Abstract
Polo-like protein kinase 3 (Plk3) has been proposed to regulate entry into S phase and promote apoptosis in response to oxidative stress. Its mRNA contains three AU-rich elements (AREs) in its 3′ untranslated region (3′-UTR) that can contribute to the rapid degradation of labile transcripts. We investigated the possibility that tristetraprolin (TTP), a tandem CCCH zinc finger protein, could promote the decay of Plk3 transcripts. TTP is known to stimulate the deadenylation and decay of mRNAs possessing one or more copies of the consensus nonamer motif UUAUUUAUU. In stable mouse fibroblast cell lines derived from wild-type and TTP knockout littermates, the decay of Plk3 transcripts after serum stimulation was slowed in the absence of TTP. The specificity of TTP for promoting the degradation of Plk3 was demonstrated by the unaltered decay of Plk3 mRNA in cell lines deficient in the TTP family members ZFP36L1 and ZFP36L2. We also found that the AREs present in the Plk3 transcript were essential for both the binding of TTP to the 3′-UTR and promoting the destruction of target transcripts in cotransfection experiments. The regulation of Plk3 mRNA stability by TTP may influence the control of the cell cycle by this protein kinase.
AU-rich elements (AREs), located in the 3′ untranslated regions (3′-UTRs) of certain mRNAs, facilitate transcript decay and provide a mechanism for attenuating protein synthesis. The rate of ARE-dependent mRNA decay is determined in part by proteins that interact with these AREs (53). The tristetraprolin (TTP) family of CCCH tandem zinc finger proteins, consisting of TTP (ZFP36), ZFP36L1, and ZFP36L2 in humans as well as a fourth family member, ZFP36L3, that is expressed only in rodents, can all bind to AREs at a consensus nonamer site, UUAUUUAUU (3). TTP, the best-characterized member of this protein family, can promote mRNA deadenylation and degradation after binding to such AREs contained within the 3′-UTRs of certain mRNAs (27). Validated physiological target transcripts of TTP include those encoding tumor necrosis factor alpha (TNF-α) (6, 8), granulocyte-macrophage colony-stimulating factor (7), interleukin-2 (38), immediate-early response 3 (Ier3) (31), and others. In these examples, the inclusion criteria for being an authentic target transcript of TTP include enhanced mRNA stability in TTP knockout (KO) cell lines, specificity of TTP toward ARE binding sites in the transcript, and TTP-mediated decay of transcripts in cell transfection experiments. Although ZFP36L1, ZFP36L2, and ZFP36L3 share characteristics with TTP in overexpression experiments (4, 27), their cellular targets are unknown, and they were proposed to regulate physiological processes that are distinct from those regulated by TTP (9, 27).
Recently, global analysis of mRNA turnover in fibroblasts derived from TTP KO mice identified polo-like kinase 3 (Plk3/Frk) as a novel potential mRNA target of TTP (31). Originally identified as an inducible immediate-early response gene (16), Plk3 mRNA is transiently expressed in NIH 3T3 fibroblasts in response to growth factors and mitogens such as fibroblast growth factor 1, fibroblast growth factor 2, platelet-derived growth factor BB, phorbol myristate acetate, and serum (16). Plk3 transcripts are also induced in Mo7e cells, a hematopoietic cell line, by cytokines such as thrombopoietin and interleukin-3 (34). Plk3 belongs to a highly conserved family of serine-threonine kinases originally identified as polo in Drosophila melanogaster (36) and later identified as Cdc5 in Saccharomyces cerevisiae (24), Plo1 in Schizosaccharomyces pombe (24, 39), Plc1 and Plc2 in Caenorhabditis elegans (41), and Plx1-3 in Xenopus laevis (17, 26). In addition to Plk3, the mammalian Plk family consists of Plk1/Plk (12), Plk2/Snk (43), and Plk4/Sak (21). Plk family members are highly related in their catalytic domains, and Plk1-3 possess two conserved “polo box” motifs (18, 48). The two polo boxes of Plk1, which comprise the polo box domain (PBD), have been reported to coordinate protein-protein interactions and subcellular localization (19). Similar to Plk1, the PBDs of Plk2 and Plk3 preferentially bind phospho-serine and phospho-threonine motifs (20). Plk4 is a divergent member of the Plk family and possesses only one of the two bipartite polo box motifs present among other Plk family members (20, 33). Through their localization to mitotic structures and phosphorylation of specific substrates via their PBD and catalytic domains, respectively, the Plks were proposed to regulate entry into mitosis, cell cycle progression, cytokinesis, and the cellular response to DNA damage (50).
The short half-life of the Plk3 transcript is consistent with the presence of conserved AREs in its 3′-UTR (16, 31, 34). Unlike Plk1, which is highly expressed in proliferating tissues (50), Plk3 mRNA was previously reported to be expressed in a tissue-specific and developmentally regulated manner in mice (16) and humans (34). Despite the diverse physiological roles and contradictory reports regarding the half-life of the Plk3 protein, the coordinated regulation of Plk3 transcript and protein appears to be critical for cell division and viability.
In this report, we describe the ability of TTP to regulate the turnover of Plk3 transcripts by comparing the decay of Plk3 mRNA in TTP-deficient and wild-type (WT) fibroblasts. No equivalent role was found for the TTP family members ZFP36L1 and ZFP36L2 by using fibroblasts derived from the respective KO mice. We further demonstrate that the 3′-UTR of Plk3 contains several TTP binding sites. Finally, we show that the AREs within the Plk3 3′-UTR are sufficient to promote TTP-dependent mRNA degradation in transiently transfected HEK293 cells. These studies establish Plk3 mRNA as a physiological target of TTP and suggest that the TTP regulation of Plk3 transcript stability may modulate the effect of this protein kinase on the cell cycle.
MATERIALS AND METHODS
Cell culture.
Stable mouse fibroblast cell lines were derived from mouse embryonic fibroblasts from Zfp36+/+ (WT) and Zfp36−/− (KO) paired littermates (31), Zfp36l1+/+ and Zfp36l1−/− paired littermates (46), or Zfp36l2+/+ and Zfp36l2−/− paired littermates (D. J. Stumpo, unpublished data). Mouse fibroblasts were grown to approximately 70% confluence in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) containing 10% (vol/vol) fetal bovine serum (FBS) (Invitrogen), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine, as described previously (31). HEK293 cells were maintained in minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. All mouse experiments were conducted according to the U.S. Public Health Services Policy on Humane Care and Use of Laboratory Animals. The National Institute of Environmental Health Sciences Institutional Animal Care and Use Committee approved all animal procedures used in this study.
TTP-mediated decay of Plk3 transcripts.
The stability of Plk3 mRNA in mouse fibroblasts deficient in TTP (Zfp36−/−) was compared with that in fibroblasts derived from paired littermate controls (Zfp36+/+) as previously described (31). Cells were grown to approximately 60 to 70% confluence and were subsequently incubated in 0.5% FBS for 16 h. FBS (HyClone, Logan, UT) was then added to a final concentration of 10% (vol/vol) for 90 min. Actinomycin D (Sigma Chemical Co., St. Louis, MO) was added (5-μg/ml final concentration) for the indicated times, and RNA was isolated with the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. For each independent experiment, time point assays were performed in triplicate in 10-cm dishes and pooled for Northern blot analysis. An expressed-sequence-tag clone for Plk3 (GenBank accession number BU523505.1) was obtained from the IMAGE consortium through the American Type Culture Collection (ATCC, Manassas, VA) and was sequence verified. The RNA probed by Northern blotting was quantified using a phosphorimager (Typhoon; GE Healthcare, Piscataway, NJ) and ImageQuant software.
