Abstract
Noroviruses (NVs) are recognized as a major cause of nonbacterial gastroenteritis in humans. Studies of the human NVs continue to be hampered by the inability to propagate them in any cell culture system. Until recently, most data concerning NV replication were derived from studies of feline calicivirus and rabbit hemorrhagic disease virus, which are cultivable members of the family Caliciviridae. From such studies, it was proposed that caliciviruses induce apoptosis to facilitate the dissemination of viral progeny in the host. The discovery that MNV type 1 (MNV-1) grows in RAW264.7 cells provided the first cell culture system for use in studying the role of apoptosis in NV infection. We first showed that MNV-1 replication triggered apoptosis in infected RAW264.7 cells and then demonstrated that cell death was associated with activation of caspase-9 and caspase-3 through the mitochondrial pathway. This process was dependent on virus replication, since inactivated virus failed to induce signs of apoptosis. In order to better understand the apoptotic process induced by MNV-1 infection of RAW264.7 cells, we investigated the expression profiles of MNV-1-infected versus mock-infected cells. Survivin, a member of the inhibitor of apoptosis protein family, was found to be significantly downregulated in an inverse relationship with the virus genome replication. This study showed that, unlike other viruses that upregulate survivin, MNV-1 is the first virus found to downregulate the levels of survivin. We observed that MNV-1 replication in RAW264.7 cells activated caspases, resulting in apoptosis through the mitochondrial pathway, possibly as a result of downregulation of survivin.
Noroviruses (NVs) are recognized as a major cause of nonbacterial gastroenteritis in humans. It has been estimated that NVs may cause up to 1.1 million hospitalizations and 218,000 deaths each year for children under 5 years of age in developing countries, ranking them second only to rotaviruses as a health threat (38). NVs are highly infectious, with attack rates reaching 56%. In institutional settings, the infection results in large-scale outbreaks, leading to considerable economic losses, especially in hospitals (24).
NVs are members of the genus Norovirus of the family Caliciviridae, which also includes viruses from three other genera: Sapovirus, Vesivirus, and Lagovirus. The nonenveloped 30- to 35-nm-diameter virions have icosahedral symmetry and contain a 7.7-kb-long single-stranded RNA genome of positive polarity. The RNA genome is organized into three open reading frames (ORF1, ORF2, and ORF3). ORF1 encodes a large nonstructural polyprotein that is processed by the virus-encoded proteinase into the mature nonstructural proteins and their precursors. ORF2 and ORF3 encode major (VP1) and minor (VP2) capsid proteins, respectively. The NVs have been divided into five major genogroups (designated GI to GV), with the human pathogens characteristically found in genogroups GI, GII, and GIV (52).
Replication of human NVs is poorly understood due to the lack of both a suitable animal model and a susceptible cell culture system. Until recently, the molecular mechanisms of NV replication were studied in vitro or derived from replication studies of two other caliciviruses, feline calicivirus (FCV) in the genus Vesivirus and rabbit hemorrhagic disease virus (RHDV) in the genus Lagovirus. Murine NV type 1 (MNV-1), a member of the genus Norovirus (classified in genogroup GV) that grows in mouse bone marrow-derived macrophages and dendritic cells as well as in a murine macrophage-like cell line (RAW264.7), provided the first cell culture system to study the pathogenesis and molecular mechanisms of NV replication (31, 50).
MNV replication in RAW 264.7 cells results in extensive cytopathic effect (CPE) followed by cell death reminiscent of apoptosis (46, 50). Apoptosis is a genetically programmed cell death pathway that provides a mechanism for removal of damaged or unwanted cells in multicellular organisms. Several studies reported the occurrence of apoptotic changes in calicivirus-infected cells and, specifically, the triggering of the apoptotic pathway in FCV-infected cells (2, 3, 28, 42, 47). Increased numbers of apoptotic cells were also observed in vivo in the small intestine tissues of pigs inoculated with human NV, and progressive histopathological lesions were detected in immunodeficient mice naturally infected with MNV (13, 49). In addition, infected cells dying by apoptosis were detected in multiple organs of the RHDV-infected rabbits. The finding of apoptotic cells at sites with pronounced tissue damage suggested involvement of apoptosis in pathogenesis of the RHDV-induced disease (3, 28). Although the functional importance of apoptosis in calicivirus replication is not understood, it has been suggested that caliciviruses use this self-destructive cellular process to facilitate the dissemination of viral progeny in the host (3, 47). The molecular mechanisms employed by caliciviruses to trigger the apoptotic cascade still remain unknown; however, it has been shown that induction of apoptosis in FCV-infected cells involves the mitochondrial pathway (35).
The intrinsic or mitochondrial apoptotic pathway is dependent on the release into the cytosol of apoptotic factors sequestered by the mitochondria, such as cytochrome c and Smac/DIABLO, and is directly linked to permeability of the organelle outer membrane. Loss of the mitochondrial membrane integrity and cytochrome c release result in apoptosome formation and processing of procaspase-9 followed by downstream activation of executioner caspase-3. The catalytic activity of caspase-9 can be inhibited by a group of proteins known as inhibitors of apoptosis (IAPs). The 16.5-kDa mouse survivin protein encoded by the BIRC5 gene is the smallest member of the IAP family. Survivin interacts with cofactor molecules, such as the X-linked mammalian IAP protein (XIAP) and the hepatitis B virus X-interacting protein (HBXIP), to specifically inhibit caspase-9 activation (4, 6). It has been shown that a number of viruses are capable of upregulating survivin in order to delay apoptosis in infected cells (9, 11, 39, 51, 53, 54).
In this study, we first established that replication-dependent apoptosis occurred in MNV-1-infected RAW264.7 cells. Then, in order to study the apoptotic cascade in detail, we investigated the expression profiles of MNV-1-infected versus mock-infected RAW264.7 cells. Here, we report that the replication of a virus (MNV-1) in cultured cells is associated with downregulation of survivin expression and results in induction of apoptosis through the mitochondrial pathway.
MATERIALS AND METHODS
Cells and virus.
The murine macrophage-like cell line RAW264.7 was obtained from ATCC (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) containing penicillin (250 units/ml) and streptomycin (250 μg/ml) and supplemented with 5% heat-inactivated fetal bovine serum. A previously characterized plaque-purified isolate of MNV-1, designated MNV-1.CW1P3, was used as the source of virus in this study (46). Propagation and plaque titration assays of MNV-1 in RAW264.7 cells were carried out as described previously (46). The MNV-1 virions were purified using ultracentrifugation in a CsCl gradient according to protocols published elsewhere (50). The purified virus was dialyzed against 1,000 volumes of phosphate-buffered saline (PBS) overnight.
For time course experiments, the viral stock was serially diluted in DMEM to obtain the desired inocula for a multiplicity of infection (MOI) of 4. RAW264.7 cells (5 × 107) were inoculated with either diluted MNV-1 or DMEM (mock infection control), incubated at 37°C, and collected at each time point for further Western blot or RNA expression analyses.
DAPI staining.
To visualize nuclear morphology, mock- and MNV-1-infected RAW264.7 cells were fixed with methanol at 28 h postinfection (h p.i.) and incubated with 4′,6-diamidino-2-phenylindole (DAPI; Roche Molecular Biochemicals, Indianapolis, IN) staining solution (1 μg/ml in PBS buffer) for 20 min at room temperature. After several washes with PBS buffer, stained cells were visualized using fluorescence microscopy.
