Abstract
Human immunodeficiency virus type 1 (HIV-1) gene expression and replication are regulated by the promoter/enhancer located in the U3 region of the proviral 5′ long terminal repeat (LTR). The binding of cellular transcription factors to specific regulatory sites in the 5′ LTR is a key event in the replication cycle of HIV-1. Since transcriptional activity is regulated by the posttranslational modification of transcription factors with the monosaccharide O-linked N-acetyl-d-glucosamine (O-GlcNAc), we evaluated whether increased O-GlcNAcylation affects HIV-1 transcription. In the present study we demonstrate that treatment of HIV-1-infected lymphocytes with the O-GlcNAcylation-enhancing agent glucosamine (GlcN) repressed viral transcription in a dose-dependent manner. Overexpression of O-GlcNAc transferase (OGT), the sole known enzyme catalyzing the addition of O-GlcNAc to proteins, specifically inhibited the activity of the HIV-1 LTR promoter in different T-cell lines and in primary CD4+ T lymphocytes. Inhibition of HIV-1 LTR activity in infected T cells was most efficient (>95%) when OGT was recombinantly overexpressed prior to infection. O-GlcNAcylation of the transcription factor Sp1 and the presence of Sp1-binding sites in the LTR were found to be crucial for this inhibitory effect. From this study, we conclude that O-GlcNAcylation of Sp1 inhibits the activity of the HIV-1 LTR promoter. Modulation of Sp1 O-GlcNAcylation may play a role in the regulation of HIV-1 latency and activation and links viral replication to the glucose metabolism of the host cell. Hence, the establishment of a metabolic treatment might supplement the repertoire of antiretroviral therapies against AIDS.
Human immunodeficiency virus type 1 (HIV-1) gene expression is regulated by the long terminal repeat (LTR) promoter, which is composed of three regions: U3 (unique 3′ end), R (repeated), and U5 (unique 5′ end). The U3 region contains an upstream regulatory element including binding sites for several cellular transcription factors such as the nuclear factor of activated T cells 1 (NFATc1) and the activator protein 1 (AP-1), an enhancer with two binding sites for the nuclear factor κB (NF-κB), and the core promoter composed of three tandem binding sites for specificity protein 1 (Sp1) and a TATA box. While NF-κB is a strong enhancer of HIV-1 transcription (8), Sp1 is essential for basal transcription and Tat-mediated activation of HIV-1 (25, 69). This is concordant with the fact that Sp1 is upregulated in activated T cells (43), which compose the primary reservoir for HIV-1 replication (68). Importantly, deletion of all three Sp1-binding sites reduces viral replication in human T-cell cultures (58).
In eukaryotic cells, myriad cytoplasmic and nuclear proteins are posttranslationally modified at the hydroxyl groups of specific serine and threonine residues by a single monosaccharide, N-acetyl-d-glucosamine (GlcNAc) (26, 80). The O-glycosidic linkage of GlcNAc to proteins is a highly dynamic and reversible posttranslational modification and differs from other glycosylation events in that it occurs in the cytosol and the nucleus rather than in the Golgi apparatus or the endoplasmic reticulum. Protein O-linked N-acetyl-d-glucosaminylation (O-GlcNAcylation) is catalyzed by O-GlcNAc transferase (OGT) and reversed by O-GlcNAc hexosaminidase (O-GlcNAcase) (30). The substrate for protein O-GlcNAcylation is UDP-GlcNAc. Therefore, O- GlcNAcylation is regulated by the cellular levels of OGT and O-GlcNAcase and by the availability of UDP-GlcNAc (81). As UDP-GlcNAc is synthesized de novo from glucose via the hexosamine biosynthetic pathway, glucose flux through the hexosamine biosynthetic pathway increases O-GlcNAcylation of proteins. Furthermore, compounds like glucosamine (GlcN), streptozotocin, O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate (PUGNAc), and 2-deoxyglucose enhance O-GlcNAcylation of proteins either by increasing the availability of UDP-GlcNAc or by inhibiting the enzyme O-GlcNAcase (23, 37, 63).
Protein O-GlcNAcylation has been shown to modulate (i) enzyme activity, (ii) protein-protein interactions, (iii) DNA-binding affinity, (iv) subcellular localization, and (v) the half-life and proteolytic processing of proteins (81). Furthermore, O-GlcNAcylation often plays an antagonistic role in relation to phosphorylation (36). The dynamic interplay between O-GlcNAcylation and phosphorylation has therefore been proposed to control protein-protein interactions and protein functions. Furthermore, O-GlcNAc plays a pivotal role in the regulation of gene expression via modification of RNA polymerase II (12, 39) and associated transcription factors (31, 32). Thus, O-GlcNAc serves as a nutrient sensor to couple the metabolic status to cellular processes, such as protein degradation, signal transduction, and gene transcription.
Some of the transcription factors involved in HIV-1 gene regulation are modified by O-GlcNAc, including AP-1 (67), yin-yang 1 (YY1) (28), NFATc1 (21), NF-κB (21), and Sp1 (31, 62). Sp1 was the first transcription factor identified as being O-GlcNAcylated (31). Sp1 is ubiquitous and belongs to the Sp1-like/Krüppel-like family of transcription factors characterized by three C-terminal Cys2His2 zinc finger motifs (34) and a DNA-binding affinity to GC-rich sites (16). Two decades ago, Tjian and coworkers showed that Sp1 bears at least eight O-GlcNAc sites and that the O-GlcNAcylated form of Sp1 was more active than the non-O-GlcNAcylated protein (31). In the meantime, other studies revealed that O-GlcNAcylation of Sp1 can also result in a reduction of transcriptional activation (62, 77), indicating that the specific effect of O-GlcNAcylation on Sp1 activity strongly depends on the targeted promoter.
Since Sp1 is critical and important for HIV-1 transcription (25) and Sp1 activity is modulated by O-GlcNAc (42), we investigated the effect of Sp1 O-GlcNAcylation on the activity of the HIV-1 LTR promoter. Here we show that an increased O-GlcNAc level represses the HIV-1 LTR promoter activity. This required O-GlcNAcylation of Sp1 and the presence of Sp1-binding sites in the HIV-1 LTR. Thus, modulation of Sp1 O-GlcNAcylation may be useful as a potential therapeutic approach for the inhibition of HIV-1 replication.
MATERIALS AND METHODS
Cell culture.
Jurkat and T1 (174 × CEM.T1) cells were maintained in very-low-endotoxin-Roswell Park Memorial Institute (RPMI) 1640 medium (Biochrom, Berlin, Germany) supplemented with 10% fetal calf serum (FCS; Biochrom) and 2 mM l-glutamine (PAA, Cölbe, Germany) at 37°C in a humidified atmosphere at 5% CO2. CD4+ T cells were isolated using anti-CD4 magnetically activated cell sorting beads (Miltenyi, Bergisch Gladbach, Germany) according to the manufacturer's instructions. CD4+ T cells were cultured in MLPC medium consisting of RPMI 1640, 10% human serum, 2 mM l-glutamine, 20 mg/liter gentamicin, 10 mM HEPES, 1 mM sodium pyruvate, and 1% minimum essential medium nonessential amino acids (all from PAA, Cölbe, Germany) at 37°C under 5% CO2. HeLa-Tat-III/LTR/d1EGFP cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Masahiko Satoh (57). These cells stably express d1EGFP (which was derived from d2EGFP, a destabilized, redshifted variant of the enhanced green fluorescent protein [EGFP], and has a half-life of approximately 1 hour) under the control of the HIV-1 LTR promoter. HeLa-Tat-III/LTR/d1EGFP cells were cultivated in Dulbecco's modified Eagle's medium (PAA) supplemented with 10% FCS, 2 mM l-glutamine, and 1 mg/ml G418 at 37°C under 8.5% CO2. HEK 293T cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% FCS and 2 mM l-glutamine at 37°C under 8.5% CO2.
Pseudovirus production.
Pseudoviruses were produced by cotransfection of HEK 293T cells with pNL4-3LucR-E- (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Nathaniel Landau [13, 27]) and a vesicular stomatitis virus G protein (VSV-G)-encoding plasmid by using the calcium phosphate technique. Pseudovirus-containing supernatants were harvested 2 days after transfection, and cellular debris was removed by centrifugation at 300 × g for 5 min. HIV-1 p24 concentration was estimated by antigen enzyme-linked immunosorbent assay. Pseudovirus-containing supernatants were stored in 1-ml aliquots at −80°C until use.
HIV-1 infection and GlcN treatment.
Jurkat and T1 cells were cultured to a density of 1.5 × 106 cells/ml and split 1:2 on the day prior to infection. Cells were infected with VSV-G env pseudotyped HIV-1NL4-3LucR-E- for 4 h at 37°C, using a concentration of 30 ng p24 per 1 × 106 cells. Cells were centrifuged at 300 × g for 5 min to remove excessive pseudovirus and resuspended in 2 ml very-low-endotoxin-RPMI 1640 containing 2 mM l-glutamine and 10% FCS supplemented with the respective concentration of GlcN (Sigma-Aldrich, Munich, Germany) in triplicate. CD4+ T cells were stimulated for 6 days with 10 U/ml interleukin-2 (Roche, Mannheim, Germany) and 10 μg/ml phytohemagglutinin P (Sigma-Aldrich) prior to infection. Cells were infected with reporter HIV-1 for 4 h at 37°C, using a concentration of 30 ng p24 per 2 × 106 cells. Cells were centrifuged at 250 × g for 10 min to remove excessive pseudovirus and resuspended in 1.5 ml MLPC medium supplemented with the respective concentration of GlcN in triplicate. After incubation for 24 h, cells were collected, washed once with phosphate-buffered saline (PBS; Biochrom), lysed in 1× passive lysis buffer (Promega, Mannheim, Germany), and assayed for luciferase activity as described below.
Cell viability assay.
Cell viability was measured using the CellTiter 96 nonradioactive cell proliferation assay (MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay; Promega) according to the manufacturer's protocol. Briefly, 100-μl cell suspensions containing either 1 × 105 HIV-1-infected Jurkat or T1 cells or 2 × 105 HIV-1-infected CD4+ primary T lymphocytes were applied for the assay.
Plasmids.