Preparation of RNA probes.
The Plk3 ARE plasmids (bp 2269 to 2312 of GenBank accession number NM_013807.2) and the Plk3 ARE mutant plasmids (see Fig. 4A) were constructed using a double-stranded synthetic oligonucleotide that included Asp718I and XbaI sequences at its 5′ or 3′ end, respectively. This was inserted between the Asp718I and XbaI sites of vector SK(−) (Stratagene, La Jolla, CA). Sequence confirmation of the DNA inserts was performed with dRhodamine and BigDye (Applied Biosystems, Foster City, CA) terminator cycle sequencing. The templates for synthesizing RNA probes were prepared by gel purification of the XbaI-linearized plasmids. The RNA probes were transcribed in the presence of [α-32P]UTP (800 Ci/mmol) using the Promega Riboprobe (Promega, Madison, WI) in vitro transcription system protocol. The resulting RNA probes were separated from the free nucleotides using G50 columns. Cell extracts were prepared from HEK293 cells transfected with CMV.hTTP.tag or expressing a zinc finger mutant of TTP, C124R, or a truncated TTP containing only the tandem zinc finger domain, as previously described (29-31).
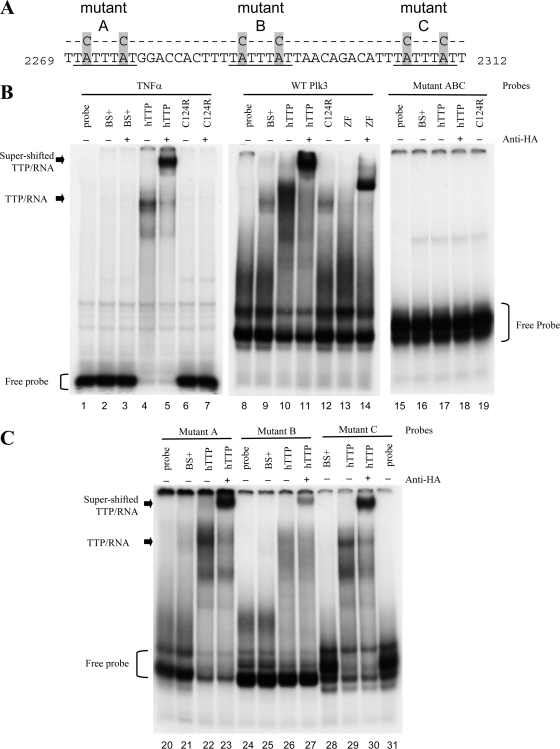
FIG. 4.
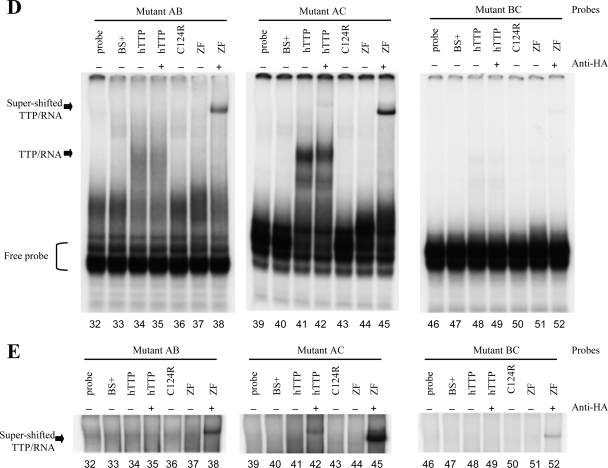
RNA gel shifts of WT and mutant Plk3 AREs with TTP. (A) The sequences of the RNA probes used are shown and include the three potential TTP binding sites within bp 2269 to 2312 of GenBank accession number NM_013807.2. The three core heptamers of the prospective TTP binding sites are underlined. The locations of the A-to-C mutations within each heptamer are highlighted. (B to E) RNA gel shift autoradiographs. The indicated RNA probes were incubated with cytosolic extracts derived from HEK293 cells transfected with either vector alone [pBS(+)], WT human TTP plasmid (hTTP), the mutant TTP plasmid with the C124R zinc finger mutation (C124R), or a fragment of TTP containing the tandem zinc finger domains (ZF). Cell extracts were incubated with (+) or without (−) anti-HA antibodies prior to the addition of RNA probes. Lanes 1 to 7 represent conditions under which the TNF ARE probe, used as a positive control, was incubated without (lane 1) or with (lanes 2 to 7) cytosolic extracts. Lanes 8 to 14 represent conditions under which the WT ARE of Plk3 was incubated without (lane 8) or with (lanes 9 to 14) cytosolic extracts. Only the presence of either unmutated hTTP or the zinc finger domains alone caused a shift of both TNF and WT Plk3 probes. Lanes 15 to 19 represent conditions in which the ABC mutant Plk3 probe was incubated without (lane 15) or with (lanes 16 to 19) the same cytosolic extracts. The unbound probes are denoted with brackets, and the complexes formed with hTTP are indicated with arrows. The supershifted TTP/RNA complex was observed when anti-HA monoclonal antibody was incubated with the indicated cell extracts for 10 min prior to the addition of probe. (C) Plk3 RNA probes with mutations in first (mutant A) (lanes 20 to 23), second (mutant B) (lanes 24 to 27), or third (mutant C) (lanes 28 to 31) ARE were incubated with or without cytosolic extracts. (D and E) Plk3 mutant probes AB (lanes 32 to 38), AC (lanes 39 to 45), and BC (lanes 46 to 52) were incubated without or with the same cytosolic extracts as in B. (E) Longer exposure of the same experiment above (D) in order to visualize the supershifted TTP/RNA complex.
RNA electrophoretic mobility shift assay.
RNA mobility gel shift assays were performed with 5 μg of extract protein in cytosolic extracts prepared from HEK293 cells. Cell extracts were incubated with 5 × 105 cpm of RNA probes containing the WT or mutant Plk3 ARE (without RNase T1 treatment) or the TNF ARE probe (pTNF bp 1309 to 1332 of GenBank accession no. X02611) treated with RNase TI (10 U/reaction; Epicentre, Madison, WI) in 25 μl buffer containing 10 mM HEPES (pH 7.6), 40 mM KCl, 3 mM MgCl2, 50 μg heparin, and 1.2 μg yeast tRNA, as previously described (29), and the protein-RNA complexes were resolved on 8% (wt/vol) nondenaturing acrylamide (37.5:1) gels.
Plk3 expression plasmids.