DNA fragmentation assay and PARP cleavage analysis.
Isolation and gel electrophoresis of equivalent quantities of total DNA samples were carried out as described previously (25). Briefly, mock-infected or MNV-1-infected RAW264.7 cells were harvested at 4, 8, 12, 16, 20, 24, and 28 h p.i., and total DNA was extracted and analyzed by electrophoresis using a 1.5% agarose gel. Mouse poly(ADP-ribose) polymerase 1 (PARP-1) cleavage was studied by Western blot analysis of the collected cell samples. Cleaved PARP-1 was detected at 4, 8, 12, 16, 20, and 24 h p.i. using an antibody that specifically binds the large (89-kDa) fragment of the caspase-cleaved protein (Cell Signaling Technology, Danvers, MA).
Immunodetection assays.
Western blot analysis was carried out using standard techniques (44). Briefly, MNV-1-infected RAW264.7 cells collected at different time points and mock-infected cells (24 h p.i.) were resuspended in PBS buffer, incubated for 10 min at 95°C with equal volumes of 2× sodium dedecyl sulfate-polyacrylamide gel electrophoresis sample buffer (Invitrogen, Carlsbad, CA) containing 5% ß-mercaptoethanol, and subjected to sodium dedecyl sulfate-polyacrylamide gel electrophoresis separation in a 4 to 20% Tris-glycine gel (Invitrogen). The separated proteins were electroblotted onto a nitrocellulose membrane and probed with antibodies specific to mouse and MNV-1 proteins. Anti-caspase-3, anti-caspase-9, and anti-survivin antibodies were obtained from Cell Signaling Technology. Anti-caspase-8 antibody was obtained from Alexis Biochemicals, San Diego, CA. Anti-PCNA and anti-ß-actin antibodies were obtained from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, and Sigma-Aldrich, St. Louis, MO, respectively. Anti-MNV-1 protein antibodies were described previously (46). Horseradish peroxidase-labeled secondary antibodies purchased from Kirkegaard & Perry Laboratories, Gaithersburg, MD, were used for detection of bound primary antibodies. After 1 h of incubation with secondary antibodies, blot membranes were developed using a chemiluminescent detection system (Thermo Fisher Scientific Inc., Rockford, IL).
Fluorometric analysis of caspase activity.
The activities of caspase-3, caspase-8, and caspase-9 were measured using the caspase-specific fluorometric substrates Ac-DEVD-AFC, Ac-IETD-AFC, and Ac-LEHD-AFC, respectively (BioVision Research Products, Mountain View, CA), according to the manufacturer's protocol. Briefly, 2 × 106 mock- or MNV-1-infected (at an MOI of 4) RAW cells were collected at different times p.i. (both mock- and virus-infected samples were collected for every time point). Cell pellets were resuspended in cell lysis buffer (BioVision Research Products), incubated on ice for 10 min, mixed with equal amounts of 2× reaction buffer (BioVision Research Products), and incubated with individual caspase substrates (50 μM) at 37°C for 2 h. Fluorescence of the samples was measured at 400-nm excitation and 505-nm emission using a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA).
Cell fractionation assay.
RAW264.7 cells that were either mock infected (16 h p.i.) or MNV-1 infected (8 and 16 h p.i.) were harvested. The collected cells were processed using an ApoAlert cell fractionation kit (Clontech, Mountain View, CA) according to the manufacturer's instructions, and the isolated protein fractions were analyzed by Western blotting. First, the collected cytosolic fractions were examined for contamination with mitochondria by the use of cytochrome c oxidase subunit IV (COX IV) antibodies. The amounts of protein loaded on gel were normalized using protein content and verified by Western blotting using antibodies specific to ß-actin (Sigma). Finally, the presence of cytochrome c in normalized samples was detected using cytochrome c antibodies provided by the manufacturer. The chemiluminescence was detected by exposing blot membranes to X-ray film, and the densities of the observed bands were quantified using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
UV inactivation of MNV-1 and inhibition of MNV-1 replication.
UV treatment of MNV-1 was conducted as previously described for other viruses (29, 37). Virus stock aliquots (5 × 107 PFU/ml) were added to a petri dish placed on ice and irradiated with UV (254 nm) light for 15 min. The source of UV was a Spectroline UV lamp (Spectronics Inc., Mobile, AL) positioned at a distance of 3 cm above the dish. The efficiency of the UV inactivation treatment was assessed by an infectivity assay that measured residual virus in the treated stock. Only preparations shown to be fully inactivated were used in experiments that employed UV-inactivated virus. RAW264.7 monolayers were infected with UV-inactivated viruses by following the same protocol used for active MNV-1 (MOI of 4).
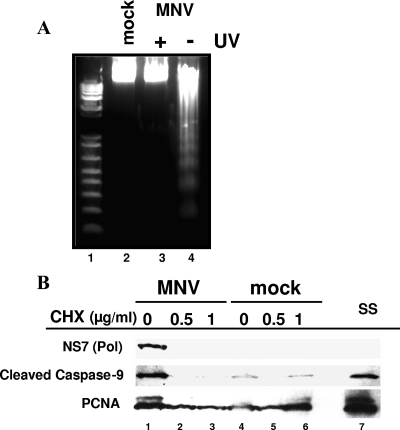
In order to inhibit protein synthesis, the MNV-1-infected RAW264.7 cells were incubated with cycloheximide (CHX; Sigma) at 0, 0.5, and 1 μg/ml for 16 h. In addition, treatment of RAW264.7 cells with 2 μM staurosporine (Sigma) served as a positive control for apoptosis induction. Equivalent amounts of the corresponding cell lysate samples were subjected to Western blot analysis to detect viral protein synthesis and activation of caspase-9.
Affymetrix GeneChip hybridization and analysis.
Total cellular RNA from mock- and MNV-1-infected RAW264.7 cells (12 h p.i.) was isolated using an RNeasy kit (Qiagen, Valencia, CA), labeled with biotin, and hybridized to mouse oligonucleotide microarrays following procedures provided by the manufacturer (mouse genome array 430 2.0; Affymetrix, Santa Clara, CA). Microarray data were analyzed using a GeneSpring GX 7.3.1 system (Agilent, Santa Clara, CA). Signal values from the probe sets were calculated using microarray suite 5. Each chip was independently normalized to the median expression level of all the genes on the chip. Each gene was then normalized to the median expression levels of that gene. Each of the 45,101 genes was filtered by identification using a “present or marginal” Affymetrix flag in three of the sample replicates. The remaining genes were analyzed by one-way analysis of variance to identify those with statistically significant differences between samples infected with MNV-1 (three replicates) and uninfected samples (three replicates). Variances were calculated using a parametric test with a P-value cutoff of 0.05. Gene ontology was used to identify changes in gene expression associated with apoptosis that were twofold or greater. Ingenuity pathway analysis (Ingenuity Systems, Redwood City, CA) was employed to identify groups of genes involved in apoptotic canonical pathways.
SuperArray analysis.