The coding sequence of OGT was cloned by amplifying 820 bp corresponding to the N terminus of nucleocytoplasmic OGT, GenBank accession number NM_181672 (24, 41), from human cDNA with specific primers containing the restriction sites for BamHI and EcoRV (underlined) and a Kozak sequence (italics) (forward primer, 5′-TAGGGATCCGATATCGCCACCATGGCGTCTTCCGTGGGCAACG-3′) and the restriction site for StuI (underlined) (reverse primer, 5′-AGATCTATCAGGCCTTGCTCATAG-3′). The PCR fragment was then ligated with the StuI restriction site to the part of the Lv4F fragment (also termed mitochondrial OGT; GenBank accession number U77413 [46, 47]) which corresponds to the C terminus of nucleocytoplasmic OGT and harbors a NotI restriction site at the 3′ end. The complete sequence for the human nucleocytoplasmic OGT was subsequently inserted into pcDNA4/myc-His B (Invitrogen, Karlsruhe, Germany) at the EcoRV and NotI sites. The rescue mutant resOGT was created by mutagenesis PCR using pcDNA-OGT as template and the QuikChange XL site-directed mutagenesis kit (Stratagene, Amsterdam, The Netherlands) according to the manufacturer's instructions. The mutagenic primers were designed by the QuikChange primer design program: forward primer, 5′-GAGGCACGGCAACCTGTGCTTAGATAAGATCAACGTGCTGCACAAGCCACCATATGAACATCCAAAAGA-3′; reverse primer, 5′-TCTTTTGGATGTTCATATGGTGGCTTGTGCAGCACGTTGATCTTATCTAAGCACAGGTTGCCGTGCCTC-3′ (nucleotide changes compared to the wild-type sequence are in bold). The expression plasmid for Sp1 was generated by subcloning the XbaI/SmaI fragment of the pBS-Sp1-f1 vector (GenBank accession number AF252284 [35]), representing the open reading frame of Sp1, into the XbaI/EcoRV-digested pcDNA3.1(−) vector (Invitrogen). The reporter constructs pXP1-LTR-κB-Sp1wt-Luc, pXP1-LTR-κB-Sp1mut-Luc, and pXP1-LTR-Sp1wt-Luc were kindly provided by Manuel López-Cabrera (2, 22). The plasmid pcDNA4-LTRwt-d2EGFP (destabilized, redshifted variant of the EGFP with a half-life of approximately 2 hours) was cloned by insertion of d2EGFP into pcDNA4/myc-His B plasmid (Invitrogen) via XhoI and XbaI. HIV-1 wild-type LTR (LTRwt) was amplified from pNL4-3, obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Malcolm Martin (1), using the following primers: forward primer, 5′-GACAATTGAAGAAAAGGGGGGACTGGAAGGGCTAATTCACTC-3′; reverse primer, 5′-ACCTCGAGTTGGCTCACTGCAACCTCTACCTCCTGGGTGCT-3′. The amplified LTRwt DNA was digested with MfeI and XhoI (underlined) and ligated into pcDNA4-d2EGFP. The plasmid pcDNA4-LTRmutSp1-d2EGFP contained the Sp1-binding-site-mutated LTR (LTRmutSp1) and was created by QuikChange XL site-directed mutagenesis PCR using the forward primer 5′-GGGGACTTTCCAGGGATTCGTGGCCTGTTCGGGACTGGTTAGTGGCGA-3′ and the reverse primer 5′-CTCGCCACTAACCAGTCCCGAACAGGCCACGAATCCCTGGAAAGTCCCC-3′ (nucleotide changes compared to the wild-type sequence are in bold). The Tat expression construct pCEP4-Tat was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Lung-Ji Chang (9). The control reporter plasmid pEF1α-Luc was constructed by subcloning the elongation factor 1 alpha (EF1α) promoter from pEF1/Myc-His vector (Invitrogen) into the promoterless pGL3-Basic vector (Promega). pcDNA4-EF1α-d2EGFP was generated by replacing the cytomegalovirus promoter in the plasmid pcDNA4-d2EGFP with the EF1α promoter. All cloned constructs were confirmed by full-length sequencing and expression experiments. The plasmid pGEM4Z-OGT-64A was generated by subcloning the OGT gene from pcDNA4-OGT into the RNA production vector pGEM4Z-5′UTR-sig-huSurvivin-DC.LAMP-3′UTR-64A (kindly provided by Kris Thielemans [4]), replacing the huSurvivin-DC.LAMP. The pGEM4Z-EGFP-64A vector was provided by I. Tcherepanova, Argos Therapeutics, Durham, NC (66). The constructs were confirmed by restriction digest and expression experiments.
Flow cytometry analyses.
HeLa-Tat-III/LTR/d1EGFP cells were treated with the respective GlcN concentrations for 5 h in triplicate. HEK 293T-LTRwt-d2EGFP and HEK 293T-LTRmutSp1-d2EGFP cells were treated with 16 mM GlcN for 20 h in duplicate. The cells were harvested with trypsin-EDTA (PAA), washed twice with FACS-PBS (PBS containing 5% FCS and 0.1% sodium azide [Sigma-Aldrich]) and resuspended in FACS-PBS. Viable GFP-positive cells (1 × 104) were analyzed for each sample using a FACSCalibur flow cytometer with CellQuest Pro software (both from BD Biosciences, Heidelberg, Germany). The relative fluorescence intensity is given as a ratio of the geometric mean of untreated cells to that of GlcN-treated cells. The geometric mean of untreated cells was defined as 100%.
In vitro transcription of mRNA.
In vitro transcription of RNA was performed as described previously (65). In short, the pGEM4Z-OGT-64A and pGEM4Z-EGFP-64A vectors were linearized with NotI and SpeI enzyme, respectively, purified by phenol-chloroform extraction and ethanol precipitation, and used as DNA templates. The in vitro transcription was performed with T7 RNA polymerase (mMessage mMachine T7 Ultra kit; Ambion/Applied Biosystems, Austin, TX) according to the manufacturer's instructions. After DNase I (Ambion) digestion and poly(A) tailing (Ambion), the transcribed RNA was recovered on RNeasy columns (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. RNA quality was verified by agarose gel electrophoresis, and the RNA concentration was measured spectrophotometrically.
Determination of protein concentration.
The protein concentration was determined in a microplate reader model 680 (Bio-Rad, Munich, Germany) at 750 nm using a detergent-compatible (DC) protein assay kit (Bio-Rad) according to the manufacturer's protocol.
Western blotting.
Cells were lysed as described under “Cell transfection” and “HIV-1 infection and GlcN treatment.” Samples were prepared in 6× sodium dodecyl sulfate (SDS) buffer (350 mM Tris-HCl, pH 6.8, 30% glycerol, 10% SDS, 0.6 M dithiothreitol, 0.012% bromophenol blue), boiled for 10 min at 99°C, and loaded on 7.5% acrylamide-bisacrylamide gels or 4 to 20% Tris-glycine gradient gels (Anamed, Gross-Bieberau, Germany). Gel-separated proteins were transferred to a polyvinylidene fluoride membrane (Roth, Karlsruhe, Germany), which was blocked with 5% nonfat dry milk in PBS-Tween (PBS containing 0.1% Tween 20) for 2 h, washed with PBS-Tween, and incubated with the primary antibody in 2.5% nonfat dry milk in PBS-Tween at room temperature for 2 h. After washing, the membranes were incubated for 45 min at room temperature with the secondary antibody in 2.5% nonfat dry milk in PBS-Tween (71, 76). In order to prevent cross-reactions with milk proteins, Western blot staining with the O-GlcNAc-recognizing antibody was performed as follows: the membranes were blocked with 10% Western blocking reagent (Roche) in PBS-Tween and the antibody was diluted in 0.5% Western blocking reagent (Roche). Subsequent to the final washing step, chemiluminescence was detected according to the manufacturer's protocol (Pierce Biotechnology, Rockford, IL).
Antibodies.
The following primary antibodies were used: polyclonal rabbit anti-human Sp1 Pep2 (1:500; Santa Cruz Biotechnology, Heidelberg, Germany), polyclonal goat anti-human lamin A/C N-18 (1:1,000; Santa Cruz Biotechnology), polyclonal rabbit anti-human OGT TI-14 (1:1,000; Sigma-Aldrich), polyclonal rabbit anti-human actin (1:1,000; Sigma-Aldrich), monoclonal mouse anti-human histone H1 (1:1,000; Santa Cruz), monoclonal mouse anti-GFP (1:10,000, Roche); monoclonal mouse anti-eukaryote O-GlcNAc CTD110.6 (1:1,000; Hiss Diagnostics, Freiburg, Germany), and monoclonal mouse anti-human glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:70,000, Millipore, Schwalbach, Germany). The monoclonal rat anti-Tat immunoglobulin G2a (IgG2a) antibody (clone 1C9; 1:150) was produced by immunization of LOU/C rats with His-tagged purified recombinant Tat protein (50 μg) according to a previously described procedure (48). The secondary, horseradish peroxidase-coupled antibodies goat anti-rat IgG, donkey anti-rabbit IgG, sheep anti-mouse IgG, and rabbit anti-goat IgG (all from Dako, Hamburg, Germany) were diluted 1:5,000.
Nuclear/cytosol fractionation.
Fractionation was performed with a nuclear/cytosol fractionation kit (BioVision, Wiesbaden, Germany) according to the manufacturer's protocol (64). For gel shift experiments, cells were counted, and the nuclear fractions were lysed in 10 μl nuclear extraction buffer per 1 × 106 HEK 293T cells in order to concentrate the fractions. The protein concentration was determined as described above. Equal protein amounts of each fraction were loaded onto the gel and analyzed by Western blotting or applied to gel shift experiments.
Immunoprecipitation.
HEK 293T cells (2 × 106) were seeded in 9 ml medium in 10-cm cell culture dishes. The cells were transfected 24 h later (via the calcium phosphate method) with pcDNA4, pcDNA-Sp1, or pcDNA-Sp1 and pcDNA-OGT. The total transfected DNA amount was adjusted to 30 μg with the control plasmid pcDNA4. At 24 h after transfection, cells were washed with PBS and subsequently lysed in IP lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 1% Igepal [all purchased from Sigma-Aldrich]), supplemented with one tablet of Complete Mini, EDTA-free protease inhibitor cocktail (Roche) per 10 ml. The protein concentration was determined, and 1 mg total protein in a maximal volume of 1 ml was used for the immunoprecipitation. The lysates were precleared by incubation with 100 μl Sepharose CL-6B (Sigma-Aldrich) at 4°C for 1 h. The Sp1 protein complex was immunoprecipitated at 4°C for 3 h with 30 μg anti-Sp1 Pep2 antibody covalently coupled to agarose beads (Santa Cruz Biotechnology). Bead-coupled protein complexes were washed twice with 1 ml IP lysis buffer and three times with IP wash buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 0.1% Igepal). The immunoprecipitates were analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting as described above.
Cell transfection.
HEK 293T cells were seeded 24 h prior to transfection in 1.5 ml medium at a density of 3 × 105 cells per well in a six-well plate. Cells were transiently cotransfected (via the calcium phosphate method) in triplicate with 0.5 μg pXP1-LTR-Sp1wt-Luc or 0.5 μg pEF1α-Luc, along with 0.75 μg pcDNA-Sp1 and/or 0.25 μg pCEP4-Tat and 1 μg pcDNA-OGT (unless otherwise indicated). The total DNA amount was adjusted to 2.5 μg with the control plasmid pcDNA4. For RNA interference experiments, 0.22 μg (amount corresponds to 10 nM) short interfering RNA (siRNA) oligonucleotides specific for human OGT (hsOGT_7 SI02665131; Qiagen), human Sp1 (hsSP1_1 SI00150976; Qiagen), or human GAPDH (Silencer GAPDH siRNA; Ambion, Darmstadt, Germany), or a nontargeting control siRNA (Silencer Negative Control no. 1 siRNA; Ambion) was cotransfected. Twenty-four hours later, cells were lysed in 200 μl 1× passive lysis buffer (Promega) and assayed for luciferase activity as described below. Stable transfection of HEK 293T cells with pcDNA4-LTRwt-d2EGFP and pcDNA4-LTRmutSp1-d2EGFP was performed via the calcium phosphate method. Stably transfected cells were selected with 600 μg/ml Zeocin (Invitrogen). After 10 days, single colonies were transferred to new culture plates. Fluorescence intensity of clones was analyzed by flow cytometry. For the final experiment, three clones of each construct, which expressed comparable fluorescence intensities, were selected. Electroporation of Jurkat cells was performed with the Nucleofector II (Amaxa/Lonza, Cologne, Germany). Therefore, cells were resuspended in 100 μl Nucleofector solution V at a final concentration of 2 × 107 cells/ml and mixed with 50 μg/ml DNA (pcDNA4-EF1α-d2EGFP or pcDNA4-OGT). Electroporation was carried out with 100 μl cell DNA suspension using program X-001. Immediately after transfection, cells were transferred to prewarmed RPMI 1640 medium supplemented with 10% FCS and 4 mM l-glutamine. At 48 h posttransfection, stably transfected Jurkat cells were selected with 400 μg/ml Zeocin (Invitrogen). After initial selection, cells were singularized by limited dilution into 96-well plates. After 13 days, d2EGFP-expressing Jurkat clones were pooled, while OGT-expressing Jurkat clones were cultivated separately.
