was 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei. Images were taken by an Eclipse C1 Plus confocal microscope (Nikon) at a ×40 magnification.
The transcription profiles generated by both MVC infection and transfection of the infectious clone.
Total RNA was isolated from MVC-infected WRD cells, and reverse transcription-PCR (RT-PCR), 3′ rapid amplification of cDNA ends, and 5′ rapid amplification of cDNA ends were performed using various primers. Transcription units shown in Fig. 3B were identified by sequencing the amplified PCR products and confirmed by RNase protection assay (data not shown).
Initially, to determine the relative abundance of various species of MVC mRNA, we assayed both mRNA isolated from MVC-infected cells and pIMVC-transfected WRD cells by Northern blot analysis using the three probes shown in Fig. 3B. The NS probe detected two species of mRNAs at 2.9 kb (R1) and 4.9 kb (R2) (shown in Fig. 3A, lanes 4 and 5, respectively), which were consistent with being unspliced mRNA at the first donor site (1D). Since the first exon was short, mRNAs spliced at 1D were not detected by the NS probe (54). The Cap probe hybridized to mRNAs that ended only at the (pA)d and detected four mRNA bands (Fig. 3A, lanes 2 and 3). They were 4.9-kb (R2), 3.0-kb (R4), 2.5-kb (R5), and 2.2-kb (R6) mRNA. The NSCap probe detected all six major MVC mRNAs, including the 1.0-kb (R3) mRNA that was polyadenylated at the internal polyadenylation site [(pA)p] (Fig. 3A, lanes 6 and 7). The 3.0-kb band of R4 mRNA was barely seen in virus infection (Fig. 3A, lanes 3 and 7), which could be generated less than that in transfection (Fig. 3A, lanes 2 and 6). The overall transcription map suggested by Northern analysis was confirmed by the RNase protection assay (data not shown) and is presented in Fig. 3B. No significant difference was seen between MVC mRNAs generated following infection or transfection of the infectious clone (Fig. 3A, compare lanes 2, 4, and 6 with lanes 3, 5, and 7, respectively). Similar to the map of BPV (54), mid-ORF-encoded mRNAs R3 and R4 were generated that were either polyadenylated at (pA)p or at (pA)d. This is a unique feature of viruses now classified in the genus Bocavirus. A band of mRNA at 3.7 kb that potentially encodes NS2 of BPV was not detected in RNAs isolated from MVC-infected and pIMVC-transfected WRD cells by RT-PCR (data not shown) and Northern blot analysis (Fig. 3A).
MVC NP1 was essential for MVC DNA replication in WRD cells.
To test the function of NP1 in MVC infection, we prematurely terminated the mid-ORF by introducing a TAG stop codon at nt 2735 with a single T mutation in pIMVC [NP1(−)] (Fig. 4B). Transfection of this NP1 knockout mutant, NP1(−), did not produce significant DpnI-resistant MVC replicative form (RF) DNA (Fig. 4A, lane 5), although NS1 was expressed at a level similar to that from the VP1(−) mutant as seen by immunofluorescence (Fig. 4C). Similar results were also seen when the AUG of NP1 was mutated to ACG, which still left the NS1 ORF unchanged (data not shown). For a comparison, the NS1 ORF was also terminated early in pIMVC [NS1(−)] (Fig. 4B). Replication of MVC DNA of the NS1(−) mutant was totally abolished as shown by immunofluorescence (Fig. 4C). No DpnI-resistant RF DNA was detected (as shown in Fig. 4A, lane 2). Parvovirus NS1 is a multifunctional polypeptide that is essential for parvoviral replication (20). Consistent with this requirement of NS1, our current findings show that MVC NS1 was absolutely required for MVC DNA replication. Surprisingly, NP1 was also essential for MVC DNA replication. Without NP1, replication of MVC DNA was reduced by 320-fold (Fig. 4A, compare lane 1 with lane 5).