SuperArray technology (SABiosciences, Frederick, MD) was utilized to compare the relative levels of mRNA from apoptosis-related genes expressed in RAW 264.7 cells with or without MNV-1 infection at 12 h p.i. A PCR array (catalog no. PAMM-012) contained 84 primer pairs that amplified genes involved in mouse apoptosis. Reactions for the infected and the mock-infected control cells were repeated in three independent experiments following the manufacturer's instructions. The quantitative PCR was run on an ABI 7900 HT instrument (Applied Biosystems, Foster City, CA). For each set of triplicates, the mean value for each gene was determined and used to calculate the changes in levels (infected versus uninfected control). With the use of appropriate cutoff criteria, a fourfold induction or repression of expression, with a P value of ≤0.05, was considered to represent significantly up- or downregulated gene expression.
Quantitative RT-PCR detection of BIRC5.
Downregulation of expression of the survivin BIRC5 gene in MNV-1-infected RAW 264.7 cells was confirmed using relative quantification of gene transcript 1 and expression of the ß-actin gene as a housekeeping gene control. Briefly, six-well cell culture plates were infected with MNV-1 at an MOI of 4. After 1 h of adsorption at 37°C, wells were rinsed with PBS, and the cells in each well were collected by scraping them into 500 μl of PBS at 0, 4, 8, 12, 16, 20, and 24 h p.i. The RNA in 200 μl of each cell suspension was extracted two consecutive times, and for each of those times, the sample was treated with DNase while adsorbed in the column (Qiagen). The resulting RNA quantity and quality were assayed using a NanoDrop spectrophotometer (Thermo Fisher Scientific), and 1 μg of the purified RNA was used to carry out the reaction with reagents and primers according to the manufacturer's protocol (SABiosciences, Frederick, MD). Nontemplate controls as well as non-reverse transcriptase (non-RT) controls were included for each time point and for each gene transcript to be detected. The reaction was performed in triplicate using an ABI 7900 HT instrument (Applied Biosystems), and the results were analyzed using relative quantification software (RQ Manager 1.2; Applied Biosystems).
MNV-1 genomic RNA quantification.
In order to determine the level of MNV-1 RNA for each of the time points described above, MNV-1 genomic RNA present in infected samples was quantified using single-step quantitative real-time RT-PCR. One TaqMan probe and two primers designed to specifically amplify the MNV-1 capsid-encoding sequence were utilized (E. L. Barron, S. V. Sosnovtsev, C. R. Rhodes, K. Bok, V. Prikhodko, K. Hasenkrug, A. B. Carmody, J. Ward, K. Perdue, and K. Y. Green, unpublished data). Briefly, 5 μl of RNA extracted from each well by use on an RNeasy kit (Qiagen) were added to an RT-PCR mix (Brilliant II quantitative RT-PCR core reagent kit; Stratagene, La Jolla, CA). The reaction was then incubated at 45°C for 30 min and for 95°C for 10 min, followed by 45 cycles at 95°C for 30 s, 50°C for 1 min, and 72°C for 30 s, using an ABI 7900 HT instrument (Applied Biosystems). Each sample was tested in duplicate, and an appropriate triplicate standard curve for absolute quantification was also included for each run.
Microarray data accession number.
The complete data set obtained from the microarray analysis has been submitted to the GEO database (NCBI) under accession number GSE12518.
RESULTS
Apoptotic changes in MNV-1-infected RAW264.7 cells.
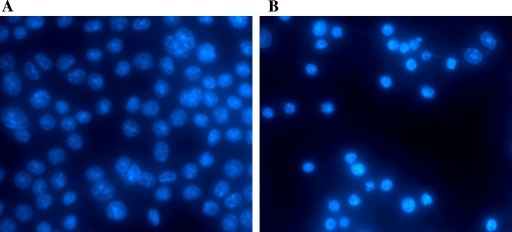
Induction of apoptotic changes in calicivirus-infected cells has been previously reported for vesiviruses and lagoviruses, and it was proposed as a putative mechanism to release viral progeny (3, 47). To investigate whether NV replication could also result in the apoptotic death of infected cells, we studied morphological and biochemical changes induced in RAW264.7 cells infected with MNV-1. It has been reported that replication of MNV-1 occurs in the cytoplasm of infected cells and is characterized by extensive rearrangement of intracellular membranes (50). Vesiculated areas could be detected as early as 12 h p.i., expanding in size as the infection progressed. An increase in the number of vesicles inside the infected cell led to displacement of the nucleus (50). One of the hallmark features of cells dying by apoptosis is nuclear chromatin condensation and irreversible oligonucleosomal fragmentation of the genomic DNA (14). We examined alterations in nuclear chromatin morphology by DAPI staining of MNV-1-infected cells fixed at different times p.i. The DAPI staining of the infected cells provided evidence of nuclear shrinkage and chromatin condensation, with changes starting between 12 and 16 h p.i. Late in the infection, the proportion of cells with collapsed nuclei gradually increased, reaching the maximum at 28 h p.i. (Fig. 1). An analysis of the cell DNA integrity showed internucleosomal fragmentation concurrent with chromatin condensation. Characteristic oligonucleosomal DNA laddering was detected beginning at 16 h p.i. (Fig. 2, lanes 5 to 8). The total DNA isolated from mock-infected RAW264.7 cells did not show signs of specific fragmentation (Fig. 2, lane 1).
FIG. 1.
Shrinkage of nuclei and chromatin condensation in MNV-1-infected RAW264.7 cells. Monolayers of mock-infected (A) or MNV-1-infected (MOI of 4) (B) RAW264.7 cells were fixed with methanol at 28 h p.i. and stained with DAPI. Nuclei were visualized by fluorescence microscopy using filters for UV excitation.
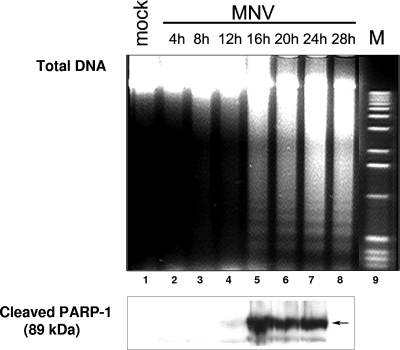
FIG. 2.
DNA degradation and PARP-1 cleavage in MNV-1-infected RAW264.7 cells. The results of isolation of total DNA and gel electrophoresis of DNA fragments over time are shown. Data represent mock-infected RAW264.7 cells (28 h p.i.; lane 1), MNV-1-infected RAW264.7 cells harvested at 4, 8, 12, 16, 20, 24, and 28 h p.i. (lanes 2 to 8), and a DNA marker (M; lane 9). An aliquot of the harvested cells was subjected to electrophoresis in an acrylamide gel and then transferred to a nitrocellulose membrane. Cleaved PARP-1 was detected in all samples (except for the samples from 28 h p.i., which were not tested) with a mouse antibody specific for the 89-kDa cleavage product.
Oligonucleosomal fragmentation of nuclear chromatin in apoptotic cells is usually accompanied by inactivation of proteins involved in DNA metabolism and DNA damage repair. PARP-1, an enzyme involved in recognition of single-stranded DNA breaks and repair signaling, has been described as one of the early targets of proteolytical degradation mediated by activated caspases (32). During apoptosis, PARP-1 is inactivated by cleavage into two fragments of 24 and 89 kDa. Western blot analysis of MNV-1-infected cell lysates collected in the same time course experiments showed that the 89-kDa caspase cleavage product of PARP-1 could be detected beginning at 12 h p.i. (Fig. 2, lanes 4 to 7).