RNA electroporation of CD4+ T lymphocytes.
T lymphocytes were electroporated as described previously (65). Briefly, cells were washed once with pure RPMI 1640 and once with OptiMEM without phenol red (Invitrogen) (all at room temperature). The cells were resuspended in OptiMEM at a concentration of 1 × 108 cells/ml. RNA was transferred to a 4-mm cuvette (Molecular Bioproducts, San Diego, CA) at a final concentration of 150 μg/ml. A volume of 100 μl of cell suspension was added and immediately pulsed in a Gene Pulser Xcell (Bio-Rad). Pulse conditions for CD4+ T lymphocytes were square-wave pulse, 500 V, and 5 ms. Immediately after electroporation, cells were transferred to prewarmed MLPC medium.
Luciferase reporter gene assays.
Cell lysates were centrifuged at 10,000 × g at 4°C for 3 min. Firefly luciferase activity of the supernatants was measured using the luciferase assay system (Promega) in a Luminoskan Ascent instrument (Thermo Fisher Scientific, Langenselbold, Germany). Each luciferase reporter gene assay was performed in triplicate (except for that shown in Fig. 3).
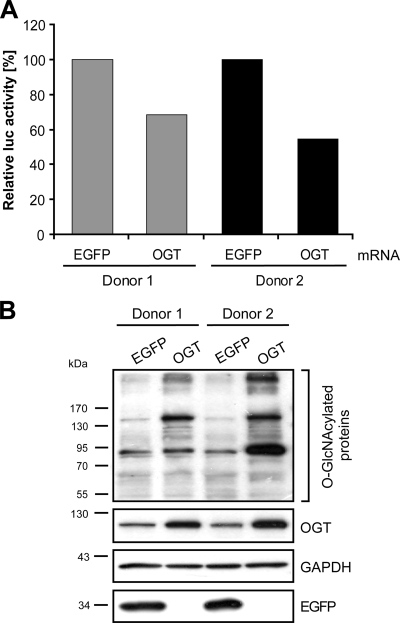
FIG. 3.
OGT inhibits HIV-1 LTR promoter activity in infected primary CD4+ T cells. Primary CD4+ T cells from two different donors were infected with VSV-G env pseudotyped HIV-1NL4-3LucR-E-. At 36 h postinfection, cells were electroporated with in vitro-transcribed polyadenylated mRNA encoding either EGFP or OGT. (A) At 8 h postelectroporation, luciferase activity was measured. (B) The O-GlcNAcylation pattern and overexpression of EGFP and OGT were verified by Western blotting. Staining of GAPDH demonstrates equal loading of proteins.
Electrophoretic mobility shift assay.
HEK 293T cells were transfected with the expression constructs for Sp1, OGT, or both, and nuclear proteins were isolated as already described. The electrophoretic mobility shift assay was essentially performed as described previously (54, 56). Briefly, binding reaction mixtures (final volume, 15 μl) contained 10 mM Tris-HCl (pH 7.5), 80 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5% glycerol, 3 μg of poly(dI-dC) · poly(dI-dC) (GE Healthcare), 10 μg of nuclear extract, and 4 × 104 cpm of the 32P-end-labeled double-stranded oligonucleotide probe. The oligonucleotides corresponded either to the wild-type HIV-1 LTR region containing the three binding sites for Sp1 (wt LTR-Sp1, 5′-GGATCGGGAGCGTGGCCTGGGCGGGACTGGGGAGTGGCGAGCCC-3′; Sp1-binding sites are underlined) or to the Sp1-binding-site-mutated HIV-1 LTR (mut LTR-Sp1, 5′-GGATCGGGATTCGTGGCCTGTTCGGGACTGGTTAGTGGCGAGCCC-3′; nucleotide substitutions in bold). After incubation for 45 min on ice, the protein-DNA complexes were resolved on nondenaturing 5% polyacrylamide gels run in 1× Tris-borate-EDTA buffer containing 89 mM Tris, 89 mM boric acid, and 2 mM EDTA (pH 8.0). For competition experiments, a 10-, 20-, or 50-fold molar excess of unlabeled competitor oligomers was added to the gel shift mixtures prior to the addition of the 32P-labeled oligonucleotide probe and incubated for 30 min at 4°C. For supershift assays, antibodies directed against Sp1, O-GlcNAc, or OGT were added to the gel shift mixture and incubated for 30 min at 4°C prior to the addition of the labeled probe. Supershift antibodies were Sp1 Pep2 sc-59X (Santa Cruz Biotechnology), O-GlcNAc CTD110.6 (Hiss Diagnostics), and OGT TI-14 (Sigma-Aldrich), as well as IgG (NF-κB p50 sc114X [Santa Cruz Biotechnology]) and IgM (heparan sulfate 370255 [Seikagaku Corporation]) isotype controls. All experiments were repeated three times. One representative gel shift of each assay is shown.
Reverse transcriptase PCR (RT-PCR).
RNA was isolated with RNeasy columns (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. RNA quality was verified by agarose gel electrophoresis, and RNA concentration was measured spectrophotometrically. Reverse transcription of total RNA and amplification of OGT and GAPDH cDNA were performed as described previously (53, 72). PCR was carried out in a total reaction volume of 25 μl with 1 μl of either undiluted (1) or diluted (1:5 and 1:10) cDNA. Primers specifically amplifying recombinant OGT were selected: the forward primer recognized specifically the vector-encoded 5′ untranslated region of the recombinant OGT mRNA (5′-TCCAGTGTGGTGGAATTCTG-3′); the reverse primer was homologous to a sequence corresponding to the N-terminal region of both endogenous and recombinant OGT (5′-TTGCGTCTCAATTGCTTTCA-3′). Amplification of GAPDH was performed using the forward primer 5′-AGCCACATCGCTCAGAACAC-3′ and the reverse primer 5′-GAGGCATTGCTGATGATCTTG-3′ as described previously (44).
Statistical analyses.
Statistical significances were calculated with the Student t test for paired samples using the SPSS 15.0 and 16.0 software for Microsoft Windows (SPSS Inc., Chicago, IL). P values smaller than 0.05 were considered statistically significant (*), P values smaller than 0.01 were considered highly significant (**), and P values smaller than 0.001 were considered the most significant (***).
RESULTS
GlcN inhibits HIV-1 transcription in lymphocytes.
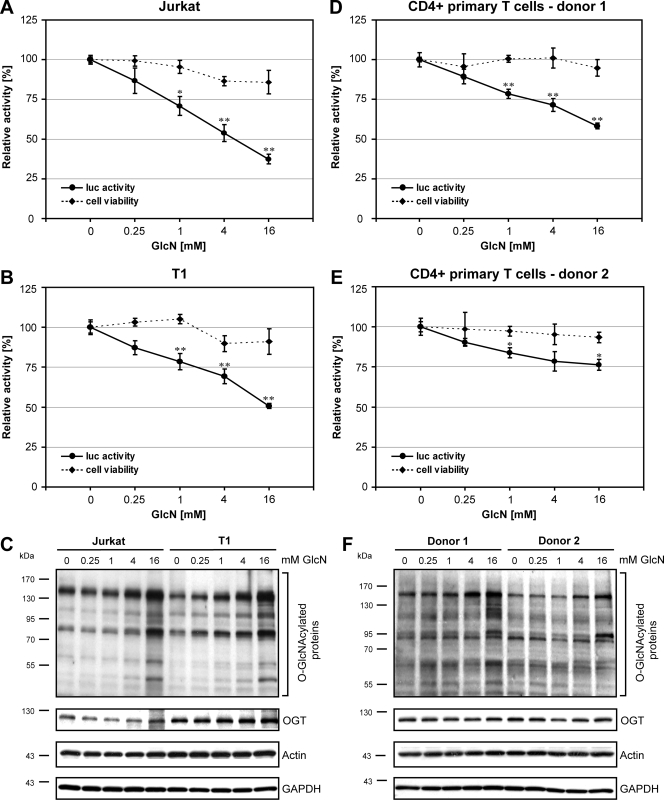
In order to evaluate whether O-GlcNAcylation affects HIV-1 replication, Jurkat, T1, and primary CD4+ T lymphocytes were infected with HIV-1NL4-3LucR-E-, a replication-deficient HIV-1 clone pseudotyped with the envelope protein VSV-G. Upon integration, this recombinant virus expresses the firefly luciferase gene driven by the HIV-1 LTR promoter. Consequently, the luciferase activity in infected cells correlates with the rate of HIV-1 gene transcription. Reporter-HIV-1-infected cells were either left untreated (0 mM) or treated for 24 h with increasing concentrations of GlcN (0.25 mM, 1 mM, 4 mM, and 16 mM), and luciferase activity was measured. GlcN significantly inhibited the HIV-1 gene transcription by more than 60% in Jurkat cells and 50% in T1 cells (Fig. 1A and B) without significantly decreasing cell viability (Fig. 1A and B). Western blot analyses using an antibody which recognizes O-GlcNAcylated proteins demonstrated increased O-GlcNAcylation upon GlcN treatment, whereas expression of OGT remained unchanged (Fig. 1C). Detection of GAPDH and actin demonstrated that equal amounts of protein were loaded. Similar results were obtained in primary CD4+ T lymphocytes from two different donors (Fig. 1D and E), although HIV-1 replication in donor 2 (Fig. 1E) was inhibited to a lesser extent than was that in donor 1 (Fig. 1D). This difference is also reflected in smaller amounts of O-GlcNAcylated proteins in the lysates of donor 2 than in lysates of donor 1 (compare donor 1 and donor 2 in Fig. 1F, upper panel).
FIG. 1.
GlcN inhibits HIV-1 transcription in lymphocytes. Jurkat cells (A), T1 cells (B), and primary CD4+ T cells from two different donors (D and E) were infected with VSV-G env pseudotyped HIV-1NL4-3LucR-E- and cultured in the absence (0 mM) or presence of different concentrations of GlcN (0.25 mM, 1 mM, 4 mM, and 16 mM) for 24 h. Subsequently, HIV-1 LTR-driven luciferase activity was measured (solid lines). The effect of GlcN on cytotoxicity and proliferation was monitored by MTT assay (dashed lines). The results are presented in terms of percent activities of untreated control cells. The means ± standard deviations from triplicate determinations are indicated. P values are calculated in comparison with control: *, P ≤ 0.05; **, P ≤ 0.01. (C and F) Western blot analyses of O-GlcNAcylated proteins and OGT in Jurkat and T1 cells (C) and in primary CD4+ T lymphocytes (F). Actin and GAPDH served as loading controls.