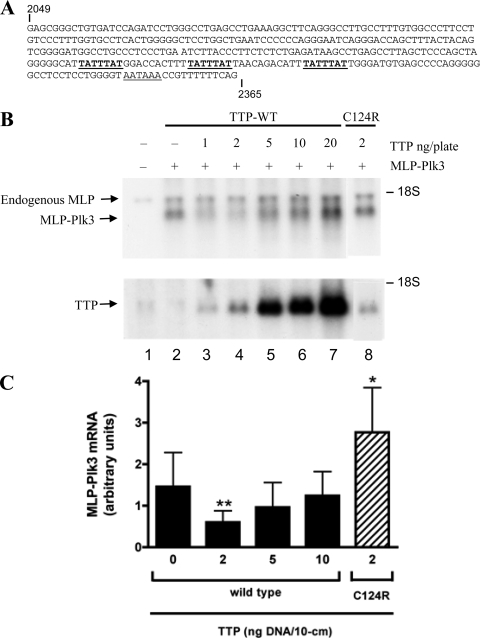
Since the overexpression of intact recombinant Plk3 is reportedly lethal in mammalian cells (13), the majority of the 3′-UTR of Plk3 (bp 2049 to 2365 of GenBank accession number NM_013807.2) was ligated 3′ of the coding region of a MARCKS-like protein (MLP/Marcksl1) (GenBank accession number NM_010807.3) to create CMV.MLP.Plk3.HA. Plk3 cDNAs were cloned by reverse transcription-PCR using total cellular RNA from either WT (67+/+) or KO (66−/−) cell lines stimulated with 10% FBS for 60 min as templates. The 5′ primer for PCR amplification of the Plk3 3′-UTR was 5′-aattgatatcGAGCGGGCTGTGATCCAGATCC-3, and the 3′ primer was 5′-tatatctagaCTGAAAAAACGGTTTATTACC-3′. The uppercase letters in the 5′ and 3′ primers contained bp 2049 to 2070 and bp 2346 to 2365, respectively, of GenBank accession number NM_013807.2. The lowercase letters in the primer sequences indicate the restriction sites for EcoRV and XbaI, respectively. The PCR products were digested with EcoRV and XbaI, and the resulting 316-bp cDNA representing the Plk3 3′-UTR was cloned into the EcoRV and XbaI sites of the pSK(−) vector containing the cytomegalovirus promoter-enhancer followed by MLP (31). Mutants within the ARE region of the Plk3 mRNA were made using the PCR primer-overlapping mutagenesis technique (32) (see Fig. 4 for the sequences). The correct sequences of the Plk3 inserts were confirmed by Big Dye terminator cycle sequencing. There were no differences in the sequences of the 3′-UTR clones from the WT or TTP KO cells, so the constructs were made using the clone from the WT cells.
Coexpression of TTP and Plk3.
The transient transfection of HEK293 cells was performed using the CaPO4 procedure (11), modified as previously described (29). MLP-Plk3 (0.5 μg) and TTP (2 to 10 ng) plasmid DNAs were transfected with carrier pBluescribe [pBS(+); Stratagene] DNA such that a total of 5 μg DNA was used per 10-cm plate. The cells were washed with minimal essential medium 16 h after the addition of DNA, and the cells were incubated for an additional 24 h in complete medium. To isolate RNA, cells were washed twice with ice-cold phosphate-buffered saline, and all liquid was removed by aspiration. RNA was isolated with the RNeasy kit (Qiagen) according to the manufacturer's instructions, and the isolated RNA from each transfection condition was used for Northern blot analysis with cDNA probes from mouse TTP (28). The cDNA probe recognizing both endogenous MLP and the MLP-Plk3 fusion transcript was a PCR product from amplification with primer CMV1F as the 5′ primer and a primer for the Plk3 3′-UTR, 5′-CTGGGGCTCACATCCCAATAAATAAATGTCTGTTAATAAATAAAAGTGGTCCATAAATAATGCCCCCTAGCTG-3′, as the 3′ primer.
Statistical analysis.
Statistical analysis was performed using Prism, version 4.0 (GraphPad Software, Inc., La Jolla, CA). One-way analysis of variance (ANOVA) was performed for multiple comparisons of WT and mutant MLP-Plk3 transcripts followed by the Bonferroni multiple-comparison test.
RESULTS
Plk3 transcript decay in TTP-deficient fibroblasts.
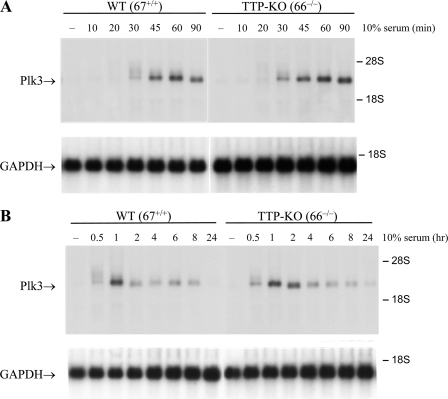
Global analysis of mRNA turnover using RNA derived from actinomycin-treated WT and TTP-deficient (KO) mouse fibroblasts suggested that Plk3 mRNA might be stabilized in TTP-deficient cells (31). This transcript contained three AREs within its 3′-UTR, conserved in several mammalian species, that are characteristic of known physiological TTP binding sites (31). To determine whether TTP promotes the degradation of endogenous Plk3 mRNA, the apparent half-lives of Plk3 mRNA in WT (67+/+) and TTP-KO (66−/−) fibroblast cell lines derived from paired littermates were compared. First, a time course after serum stimulation was performed to determine the time at which Plk3 mRNA levels were maximal and to examine differences in Plk3 expression between WT and TTP-deficient cells (Fig. 1). Plk3 mRNA was essentially undetectable in both WT and TTP-KO fibroblasts in the serum-starved state. Maximal increases in Plk3 transcript levels were observed after 1 h of serum stimulation (Fig. 1); the 2.4-kb Plk3 transcript level then declined rapidly in WT fibroblasts and approached basal levels by 2 h (Fig. 1B). These observations are consistent with previous reports that Plk3 is an immediate-early response gene in NIH 3T3 cells (16). Plk3 mRNA levels appeared to return to baseline levels more slowly in the TTP-KO cells, reaching near-basal levels after 4 h (Fig. 1B).
FIG. 1.
Time course of Plk3 mRNA expression after serum stimulation in mouse fibroblasts. Plk3 transcript levels were measured in stable fibroblast cell lines by Northern blot analysis using 10 μg of total cellular RNA and a cDNA probe for Plk3. WT (67+/+) and TTP KO (66−/−) cell lines were serum starved for 16 h in 0.5% FBS and then stimulated with 10% FBS for short (A) and long (B) time courses. The 28S and 18S ribosomal RNAs are indicated with arrows. Equal RNA loading was verified by staining the total RNA with acridine orange (data not shown) and Northern blot analysis of the original blot with a probe for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (bottom in each case).
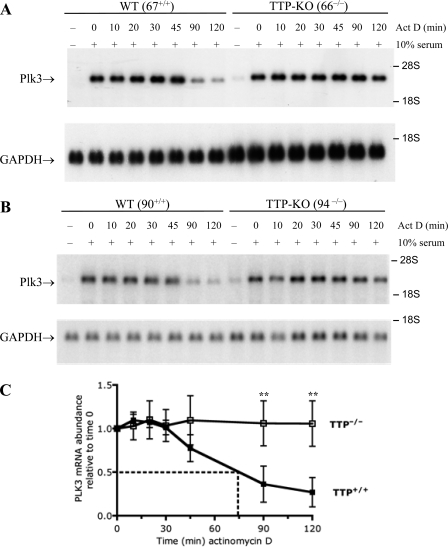
To exclude a cell line-specific effect, Plk3 mRNA decay was examined in three matched pairs of fibroblast cell lines that were derived from WT and TTP-KO littermate mice from three different litters. Fibroblasts were serum starved for 16 h and subsequently incubated with serum for 1 h, a time that resulted in peak levels of Plk3 mRNA (Fig. 1) and is known to exhibit near-maximal TTP protein accumulation in these cells (31). The decay of Plk3 mRNA was then monitored in a time course experiment after blocking transcription with actinomycin D (Fig. 2). The decay of Plk3 mRNA was considerably slower in the KO cells than in the WT cells in all three matched cell lines derived from paired littermates (Fig. 2 and data not shown). In fact, near-maximal Plk3 mRNA levels persisted in TTP KO cells for the entire 2-h actinomycin treatment (Fig. 2). When the results from four independent experiments were averaged to compare levels of Plk3 mRNA decay between one matched pair of WT and TTP-deficient fibroblasts, Plk3 mRNA levels decreased rapidly after actinomycin D treatment in the WT cells, with 50% of the Plk3 mRNA remaining after 74 min (Fig. 2C). In comparison, Plk3 transcripts remained stable in the TTP KO cells for the duration of the time course (Fig. 2C). The differences between the means after 90 and 120 min were statistically significant (Fig. 2C) (P < 0.05).