To quantify reduction of replication in the absence of NP1, we constructed mutants that could no longer express VP1 or VP2 by mutating the VP1 or VP2 AUG in pIMVC [VP1(−) and VP2(−)] (Fig. 4B). DpnI-resistant RF DNA generated from transfection of these two mutants was reduced by approximately five- to eightfold compared with that of the parent infectious clone (Fig. 4A, compare lanes 3 and 4 with lane 1). As VP1 is absolutely required for parvovirus infection (62, 67) and VP2 is the major capsid protein for assembling infectious virions, the VP1 and VP2 knockout mutants did not produce infectious progeny virions (data not shown). Therefore, RF DNA generated from transfection of VP1(−) and VP2(−) mutants resulted only from single-burst replication of transfected plasmid DNA, accounting for the decrease of five- to eightfold in the amount of RF DNA. Compared with the level of RF DNA in transfection of VP1(−) and VP2(−) mutants, transfection of NP1(−) reduced RF DNA by at least 38- to 65-fold (Fig. 4A, compare lane 5 with lanes 3 and 4). This is the second time that a second nonstructural protein of parvoviruses has been demonstrated to be essential for parvoviral monomer form DNA replication (16, 45).
To confirm the function of Bocavirus NP1 in DNA replication, we carried out NP1 cross-complementation tests by providing the NP1 of MVC, BPV, and HBoV in trans, respectively. MVC NP1 increased DNA replication by 8.4-fold, but not to the full wild-type level (Fig. 4A, lane 6). Surprisingly, BPV NP1 and HBoV NP1 were also able to recover MVC RF DNA by 8.8-fold and 5.1-fold, respectively (Fig. 4A, lanes 7 and 8). Expression of these tagged NP1 proteins was confirmed by Western blot analysis in cotransfections (data not shown). These results suggested that functions of Bocavirus NP1 could be cross-complemented among these viruses.
VP1u of MVC contained a phospholipase A2 (PLA2) motif that was critical to MVC infection.
To examine the role of the PLA2 motif in viral infection, we mutated three amino acids in the VP1 unique (VP1u) region of the infectious clone that are conserved in bocaviruses and in the HDXXY motif of secretory PLA2s (sPLA2s; VP1u35N, VP1u41A, and VP1u42N) (21, 58) (Fig. 5A). For a comparison, two random mutations in the VP1u region, which were outside of the functional PLA2 motif (67), and the nuclear localization sites that are composed of basic amino acid K/R repeats (40, 62, 64) were also made in the infectious clone (VP1u96G and Vp1u115P) (Fig. 5A). These two control mutations were at amino acids 96 and 115. As expected, levels of RF DNA generated in transfection of the PLA2 mutants, VP1u35N, VP1u41A, and VP1u42N, were reduced by approximately eightfold compared with the levels of RF DNA produced by the infectious clone (Fig. 5B, compare lanes 2 to 4 with lane 1). Similarly, the PLA2 mutants achieved the same reduced level of RF DNA as that produced by the VP1(−) mutant (Fig. 5B, compare lanes 2 to 4 with lane 7). These results indicated that, similar to the RF DNA of the VP1(−) mutant, RF DNA of the PLA2 mutants was generated only by single-burst replication of transfected MVC DNA. However, the levels of RF DNA from transfection of the two random mutants, VP1u115P and VP1u96G, of the VP1u region were not significantly reduced (Fig. 5B, compare lanes 5 and 6 with lane 1). The RF DNA still was at six- to eightfold higher than that produced by PLA2 mutants and the VP1(−) mutant (Fig. 5B, compare lanes 2 to 4 and 7 with lanes 5 and 6). Further quantification of progeny virus production from infection using transfected cell lysates showed that the PLA2 mutants, similar to the VP1(−) mutant, did not produce significant viral progeny. In contrast, the VP1u control mutants produced viral progeny at a level similar to that of the infectious clone (Fig. 5C). These results indicated that mutations in the PLA2 motif abolished production of infectious progeny; however, random mutations in the VP1u outside of the PLA2 motifs and nuclear localization sites still generated infectious progeny at a level comparable to that of the infectious clone. Thus, we conclude that the PLA2 motif in VP1u was critical in MVC infection.
DISCUSSION
In this study, we have sequenced the palindromic termini of Bocavirus MVC and have constructed four infectious clones with cloned termini. Systematic analysis of the selected infectious clone revealed common features among viruses in the genus Bocavirus and features unique to parvoviruses. Study of the animal bocaviruses, especially MVC, could help us to elucidate the pathogenesis of HBoV as both MVC and HBoV cause similar symptoms of diseases in their respective hosts (2, 10, 31, 52, 56).