GlcN represses HIV-1 LTR promoter activity in HeLa cells.
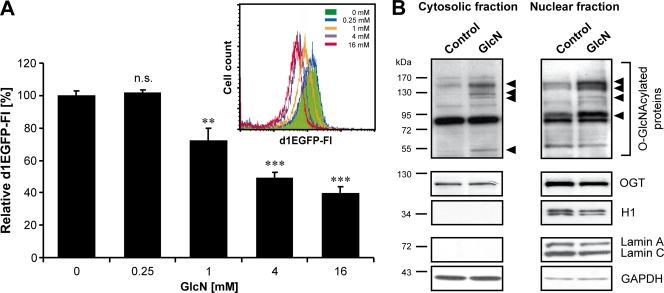
To exclude the possibility that reduced HIV-1 gene transcription is caused by impaired nuclear transport of the preintegration complex or reduced proviral integration, we evaluated the impact of GlcN treatment on the HIV-1 LTR promoter activity. Therefore, HeLa-Tat-III/LTR/d1EGFP cells, which stably express d1EGFP under the control of the HIV-1 LTR, were either left untreated (0 mM) or treated with increasing concentrations of GlcN (0.25 mM, 1 mM, 4 mM, and 16 mM). Subsequently, fluorescence intensity was measured by flow cytometry. GlcN treatment inhibited the HIV-1 LTR-triggered d1EGFP expression in a dose-dependent manner, as detected by the shift of the fluorescence emission peak to a lower fluorescence intensity (Fig. 2A, bar diagram). Quantification of the different values demonstrated that GlcN significantly decreased the HIV-1 LTR activity by more than 60% (Fig. 2A, bar diagram). Comparable results were obtained after treatment of HeLa-Tat-III/LTR/d1EGFP cells with the UDP-GlcNAc analog streptozotocin (data not shown), which inhibits the O-GlcNAcase activity and thereby increases O-GlcNAcylation (63).
FIG. 2.
GlcN inhibits HIV-1 LTR promoter activity and increases O-GlcNAcylation in HeLa cells. (A) HeLa-Tat-III/LTR/d1EGFP cells were either left untreated (0 mM) or stimulated for 5 h with increasing concentrations of GlcN (0.25 mM, 1 mM, 4 mM, and 16 mM). The HIV-1 LTR activity was assessed by measuring the d1EGFP fluorescence intensity (FI) with flow cytometry analyses. The shift of the fluorescence emission peak (inset) and the bar diagram of the respective geometric mean values compared to those for unstimulated control cells are shown. The means ± standard deviations calculated from triplicate determinations are indicated. P values are given for comparison with control: n.s., not significant; **, P ≤ 0.01; ***, P ≤ 0.001. (B) Nuclear and cytosolic fractions of unstimulated (control) or stimulated (16 mM GlcN) HeLa-Tat-III/LTR/d1EGFP cells were analyzed by Western blotting to detect O-GlcNAcylated proteins. Arrowheads mark proteins whose O-GlcNAcylation patterns are increased upon GlcN treatment. Staining of OGT (an example of an O-GlcNAcylated protein) and histone H1 (non-O-GlcNAcylated) shows that protein expression levels were not altered. Staining of GAPDH and lamin A/C demonstrates successful fractionation.
To demonstrate O-GlcNAcylation of cellular proteins in the presence of GlcN, cytosolic and nuclear fractions of control-treated versus GlcN-treated (16 mM) HeLa-Tat-III/LTR/d1EGFP cells were analyzed by Western blotting (Fig. 2B). In both fractions, several proteins showed increased O-GlcNAcylation upon GlcN treatment (Fig. 2B, upper panels), whereas the expression of OGT (example of an O-GlcNAc-modified protein) and of histone H1 (non-O-GlcNAc-modified protein) remained unchanged (Fig. 2B, middle panels). Successful fractionation of cytoplasmic and nuclear proteins and loading of equal protein amounts were verified by staining of lamin A/C and GAPDH (Fig. 2B, lower panels). Altogether, these results demonstrate that GlcN treatment enhances O-GlcNAcylation of cytoplasmic and nuclear proteins and that this correlates with decreased gene expression from the HIV-1 LTR promoter.
OGT inhibits HIV-1 transcription in primary T lymphocytes.
Since OGT is the only known enzyme to mediate O-GlcNAcylation, we investigated the effect of OGT on HIV-1 transcription in primary lymphocytes. To this goal, primary CD4+ T lymphocytes from two different donors were infected with pseudotyped reporter HIV-1. At 36 h postinfection the cells were transfected with in vitro-transcribed polyadenylated mRNA encoding either EGFP or OGT. OGT expression levels were maximal at 8 h after transfection (data not shown), and the cells were harvested. Reporter analyses showed that overexpression of OGT inhibited HIV-1 transcription to an extent similar to that of treatment with 16 mM GlcN (compare Fig. 3A with Fig. 1D and E). The inhibition was slightly stronger in the cells of donor 2 than in the cells of donor 1, which correlated well with the increased OGT expression and O-GlcNAcylation level observed in the cells of donor 2 (Fig. 3B, upper panels; compare donor 1 and donor 2).
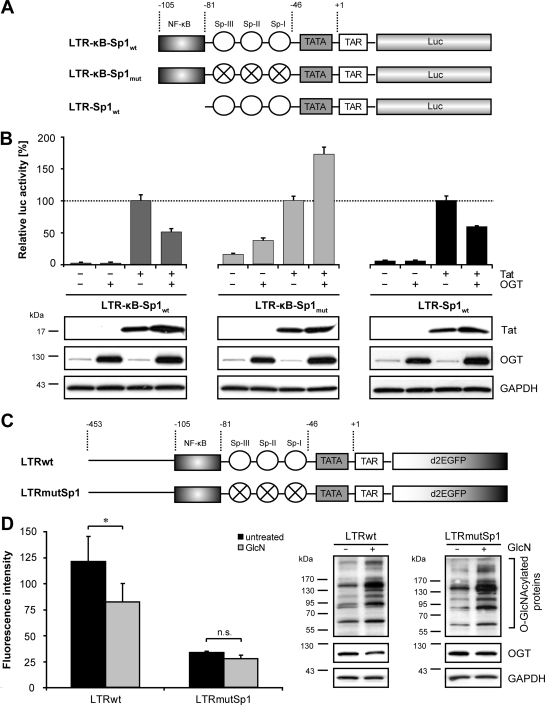
The Sp1-binding sites are necessary for inhibition of HIV-1 LTR activity by OGT.
Assuming that O-GlcNAcylation alters the activity of transcription factors, Sp1 and NF-κB were prime candidates for the O-GlcNAc-mediated inhibition of the HIV-1 LTR. Binding sites for both factors constitute the core promoter/enhancer of HIV-1, and both have been reported to be modified by O-GlcNAc (21, 31). In order to prove this hypothesis, luciferase reporter assays were performed using LTR reporters lacking the upstream modulatory region but containing the NF-κB- and/or Sp1-binding sites (either combined or isolated), as well as a TATA box and the trans-activation response (TAR) element for Tat-mediated activation (Fig. 4A, LTR-κB-Sp1wt, LTR-κB-Sp1mut, and LTR-Sp1wt). HEK 293T cells were cotransfected with the reporter constructs and a Tat-encoding plasmid, along with an OGT-encoding plasmid or a control plasmid (Fig. 4B). Overexpression of OGT inhibited the Tat-induced activity of the promoters containing functional Sp1-binding sites (Fig. 4B, LTR-κB-Sp1wt and LTR-Sp1wt). In contrast, the activity of the promoter containing mutated Sp1-binding sites was increased upon overexpression of OGT (Fig. 4B, LTR-κB-Sp1mut). The transient expression of Tat itself was not significantly affected by overexpression of OGT (Fig. 4B, lower panels). These findings suggest that the presence of functional Sp1-binding sites is crucial for the inhibitory effect of OGT on the HIV-1 LTR.
FIG. 4.
The presence of Sp1-binding sites in the HIV-1 LTR promoter is required for OGT-mediated inhibition. (A) Schematic representation of the promoter constructs used in the luciferase assay in panel B. The truncated promoters lack the upstream modulatory region but contain the NF-κB- and/or Sp1-binding sites (either combined or isolated), as well as a TATA box and the TAR element for Tat-mediated activation. Quartered circles represent mutated Sp1-binding sites. Numbers reflect the positions on the wild-type LTRLAI. (B) Promoter activities were detected by measuring the luciferase activity in the cell lysates of HEK 293T cells transiently transfected with the reporter constructs (LTR-κB-Sp1wt, LTR-κB-Sp1mut, and LTR-Sp1wt) together with plasmids coding for OGT and Tat as indicated. The total DNA amount was adjusted with pcDNA4. The values were normalized to the total amount of protein and are presented in terms of percentages of the corresponding Tat-induced promoter activity. The means ± standard deviations from triplicate determinations are shown (upper panel). Western blot analyses of Tat and OGT in transfected cells are shown in the lower panel. GAPDH staining demonstrates that equal amounts of protein were loaded. (C) Schematic representation of the full-length wild-type and full-length Sp1-mutated promoter constructs used for the stable transfection of HEK 293T cells. (D) HEK 293T cells were stably transfected with d2EGFP either under the control of the wild-type HIV-1 LTR (LTRwt) or under the control of the LTR containing mutated Sp1-binding sites in the basal promoter (LTRmutSp1). The effect of GlcN (16 mM) on the HIV-1 LTR was analyzed by flow cytometry, and the mean fluorescence intensity is represented in the bar diagram. The means ± standard deviations were calculated from duplicate determinations of three independent clones with similar fluorescence intensities. P values are given for comparison with control: *, P ≤ 0.05; n.s., not significant. Western blot analyses of one representative clone verify the increased O-GlcNAcylation pattern upon GlcN treatment and constant OGT expression. GAPDH staining demonstrates that equal amounts of protein were loaded.
In a next step, the role of the Sp1-binding sites in the full-length HIV-1 LTR integrated into the cellular genome was analyzed. Toward this goal, HEK 293T cells were stably transfected with plasmids expressing d2EGFP either under the control of the full-length wild-type HIV-1 LTR (Fig. 4C, LTRwt) or under the control of the full-length LTR containing mutated Sp1-binding sites (Fig. 4C, LTRmutSp1). Three independent HEK 293T cell clones with similar fluorescence intensities were either left untreated or stimulated with 16 mM GlcN. Subsequently, the promoter activities were measured by flow cytometry. Mutation of the Sp1-binding sites strongly decreased HIV-1 LTR activity (Fig. 4D, compare fluorescence intensities in the bar diagram). However, GlcN treatment significantly decreased the fluorescence intensity only in cells expressing d2EGFP under the control of the wild-type HIV-1 LTR and had no significant effect on cells expressing d2EGFP under the control of the Sp1-mutated HIV-1 LTR (Fig. 4D, bar diagram). Western blot analyses demonstrated that GlcN treatment similarly increased the O-GlcNAcylation pattern in the two cell types, that OGT expression was not altered, and that equal amounts of proteins were loaded (Fig. 4D, right panels). Altogether these results demonstrate that the Sp1-binding sites are required for the GlcN-mediated inhibition of the full-length HIV-1 promoter integrated into the cellular genome.