FIG. 2.
Plk3 mRNA stability in TTP-deficient fibroblasts. Plk3 mRNA decay was measured in serum-starved fibroblasts that were stimulated for 60 min with FBS and then treated with actinomycin D for the indicated times. Plk3 mRNA decay was compared in two matched pairs of cell lines: WT 67+/+ and TTP KO 66−/− (A) and WT 90+/+ and TTP KO (94−/−) (B) cells. GAPDH mRNA levels are shown for comparison. (C) The half-life of Plk3 mRNA was examined by averaging data from one of the pairs of fibroblast cell lines (67+/+ and 66−/−); shown are the means of data from four separate experiments. The mean half-life was 74 min for the WT cells in this experiment. Plk3 mRNA levels did not decline sufficiently in the TTP KO cells to calculate the half-life of the Plk3 transcript. The black and white squares represent the WT and TTP KO cell lines, respectively. Error bars represent the standard deviations (SD) derived from four separate experiments, with the Northern blots being quantitated by phosphorimager analysis and normalized for GAPDH mRNA levels. The double asterisks represent P values of <0.05 from a two-tailed Student t test.
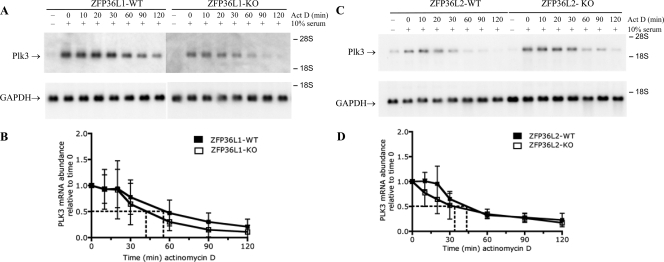
Effect of Zfp36l1 and Zfp36l2 deficiency on Plk3 mRNA decay.
The only other TTP family members expressed in mouse fibroblasts are ZFP36L1 and ZFP36L2. Fibroblast cell lines derived from either Zfp36l1 (46) or Zfp36l2 (D. J. Stumpo, unpublished data) KO mouse embryos were used to determine whether either of these proteins contributed to the turnover of Plk3 mRNA. Plk3 mRNA expression was maximal after 1.5 h of serum stimulation in both WT and ZFP36L1-deficient fibroblasts (data not shown). As described previously, the ZFP36L1 mRNA and protein levels were essentially unchanged in both serum-starved and growth factor-simulated fibroblasts (31). In addition, the TTP mRNA expression levels are similar in both WT and ZFP36L1-deficient fibroblasts (31). Plk3 mRNA decayed rapidly in both WT and ZFP36L1-deficient cell lines (Fig. 3A and B), similar to the decay of Plk3 mRNA in the WT fibroblast cell lines in Fig. 2. There was a 50% reduction of Plk3 mRNA after 55 and 42 min of actinomycin treatment for three pairs of WT and ZFP36L1-KO cells, respectively (Fig. 3B). Similarly, the decay rates of Plk3 mRNA were comparable between three pairs of WT and ZFP36L2-deficient cell lines (Fig. 3C and D). The mean apparent half-lives of Plk3 mRNA, obtained from three independent pairs of WT and ZFP36L2-KO cells, were 44 and 34 min, respectively.
FIG. 3.
Plk3 mRNA half-life in WT and ZFP36L1-and ZFP36L2-deficient fibroblasts. The half-lives of Plk3 mRNA in paired fibroblast cell lines deficient in either ZFP36L1 or ZFP36L2 were compared. Serum-starved fibroblasts were stimulated for 90 min with FBS and treated with actinomycin D (Act D) for the indicated times. (A and C) Representative Northern blots are shown for Plk3 mRNA decay and GAPDH expression for WT and ZFP36L1-KO (A) and WT and ZFP36L2-KO (C) fibroblasts. (B) The mean half-lives of Plk3 mRNA normalized for GAPDH mRNA expression, when calculated from three different paired fibroblast cell lines, were 55 and 42 min for WT and ZFP36L1-deficient fibroblasts, respectively. (D) The half-lives of Plk3 mRNA from three different paired cell lines were 44 and 34 min for WT and ZFP36L2 KO cells, respectively. The black and white squares represent the WT and KO cell lines, respectively. Error bars represent the SD derived from data for three to four separate experiments, with the Northern blots being quantitated by phosphorimager analysis and normalized for GAPDH mRNA levels.
TTP binds to AREs within the Plk3 mRNA 3′-UTR.
The 3′-UTR of Plk3 contains three conserved, closely spaced copies of the heptamer UAUUUAU, the core of the ideal TTP binding site (31). The ability of TTP to bind to these sequences was examined in RNA gel shift experiments (Fig. 4). We mutated combinations of the six core A residues to C's within the three binding sites to test the specificity of TTP for these sequences (Fig. 4A). The resulting 60-base RNA probes, transcribed in vitro, include 16 bases from the vector SK(−) (Stratagene) and 44 bases of the Plk3 3′-UTR (bp 2269 to 2312 of GenBank accession number NM_013807.2). The TNF-α ARE probe, used as a positive control, exhibited delayed migration in the gel when a cytosolic HEK293 cell extract containing recombinant TTP was incubated with the probe (Fig. 4B, lane 4). When the TTP-containing extract was incubated with an antibody recognizing the hemagglutinin (HA) epitope, there was a further delay in the migration of the TNF-α ARE probe, as evidenced by a supershifted complex (Fig. 4B, lane 5). In contrast, the TNF-α ARE probe did not exhibit delayed migration when it was incubated with extracts from cells transfected with either vector alone (BS+) (Fig. 4B, lanes 2 and 3), a plasmid encoding the nonbinding TTP C124R zinc finger mutant (lane 6), or the TTP C124R mutant coincubated with the anti-HA antibody (lane 7) (27, 31). The same series of cell extracts was used with probes derived from WT and mutant Plk3 sequences, in which the A's in one or more of the three putative TTP binding sites were mutated to C's (Fig. 4). Incubation of the WT Plk3 ARE probe with TTP-containing cell extracts resulted in the shifted, slower migration of the probe (Fig. 4B, lane 10) and was supershifted when coincubated with the anti-HA antibody (lane 11). These data confirm that the RNA-protein complex contained TTP. In comparison, the TTP C124R mutant (Fig. 4B, lane 12) and vector control-transfected extracts (lane 9) did not cause a delayed migration of the WT Plk3 ARE probe.