The MVC full-length genome consists of 5,402 nt, and it has disparate palindromic hairpins of 183 nt and 198 nt at the left-end and right-end termini of the genome, respectively. By including the two palindromic termini, we revealed a high identity of 52.1% and 52.6% in genome composition between MVC and BPV and between MVC and HBoV, respectively. An uncorrected insertion of T at nt 300 was observed in the previous NSCap sequence (accession no. NC_004442), which extended 58 amino acids at the N terminus of MVC NS1 in the full-length MVC genome (accession no. FJ214110). The fidelity of this MVC genome was confirmed by its infectivity in MVC-permissive WRD cells. Although BPV has been cloned and sequenced, the infectious clone was not available (54). Thus, the infectious clone of MVC reported here will expand our capability to deeply understand the molecular pathogenesis of bocaviruses in general. Moreover, conserved sequences identified in the rabbit ear structures in the left-end terminus and on the stem in the right-end terminus provide information that can be used to clone the termini of HBoV by designing degenerative primers.
The secondary structure of the parvoviral terminus is remarkably important for DNA replication, sometimes more than the sequence per se. It has been shown that the secondary structure of the arm of the AAV terminus is more important than its primary sequences for AAV DNA replication (37). Interestingly, the primary sequence of the rabbit ear structures on the left-end terminus of the two bocaviruses is conserved. The bubble region in the stem of the left-end hairpin BPV was confirmed as being important for BPV DNA replication. Mutation of to-be-paired nucleotides of either the top strand or bottom strand in the bubble region abolished replication in transfected cells (63). The right-end palindromic hairpin of BPV has the capability to form a secondary structure of the stem-arm (Fig. 1B). In another viral model, a similar stem-arm structure that contains the NS1 binding sites had important implications for the viral DNA replication of MVM (6, 18, 19). In comparison, MVC also has the potential to form such a structure of stem-arm at the right-end hairpin (Fig. 1B); however, the free energy in the potential stem-arm structure of MVC is probably not sufficient thermodynamically. The importance of this structure in viral DNA replication warrants further investigation. Interestingly, this stem-arm structure could be created just after the poly(A) signal. It is possible that this structure can slow down RNA polymerase II as a pausing element to terminate transcription for sufficient polyadenylation at the distal poly(A) site (23, 51). Overall, these two bocaviruses, MVC and BPV, shared extensive similarities in structure and conserved motifs on both termini. Identification of these conserved motif sequences may, in the future, be applied to clone the palindromic hairpins of HBoV.
The transcription map of MVC further confirmed that, similar to parvovirus B19 (49) and Aleutian mink disease virus (53), mRNAs from all species of bocavirus were transcribed from a single unique promoter and were processed by alternative splicing and alternative polyadenylation (54). We made silent mutations in two actively utilized poly(A) signals in viral infection as well as one AAUAAA signal that was not used in viral infection in the infectious clone. Analysis of RNAs generated by transfection of this mutant showed that MVC RNAs were still polyadenylated at the (pA)p site at a ratio similar to that of the wild type (Y. Sun and J. Qiu, unpublished data). However, these mutations significantly decreased the stabilities of MVC mRNAs, indicating that this region was important to maintain MVC mRNAs in a stable form. Further attempts to knock out the (pA)p sites were not tried since this region is in the coding region for the VP1 unique part.
In all parvoviruses except Aleutian mink disease virus, the minor structural protein, VP1, possesses motifs that have PLA2 activity shown to be essential for nuclear entry of the virus during infection (22, 27, 42, 54, 67). The HDXXY motif that was found in the catalytic site of sPLA2s (21) is also predicted to be present in the VP1u of MVC (58). The aspartic amino acid residue (35D) that is 5 amino acids before the HDXXY motif and the HD amino acids in the HDXXY motif are the most conserved ones in the parvovirus PLA2 motifs and sPLA2s (67). Mutations of these three amino acids in the PLA2 motif of the VP1u in the infectious clone confirmed the identity of this protein as VP1, and the PLA2 phospholipase motif is present in VP1 that is critical in MVC infection and may play an essential role for virus to across the endosomal membrane (26). Protein analysis of purified MVC by SDS-PAGE showed three structural proteins, VP1, VP2, and VP3, at 81 kDa, 67/63 kDa, and 61 kDa, respectively (58), which was not in agreement with our transcription map of MVC in that only R5 and R6 had capabilities to encode VP1 and VP2. However, three capsid proteins of BPV, VP1 (80 kDa), VP2 (72 kDa), and VP3 (62 kDa), were also detected on SDS-PAGE gels (32, 37). It was suggested that multiple capsid bands might be degraded or cleaved capsid proteins (32, 35). Thus, the nature of these Bocavirus capsid proteins warrants further investigation.