Apoptosis in MNV-1-infected RAW 264.7 cells is associated with activation of caspases through the mitochondrial pathway.
Activation of the DNase responsible for oligonucleosomal degradation of nuclear chromatin is dependent on cleavage of an inhibitor complexed with the DNase molecule (43). The cleavage is mediated by caspase-3, which in turn is activated by either initiator caspase-8 or caspase-9. Caspase-8 and caspase-9 represent the extrinsic and intrinsic pathways of apoptosis cascade triggering, respectively. Activation of caspase-8 occurs in a death-inducing signaling complex that assembles in response to the binding of apoptogenic ligands by death receptors. Meanwhile, activation of caspase-9 takes place in an apoptosome complex formed by the interaction of Apaf1, ATP, and cytochrome c, which are released by mitochondria in response to intracellular stimuli.
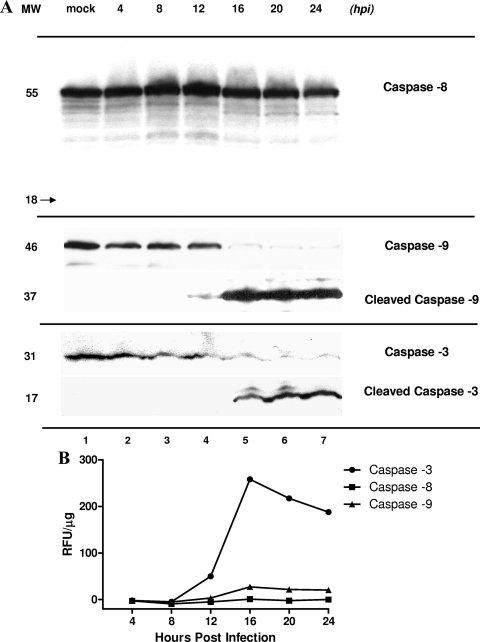
In order to characterize the pathway that triggers the apoptotic cascade during MNV-1 infection, we examined cleavage of caspase-8 and caspase-9 in samples of mock- and MNV-1-infected RAW264.7 cells collected at various time points of infection by the use of Western blotting and caspase-specific antibodies. Western blot analysis did not show any significant changes in the amounts of procaspase-8 throughout the course of infection (Fig. 3A, lanes 1 to 7). Consequently, we did not observe a 18-kDa protein band corresponding to the processed form of caspase-8 that would be detected with the same antibodies. In contrast, the appearance of a 37-kDa protein corresponding to the cleaved form of caspase-9 was observed starting at 12 h p.i. (Fig. 3A, lanes 4 to 7). Unprocessed caspase-9 at 46 kDa was the predominant form detected in samples of mock-infected cells and in cells collected at early times of infection (Fig. 3A, lanes 1 to 4).
FIG. 3.
Activation of caspase-3, caspase-8, and caspase-9 in MNV-1-infected RAW264.7 cells. (A) Equivalent amounts of protein from serial collection time points in mock (24 h p.i.)- and MNV-1-infected RAW264.7 cells were loaded onto a polyacrylamide gel and subjected to electrophoresis. Unprocessed caspase-8 (55 kDa) was detected by Western blot analysis throughout the experiment (lanes 1 to 7). A corresponding band representing cleaved caspase-8 (18 kDa) was not detected. Unprocessed caspase-3 and caspase-9 were detected at earlier time points and in mock-infected cells (lanes 1 to 4) at 31 and 46 kDa, respectively. Cleaved caspase-9 and caspase-3 were observed starting at 12 and 16 h p.i., respectively, in MNV-1-infected cells (lanes 4 to 7). MW, molecular weight (kDa). (B) Fluorometric analysis of caspase-3, caspase-8, and caspase-9 activities in MNV-1-infected RAW264.7 cells collected at serial time points. Caspase-3, caspase-8, and caspase-9 activities were determined by incubation of cell lysates with AFC-conjugated caspase-specific peptide substrates. Proteolytic activities were quantified by measuring fluorescence at 505 nm. After normalization for protein content, the final values of caspase activity were derived by subtracting background fluorescence from the corresponding mock-infected RAW cell lysates. Data represent the mean values obtained in two replicate experiments. RFU, relative fluorescence units.
Once processed, caspase-9 further cleaves procaspase-3, activating caspase-3. A cleaved caspase-3 fragment corresponding to the 17-kDa protein band was detected starting at 16 h p.i. (Fig. 3A, lanes 5 to 7), while unprocessed procaspase-3 was observed as a major 31-kDa band in mock-infected cells and during the first 12 h of infection (Fig. 3A, lanes 1 to 4). Analysis of caspase proteolytic activity based on use of fluorogenic caspase-specific peptide substrates confirmed preferential activation of the caspase-9/caspase-3 pathway (Fig. 3B). We observed a 46.5-fold increase in the activity of caspase-3 and a 5-fold increase in the activities of caspase-9 between 8 and 16 h p.i. The activity level of caspase-8 remained low throughout the experiment.
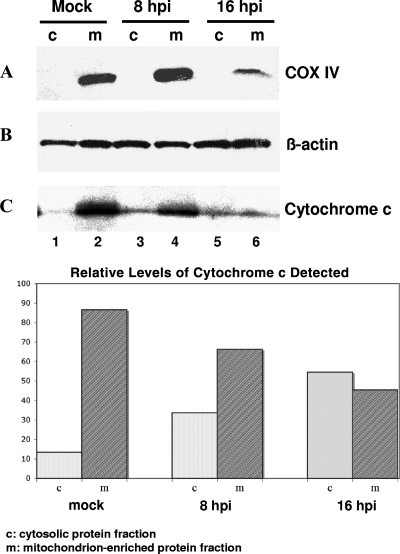
Additional data supporting the induction of the mitochondrial (intrinsic) pathway were obtained from an analysis of the distribution of cytochrome c in infected cells. Extraction of cytosolic and mitochondrion-enriched protein fractions from mock- and MNV-1-infected RAW264.7 cells followed by Western blot analysis showed the release of cytochrome c into the cytosol at between 8 and 16 h p.i. (Fig. 4C, lanes 3 and 5). While most of the cytochrome c was detected in the mitochondrial fraction of the mock-infected cells (Fig. 4C, lane 2), more than 50% of the cytochrome c was found in the cytosolic fraction of the infected cells (16 h p.i.), consistent with loss of the mitochondrial outer membrane integrity (Fig. 4C). Western blot analysis of the separated protein fractions with COX IV-specific antibodies showed confinement of this protein to the mitochondrion-enriched fraction, indicating the absence of general contamination of the cytosolic fractions with mitochondrial proteins during the fractionation procedure (Fig. 4A, lanes 2, 4, and 6).
FIG. 4.