Further evidence that the tandem zinc finger domain of TTP was specifically required for binding of the WT Plk3 probe was observed using a truncated version of TTP that contains the tandem zinc finger domain located between amino acids 97 and 173 (29). The migration of the WT Plk3 ARE probe was delayed when extracts containing the zinc finger domain fragment (Fig. 4B, lane 14) were coincubated with the anti-HA antibody.
To investigate the relative contributions of each ARE to TTP binding, three individual (mutants A, B, and C) and three paired (mutants AB, AC, and BC) Plk3 ARE mutants were evaluated and compared to a mutant Plk3 ARE probe (mutant ABC) that contained A-to-C mutations in all three potential binding heptamers (Fig. 4B to D). Unlike the WT Plk3 probe, TTP did not delay the migration of the mutant Plk3 ARE probe that contained A-to-C mutations in all three heptamers (Fig. 4B, lanes 17 and 18). In addition, extracts from either the BS vector control or the TTP C124R mutant did not cause a delayed migration of the mutant ABC probe (Fig. 4B, lanes 16 and 19, respectively).
Since mutating all three of the potential TTP binding sites eliminated binding to TTP, each potential binding site was examined to determine its relative contribution to TTP binding. Plk3 RNA probes with mutations in individual putative binding sites (mutants A, B, and C) still permitted the binding of TTP to the probes (Fig. 4C, lanes 22, 26, and 29, respectively). All three of these individual Plk3 mutants were supershifted when incubated with the anti-HA antibody, indicating the presence of TTP in the RNA-protein complexes (Fig. 4C, lanes 23, 27, and 30). The TTP C124R mutant failed to bind to any of the three individual binding-site mutants (data not shown). The binding of TTP to probes containing mutations in two of the three putative binding sites suggests a hierarchy of these binding sites (Fig. 4D). For example, mutant AC was appreciably shifted in the presence of TTP (Fig. 4D, lane 41), whereas mutant AB demonstrated less of a mobility shift (lane 34). Both of these mutant probes were supershifted when coincubated with the zinc finger TTP fragment and the anti-HA antibody (Fig. 4D and E, lanes 38 and 45, respectively). A longer exposure of the gels from the same experiments revealed that intact TTP-containing extracts incubated with anti-HA antibodies caused a supershift of the protein complex with the AC mutant but not with the AB mutant probe (Fig. 4E, lanes 42 and 35, respectively). Among the three paired ARE mutants, mutant BC showed the weakest binding of TTP (Fig. 4D and E, lanes 48 and 49) and exhibited only a modest supershift when coincubated with the zinc finger domain of TTP and anti-HA antibodies (Fig. 4E, lane 52).
As an alternative measure of RNA-protein interactions, we performed immunoprecipitation of TTP from cells deprived of serum overnight and then stimulated for 90 min with 10% fetal calf serum. At this time point of stimulation, both TTP protein levels (31) and Plk3 mRNA levels (Fig. 1A) were near maximal. We used a well-validated antibody that reacts with mouse TTP (5, 31), along with its corresponding preimmune serum, and followed a widely used immunoprecipitation and reverse transcription-PCR protocol (23). As shown in Fig. S1 in the supplemental material, in one experiment (experiment A), there was an average sevenfold enrichment of Plk3 mRNA in the TTP immunoprecipitates compared to the pellets from the preimmune serum. In a second experiment, the enrichment was approximately 2.5-fold. Although this type of experiment has been criticized (37), particularly for the possibility of posthomogenization protein-RNA associations, these findings support an association of TTP with Plk3 mRNA in the intact fibroblasts after serum stimulation. In addition, a recent global analysis of TTP immunoprecipitates in macrophages did not identify Plk3 mRNA as an enriched mRNA, but it also did not pick up the best-validated TTP binding target in these cells, TNF itself (45). Finally, we noted that both the immune beads and the control beads brought down a variety of control mRNAs in a nonspecific fashion (see Fig. S1 in the supplemental material).
Concentration-dependent TTP-mediated decay of a fusion MLP-Plk3 transcript in HEK293 cells.
Since each of the three conserved AREs within the 3′-UTR of Plk3 could bind TTP in a cell-free system, we next examined whether these sequences conferred TTP sensitivity to a transcript consisting of the MARCKS-like protein (MLP or Marcksl1)-coding region fused to part of the Plk3 3′-UTR. We used this fusion transcript as a potential TTP “target” since previous reports noted that the overexpression of Plk3 can be lethal in mammalian cells (13). Since TTP has been shown to promote mRNA degradation in a concentration-dependent manner in this transfection system (31), HEK293 cells were transiently cotransfected with various amounts of plasmids encoding TTP and a fixed amount of MLP-Plk3-encoding DNA to determine the optimal concentration of TTP to promote decreased accumulation of MLP-Plk3 mRNA. Endogenous MLP mRNA was present in mock-transfected cells (Fig. 5B, lane 1) and served as a loading control for normalization, whereas TTP is not expressed in HEK293 cells. Compared to the MLP-Plk3 plasmid transfected alone, the smallest amounts of transfected TTP DNA (1 and 2 ng DNA per 10-cm dish) caused a substantial reduction in steady-state levels of the MLP-Plk3 transcripts (Fig. 5B, lanes 3 and 4). With increasing amounts of transfected TTP DNA (Fig. 5B, lanes 5 to 7), MLP-Plk3 transcripts became less sensitive to degradation, as noted previously with several other target transcripts in this experimental system (30, 31). Subsequent experiments used the most effective concentration of TTP plasmid (2 ng/plate) for cotransfection experiments with the MLP-Plk3 plasmid. In contrast to the potent reduction in mRNA levels seen with small amounts of WT TTP (Fig. 5B, lane 4), the same amount of the C124R TTP mutant did not reduce the steady-state levels of MLP-Plk3 transcripts (lane 8).
FIG. 5.
Effect of TTP on steady-state levels of MLP-Plk3 fusion mRNA. A portion of the Plk3 3′-UTR (bp 2049 to 2365 of GenBank accession number NM_013807.2) was fused 3′ of the coding region of MLP to create the MLP-Plk3 fusion transcript for cotransfection experiments with TTP DNA. (A) The three potential TTP binding sites within the 3′-UTR of the Plk3 transcript are underlined. (B) Various amounts of TTP DNA were cotransfected into HEK293 cells along with 0.5 μg of the plasmid coding for the MLP-Plk3 fusion transcript. Total plasmid DNA was kept constant (5 μg) by the addition of the pBS(+) vector. RNA was isolated 48 h after DNA was introduced into the cells. A total of 10 μg of total cellular RNA was loaded per gel lane. The results of a Northern blot from such an experiment probed for MLP-Plk3 and TTP are shown at the top and bottom, respectively. Cells were transfected with the pBS(+) vector alone (mock) (lane 1), MLP-Plk3 DNA alone (lane 2), MLP-Plk3 DNA plus various amounts of TTP DNA (lanes 3 to 7), and the TTP mutant C124R (lane 8). The native MLP transcript migrates above the MLP-Plk3 fusion transcript, as indicated. (C) The amount of MLP-Plk3 mRNA shown by Northern blotting was quantified using a phosphorimager and ImageQuant software. The means represent the amounts of MLP-Plk3 mRNA normalized against endogenous MLP, and error bars represent the SD derived from between 3 and 12 experiments (arbitrary units). Comparisons of MLP-Plk3 transcripts in the absence of TTP and various amounts of TTP were determined by one-way ANOVA and the Bonferroni multiple-comparisons test (* represents a P value of <0.05, and ** represents a P value of <0.01).