Sp1 is necessary for the inhibitory effect of OGT on the HIV-1 LTR.
Since the presence of the Sp1-binding sites is crucial for the inhibitory effect of OGT on the HIV-1 LTR promoter, we investigated whether this is also the case for the Sp1 protein itself. To this goal, the sensitivity of the Tat-induced LTR-Sp1wt promoter to OGT was analyzed in HEK 293T cells in the absence and presence of Sp1. Sp1 was recombinantly overexpressed, and its expression was inhibited with a specific siRNA. A nontargeting siRNA was used as control. Cotransfection of the control siRNA did not affect the OGT-mediated inhibition of the LTR-Sp1wt (Fig. 5A, compare bars 3 and 4). Tat-mediated activation of the LTR-Sp1wt was less pronounced after knockdown of Sp1 (Fig. 5A, compare protein content-normalized relative light unit values given above bars 3 and 7). However, OGT clearly did not inhibit LTR-Sp1wt promoter activity under these conditions (Fig. 5A, compare bars 7 and 8). The expression of OGT and Tat was not altered upon depletion of Sp1 expression (Fig. 5B). Thus, in addition to the Sp1-binding sites, the presence of the Sp1 protein itself is also necessary for the OGT-mediated inhibition of the HIV-1 LTR activity.
FIG. 5.
Sp1 is crucial for the inhibitory effect of OGT on the HIV-1 LTR. (A) HIV-1 LTR activity was measured by luciferase assay after cotransfection of HEK 293T cells with an LTR-Sp1wt reporter construct along with Tat-and Sp1-encoding plasmids in the absence or presence of an OGT-encoding vector together with 10 nM control siRNA or an siRNA specifically targeting Sp1. Total DNA amount was adjusted with pcDNA4. The relative light units normalized to the total protein content indicate the means ± standard deviations from triplicate determinations (values above the bars). The results were normalized for each siRNA data set individually and are presented in terms of percentages of control (Tat- and Sp1-induced LTR-Sp1wt activity). P values are given for comparison with control: **, P ≤ 0.01; n.s., not significant. (B) Western blot analyses of the lysates verify the knockdown of Sp1 by siRNA and prove that the expression of OGT and Tat remains unaffected upon silencing of Sp1. The GAPDH staining demonstrates that equal amounts of protein were loaded.
O-GlcNAcylation of Sp1 selectively inhibits the HIV-1 LTR promoter in a dose-dependent manner.
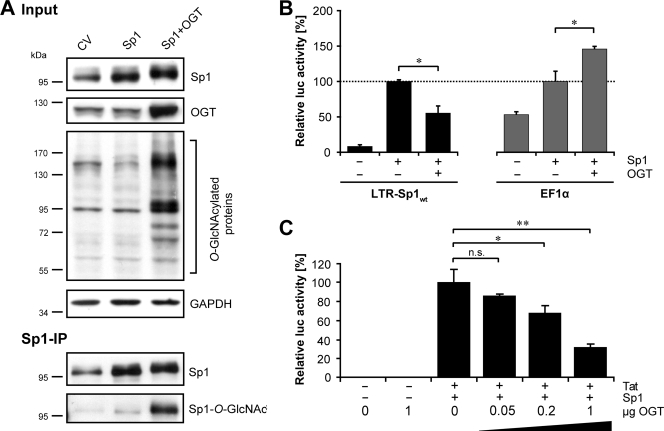
To investigate the role of Sp1 in the OGT-mediated inhibition of the HIV-1 LTR in more detail, we explored whether overexpression of OGT enhances O-GlcNAcylation of Sp1 (Fig. 6A). For this purpose, Sp1 was expressed alone or in combination with OGT (Fig. 6A, input). Immunoprecipitation experiments demonstrated that O-GlcNAcylation of Sp1 was substantially increased when OGT was overexpressed (Fig. 6A, Sp1-IP). Coimmunoprecipitations revealed that OGT physically interacts with Sp1 under these conditions (data not shown).
FIG. 6.
O-GlcNAcylation of Sp1 selectively inhibits HIV-1 LTR promoter activity in a dose-dependent manner. (A) HEK 293T cells were transfected with control vector (CV), Sp1, or Sp1- and OGT-expressing vectors. Total DNA amount was adjusted with pcDNA4. Expression of transfected plasmids and O-GlcNAcylation were assessed via Western blotting of cell lysates (input). Immunoprecipitation of Sp1 (Sp1-IP) and O-GlcNAcylation of Sp1 were verified by Western blotting of Sp1-IPs with anti-Sp1 and anti-O-GlcNAc antibodies, respectively. (B) The effects of OGT on the LTR-Sp1wt and the control promoter EF1α in HEK 293T cells cotransfected with the reporter and an Sp1-encoding plasmid were analyzed by luciferase assay. (C) Luciferase activity of HEK 293T cells transiently cotransfected with the LTR-Sp1wt reporter construct along with Tat- and Sp1-encoding plasmids and increasing concentrations of an OGT-encoding vector (0.05 μg, 0.2 μg, and 1 μg). The total DNA amount in panels B and C was adjusted with pcDNA4. The values were normalized to the total amount of protein and are presented in terms of percentages of control (Sp1 induced [B] and Tat- and Sp1-induced [C] promoter activity). The means ± standard deviations from triplicate determinations are indicated. P values are given for comparison with control: n.s., not significant; *, P ≤ 0.05; **, P ≤ 0.01.
In order to determine whether O-GlcNAcylation of Sp1 inhibits solely HIV-1 LTR activity or transcription in general, luciferase reporter assays were performed using LTR-Sp1wt and an EF1α promoter construct. The latter triggers the expression of the housekeeping gene EF1α and harbors Sp1-binding sites. Unlike the LTR-Sp1wt promoter, the EF1α promoter has no TAR element. In order to ensure comparable induction levels, both reporter promoters were activated with Sp1 alone (Fig. 6B). In agreement with the results above, overexpression of OGT significantly inhibited the Sp1-triggered expression of LTR-Sp1wt (Fig. 6B, LTR-Sp1wt). In contrast, the activity of the EF1α promoter was not repressed but significantly increased by OGT (Fig. 6B, EF1α), suggesting that OGT selectively inhibits the HIV-1 LTR promoter and does not generally repress Sp1-regulated gene expression.
Furthermore, we evaluated by luciferase reporter assay whether the inhibitory effect of OGT on the Sp1-regulated HIV-1 LTR promoter is dose dependent. To this end, the reporter LTR-Sp1wt was cotransfected with Sp1- and Tat-encoding constructs in HEK 293T cells together with increasing amounts of OGT-encoding plasmid (0.05 μg, 0.2 μg, and 1 μg). OGT clearly inhibited dose dependently the HIV-1 LTR promoter activity (Fig. 6C).
OGT does not inhibit expression and DNA-binding affinity of Sp1.
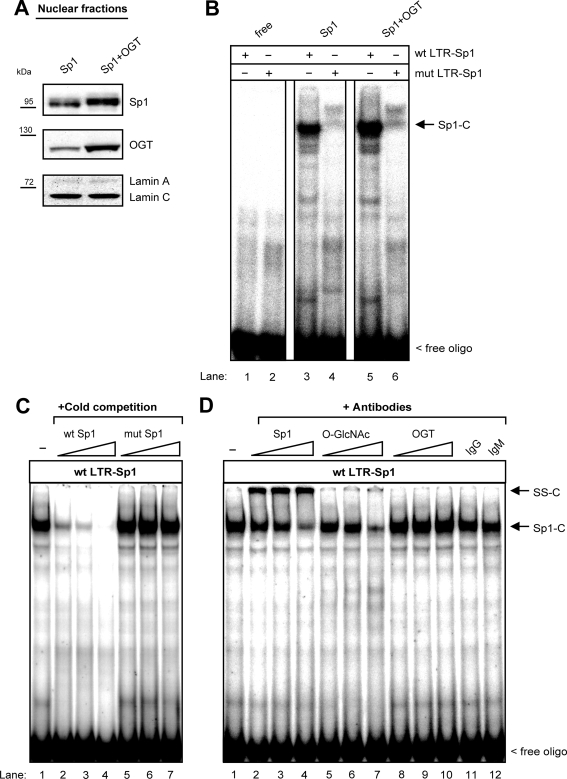
The inhibitory effect of OGT on the HIV-1 LTR may rely on impaired nuclear translocation or decreased DNA-binding affinity of Sp1 upon O-GlcNAcylation. The impact of OGT on the nuclear translocation of Sp1 was investigated by comparing the nuclear amounts of Sp1 in cells expressing endogenous or increased levels of OGT. The amount of nuclear Sp1 was not decreased upon overexpression of OGT (Fig. 7A) but rather increased, suggesting that the observed decrease in the HIV-1 LTR promoter activity was not due to impaired nuclear translocation of Sp1.
FIG. 7.
OGT does not interfere with Sp1 expression and DNA binding of Sp1. (A) HEK 293T cells were transfected with an Sp1-encoding plasmid, along with an OGT-encoding plasmid or control plasmid. Protein expression was detected in nuclear protein extracts via Western blotting. Staining of lamin A/C was used as a loading control. (B) Electrophoretic mobility shift assays were carried out with 32P-end-labeled double-stranded oligonucleotides corresponding to the Sp1-binding sites in the HIV-1 LTR promoter (wt LTR-Sp1) or with oligonucleotides containing mutated Sp1-binding sites in order to prevent binding (mut LTR-Sp1). One representative gel shift assay out of three is shown. Reactions were performed either without nuclear extracts (free) or with lysates from cells transfected with an Sp1-encoding plasmid alone (Sp1) or in combination with an OGT-encoding plasmid (Sp1+OGT). The Sp1-oligonucleotide complex is indicated as Sp1-C. (C) Competition experiments were carried out with 10-, 20-, and 50-fold molar excesses of unlabeled wild-type (wt LTR-Sp1) or mutated (mut LTR-Sp1) oligonucleotides. (D) Supershift analyses were performed using 1 μg, 2 μg, or 5 μg anti-Sp1, anti-O-GlcNAc, or anti-OGT antibodies as well as 5 μg anti-IgG or anti-IgM as an isotype control.
To investigate whether OGT interferes with the DNA-binding affinity of Sp1, gel shift assays were carried out with templates corresponding to the Sp1-binding sites in the HIV-1 LTR promoter (Fig. 7B, wt LTR-Sp1). No decrease in the formation of protein-oligonucleotide complexes was detected upon overexpression of OGT (Fig. 7B, compare lane 3 and 5). As a specificity control, gel shift experiments were performed using templates containing mutated Sp1-binding sites (Fig. 7B, mut LTR-Sp1). The specific Sp1 complex (Fig. 7B, Sp1-C) was not detectable with these oligonucleotides (Fig. 7B, lanes 4 and 6).