MNV-1 infection triggers the mitochondrial pathway of apoptosis. Cytosolic and mitochondrion-enriched protein fractions were isolated from mock- and MNV-1-infected RAW264.7 cells collected at 16 h p.i. and at 8 and 16 h p.i., respectively. (A) The collected cytosolic proteins were probed with antibodies specific to mitochondrial marker protein COX IV to verify the absence of contamination with mitochondrial proteins. (B) The loading amounts of mitochondrial and cytosolic protein samples were normalized using total protein content and verified by probing with ß-actin-specific antibodies. (C) Cytochrome c release from the mitochondria was examined using normalized amounts of cytosolic (lanes 1, 3, and 5) and mitochondrial (lanes 2, 4, and 6) protein samples and cytochrome c-specific antibodies. Quantification of the cytochrome c release was performed using densitometry with the help of ImageQuant software (Molecular Dynamics).
MNV-1 replication is required to trigger apoptosis in RAW264.7 cells.
We examined whether active viral replication was required to trigger apoptosis in MNV-1-infected RAW264.7 cells. Evidence that active virus replication was required for the induction of apoptosis in the MNV-1-infected RAW264.7 cells was obtained in experiments with UV-inactivated virus. We found that a 15-min UV treatment of the virus, under conditions similar to those described previously, resulted in a more than 5-log reduction of the virus titer (data not shown). Cells inoculated with UV-inactivated virus showed no signs of characteristic DNA fragmentation, indicating that the interaction of the virus with the cells (Fig. 5A, lane 3) is not sufficient to trigger the apoptotic pathway. Alternatively, virus-infected RAW264.7 cells showed the characteristic DNA fragmentation observed at 16 h p.i. (Fig. 5A, lane 4) compared to mock-infected cells (Fig. 5A, lane 2).
FIG. 5.
Induction of apoptosis in RAW264.7 cells is dependent on virus replication. (A) DNA laddering in RAW264.7 cells infected with UV-treated MNV-1 virions. The results of electrophoresis of extracted DNA from UV-treated or nontreated MNV-1-infected RAW264.7 cells are shown and represent MNV-1 infection at 16 h p.i. (lane 4), UV-treated MNV-1-infected RAW264.7 cells (lane 3), mock-infected RAW264.7 cells (lane 2), and a DNA marker (lane 1). (B) Effect of CHX on MNV-1 replication in infected RAW264.7 cells. Mock- and MNV-1-infected RAW264.7 cells were treated with no CHX (lanes 1 and 4), with 0.5 μg of CHX/ml (lanes 2 and 5), with 1 μg of CHX/ml (lanes 3 and 6), and with staurosporine (ss; lane 7). Equivalent amounts of protein collected at 16 h p.i. were loaded in each gel and detected with α-PCNA antibody in infected and mock-infected cultures with or without CHX (lanes 1 to 7). The virus polymerase (NS7) was observed in virus-infected cells without CHX in the growth medium (lane 1). Cleaved caspase-9 was observed in MNV-1-infected cells without CHX (lane 1) and in RAW264.7 cells in the presence of an apoptosis inductor, staurosporine (ss) (lane 7).
Additional evidence supported the requirement of virus replication in order to induce apoptosis. Incubation of the infected cells in the presence of 0.5 or 1 μg of CHX/ml resulted in the inhibition of viral protein synthesis. When protein synthesis was inhibited, Western blot analysis of lysates of MNV-1-infected cells showed no expression of the viral polymerase (NS7) at 16 h p.i. (Fig. 5B, lanes 2 and 3). Consequently, cleaved caspase-9 was detected only at background levels of its activation in these samples. In contrast, processed caspase-9 was readily detected in the control MNV-1-infected cell lysate and in the lysate of cells treated with the apoptosis inducer staurosporine (Fig. 5B, lanes 1 and 7), showing that MNV-1 replication was required to induce apoptosis in RAW264.7 cells.
Microarray and PCR array expression analysis of MNV-1- and mock-infected RAW264.7 cells.
In order to study the apoptotic cascade in MNV-1-infected RAW264.7 cells, we profiled the transcription level of the whole mouse genome by the use of microarray analysis. The infection (12 h p.i.) was performed with purified virus to account for transcriptional changes due to viral infection only. A total of 1,781 genes were significantly (P ≤ 0.005) differentially expressed, and 1,322 genes presented results representing a twofold or greater up- or downregulation (data not shown). Ingenuity pathway analysis identified 82 differentially expressed genes associated with cell death. Eight apoptosis-associated genes found up- or downregulated by microarray analysis were further confirmed by PCR array analysis (Table 1). BIRC5 (survivin) and Hells (a lymphoid-specific helicase) were the only downregulated genes detected in this analysis, while several upregulated genes associated with apoptotic functions were also found. The PCR array analysis confirmed the striking downregulation of BIRC5 in MNV-1-infected RAW264.7 cells detected in the microarray experiments. This gene transcript presented the highest negative change, −22.8-fold.
TABLE 1.
Analysis of apoptosis-related genes by microarrays and PCR arrays in MNV-1-infected RAW264.7 cells
| Gene designation | UniGene designation | Protein designation or description | Fold changea
|
P | |
|---|---|---|---|---|---|
| PCR array | Microarray | ||||
| Birc5 | Mm.8552 | Baculoviral IAP repeat-containing 5 | −22.8 | −9.5 | 0.0001 |
| Casp11 | Mm.1569 | Caspase-11, apoptosis-related cysteine peptidase | 8.6 | 2.6 | 0.0001 |
| Hells | Mm.57223 | Helicase, lymphoid specific | −4 | −10 | 0.002 |
| Pycard | Mm.24163 | PYD and CARD domain containing | 25 | 2.7 | 0.0001 |
| Cd40 | Mm.271833 | CD40 antigen | 29.9 | 2.94 | 0.0000 |
| Tnf | Mm.1293 | TNF | 14.9 | 2.7 | 0.002 |
| Tnfsf10 | Mm.1062 | TNF-SF10 | 128.6 | 10.36 | 0.0000 |
| Traf1 | Mm.239514 | TNF receptor-associated factor 1 | 17.8 | 9.02 | 0.0005 |
Genes were differentially expressed by the microarrays and PCR arrays. Data represent changes in levels of specified genes in MNV-1-infected RAW 264.7 cells compared to mock-infected cells.
Of interest among upregulated proapoptotic protein genes were caspase-11 and Pycard protein genes, as well as several other proapoptotic genes of proteins regularly found expressed in blood line-derived cells, such as macrophages (CD40, tumor necrosis factor [TNF], and TNF [ligand] superfamily, member 10 [TNF-SF10]).
Downregulation of survivin over time.