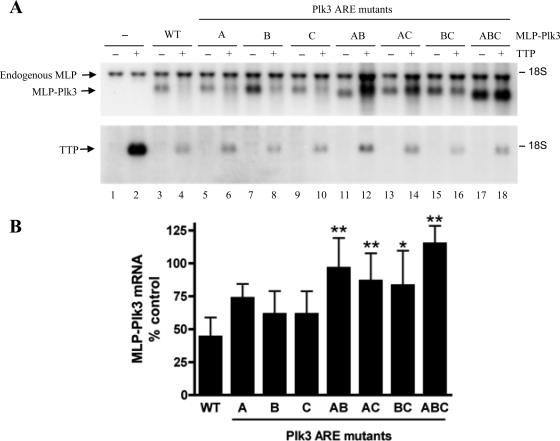
Intact AREs determine the steady state of MLP-Plk3 mRNA.
The Plk3 3′-UTR, shown in Fig. 5A, is more than 300 bp in length and contains three closely spaced AREs. Similar to the experiment shown in Fig. 4, the six core A residues within the three AREs were converted to C's to examine the importance of these potential TTP binding sites for the steady-state levels of the MLP-Plk3 transcript in this cotransfection system. HEK293 cells were transfected with MLP-Plk3 DNA containing either WT or mutant Plk3 AREs and cotransfected with or without the WT TTP plasmid. Compared to transcript levels in the absence of TTP, the steady-state levels of the WT MLP-Plk3 transcript were reduced by 56% when TTP was present (Fig. 6A, lanes 3 and 4, and B). In the presence of TTP, mutations in individual AREs (mutant B and mutant C) (Fig. 6A, lanes 7 and 8 and lanes 9 and 10, respectively) resulted in decreases in MLP-Plk3 transcript levels by approximately 39%. Steady-state levels of MLP-Plk3 transcripts lacking the first TTP binding sequence (mutant A) (Fig. 6A, lanes 5 and 6) were decreased by 27%. However, mutating any two of the three binding sites reduced the TTP-dependent destabilization of the transcript still further (lanes 11 to 16). In the presence of TTP, MLP-Plk3 mutant AB exhibited a 4% decrease in steady-state levels, and levels of mutant AC and BC transcripts were decreased by 13% and 17%, respectively. Mutation of all three AREs resulted in the complete insensitivity of MLP-Plk3 to TTP (mutant ABC) (Fig. 6A, lanes 17 and 18). The mean differences between the TTP-dependent decays of WT MLP-PLK3 and the double MLP-Plk3 mutants were statistically significant (P < 0.001 for mutants AB and AC and P < 0.01 for mutant BC). Similarly, there was a statistically significant difference between the steady-state levels of WT MLP-Plk3 and the triple ARE mutant (P < 0.001 for mutant ABC). Collectively, these data suggest that the three AREs within the 3′-UTR of Plk3 mediate TTP-dependent destabilization and rapid turnover of the transcript.
FIG. 6.
Effect of TTP on steady-state levels of MLP-Plk3 transcript mutated in the ARE. The WT human TTP plasmid was cotransfected into HEK293 cells with plasmids expressing MLP-Plk3 constructs with WT or mutant AREs. The MLP-Plk3 mutants consist of A-to-C mutations within individual AREs, a combination of two AREs, or all three AREs, as depicted in Fig. 4A. (A) Cells were transfected with the pBS(+) vector alone (lane 1), the pBS(+) vector plus TTP (lane 2), MLP-Plk3 DNA alone (lane 3), or MLP-Plk3 DNA plus TTP (lane 4). The steady-state levels of MLP-Plk3 mutant constructs were compared without or with TTP coexpression for MLP-Plk3 mutant A (lanes 5 and 6), mutant B (lanes 7 and 8), mutant C (lanes 9 and 10), mutant AB (lanes 11 and 12), mutant AC (lanes 13 and 14), mutant BC (lanes 15 and 16), and mutant ABC (lanes 17 and 18). Shown are representative Northern blots for the MLP-Plk3 fusion transcript (top) and TTP (bottom) mRNAs. RNA was isolated 48 h after transfection, and 10 μg of total cellular RNA was loaded into each gel lane for Northern blotting. The amount of MLP-Plk3 mRNA probed by Northern blotting was quantified using a phosphorimager and ImageQuant software, with normalization against endogenous MLP. The data shown in B reflect the means of the ratio of the level of MLP-Plk3 in the presence of TTP divided by that in the absence of TTP (percent control). Error bars represent the SD derived from data from between 3 and 12 experiments. Comparisons between WT and mutant Plk3 ARE transcripts were determined by one-way ANOVA and the Bonferroni multiple-comparisons test (* represents a P value of <0.01, and ** represents a P value of <0.001).
DISCUSSION
TTP is a zinc finger protein that binds certain class II AREs, thereby promoting the removal of the poly(A) tail and increasing the rate of mRNA turnover. Unlike the class I AREs that do not necessarily confer transcript instability under physiological conditions, class II AREs are major determinants of mRNA instability under physiological conditions (53). Despite this, the physiological relevance of class II AREs has been described for only a few transcripts. Recently, Lai et al. provided preliminary data suggesting that Plk3 mRNA and several additional transcripts were stabilized in TTP-deficient fibroblasts (31). The three potential TTP binding sites within the 3′-UTR of the Plk3 transcript have been proposed to play a role in the rapid degradation of this inducible early-response gene (16, 31, 34). Since the three canonical UAUUUAU heptamers representing the core sequence of the TTP binding sites were highly conserved within the Plk3 3′-UTR of mammals (31), we investigated in detail whether TTP was involved in the physiological regulation of Plk3 transcript degradation.
Our results from three pairs of mouse fibroblast cell lines, with each pair derived from WT and TTP-KO littermates, demonstrated that the absence of TTP results in the enhanced stability of the Plk3 transcript. Furthermore, Plk3 mRNA was not stabilized in mouse fibroblasts lacking the TTP-related proteins ZFP36L1 and ZFP36L2. These data establish Plk3 mRNA as a physiological target for TTP, one that appears to be specific for this family member, at least in this cell type after serum stimulation. As predicted from the RNA sequence and as demonstrated by RNA gel shift experiments, TTP bound to each of the individual AREs within the 3′-UTR of Plk3. When placed back in the context of the entire 3′-UTR of Plk3, the single ARE mutants each caused the mutant transcripts to be less sensitive to TTP than the transcript containing all intact AREs. Among RNA probes lacking two of the putative TTP binding sites, TTP bound most readily to the RNA probe containing an intact ARE in the second position (mutant AC) (Fig. 4D and E). TTP binding to the mutant AB probe was less, and binding to the BC probe was nearly undetectable. The conversion of the two adenylate residues to cytidines in all three putative TTP binding sites completely abrogated TTP binding. Similarly, in cotransfection experiments, transcripts containing the WT Plk3 3′-UTR were inherently less stable in the presence of TTP than those transcripts with the mutant AREs. MLP-Plk3 mutant transcripts became more resistant to TTP-promoted transcript decay with the progressive loss of each TTP binding site. Finally, the TTP C124R zinc finger mutant did not bind the Plk3 probe in the gel shift assay, nor did it promote Plk3 transcript destabilization in HEK293 cells, confirming the tandem zinc finger domain as the Plk3 mRNA binding domain within TTP.