The specificity of Sp1-C was confirmed by competition experiments with increasing molar excess of unlabeled wt LTR-Sp1 (Fig. 7C, lanes 2 to 4) and mut LTR-Sp1 oligonucleotides (Fig. 7C, lanes 5 to 7). In addition, the presence of O-GlcNAc-modified Sp1 in Sp1-C was confirmed by supershift analyses using increasing amounts of anti-Sp1 (Fig. 7D, lanes 2 to 4) and anti-O-GlcNAc (Fig. 7D, lanes 5 to 7) antibodies. Both antibodies shifted Sp1-C almost completely (Fig. 7D, lanes 4 and 7), suggesting that most of the DNA-bound Sp1 is O-GlcNAcylated in OGT-overexpressing cells. Addition of an anti-OGT antibody did not shift Sp1-C (Fig. 7D, lanes 8 to 10). Isotype control anti-IgG (Fig. 7D, lane 11) and anti-IgM (Fig. 7D, lane 12) antibodies had no effect on Sp1-C. These results demonstrate that increased O-GlcNAcylation of Sp1 inhibits neither the ability of Sp1 to translocate into the nucleus nor the DNA-binding affinity of Sp1.
Sp1 O-GlcNAcylation is necessary for the inhibition of the HIV-1 LTR.
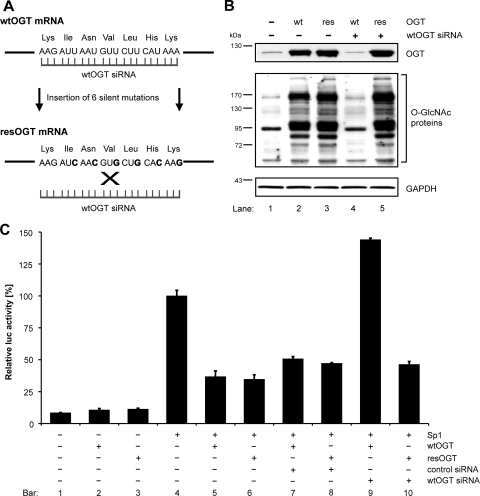
To confirm that the inhibition of the HIV-1 LTR is dependent on Sp1 O-GlcNAcylation, a specific siRNA was used to deplete the expression of OGT. The specificity of the siRNA was verified with an OGT rescue mutant (resOGT)-encoding plasmid, which contains six silent nucleotide exchanges in the siRNA-binding site (Fig. 8A). Western blot analyses proved that overexpression of wild-type OGT (wtOGT) was inhibited in HEK 293T cells cotransfected with the wtOGT-specific siRNA (Fig. 8B, top panel, lanes 2 and 4), whereas expression of resOGT was not affected (Fig. 8B, top panel, lanes 3 and 5). Staining of O-GlcNAc-modified proteins served as an additional control for the functionality of resOGT (Fig. 8B, middle panel).
FIG. 8.
Sp1 O-GlcNAcylation is necessary for the inhibition of the HIV-1 LTR. (A) Generation of a plasmid encoding an OGT rescue mutant in order to escape silencing by the siRNA targeting wild-type OGT: six silent mutations (bold) were introduced into the siRNA-binding sequence of OGT. (B) Western blot analyses of wild-type (wt) and rescue mutant (res) OGT were performed after transfection with the siRNA specifically targeting wtOGT. Detection of O-GlcNAcylation levels was used as a control for the functionality of the rescue mutant. Immunodetection of GAPDH demonstrates that equal amounts of protein were loaded. (C) HEK 293T cells were transfected with the reporter construct LTR-Sp1wt along with plasmids encoding Sp1 and OGT (wild type or rescue mutant). Total DNA amount was adjusted with pcDNA4. Control siRNA or siRNA targeting wtOGT (10 nM) was cotransfected, and the luciferase activity was measured. The values were adjusted to the total amount of protein and indicate the means ± standard deviations from triplicate determinations. The results are presented in terms of percentages of control (Sp1-induced LTR activity).
Applying these tools to test the effect of Sp1 O-GlcNAcylation on HIV-1 LTR, we showed that the promoter activity (Fig. 8C, bar 4) was strongly reduced after overexpression of wtOGT or resOGT (Fig. 8C, bars 5 and 6). Cotransfection of a control siRNA targeting GAPDH had no impact on the HIV-1 LTR activity (Fig. 8C, bars 7 and 8). Knockdown of wtOGT restored the HIV-1 LTR activity (Fig. 8C, bar 9), whereas resOGT escaped silencing and was still able to suppress the promoter activity (Fig. 8C, bar 10). These findings demonstrate that O-GlcNAcylation of Sp1 is required for the inhibition of the HIV-1 LTR promoter by OGT.
Increased OGT expression before infection intensifies OGT-inhibitory effects on HIV-1 replication.
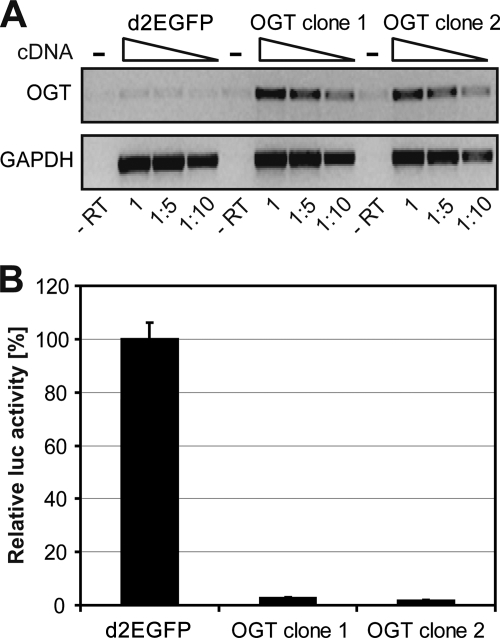
In all previous experiments the O-GlcNAcylation level was increased after infection with HIV-1. Thus, we aimed to investigate whether increased OGT expression prior to infection may amplify the inhibitory effects on HIV-1 transcription in lymphocytes. Jurkat cells were stably transfected with OGT or, as a control, with d2EGFP. Expression of recombinant OGT in selected clones was confirmed by RT-PCR (Fig. 9A). The stably transfected cells were infected with VSV-G env pseudotyped HIV-1NL4-3LucR-E-, and HIV-1 LTR activity was determined by luciferase assay. Of note, HIV-1 LTR activity was 40-fold lower in both OGT-overexpressing Jurkat cell clones than in control cells expressing d2EGFP (Fig. 9B). These results demonstrate that OGT is a potent inhibitor of HIV-1 LTR activity.
FIG. 9.
OGT overexpression prior to infection amplifies the inhibitory effect on HIV-1 replication. (A) Jurkat cells were stably transfected with plasmids encoding either d2EGFP or OGT. Expression of recombinantly expressed OGT mRNA was confirmed by RT-PCR (upper panel) using specific primers. Amplification of GAPDH served as a loading control (lower panel). Cellular cDNA was subjected to the amplification reactions in increasing dilutions: undiluted (1), 1:5, and 1:10. Control reactions were carried out in the absence of reverse transcriptase (−RT) with undiluted cDNA template. (B) Jurkat cells stably expressing d2EGFP or OGT were infected with VSV-G env pseudotyped HIV-1NL4-3LucR-E-. Luciferase activity was measured 48 h after infection.
DISCUSSION
Regulation of gene expression by nutrients like glucose and glucosamine is well established (20, 37, 52, 74) and demonstrates that cellular transcription adapts to environmental and metabolic changes. Viruses critically depend on the host cell metabolism (29). This directed us to investigate the impact of the monosaccharidic metabolite O-GlcNAc on HIV-1 gene expression.
We showed that increased O-GlcNAc levels inhibit HIV-1 transcription in human T-cell lines as well as in human primary CD4+ T lymphocytes. This effect appeared to be mediated by the transcription factor Sp1, as supported by several lines of experimental evidence. First, we demonstrated that O- GlcNAc-mediated inhibition of HIV-1 transcription required the presence of Sp1-binding sites in the HIV-1 LTR promoter. The Sp1-binding sites are well conserved throughout the lentiviral HIV-1 LTRs (18), and mutation of one or more Sp1-binding sites in the basal HIV-1 LTR promoter leads to a strong delay in replication in human peripheral blood lymphocytes and in T-cell lines (45, 58). This is also reflected in our observations that mutation of the Sp1-binding sites decreases HIV-1 LTR activity in transiently and stably transfected cells. But although Sp1-mutated HIV-1 LTR activity was still measurable and inducible, no significant O-GlcNAc-mediated inhibition of promoter activity was observed.
Second, the presence of the Sp1 protein was crucial for the inhibitory effect of O-GlcNAc on the HIV-1 LTR. Inhibition of Sp1 expression with siRNA completely abolished OGT-mediated inhibition of HIV-1 LTR activity. Interestingly, although Sp1 is implicated in the activation of a large number of genes, depletion of the ubiquitous transcription factor did not induce cytotoxicity. This is in agreement with the observation of Philipsen and coworkers, who disrupted the mouse Sp1 gene and found that Sp1-deficient embryonic stem cells were viable and showed normal growth characteristics (50). This might be attributed to the compensation of Sp1-triggered gene transcription by other Sp family members, such as Sp3, which is also ubiquitously expressed and has the potential to activate transcription of certain otherwise Sp1-regulated promoters (70).
Third, Sp1 was found to be O-GlcNAcylated by OGT, and this decreased its ability to activate the HIV-1 LTR promoter. These findings are in accordance with the results of Kudlow and colleagues, who showed that O-GlcNAcylation decreases the capability of Sp1 to activate GC-box-containing promoters (77). But it has to be emphasized that O-GlcNAcylation of Sp1 does not generally inhibit transcription. We have shown here that the human EF1α promoter, which also contains Sp1-binding sites (55, 73, 75), is activated by O-GlcNAcylation of Sp1. Additionally, others have shown that the expression of plasminogen activator inhibitor 1 (14, 19, 20), calmodulin (38), and argininosuccinate synthetase (6, 7) is also increased by Sp1 O-GlcNAcylation. Thus, O-GlcNAcylation of Sp1 differentially modulates gene expression. These differential effects may depend not only on the individual composition of the transcription complexes at different promoters but also on differential Sp1 O-GlcNAcylation patterns in different transcription complexes. The latter is supported by the fact that transcriptional corepressors recruit OGT to promoters (78), indicating that O-GlcNAcylation is modulated directly at the promoter site. Furthermore, it has been shown that phosphorylation of distinct residues differentially regulates Sp1 activity (5, 10, 11). As O-GlcNAcylation appears to have a yin-yang relationship with phosphorylation, the same may apply for O-GlcNAcylation.
Several mechanisms have been suggested for how O-GlcNAcylation of Sp1 inhibits promoter activity. For example, O-GlcNAcylation reduces the ability of Sp1 to homomultimerize and to interact with transcriptional coactivator TATA-binding protein-associated factor II 110 (TAFII110) (62, 77). These mechanisms are in accordance with our findings that O-GlcNAcylated Sp1 localizes at the HIV-1 LTR promoter and does not exhibit a decreased DNA-binding activity. Furthermore, O-GlcNAcylated Sp1 seems to act additionally as a repressor of HIV-1 transcription, as the presence of the Sp1-binding sites in the HIV-1 LTR is crucial for the inhibitory effect of OGT. The HIV-1 LTR promoter containing only functional NF-κB-binding sites but mutated Sp1-binding sites (LTR-κB-Sp1mut) was activated by OGT overexpression, indicating that O-GlcNAcylation of NF-κB increases the transcription rate from the LTR-κB-Sp1mut promoter. Accordingly, O-GlcNAcylation of NF-κB has been described and it was suggested to enhance the nuclear translocation of NF-κB (21). However, the transcription of the wild-type HIV-1 LTR (LTR-κB-Sp1wt) containing both NF-κB- and Sp1-binding sites was inhibited by O-GlcNAcylation of Sp1. This indicates that O-GlcNAcylation of Sp1 inhibits the activity of the HIV-1 LTR in a dominant manner. Unlike the NF-κB-binding sites, the Sp1-binding sites are well conserved throughout primate lentiviral LTRs (18, 79), suggesting a conserved regulation of Sp1-mediated gene transcription for lentiviral LTRs, which is probably dominant over NF-κB regulation. Furthermore, the dominant effect of Sp1 over NF-κB might be attributed to the synergistic mode of action of both transcription factors on the HIV-1 LTR (59, 60).