We have demonstrated that MVC NS1, similar to the NS1 of MVM (14), is a multifunctional protein that is essential for viral DNA replication. Parvoviruses express more than one nonstructural protein. The mid-ORF NP1 protein is a bocavirus-unique protein among parvoviruses, which is translated from a continuous ORF located in the middle of the genome. The N terminus of the HBoV NP1 shares the same coding sequence (nt 2537 to 2724) with the C terminus of the NS1 but in a different reading frame. The rest of the coding region of MVC NP1, 369 nt, contained unique coding sequences that lay mostly in the third intron, which was spliced from donor site 3D to acceptor site 3A, in front of the VP1 coding region (Fig. 3B). NP1 has a predicted size of approximately 20 kDa. Similar proteins are not present in other parvoviruses of other genera, setting bocaviruses apart from all other known parvoviruses. Furthermore, NP1 homologous proteins were also not found by BLAST databases. MVC NP1 protein shares an identity of 39.9% and 41.4% with HBoV NP1 and BPV NP1, respectively.
For MVM, the N-terminal exon of NS2 shares its start site and the same reading as NS2 and then after splicing shifts to an alternative ORF (45). The MVM NS2 has a molecular mass of 25 kDa, and the 84-amino-acid N terminus is shared with NS1 and the C terminus is expressed from spliced mRNA with an alternative frame of the NS1-encoding mRNA (17). MVM NS2 is dispensable for productive replication in a variety of nonmurine cells, such as transformed human 324 K cells, but it is required for maximum viral DNA replication in murine cells. The NS2 early-terminated mutants reduced MVM DNA replication maximally by fivefold compared with the wild-type level in a single-burst cycle of replication (16, 45). However, MVC DNA replication was extremely dependent on the function of NP1. An NP1 knockout mutant reduced RF DNA by nearly 50-fold in a single-burst cycle of replication. The NS2 of canine parvovirus (CPV), similar to the MVM NS2, is translated from spliced NS1 ORF and contains 87 amino-terminal amino acids in common with NS1 joined to 78 amino acids from an alternative ORF (65). NS2 knockout mutants in the infectious clone of CPV show no obvious differences in viral DNA replication in cell culture or in dogs (65). However, recent studies examining the within-host genetic diversity of CPV infection showed that NS2 might play a role in host adaptation of CPV (30). MVM NS2 is a multifunctional protein; it also has a role in ssDNA production, capsid assembly, and nuclear export of virions (16, 24, 43, 45, 55). Whether MVC NP1 has multiple roles in MVC infection warrants further investigation. Importantly, BPV NP1 can rescue MVC DNA replication to a similar level as the MVC NP1. Whether the Bocavirus NP1 unique coding sequence can be exchangeable is currently under investigation by replacing the MVC NP1 with BPV or HBoV NP1 in the MVC infectious clone. Thus, we can study the function of HBoV NP1 within the context of MVC.
Acknowledgments
This work was supported by PHS grant no. RO1 AI070723 from NIAID and grant no. P20 RR016443 from the NCRR COBRE program.
We thank Colin Parrish at the James A. Baker Institute of Cornell University and David Pintel at the Life Sciences Center of the University of Missouri-Columbia for valuable reagents. We thank Martha Montello and Heather McNeill for editing the manuscript.
Footnotes
Published ahead of print on 11 February 2009.