Consistent with their diverse proposed roles within the cell, the Plks are differentially expressed, exhibit distinct subcellular localization patterns, and are subject to different types of posttranslational modifications (48). The mRNA and protein levels of Plk1 and Plk2 appear to be coordinately expressed during the cell cycle (50). In contrast, there is little reported concordance regarding the expression of Plk3 mRNA and protein. Several contradictory reports have proposed that Plk3 protein levels (i) remain constant throughout the cell cycle (2, 16), (ii) increase as cells enter S phase and mitosis (10), or (iii) peak during the G1 stage and become undetectable during mitosis (54). Others have suggested that an inactive form of Plk3 is maintained at a steady state in the cytosol throughout the cell cycle, whereas an activated form of Plk3 is localized in the nucleus, where it undergoes rapid proteolysis via a ubiquitin-mediated pathway (1). Among other factors, these apparently contradictory data may be due to differences between antibody sources and methods of protein extraction.
The various proposed intracellular localizations of Plk3 have implicated this kinase in a wide range of cellular functions. For example, Plk3 was reportedly found in the Golgi apparatus and the centrosome, where it could play a role in Golgi disassembly (42) or microtubule dynamics (14, 15, 49). In addition, Plk3 has been described to be located at the spindle pole, at spindle microtubules, within actin-containing plaques, and within the nucleus. A recent report of a study that employed RNA interference to knock down Plk3 expression found Plk3 solely in the nucleolus and absent from cytoplasmic structures, the centrosome, and the spindle pole (54). Although numerous Plk3 substrates have been identified in vitro, including p53, Cdc25A, Cdc25B, Cdc25C (40), SPAR, DNA polymerase δ (48), and topoisomerase IIα (22), the majority of these putative substrates require further validation in vivo.
Alterations in transcript stability have been associated with human diseases ranging from cancer, inflammatory disease, α-thalassemia, and Alzheimer's disease (35). The consequences of prolonging the half-lives of labile transcripts are illustrated by the TTP-deficient mouse, which exhibits arthritis, autoimmunity, skin lesions, myeloid hyperplasia, and weight loss (47), all apparently secondary to the stabilization of a TTP target transcript, that encoding TNF-α (6, 8). A similar but more severe inflammatory disease was caused by the complete removal of the TNF ARE (25). Using a similar approach, we are exploring the consequences of increasing the stability of Plk3 transcripts using a mouse “knock-in” strategy, in which the TTP binding sites within the Plk3 3′-UTR are removed.
Although Plk1 and Plk3 share some substrates in common in vitro and can rescue the Cdc5 temperature-sensitive mutant in S. cerevisiae (40), the overexpression of recombinant Plk1 and Plk3 in mammalian cells results in cell transformation (44) and cell death, respectively (13). Plk3 has been proposed to regulate apoptosis by, among others, the p53 pathway (51), and characterization of Plk3 KO mice has implicated Plk3 as a tumor suppressor (52). In aging mice, a Plk3 deficiency caused accelerated tumor development, larger tumor size, and more-pronounced angiogenesis in the KO mice than in wild-type litter mates. An understanding of the situations in which TTP attenuates Plk3 expression will be critical for unraveling the role of this protein kinase during normal and malignant cell proliferation and apoptosis.
Supplementary Material
Acknowledgments
We thank Douglas Bell and Farhad Imani for critical comments on the manuscript and Elizabeth Kennington for her thoughtful review.
This research was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH.
Footnotes
Published ahead of print on 2 February 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alberts, G. F., and J. A. Winkles. 2004. Murine FGF-inducible kinase is rapidly degraded via the nuclear ubiquitin-proteosome system when overexpressed in NIH 3T3 cells. Cell Cycle 3678-684. [PubMed] [Google Scholar]
- 2.Bahassi el, M., C. W. Conn, D. L. Myer, R. F. Hennigan, C. H. McGowan, Y. Sanchez, and P. J. Stambrook. 2002. Mammalian Polo-like kinase 3 (Plk3) is a multifunctional protein involved in stress response pathways. Oncogene 216633-6640. [DOI] [PubMed] [Google Scholar]
- 3.Blackshear, P. J. 2002. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30945-952. [DOI] [PubMed] [Google Scholar]
- 4.Blackshear, P. J., R. S. Phillips, S. Ghosh, S. B. Ramos, E. K. Richfield, and W. S. Lai. 2005. Zfp3613, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol. Reprod. 73297-307. [DOI] [PubMed] [Google Scholar]
- 5.Cao, H., J. S. Tuttle, and P. J. Blackshear. 2004. Immunological characterization of tristetraprolin as a low abundance, inducible, stable cytosolic protein. J. Biol. Chem. 27921489-21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carballo, E., G. S. Gilkeson, and P. J. Blackshear. 1997. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (−/−) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFalpha overproduction. J. Clin. Investig. 100986-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carballo, E., W. S. Lai, and P. J. Blackshear. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 951891-1899. [PubMed] [Google Scholar]
- 8.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 2811001-1005. [DOI] [PubMed] [Google Scholar]
- 9.Carrick, D. M., and P. J. Blackshear. 2007. Comparative expression of tristetraprolin (TTP) family member transcripts in normal human tissues and cancer cell lines. Arch. Biochem. Biophys. 462278-285. [DOI] [PubMed] [Google Scholar]
- 10.Chase, D., Y. Feng, B. Hanshew, J. A. Winkles, D. L. Longo, and D. K. Ferris. 1998. Expression and phosphorylation of fibroblast-growth-factor-inducible kinase (Fnk) during cell-cycle progression. Biochem. J. 333655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 72745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clay, F. J., S. J. McEwen, I. Bertoncello, A. F. Wilks, and A. R. Dunn. 1993. Identification and cloning of a protein kinase-encoding mouse gene, Plk, related to the polo gene of Drosophila. Proc. Natl. Acad. Sci. USA 904882-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conn, C. W., R. F. Hennigan, W. Dai, Y. Sanchez, and P. J. Stambrook. 2000. Incomplete cytokinesis and induction of apoptosis by overexpression of the mammalian polo-like kinase, Plk3. Cancer Res. 606826-6831. [PubMed] [Google Scholar]
- 14.Dai, W. 2002. When duplicated centrosomes won't split. Cell Cycle 1181-182. [PubMed] [Google Scholar]
- 15.Dai, W., Q. Wang, and F. Traganos. 2002. Polo-like kinases and centrosome regulation. Oncogene 216195-6200. [DOI] [PubMed] [Google Scholar]
- 16.Donohue, P. J., G. F. Alberts, Y. Guo, and J. A. Winkles. 1995. Identification by targeted differential display of an immediate early gene encoding a putative serine/threonine kinase. J. Biol. Chem. 27010351-10357. [DOI] [PubMed] [Google Scholar]