A potential molecular mechanism for the dominant repressive effect of O-GlcNAcylated Sp1 on the HIV-1 LTR relies on the ability of Sp1 to recruit corepressors to the promoter. It has been reported that Sp1 can act as an anchor site for a repressor complex consisting of histone deacetylases and mSin3a (82). Intriguingly, Sp1 has also been detected in a DNA-protein complex with c-Myc and histone deacetylase 1 at the HIV-1 LTR, and it has been shown that this complex is involved in the establishment of proviral latency (33). Thus, O-GlcNAcylation of Sp1 might quickly put initiated viral replication into a state of latency. Most importantly, increased O-GlcNAcylation of Sp1 prior to HIV-1 infection might efficiently prevent the onset of viral replication. This is supported by the fact that stable overexpression of OGT strongly inhibited replication of HIV-1 in de novo-infected lymphocytes. Altogether, these results suggest that O-GlcNAcylated Sp1 efficiently inhibits gene expression from the HIV-1 LTR and may be involved in the regulation of the viral life cycle.
Many reports have evaluated the effect of highly active antiretroviral therapy (HAART) on glucose metabolism (15, 17, 49) and the appearance of diabetes as a consequence of HAART (40, 51). But up to now, no studies have investigated the effect of glucose metabolism on HIV-1 replication. Our results indicate that the O-GlcNAc level, and thus the glucose metabolism, may influence the life cycle of HIV-1. Accordingly, inducers of Sp1 O-GlcNAcylation such as GlcN and 2-deoxyglucose, which are approved in clinical treatment of osteoarthritis and human genital herpes infections, respectively (3, 61), may support HAART. The establishment of a metabolic treatment may supplement the repertoire of antiretroviral therapies against AIDS.
Acknowledgments
We thank Gertrud Hoffmann, Mahimaidos Manoharan, and Melanie Nurtsch for excellent technical assistance (all Division of Molecular and Experimental Surgery); Manuel López-Cabrera (University of Molecular Biology, University Hospital of Princesa, Madrid, Spain) for kindly providing us with the plasmids pXP1-LTR-κB-Sp1wt-Luc, pXP1-LTR-κB Sp1mut-Luc, and pXP1-LTR-Sp1wt-Luc; and Christoph Garlichs (Department of Medicine II, Cardiology and Angiology, University of Erlangen) for kindly providing the FACSCalibur. We further thank Jan Dörrie (Department of Dermatology, University Hospital Erlangen) for helping us with the transfection of primary CD4+ T cells and Ulrich Schubert (Institute of Clinical and Molecular Virology, University of Erlangen-Nuremberg) for providing us with the Gene Pulser XCell. The generous support of Werner Hohenberger (Director of the Department of Surgery) is gratefully acknowledged.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG-SPP 1130; DFG-GK 1071, project A2; and DFG 317/2-1), German Cancer Aid (Deutsche Krebshilfe, Apoptose-Schwerpunktprogramm), and the Interdisciplinary Center for Clinical Research (IZKF) of the University Hospital of the University of Erlangen-Nuremberg to M.S.
We have no conflicting financial interests.
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkhout, B., and K. T. Jeang. 1992. Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 66139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blough, H. A., and R. L. Giuntoli. 1979. Successful treatment of human genital herpes infections with 2-deoxy-D-glucose. JAMA 2412798-2801. [PubMed] [Google Scholar]
- 4.Bonehill, A., C. Heirman, S. Tuyaerts, A. Michiels, K. Breckpot, F. Brasseur, Y. Zhang, P. Van Der Bruggen, and K. Thielemans. 2004. Messenger RNA-electroporated dendritic cells presenting MAGE-A3 simultaneously in HLA class I and class II molecules. J. Immunol. 1726649-6657. [DOI] [PubMed] [Google Scholar]
- 5.Bonello, M. R., and L. M. Khachigian. 2004. Fibroblast growth factor-2 represses platelet-derived growth factor receptor-alpha (PDGFR-alpha) transcription via ERK1/2-dependent Sp1 phosphorylation and an atypical cis-acting element in the proximal PDGFR-alpha promoter. J. Biol. Chem. 2792377-2382. [DOI] [PubMed] [Google Scholar]
- 6.Brasse-Lagnel, C., A. Fairand, A. Lavoinne, and A. Husson. 2003. Glutamine stimulates argininosuccinate synthetase gene expression through cytosolic O-glycosylation of Sp1 in Caco-2 cells. J. Biol. Chem. 27852504-52510. [DOI] [PubMed] [Google Scholar]
- 7.Brasse-Lagnel, C., A. Lavoinne, D. Loeber, A. Fairand, C. Bole-Feysot, N. Deniel, and A. Husson. 2007. Glutamine and interleukin-1beta interact at the level of Sp1 and nuclear factor-kappaB to regulate argininosuccinate synthetase gene expression. FEBS J. 2745250-5262. [DOI] [PubMed] [Google Scholar]
- 8.Calman, A. F., M. P. Busch, G. N. Vyas, T. M. McHugh, D. P. Stites, and B. M. Peterlin. 1988. Transcription and replication of human immunodeficiency virus-1 in B lymphocytes in vitro. AIDS 2185-193. [PubMed] [Google Scholar]
- 9.Chang, L. J., V. Urlacher, T. Iwakuma, Y. Cui, and J. Zucali. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 6715-728. [DOI] [PubMed] [Google Scholar]
- 10.Chu, S., and T. J. Ferro. 2005. Sp1: regulation of gene expression by phosphorylation. Gene 3481-11. [DOI] [PubMed] [Google Scholar]
- 11.Chun, R. F., O. J. Semmes, C. Neuveut, and K. T. Jeang. 1998. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol. 722615-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comer, F. I., and G. W. Hart. 2001. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry 407845-7852. [DOI] [PubMed] [Google Scholar]
- 13.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206935-944. [DOI] [PubMed] [Google Scholar]
- 14.Du, X. L., D. Edelstein, L. Rossetti, I. G. Fantus, H. Goldberg, F. Ziyadeh, J. Wu, and M. Brownlee. 2000. Hyperglycemia-induced mitochondrial superoxide overproduction activates the hexosamine pathway and induces plasminogen activator inhibitor-1 expression by increasing Sp1 glycosylation. Proc. Natl. Acad. Sci. USA 9712222-12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dube, M. P. 2000. Disorders of glucose metabolism in patients infected with human immunodeficiency virus. Clin. Infect. Dis. 311467-1475. [DOI] [PubMed] [Google Scholar]
- 16.Dynan, W. S., and R. Tjian. 1983. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 3579-87. [DOI] [PubMed] [Google Scholar]
- 17.El-Sadr, W. M., C. M. Mullin, A. Carr, C. Gibert, C. Rappoport, F. Visnegarwala, C. Grunfeld, and S. S. Raghavan. 2005. Effects of HIV disease on lipid, glucose and insulin levels: results from a large antiretroviral-naive cohort. HIV Med. 6114-121. [DOI] [PubMed] [Google Scholar]
- 18.Frech, K., R. Brack-Werner, and T. Werner. 1996. Common modular structure of lentivirus LTRs. Virology 224256-267. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg, H. J., C. I. Whiteside, and I. G. Fantus. 2002. The hexosamine pathway regulates the plasminogen activator inhibitor-1 gene promoter and Sp1 transcriptional activation through protein kinase C-beta I and -delta. J. Biol. Chem. 27733833-33841. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg, H. J., C. I. Whiteside, G. W. Hart, and I. G. Fantus. 2006. Posttranslational, reversible O-glycosylation is stimulated by high glucose and mediates plasminogen activator inhibitor-1 gene expression and Sp1 transcriptional activity in glomerular mesangial cells. Endocrinology 147222-231. [DOI] [PubMed] [Google Scholar]
- 21.Golks, A., T. T. Tran, J. F. Goetschy, and D. Guerini. 2007. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 264368-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez-Gonzalo, M., M. Carretero, J. Rullas, E. Lara-Pezzi, J. Aramburu, B. Berkhout, J. Alcami, and M. Lopez-Cabrera. 2001. The hepatitis B virus X protein induces HIV-1 replication and transcription in synergy with T-cell activation signals: functional roles of NF-kappaB/NF-AT and SP1-binding sites in the HIV-1 long terminal repeat promoter. J. Biol. Chem. 27635435-35443. [DOI] [PubMed] [Google Scholar]
- 23.Haltiwanger, R. S., K. Grove, and G. A. Philipsberg. 1998. Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O-GlcNAc-beta-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino-N-phenylcarbamate. J. Biol. Chem. 2733611-3617. [DOI] [PubMed] [Google Scholar]
- 24.Hanover, J. A., S. Yu, W. B. Lubas, S.-H. Shin, M. Ragano-Caracciola, J. Kochran, and D. C. Love. 2003. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409287-297. [DOI] [PubMed] [Google Scholar]
- 25.Harrich, D., J. Garcia, F. Wu, R. Mitsuyasu, J. Gonzalez, and R. Gaynor. 1989. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 632585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hart, G. W. 1997. Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu. Rev. Biochem. 66315-335. [DOI] [PubMed] [Google Scholar]
- 27.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 696705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiromura, M., C. H. Choi, N. A. Sabourin, H. Jones, D. Bachvarov, and A. Usheva. 2003. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation). J. Biol. Chem. 27814046-14052. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda, M., and N. Kato. 2007. Modulation of host metabolism as a target of new antivirals. Adv. Drug Deliv. Rev. 591277-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer, S. P., and G. W. Hart. 2003. Dynamic nuclear and cytoplasmic glycosylation: enzymes of O-GlcNAc cycling. Biochemistry 422493-2499. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, S. P., and R. Tjian. 1988. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell 55125-133. [DOI] [PubMed] [Google Scholar]
- 32.Jackson, S. P., and R. Tjian. 1989. Purification and analysis of RNA polymerase II transcription factors by using wheat germ agglutinin affinity chromatography. Proc. Natl. Acad. Sci. USA 861781-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, G., A. Espeseth, D. J. Hazuda, and D. M. Margolis. 2007. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 8110914-10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadonaga, J. T., K. R. Carner, F. R. Masiarz, and R. Tjian. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 511079-1090. [DOI] [PubMed] [Google Scholar]
- 35.Kadonaga, J. T., A. J. Courey, J. Ladika, and R. Tjian. 1988. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science 2421566-1570. [DOI] [PubMed] [Google Scholar]
- 36.Kamemura, K., and G. W. Hart. 2003. Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: a new paradigm for metabolic control of signal transduction and transcription. Prog. Nucleic Acid Res. Mol. Biol. 73107-136. [DOI] [PubMed] [Google Scholar]