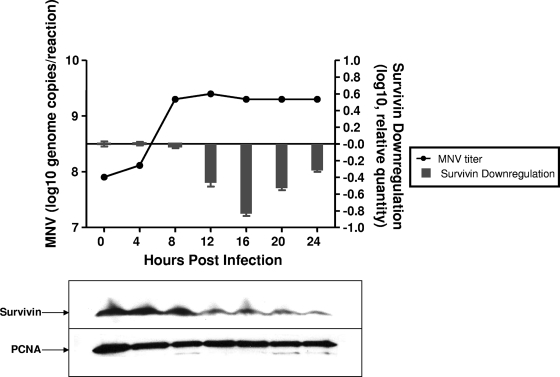
Survivin is part of a limited group of molecules that are known to be differentially expressed in several types of tumors, and its role in the apoptotic pathway and cell cycle regulation has been extensively described (4-6). Survivin functions as both a regulator of mitosis and a protector from apoptosis through modulation of caspase-9 activation (5, 6). Several viruses, including RNA viruses, take advantage of survivin's antiapoptotic properties by enhancing its expression in infected cells in order to prolong the time for replication (10, 39, 51, 53, 54). It was therefore surprising to discover that MNV-1 infection resulted in the downregulation of BIRC5 transcription in RAW264.7 cells. In order to correlate this effect with the growth cycle of MNV-1, we examined the levels of viral RNA and BIRC5 expression over time. The decrease in BIRC5 transcription became evident at 12 h p.i. and reached a maximum of inhibition at 16 h p.i., when CPE in the cell monolayer was visible (data not shown, Fig. 6). The downregulatory effect was observed at 24 h p.i., when extensive CPE was detected in most of the cell monolayer. The observed BIRC5 downregulation was preceded by an increase in MNV-1 RNA synthesis. The viral RNA level reached a maximum at 8 h p.i. and remained constant after 12 h p.i., when the inhibitory effect on survivin transcription became evident (Fig. 6).
FIG. 6.
Downregulation of BIRC5 (survivin) over time in MNV-1-infected RAW264.7 cells. The levels of BIRC5 and survivin, as well as the level of genomic MNV-1 RNA, are shown. Survivin transcription in MNV-1-infected cells compared to mock-infected cells (24 h p.i.) was normalized to the level of ß-actin transcription. MNV-1 genomic RNA detection by quantitative real-time PCR is plotted in comparison to the decreasing levels of BIRC5. Survivin was detected by Western blot analyses of infected cells corresponding to the same time points of MNV-1 infection. Equivalent protein amounts were loaded in each gel and detected with anti-PCNA antibody.
The inhibitory effect of MNV-1 on BIRC5 transcription was confirmed by immunoblot detection of survivin at the same time points as described above (Fig. 6). As is consistent with inhibition of the BIRC5 gene transcription, the amount of survivin protein began to decrease, starting at 12 h p.i. The decrease in survivin expression was also observed for up to 24 h p.i.
Effect of MNV-1 proteins on downregulation of survivin.
The MNV-1 genome encodes six nonstructural proteins in ORF1 (NS1-2, NS3, NS4, NS5, NS6, and NS7), as well as two structural capsid proteins in ORF2 and ORF3 (VP1 and VP2, respectively). In order to determine whether the effect of MNV-1 infection on survivin expression could be attributed to any individual MNV-1 protein, RAW264.7 cells were transfected with individual plasmid constructs encoding each of the MNV-1 proteins (46). At 12 h p.i., the monolayer was harvested and the RNA was extracted to evaluate the variation in BIRC5 (survivin) transcription levels. Normalization of samples was performed using ß-actin gene expression as an internal control. We did not observe a significant change in BIRC5 transcription levels in transfected cells compared to the results obtained with a control transfected with plasmid only (data not shown), indicating that the downregulation of survivin could not be attributed to any individual MNV-1 protein.
DISCUSSION
Several viruses trigger apoptosis in infected cells at an early stage of infection when virus particles interact with receptors on the cell surface or at the time of fusion with cell membrane and disassembly (8, 15, 26, 41). In addition, certain viruses induce apoptosis at late stages of replication, providing a mechanism for dissemination of progeny virus. The use of apoptosis as an exit strategy for the virus has also been suggested for caliciviruses (3, 47). Apoptotic changes in calicivirus-infected cells had been documented for FCV and RHDV (2, 28, 40, 47), and we examined whether infection with another calicivirus, MNV-1, triggered apoptosis in RAW264.7 cells. Changes were observed in nuclear chromatin, DNA fragmentation, and caspase activation consistent with apoptosis induced by MNV-1. Moreover, the induction of apoptosis was dependent on viral replication, as evidenced by the lack of DNA fragmentation in cells infected with the UV-inactivated virus. The finding of apoptosis in the NVs suggests that this process may be part of a conserved strategy among the caliciviruses to release viral progeny.
Triggering of apoptosis and downstream caspase activation can be initiated through two main pathways. The “intrinsic or mitochondrial pathway” involves changes in mitochondrial membrane permeability that are regulated by a family of Bcl-2-related proteins. Activated proapoptotic members of this group (Bax, Bak) can homo-oligomerize, forming pores in the mitochondrial membrane and promoting leakage of cytochrome c into the cytoplasm (7, 20). Cytochrome c release from the mitochondria results in apoptosome formation and processing of procaspase-9 (27). The “extrinsic pathway” is a sensor for extracellular signals and activates caspase-8 (18). Caspase-8 and caspase-9 converge on caspase-3 and caspase-7, the executioner caspases, which cleave proteins involved in maintaining the filamentous network of the cytoplasm and in retaining the structural organization of nuclear membranes and chromatin as well as proteins involved in DNA replication and repair (17). The marked increase in processed caspase-3 levels in MNV-1-infected cells was noteworthy, because caspase-3 is directly involved in the execution of cellular disassembly, which would likely facilitate the release of viral progeny from an infected cell. In addition, cleavage of caspase-3 was preceded by the appearance of the processed form of caspase-9, indicating involvement of the mitochondrial or intrinsic pathway, which has also been suggested for another calicivirus, FCV (35, 47).
Several RNA viruses (measles virus, vesicular stomatitis virus, Sindbis virus, and vaccinia virus, among others) induce apoptosis via mechanisms involving receptor binding and internalization or other replication events (8, 12, 15, 19, 21, 22, 26, 41, 48). Alternatively, some DNA viruses (Epstein-Barr virus and papillomavirus 16) inhibit programmed cell death by inducing the expression of cellular IAP (1, 10). MNV-1 inhibits expression of the antiapoptotic protein survivin, which may be a previously unrecognized mechanism used by viruses to actively induce programmed cell death. Our finding of downregulation of survivin by a replicating virus is the first evidence for a pathogen-induced mechanism to reduce the levels of this antiapoptotic protein. Decreased levels of survivin have previously been observed only in the presence of molecular antagonists of survivin such as antisense compounds, ribozymes, and small interfering RNA sequences or with dominant-negative mutants at the BIRC5 gene level. Downregulation of survivin always results in typical caspase-dependent cell death accompanied by cytochrome c release and caspase-9 activation (16, 33, 34). The cellular changes (e.g., cytochrome c release and caspase-9 activation) resulting from the nonviral downregulation of survivin expression are consistent with the apoptotic effects observed during replication of MNV-1 in RAW264.7 cells. It will be of interest to examine whether other viruses that induce apoptosis are associated with the downregulation of BIRC5 transcription. Ultimately, MNV-1 might prove to cause apoptosis by the use of a complex mechanism that involves the regulation of several genes. Because of the similarities observed between the apoptosis triggered by induced survivin inhibition and cell death caused by MNV-1 infection of RAW264.7 cells, we propose that cell death triggered by MNV-1 might be a direct consequence of BIRC5 transcription inhibition during virus replication.