REFERENCES
- 1.Abinanti, F. R., and M. S. Warfield. 1961. Recovery of a hemadsorbing virus (HADEN) from the gastrointestinal tract of calves. Virology 14288-289. [DOI] [PubMed] [Google Scholar]
- 2.Allander, T. 2008. Human bocavirus. J. Clin. Virol. 4129-33. [DOI] [PubMed] [Google Scholar]
- 3.Allander, T., T. Jartti, S. Gupta, H. G. Niesters, P. Lehtinen, R. Osterback, T. Vuorinen, M. Waris, A. Bjerkner, A. Tiveljung-Lindell, B. G. van den Hoogen, T. Hyypia, and O. Ruuskanen. 2007. Human bocavirus and acute wheezing in children. Clin. Infect. Dis. 44904-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 10212891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astell, C. R. 1990. Terminal hairpins of parvovirus genomes and their role in DNA replication, p. 59-80. In P. Tijssen (ed.), Handbook of parvoviruses, vol. I. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 6.Astell, C. R., M. Thomson, M. B. Chow, and D. C. Ward. 1983. Structure and replication of minute virus of mice DNA. Cold Spring Harb. Symp. Quant. Biol. 47751-762. [DOI] [PubMed] [Google Scholar]
- 7.Bastien, N., N. Chui, J. L. Robinson, B. E. Lee, K. Dust, L. Hart, and Y. Li. 2007. Detection of human bocavirus in Canadian children in a 1-year study. J. Clin. Microbiol. 45610-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binn, L. N., E. C. Lazar, G. A. Eddy, and M. Kajima. 1970. Recovery and characterization of a minute virus of canines. Infect. Immun. 1503-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binn, L. N., R. H. Marchwicki, E. H. Eckermann, and T. E. Fritz. 1981. Viral antibody studies of laboratory dogs with diarrheal disease. Am. J. Vet. Res. 421665-1667. [PubMed] [Google Scholar]
- 10.Carmichael, L. E., D. H. Schlafer, and A. Hashimoto. 1991. Pathogenicity of minute virus of canines (MVC) for the canine fetus. Cornell Vet. 81151-171. [PubMed] [Google Scholar]
- 11.Carmichael, L. E., D. H. Schlafer, and A. Hashimoto. 1994. Minute virus of canines (MVC, canine parvovirus type-1): pathogenicity for pups and seroprevalence estimate. J. Vet. Diagn. Invest. 6165-174. [DOI] [PubMed] [Google Scholar]
- 12.Chen, K. C., B. C. Shull, M. Lederman, E. R. Stout, and R. C. Bates. 1988. Analysis of the termini of the DNA of bovine parvovirus: demonstration of sequence inversion at the left terminus and its implication for the replication model. J. Virol. 623807-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, K. C., B. C. Shull, E. A. Moses, M. Lederman, E. R. Stout, and R. C. Bates. 1986. Complete nucleotide sequence and genome organization of bovine parvovirus. J. Virol. 601085-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen, J., S. F. Cotmore, and P. Tattersall. 1995. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 695422-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung, J. Y., T. H. Han, S. W. Kim, C. K. Kim, and E. S. Hwang. 2007. Detection of viruses identified recently in children with acute wheezing. J. Med. Virol. 791238-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore, S. F., A. M. D'Abramo, Jr., L. F. Carbonell, J. Bratton, and P. Tattersall. 1997. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology 231267-280. [DOI] [PubMed] [Google Scholar]
- 17.Cotmore, S. F., L. J. Sturzenbecker, and P. Tattersall. 1983. The autonomous parvovirus MVM encodes two nonstructural proteins in addition to its capsid polypeptides. Virology 129333-343. [DOI] [PubMed] [Google Scholar]
- 18.Cotmore, S. F., and P. Tattersall. 1998. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J. Virol. 728477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotmore, S. F., and P. Tattersall. 2005. Structure and organization of the viral genome, p. 73-94. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 20.Cotmore, S. F., and P. Tattersall. 2006. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses, p. 171-181. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 21.Dennis, E. A. 1997. The growing phospholipase A2 superfamily of signal transduction enzymes. Trends Biochem. Sci. 221-2. [DOI] [PubMed] [Google Scholar]
- 22.Dorsch, S., G. Liebisch, B. Kaufmann, P. von Landenberg, J. H. Hoffmann, W. Drobnik, and S. Modrow. 2002. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J. Virol. 762014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dye, M. J., and N. J. Proudfoot. 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105669-681. [DOI] [PubMed] [Google Scholar]