- 37.Kang, H. T., J. W. Ju, J. W. Cho, and E. S. Hwang. 2003. Down-regulation of Sp1 activity through modulation of O-glycosylation by treatment with a low glucose mimetic, 2-deoxyglucose. J. Biol. Chem. 27851223-51231. [DOI] [PubMed] [Google Scholar]
- 38.Keembiyehetty, C. N., R. P. Candelaria, G. Majumdar, R. Raghow, A. Martinez-Hernandez, and S. S. Solomon. 2002. Paradoxical regulation of Sp1 transcription factor by glucagon. Endocrinology 1431512-1520. [DOI] [PubMed] [Google Scholar]
- 39.Kelly, W. G., M. E. Dahmus, and G. W. Hart. 1993. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J. Biol. Chem. 26810416-10424. [PubMed] [Google Scholar]
- 40.Koster, J. C., M. S. Remedi, H. Qiu, C. G. Nichols, and P. W. Hruz. 2003. HIV protease inhibitors acutely impair glucose-stimulated insulin release. Diabetes 521695-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreppel, L. K., M. A. Blomberg, and G. W. Hart. 1997. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 2729308-9315. [DOI] [PubMed] [Google Scholar]
- 42.Kudlow, J. E. 2006. Post-translational modification by O-GlcNAc: another way to change protein function. J. Cell. Biochem. 981062-1075. [DOI] [PubMed] [Google Scholar]
- 43.Lacroix, I., C. Lipcey, J. Imbert, and B. Kahn-Perles. 2002. Sp1 transcriptional activity is up-regulated by phosphatase 2A in dividing T lymphocytes. J. Biol. Chem. 2779598-9605. [DOI] [PubMed] [Google Scholar]
- 44.Lehmann, M. H., J. Weber, O. Gastmann, and H. H. Sigusch. 2002. Pseudogene-free amplification of human GAPDH cDNA. BioTechniques 33766, 769-770. [DOI] [PubMed] [Google Scholar]
- 45.Leonard, J., C. Parrott, A. J. Buckler-White, W. Turner, E. K. Ross, M. A. Martin, and A. B. Rabson. 1989. The NF-κB binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J. Virol. 634919-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lubas, W. A., D. W. Frank, M. Krause, and J. A. Hanover. 1997. O-linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 2729316-9324. [DOI] [PubMed] [Google Scholar]
- 47.Lubas, W. A., and J. A. Hanover. 2000. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J. Biol. Chem. 27510983-10988. [DOI] [PubMed] [Google Scholar]
- 48.Lubeseder-Martellato, C., E. Guenzi, A. Jörg, K. Töpolt, E. Naschberger, E. Kremmer, C. Zietz, E. Tschachler, P. Hutzler, M. Schwemmle, K. Matzen, T. Grimm, B. Ensoli, and M. Stürzl. 2002. Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am. J. Pathol. 1611749-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lutz, N. W., N. Yahi, J. Fantini, and P. J. Cozzone. 1997. Perturbations of glucose metabolism associated with HIV infection in human intestinal epithelial cells: a multinuclear magnetic resonance spectroscopy study. AIDS 11147-155. [DOI] [PubMed] [Google Scholar]
- 50.Marin, M., A. Karis, P. Visser, F. Grosveld, and S. Philipsen. 1997. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 89619-628. [DOI] [PubMed] [Google Scholar]
- 51.Mathe, G. 1999. Human obesity and thinness, hyperlipidemia, hyperglycemia, and insulin resistance associated with HIV1 protease inhibitors. Prevention by alternating several antiproteases in short sequences. Biomed. Pharmacother. 53449-451. [DOI] [PubMed] [Google Scholar]
- 52.McClain, D. A., A. J. Paterson, M. D. Roos, X. Wei, and J. E. Kudlow. 1992. Glucose and glucosamine regulate growth factor gene expression in vascular smooth muscle cells. Proc. Natl. Acad. Sci. USA 898150-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naschberger, E., R. S. Croner, S. Merkel, A. Dimmler, P. Tripal, K. U. Amann, E. Kremmer, W. M. Brueckl, T. Papadopoulos, C. Hohenadl, W. Hohenberger, and M. Stürzl. 2008. Angiostatic immune reaction in colorectal carcinoma: impact on survival and perspectives for antiangiogenic therapy. Int. J. Cancer 1232120-2129. [DOI] [PubMed] [Google Scholar]
- 54.Naschberger, E., T. Werner, A. B. Vicente, E. Guenzi, K. Topolt, R. Leubert, C. Lubeseder-Martellato, P. J. Nelson, and M. Stürzl. 2004. Nuclear factor-kappaB motif and interferon-alpha-stimulated response element co-operate in the activation of guanylate-binding protein-1 expression by inflammatory cytokines in endothelial cells. Biochem. J. 379409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen, S. J., M. Praestegaard, H. F. Jorgensen, and B. F. Clark. 1998. Different Sp1 family members differentially affect transcription from the human elongation factor 1 A-1 gene promoter. Biochem. J. 333511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ortiz, B. D., A. M. Krensky, and P. J. Nelson. 1996. Kinetics of transcription factors regulating the RANTES chemokine gene reveal a developmental switch in nuclear events during T-lymphocyte maturation. Mol. Cell. Biol. 16202-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parent, M., T. M. C. Yung, A. Rancourt, E. L. Y. Ho, S. Vispe, F. Suzuki-Matsuda, A. Uehara, T. Wada, H. Handa, and M. S. Satoh. 2005. Poly(ADP-ribose) polymerase-1 is a negative regulator of HIV-1 transcription through competitive binding to TAR RNA with Tat-positive transcription elongation factor b (p-TEFb) complex. J. Biol. Chem. 280448-457. [DOI] [PubMed] [Google Scholar]
- 58.Parrott, C., T. Seidner, E. Duh, J. Leonard, T. S. Theodore, A. Buckler-White, M. A. Martin, and A. B. Rabson. 1991. Variable role of the long terminal repeat Sp1-binding sites in human immunodeficiency virus replication in T lymphocytes. J. Virol. 651414-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perkins, N. D., A. B. Agranoff, E. Pascal, and G. J. Nabel. 1994. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol. Cell. Biol. 146570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perkins, N. D., N. L. Edwards, C. S. Duckett, A. B. Agranoff, R. M. Schmid, and G. J. Nabel. 1993. A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. EMBO J. 123551-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pujalte, J. M., E. P. Llavore, and F. R. Ylescupidez. 1980. Double-blind clinical evaluation of oral glucosamine sulphate in the basic treatment of osteoarthrosis. Curr. Med. Res. Opin. 7110-114. [DOI] [PubMed] [Google Scholar]
- 62.Roos, M. D., K. Su, J. R. Baker, and J. E. Kudlow. 1997. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol. Cell. Biol. 176472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roos, M. D., W. Xie, K. Su, J. A. Clark, X. Yang, E. Chin, A. J. Paterson, and J. E. Kudlow. 1998. Streptozotocin, an analog of N-acetylglucosamine, blocks the removal of O-GlcNAc from intracellular proteins. Proc. Assoc. Am. Physicians 110422-432. [PubMed] [Google Scholar]
- 64.Sander, G., A. Konrad, M. Thurau, E. Wies, R. Leubert, E. Kremmer, H. Dinkel, T. Schulz, F. Neipel, and M. Stürzl. 2008. Intracellular localization map of human herpesvirus 8 proteins. J. Virol. 821908-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schaft, N., J. Dorrie, I. Muller, V. Beck, S. Baumann, T. Schunder, E. Kampgen, and G. Schuler. 2006. A new way to generate cytolytic tumor-specific T cells: electroporation of RNA coding for a T cell receptor into T lymphocytes. Cancer Immunol. Immunother. 551132-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaft, N., J. Dorrie, P. Thumann, V. E. Beck, I. Muller, E. S. Schultz, E. Kampgen, D. Dieckmann, and G. Schuler. 2005. Generation of an optimized polyvalent monocyte-derived dendritic cell vaccine by transfecting defined RNAs after rather than before maturation. J. Immunol. 1743087-3097. [DOI] [PubMed] [Google Scholar]
- 67.Srinivasan, S., M. E. Hatley, D. T. Bolick, L. A. Palmer, D. Edelstein, M. Brownlee, and C. C. Hedrick. 2004. Hyperglycaemia-induced superoxide production decreases eNOS expression via AP-1 activation in aortic endothelial cells. Diabetologia 471727-1734. [DOI] [PubMed] [Google Scholar]
- 68.Stevenson, M., T. L. Stanwick, M. P. Dempsey, and C. A. Lamonica. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 91551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suñé, C., and M. A. García-Blanco. 1995. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 696572-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suske, G. 1999. The Sp-family of transcription factors. Gene 238291-300. [DOI] [PubMed] [Google Scholar]
- 71.Thurau, M., G. Marquardt, N. Gonin-Laurent, K. Weinländer, E. Naschberger, R. Jochmann, K. R. Alkharsah, T. F. Schulz, M. Thome, F. Neipel, and M. Stürzl. 2009. Viral inhibitor of apoptosis vFLIP/K13 protects endothelial cells against superoxide-induced cell death. J. Virol. 83598-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tripal, P., M. Bauer, E. Naschberger, T. Mörtinger, C. Hohenadl, E. Cornali, M. Thurau, and M. Stürzl. 2007. Unique features of different members of the human guanylate-binding protein family. J. Interferon Cytokine Res. 2744-52. [DOI] [PubMed] [Google Scholar]
- 73.Uetsuki, T., A. Naito, S. Nagata, and Y. Kaziro. 1989. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1 alpha. J. Biol. Chem. 2645791-5798. [PubMed] [Google Scholar]
- 74.Vanderford, N. L., S. S. Andrali, and S. Özcan. 2007. Glucose induces MafA expression in pancreatic beta cell lines via the hexosamine biosynthetic pathway. J. Biol. Chem. 2821577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wakabayashi-Ito, N., and S. Nagata. 1994. Characterization of the regulatory elements in the promoter of the human elongation factor-1 alpha gene. J. Biol. Chem. 26929831-29837. [PubMed] [Google Scholar]
- 76.Weinländer, K., E. Naschberger, M. H. Lehmann, P. Tripal, W. Paster, H. Stockinger, C. Hohenadl, and M. Stürzl. 2008. Guanylate binding protein-1 inhibits spreading and migration of endothelial cells through induction of integrin alpha4 expression. FASEB J. 224168-4178. [DOI] [PubMed] [Google Scholar]
- 77.Yang, X., K. Su, M. D. Roos, Q. Chang, A. J. Paterson, and J. E. Kudlow. 2001. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. USA 986611-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang, X., F. Zhang, and J. E. Kudlow. 2002. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 11069-80. [DOI] [PubMed] [Google Scholar]
- 79.Yedavalli, V. S., M. Benkirane, and K. T. Jeang. 2003. Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J. Biol. Chem. 2786404-6410. [DOI] [PubMed] [Google Scholar]
- 80.Zachara, N. E., and G. W. Hart. 2006. Cell signaling, the essential role of O-GlcNAc! Biochim. Biophys. Acta 1761599-617. [DOI] [PubMed] [Google Scholar]
- 81.Zachara, N. E., and G. W. Hart. 2004. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 167313-28. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, Y., and M. L. Dufau. 2002. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem. 27733431-33438. [DOI] [PubMed] [Google Scholar]