Although we showed an association between MNV-1 infection and downregulation of survivin, it is not yet clear how MNV-1 affects other steps of the apoptotic pathway. We identified eight different genes (including the gene encoding survivin) in MNV-1-infected RAW264.7 cells that differed significantly in their transcription levels compared to those in mock-infected cells. Some of these genes may also contribute to the signs observed in apoptotic MNV-1-infected cells. Pycard, which was upregulated 25-fold in virus-infected cells, has been shown to associate with Bax, a key protein in the apoptotic cascade that induces cytochrome c release from the mitochondria (36). This effect ultimately leads to activation of caspase-9 and caspase-3, a result that was also observed in MNV-1-infected cells. In addition, the importance of caspase-11 for the activation of caspase-3 and caspase-1 has been demonstrated in experiments using caspase-11-knockout mice (30, 45). Therefore, the increase (8.6-fold) in transcription levels of caspase-11 in MNV-1-infected cells, together with the downregulation of survivin, might have also contributed to the more than 40-fold increase in caspase-3 activation due to MNV-1 infection.
A number of viral proteins have been reported to be directly involved in the modulation of apoptotic pathways, in particular, in the control of the mitochondrial apoptosis pathway (23). We attempted to identify an MNV-1 protein that would individually induce downregulation of survivin. However, we have not been able to associate a single MNV-1 protein with induction of apoptosis through regulation of BIRC5. The downregulation of survivin in MNV-1-infected cells might involve multiple factors, such as the interaction of several proteins, the presence of viral protein precursors, a cascade of events, or simply active RNA replication. Alternatively, it is also possible that downregulation of survivin is mediated via an indirect effect of other cellular changes induced by MNV-1 infection.
This report shows that, in contrast to viruses that increase survivin levels during replication, MNV-1 is the first agent found to decrease the levels of survivin in cells. We demonstrated that MNV-1 replication induces apoptosis in RAW264.7 cells, activating caspases through the mitochondrial pathway, possibly as a result of downregulation of survivin. There is a consensus that the gene encoding survivin is an essential cancer gene and an appropriate target for anticancer drug discovery (5), and the implications of our finding that certain viruses may naturally target this gene will need to be assessed in the future.
Acknowledgments
We thank Tanaji Mitra for technical support.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 11 February 2009.
REFERENCES
- 1.Ai, M. D., L. L. Li, X. R. Zhao, Y. Wu, J. P. Gong, and Y. Cao. 2005. Regulation of survivin and CDK4 by Epstein-Barr virus encoded latent membrane protein 1 in nasopharyngeal carcinoma cell lines. Cell Res. 15777-784. [DOI] [PubMed] [Google Scholar]
- 2.Al-Molawi, N., V. A. Beardmore, M. J. Carter, G. E. Kass, and L. O. Roberts. 2003. Caspase-mediated cleavage of the feline calicivirus capsid protein. J. Gen. Virol. 841237-1244. [DOI] [PubMed] [Google Scholar]
- 3.Alonso, C., J. M. Oviedo, J. M. Martin-Alonso, E. Diaz, J. A. Boga, and F. Parra. 1998. Programmed cell death in the pathogenesis of rabbit hemorrhagic disease. Arch. Virol. 143321-332. [DOI] [PubMed] [Google Scholar]
- 4.Altieri, D. C. 2006. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr. Opin. Cell Biol. 18609-615. [DOI] [PubMed] [Google Scholar]
- 5.Altieri, D. C. 2008. Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer 861-70. [DOI] [PubMed] [Google Scholar]
- 6.Altieri, D. C. 2003. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene 228581-8589. [DOI] [PubMed] [Google Scholar]
- 7.Annis, M. G., E. L. Soucie, P. J. Dlugosz, J. A. Cruz-Aguado, L. Z. Penn, B. Leber, and D. W. Andrews. 2005. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J. 242096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton, E. S., J. D. Chappell, J. L. Connolly, J. C. Forrest, and T. S. Dermody. 2001. Reovirus receptors and apoptosis. Virology 290173-180. [DOI] [PubMed] [Google Scholar]
- 9.Bokarewa, M., A. Tarkowski, and M. Magnusson. 2007. Pathological survivin expression links viral infections with pathogenesis of erosive rheumatoid arthritis. Scand. J. Immunol. 66192-198. [DOI] [PubMed] [Google Scholar]
- 10.Borbély, A. A., M. Murvai, K. Szarka, J. Konya, L. Gergely, Z. Hernadi, and G. Veress. 2007. Survivin promoter polymorphism and cervical carcinogenesis. J. Clin. Pathol. 60303-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branca, M., C. Giorgi, D. Santini, L. Di Bonito, M. Ciotti, S. Costa, A. Benedetto, E. A. Casolati, C. Favalli, P. Paba, P. Di Bonito, L. Mariani, S. Syrjanen, D. Bonifacio, L. Accardi, F. Zanconati, and K. Syrjanen. 2005. Survivin as a marker of cervical intraepithelial neoplasia and high-risk human papillomavirus and a predictor of virus clearance and prognosis in cervical cancer. Am. J. Clin. Pathol. 124113-121. [DOI] [PubMed] [Google Scholar]
- 12.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and J. A. Young. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87845-855. [DOI] [PubMed] [Google Scholar]
- 13.Cheetham, S., M. Souza, T. Meulia, S. Grimes, M. G. Han, and L. J. Saif. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 8010372-10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Compton, M. M. 1992. A biochemical hallmark of apoptosis: internucleosomal degradation of the genome. Cancer Metastasis Rev. 11105-119. [DOI] [PubMed] [Google Scholar]
- 15.Connolly, J. L., and T. S. Dermody. 2002. Virion disassembly is required for apoptosis induced by reovirus. J. Virol. 761632-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway, E. M., S. Pollefeyt, M. Steiner-Mosonyi, W. Luo, A. Devriese, F. Lupu, F. Bono, N. Leducq, F. Dol, P. Schaeffer, D. Collen, and J. M. Herbert. 2002. Deficiency of survivin in transgenic mice exacerbates Fas-induced apoptosis via mitochondrial pathways. Gastroenterology 123619-631. [DOI] [PubMed] [Google Scholar]
- 17.Cryns, V., and J. Yuan. 1998. Proteases to die for. Genes Dev. 121551-1570. [DOI] [PubMed] [Google Scholar]
- 18.Debatin, K. M., and P. H. Krammer. 2004. Death receptors in chemotherapy and cancer. Oncogene 232950-2966. [DOI] [PubMed] [Google Scholar]
- 19.Deng, L., T. Adachi, K. Kitayama, Y. Bungyoku, S. Kitazawa, S. Ishido, I. Shoji, and H. Hotta. 2008. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase-3-dependent pathway. J. Virol. 8210375-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Epand, R. F., J. C. Martinou, S. Montessuit, R. M. Epand, and C. M. Yip. 2002. Direct evidence for membrane pore formation by the apoptotic protein Bax. Biochem. Biophys. Res. Commun. 298744-749. [DOI] [PubMed] [Google Scholar]