- 24.Eichwald, V., L. Daeffler, M. Klein, J. Rommelaere, and N. Salome. 2002. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J. Virol. 7610307-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo, R., N. Ishiguro, H. Kikuta, S. Teramoto, R. Shirkoohi, X. Ma, T. Ebihara, H. Ishiko, and T. Ariga. 2007. Seroepidemiology of human bocavirus in Hokkaido Prefecture, Japan. J. Clin. Microbiol. 453218-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farr, G. A., L. G. Zhang, and P. Tattersall. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. USA 10217148-17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83973-978. [DOI] [PubMed] [Google Scholar]
- 28.Guan, W., F. Cheng, Y. Yoto, S. Kleiboeker, S. Wong, N. Zhi, D. J. Pintel, and J. Qiu. 2008. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J. Virol. 829951-9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison, L. R., E. L. Styer, A. R. Pursell, L. E. Carmichael, and J. C. Nietfeld. 1992. Fatal disease in nursing puppies associated with minute virus of canines. J. Vet. Diagn. Invest. 419-22. [DOI] [PubMed] [Google Scholar]
- 30.Hoelzer, K., L. A. Shackelton, E. C. Holmes, and C. R. Parrish. 2008. Within-host genetic diversity of endemic and emerging parvoviruses of dogs and cats. J. Virol. 8211096-11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Järplid, B., H. Johansson, and L. E. Carmichael. 1996. A fatal case of pup infection with minute virus of canines (MVC). J. Vet. Diagn. Invest. 8484-487. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, F. B., and M. D. Hoggan. 1973. Structural proteins of HADEN virus. Virology 51129-137. [DOI] [PubMed] [Google Scholar]
- 33.Kahn, J. S., D. Kesebir, S. F. Cotmore, A. D'Abramo, Jr., C. Cosby, C. Weibel, and P. Tattersall. 2008. Seroepidemiology of human bocavirus defined using recombinant virus-like particles. J. Infect. Dis. 19841-50. [DOI] [PubMed] [Google Scholar]
- 34.Kantola, K., L. Hedman, T. Allander, T. Jartti, P. Lehtinen, O. Ruuskanen, K. Hedman, and M. Soderlund-Venermo. 2008. Serodiagnosis of human bocavirus infection. Clin. Infect. Dis. 46540-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lederman, M., R. C. Bates, and E. R. Stout. 1983. In vitro and in vivo studies of bovine parvovirus proteins. J. Virol. 4810-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lederman, M., J. T. Patton, E. R. Stout, and R. C. Bates. 1984. Virally coded noncapsid protein associated with bovine parvovirus infection. J. Virol. 49315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefebvre, R. B., S. Riva, and K. I. Berns. 1984. Conformation takes precedence over sequence in adeno-associated virus DNA replication. Mol. Cell. Biol. 41416-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, F., W. Guan, F. Cheng, N. Yang, D. Pintel, and J. Qiu. 2008. ELISAs using human bocavirus VP2 virus-like particles for detection of antibodies against HBoV. J. Virol. Methods 149110-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, F., A. Zeng, N. Yang, H. Lin, E. Yang, S. Wang, D. Pintel, and J. Qiu. 2007. Quantification of human bocavirus in lower respiratory tract infections in China 161. Infect. Agents Cancer 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lombardo, E., J. C. Ramirez, J. Garcia, and J. M. Almendral. 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 767049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lubeck, M. D., and F. B. Johnson. 1976. Multiplication of bovine parvovirus in two cell strains. Infect. Immun. 131289-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupescu, A., C. T. Bock, P. A. Lang, S. Aberle, H. Kaiser, R. Kandolf, and F. Lang. 2006. Phospholipase A2 activity-dependent stimulation of Ca2+ entry by human parvovirus B19 capsid protein VP1. J. Virol. 8011370-11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, C. L., and D. J. Pintel. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 763257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mochizuki, M., M. Hashimoto, T. Hajima, M. Takiguchi, A. Hashimoto, Y. Une, F. Roerink, T. Ohshima, C. R. Parrish, and L. E. Carmichael. 2002. Virologic and serologic identification of minute virus of canines (canine parvovirus type 1) from dogs in Japan. J. Clin. Microbiol. 403993-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naeger, L. K., J. Cater, and D. J. Pintel. 1990. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J. Virol. 646166-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naeger, L. K., R. V. Schoborg, Q. Zhao, G. E. Tullis, and D. J. Pintel. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 61107-1119. [DOI] [PubMed] [Google Scholar]
- 47.Naghipour, M., L. E. Cuevas, T. Bakhshinejad, W. Dove, and C. A. Hart. 2007. Human bocavirus in Iranian children with acute respiratory infections. J. Med. Virol. 79539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohshima, T., M. Kishi, and M. Mochizuki. 2004. Sequence analysis of an Asian isolate of minute virus of canines (canine parvovirus type 1). Virus Genes 29291-296. [DOI] [PubMed] [Google Scholar]
- 49.Ozawa, K., J. Ayub, Y. S. Hao, G. Kurtzman, T. Shimada, and N. Young. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J. Virol. 612395-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pintel, D., D. Dadachanji, C. R. Astell, and D. C. Ward. 1983. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Res. 111019-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plant, K. E., M. J. Dye, C. Lafaille, and N. J. Proudfoot. 2005. Strong polyadenylation and weak pausing combine to cause efficient termination of transcription in the human Gγ-globin gene. Mol. Cell. Biol. 253276-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pratelli, A., D. Buonavoglia, M. Tempesta, F. Guarda, L. Carmichael, and C. Buonavoglia. 1999. Fatal canine parvovirus type-1 infection in pups from Italy. J. Vet. Diagn. Invest. 11365-367. [DOI] [PubMed] [Google Scholar]
- 53.Qiu, J., F. Cheng, L. R. Burger, and D. Pintel. 2006. The transcription profile of Aleutian mink disease virus (AMDV) in CRFK cells is generated by alternative processing of pre-mRNAs produced from a single promoter. J. Virol. 80654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu, J., F. Cheng, F. B. Johnson, and D. Pintel. 2007. The transcription profile of the bocavirus bovine parvovirus is unlike those of previously characterized parvoviruses. J. Virol. 8112080-12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruiz, Z., A. D'Abramo, Jr., and P. Tattersall. 2006. Differential roles for the C-terminal hexapeptide domains of NS2 splice variants during MVM infection of murine cells. Virology 349382-395. [DOI] [PubMed] [Google Scholar]
- 56.Schildgen, O., A. Muller, T. Allander, I. M. Mackay, S. Volz, B. Kupfer, and A. Simon. 2008. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin. Microbiol. Rev. 21291-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 18122-34. [DOI] [PubMed] [Google Scholar]
- 58.Schwartz, D., B. Green, L. E. Carmichael, and C. R. Parrish. 2002. The canine minute virus (minute virus of canines) is a distinct parvovirus that is most similar to bovine parvovirus. Virology 302219-223. [DOI] [PubMed] [Google Scholar]
- 59.Shull, B. C., K. C. Chen, M. Lederman, E. R. Stout, and R. C. Bates. 1988. Genomic clones of bovine parvovirus: construction and effect of deletions and terminal sequence inversions on infectivity. J. Virol. 62417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sloots, T. P., P. McErlean, D. J. Speicher, K. E. Arden, M. D. Nissen, and I. M. Mackay. 2006. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J. Clin. Virol. 3599-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tattersall, P. 2006. The evolution of parvovirus taxonomy, p. 5-14. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 62.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1993. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J. Virol. 67131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Via, L. E., C. Mainguy, S. Naides, and M. Lederman. 2006. Bovine parvovirus, p. 479-486. In J. Kerr, S. F. Cotmore, M. E. Bloom, M. E. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 64.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 761884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, D., W. Yuan, I. Davis, and C. R. Parrish. 1998. Nonstructural protein-2 and the replication of canine parvovirus. Virology 240273-281. [DOI] [PubMed] [Google Scholar]
- 66.Word, P. 2006. Replication of adeno-associated virus DNA, p. 189-212. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 67.Zádori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1291-302. [DOI] [PubMed] [Google Scholar]
- 68.Zhi, N., Z. Zadori, K. E. Brown, and P. Tijssen. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318142-152. [DOI] [PubMed] [Google Scholar]