- 21.Esolen, L. M., S. W. Park, J. M. Hardwick, and D. E. Griffin. 1995. Apoptosis as a cause of death in measles virus-infected cells. J. Virol. 693955-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gadaleta, P., M. Vacotto, and F. Coulombie. 2002. Vesicular stomatitis virus induces apoptosis at early stages in the viral cycle and does not depend on virus replication. Virus Res. 8687-92. [DOI] [PubMed] [Google Scholar]
- 23.Galluzzi, L., C. Brenner, E. Morselli, Z. Touat, and G. Kroemer. 2008. Viral control of mitochondrial apoptosis. PLoS Pathog 4e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green, K. Y. 2007. Caliciviridae: the noroviruses, p. 949-979. In D. M. Knipe, Peter M. Howley, Diane E. Griffin, Robert A. Lamb, and Malcolm A. Martin (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 25.Ikeda, Y., J. Shinozuka, T. Miyazawa, K. Kurosawa, Y. Izumiya, Y. Nishimura, K. Nakamura, J. Cai, K. Fujita, K. Doi, and T. Mikami. 1998. Apoptosis in feline panleukopenia virus-infected lymphocytes. J. Virol. 726932-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jan, J. T., S. Chatterjee, and D. E. Griffin. 2000. Sindbis virus entry into cells triggers apoptosis by activating sphingomyelinase, leading to the release of ceramide. J. Virol. 746425-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, X., and X. Wang. 2004. Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 7387-106. [DOI] [PubMed] [Google Scholar]
- 28.Jung, J. Y., B. J. Lee, J. H. Tai, J. H. Park, and Y. S. Lee. 2000. Apoptosis in rabbit haemorrhagic disease. J. Comp. Pathol. 123135-140. [DOI] [PubMed] [Google Scholar]
- 29.Kang, J. I., S. H. Park, P. W. Lee, and B. Y. Ahn. 1999. Apoptosis is induced by hantaviruses in cultured cells. Virology 26499-105. [DOI] [PubMed] [Google Scholar]
- 30.Kang, S. J., S. Wang, H. Hara, E. P. Peterson, S. Namura, S. Amin-Hanjani, Z. Huang, A. Srinivasan, K. J. Tomaselli, N. A. Thornberry, M. A. Moskowitz, and J. Yuan. 2000. Dual role of caspase-11 in mediating activation of caspase-1 and caspase-3 under pathological conditions. J. Cell Biol. 149613-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. Virgin IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 2991575-1578. [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann, S. H., S. Desnoyers, Y. Ottaviano, N. E. Davidson, and G. G. Poirier. 1993. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 533976-3985. [PubMed] [Google Scholar]
- 33.Li, F., and D. C. Altieri. 1999. The cancer antiapoptosis mouse survivin gene: characterization of locus and transcriptional requirements of basal and cell cycle-dependent expression. Cancer Res. 593143-3151. [PubMed] [Google Scholar]
- 34.Mesri, M., N. R. Wall, J. Li, R. W. Kim, and D. C. Altieri. 2001. Cancer gene therapy using a survivin mutant adenovirus. J. Clin. Investig. 108981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Natoni, A., G. E. Kass, M. J. Carter, and L. O. Roberts. 2006. The mitochondrial pathway of apoptosis is triggered during feline calicivirus infection. J. Gen. Virol. 87357-361. [DOI] [PubMed] [Google Scholar]
- 36.Ohtsuka, T., H. Ryu, Y. A. Minamishima, S. Macip, J. Sagara, K. I. Nakayama, S. A. Aaronson, and S. W. Lee. 2004. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat. Cell Biol. 6121-128. [DOI] [PubMed] [Google Scholar]
- 37.Parquet, M. C., A. Kumatori, F. Hasebe, E. G. Mathenge, and K. Morita. 2002. St. Louis encephalitis virus induced pathology in cultured cells. Arch. Virol. 1471105-1119. [DOI] [PubMed] [Google Scholar]
- 38.Patel, M. M., M. A. Widdowson, R. I. Glass, K. Akazawa, J. Vinje, and U. D. Parashar. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 141224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piña-Oviedo, S., K. Urbanska, S. Radhakrishnan, T. Sweet, K. Reiss, K. Khalili, and L. Del Valle. 2007. Effects of JC virus infection on anti-apoptotic protein survivin in progressive multifocal leukoencephalopathy. Am. J. Pathol. 1701291-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramiro-Ibáñez, F., J. M. Martin-Alonso, P. Garcia Palencia, F. Parra, and C. Alonso. 1999. Macrophage tropism of rabbit hemorrhagic disease virus is associated with vascular pathology. Virus Res. 6021-28. [DOI] [PubMed] [Google Scholar]
- 41.Ramsey-Ewing, A., and B. Moss. 1998. Apoptosis induced by a postbinding step of vaccinia virus entry into Chinese hamster ovary cells. Virology 242138-149. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, L. O., N. Al-Molawi, M. J. Carter, and G. E. Kass. 2003. Apoptosis in cultured cells infected with feline calicivirus. Ann. N. Y. Acad. Sci. 1010587-590. [DOI] [PubMed] [Google Scholar]
- 43.Sakahira, H., M. Enari, and S. Nagata. 1998. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 39196-99. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Shibata, M., S. Hisahara, H. Hara, T. Yamawaki, Y. Fukuuchi, J. Yuan, H. Okano, and M. Miura. 2000. Caspases determine the vulnerability of oligodendrocytes in the ischemic brain. J. Clin. Investig. 106643-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sosnovtsev, S. V., G. Belliot, K. O. Chang, V. G. Prikhodko, L. B. Thackray, C. E. Wobus, S. M. Karst, H. W. Virgin, and K. Y. Green. 2006. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 807816-7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sosnovtsev, S. V., E. A. Prikhod'ko, G. Belliot, J. I. Cohen, and K. Y. Green. 2003. Feline calicivirus replication induces apoptosis in cultured cells. Virus Res. 941-10. [DOI] [PubMed] [Google Scholar]
- 48.Urban, C., C. Rheme, S. Maerz, B. Berg, R. Pick, R. Nitschke, and C. Borner. 2008. Apoptosis induced by Semliki Forest virus is RNA replication dependent and mediated via Bak. Cell Death Differ. 151396-1407. [DOI] [PubMed] [Google Scholar]
- 49.Ward, J. M., C. E. Wobus, L. B. Thackray, C. R. Erexson, L. J. Faucette, G. Belliot, E. L. Barron, S. V. Sosnovtsev, and K. Y. Green. 2006. Pathology of immunodeficient mice with naturally occurring murine norovirus infection. Toxicol. Pathol. 34708-715. [DOI] [PubMed] [Google Scholar]
- 50.Wobus, C. E., S. M. Karst, L. B. Thackray, K. O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackenzie, K. Y. Green, and H. W. Virgin. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, X., N. Dong, L. Yin, N. Cai, H. Ma, J. You, H. Zhang, H. Wang, R. He, and L. Ye. 2005. Hepatitis B virus X protein upregulates survivin expression in hepatoma tissues. J. Med. Virol. 77374-381. [DOI] [PubMed] [Google Scholar]
- 52.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346312-323. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, Y., and G. Z. Gong. 2006. [Hepatitis C virus NS5A protein upregulates survivin gene expression]. Zhonghua Gan Zang Bing Za Zhi 14414-417. (In Chinese.) [PubMed] [Google Scholar]
- 54.Zhu, Y., M. Roshal, F. Li, J. Blackett, and V. Planelles. 2003. Upregulation of survivin by HIV-1 Vpr. Apoptosis 871-79. [DOI] [PubMed] [Google Scholar]