- 17.Duncan, P. I., N. Pollet, C. Niehrs, and E. A. Nigg. 2001. Cloning and characterization of Plx2 and Plx3, two additional Polo-like kinases from Xenopus laevis. Exp. Cell Res. 27078-87. [DOI] [PubMed] [Google Scholar]
- 18.Eckerdt, F., J. Yuan, and K. Strebhardt. 2005. Polo-like kinases and oncogenesis. Oncogene 24267-276. [DOI] [PubMed] [Google Scholar]
- 19.Elia, A. E., L. C. Cantley, and M. B. Yaffe. 2003. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 2991228-1231. [DOI] [PubMed] [Google Scholar]
- 20.Elia, A. E., P. Rellos, L. F. Haire, J. W. Chao, F. J. Ivins, K. Hoepker, D. Mohammad, L. C. Cantley, S. J. Smerdon, and M. B. Yaffe. 2003. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 11583-95. [DOI] [PubMed] [Google Scholar]
- 21.Fode, C., B. Motro, S. Yousefi, M. Heffernan, and J. W. Dennis. 1994. Sak, a murine protein-serine/threonine kinase that is related to the Drosophila polo kinase and involved in cell proliferation. Proc. Natl. Acad. Sci. USA 916388-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iida, M., M. Matsuda, and H. Komatani. 2008. Plk3 phosphorylates topoisomerase IIalpha at Thr(1342), a site that is not recognized by Plk1. Biochem. J. 41127-32. [DOI] [PubMed] [Google Scholar]
- 23.Keene, J. D., J. M. Komisarow, and M. B. Friedersdorf. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1302-307. [DOI] [PubMed] [Google Scholar]
- 24.Kitada, K., A. L. Johnson, L. H. Johnston, and A. Sugino. 1993. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol. Cell. Biol. 134445-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kontoyiannis, D., M. Pasparakis, T. T. Pizarro, F. Cominelli, and G. Kollias. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10387-398. [DOI] [PubMed] [Google Scholar]
- 26.Kumagai, A., and W. G. Dunphy. 1996. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science 2731377-1380. [DOI] [PubMed] [Google Scholar]
- 27.Lai, W. S., and P. J. Blackshear. 2001. Interactions of CCCH zinc finger proteins with mRNA: tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. J. Biol. Chem. 27623144-23154. [DOI] [PubMed] [Google Scholar]
- 28.Lai, W. S., E. Carballo, J. R. Strum, E. A. Kennington, R. S. Phillips, and P. J. Blackshear. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 194311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 27517827-17837. [DOI] [PubMed] [Google Scholar]
- 30.Lai, W. S., E. A. Kennington, and P. J. Blackshear. 2002. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 2779606-9613. [DOI] [PubMed] [Google Scholar]
- 31.Lai, W. S., J. S. Parker, S. F. Grissom, D. J. Stumpo, and P. J. Blackshear. 2006. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 269196-9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai, W. S., M. J. Thompson, and P. J. Blackshear. 1998. Characteristics of the intron involvement in the mitogen-induced expression of Zfp-36. J. Biol. Chem. 273506-517. [DOI] [PubMed] [Google Scholar]
- 33.Leung, G. C., J. W. Hudson, A. Kozarova, A. Davidson, J. W. Dennis, and F. Sicheri. 2002. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat. Struct. Biol. 9719-724. [DOI] [PubMed] [Google Scholar]
- 34.Li, B., B. Ouyang, H. Pan, P. T. Reissmann, D. J. Slamon, R. Arceci, L. Lu, and W. Dai. 1996. Prk, a cytokine-inducible human protein serine/threonine kinase whose expression appears to be down-regulated in lung carcinomas. J. Biol. Chem. 27119402-19408. [DOI] [PubMed] [Google Scholar]
- 35.Linker, K., A. Pautz, M. Fechir, T. Hubrich, J. Greeve, and H. Kleinert. 2005. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 334813-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llamazares, S., A. Moreira, A. Tavares, C. Girdham, B. A. Spruce, C. Gonzalez, R. E. Karess, D. M. Glover, and C. E. Sunkel. 1991. Polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 52153-2165. [DOI] [PubMed] [Google Scholar]
- 37.Mili, S., and J. A. Steitz. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 101692-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogilvie, R. L., M. Abelson, H. H. Hau, I. Vlasova, P. J. Blackshear, and P. R. Bohjanen. 2005. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 174953-961. [DOI] [PubMed] [Google Scholar]
- 39.Ohkura, H., I. M. Hagan, and D. M. Glover. 1995. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 91059-1073. [DOI] [PubMed] [Google Scholar]
- 40.Ouyang, B., H. Pan, L. Lu, J. Li, P. Stambrook, B. Li, and W. Dai. 1997. Human Prk is a conserved protein serine/threonine kinase involved in regulating M phase functions. J. Biol. Chem. 27228646-28651. [DOI] [PubMed] [Google Scholar]
- 41.Ouyang, B., Y. Wang, and D. Wei. 1999. Caenorhabditis elegans contains structural homologs of human prk and plk. DNA Seq. 10109-113. [DOI] [PubMed] [Google Scholar]
- 42.Ruan, Q., Q. Wang, S. Xie, Y. Fang, Z. Darzynkiewicz, K. Guan, M. Jhanwar-Uniyal, and W. Dai. 2004. Polo-like kinase 3 is Golgi localized and involved in regulating Golgi fragmentation during the cell cycle. Exp. Cell Res. 29451-59. [DOI] [PubMed] [Google Scholar]
- 43.Simmons, D. L., B. G. Neel, R. Stevens, G. Evett, and R. L. Erikson. 1992. Identification of an early-growth-response gene encoding a novel putative protein kinase. Mol. Cell. Biol. 124164-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, M. R., M. L. Wilson, R. Hamanaka, D. Chase, H. Kung, D. L. Longo, and D. K. Ferris. 1997. Malignant transformation of mammalian cells initiated by constitutive expression of the polo-like kinase. Biochem. Biophys. Res. Commun. 234397-405. [DOI] [PubMed] [Google Scholar]
- 45.Stoecklin, G., S. A. Tenenbaum, T. Mayo, S. V. Chittur, A. D. George, T. E. Baroni, P. J. Blackshear, and P. Anderson. 2008. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J. Biol. Chem. 28311689-11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stumpo, D. J., N. A. Byrd, R. S. Phillips, S. Ghosh, R. R. Maronpot, T. Castranio, E. N. Meyers, Y. Mishina, and P. J. Blackshear. 2004. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the tristetraprolin family. Mol. Cell. Biol. 246445-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, G. A., E. Carballo, D. M. Lee, W. S. Lai, M. J. Thompson, D. D. Patel, D. I. Schenkman, G. S. Gilkeson, H. E. Broxmeyer, B. F. Haynes, and P. J. Blackshear. 1996. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4445-454. [DOI] [PubMed] [Google Scholar]
- 48.van de Weerdt, B. C., and R. H. Medema. 2006. Polo-like kinases: a team in control of the division. Cell Cycle 5853-864. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Q., S. Xie, J. Chen, K. Fukasawa, U. Naik, F. Traganos, Z. Darzynkiewicz, M. Jhanwar-Uniyal, and W. Dai. 2002. Cell cycle arrest and apoptosis induced by human Polo-like kinase 3 is mediated through perturbation of microtubule integrity. Mol. Cell. Biol. 223450-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkles, J. A., and G. F. Alberts. 2005. Differential regulation of polo-like kinase 1, 2, 3, and 4 gene expression in mammalian cells and tissues. Oncogene 24260-266. [DOI] [PubMed] [Google Scholar]
- 51.Xie, S., H. Wu, Q. Wang, J. P. Cogswell, I. Husain, C. Conn, P. Stambrook, M. Jhanwar-Uniyal, and W. Dai. 2001. Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J. Biol. Chem. 27643305-43312. [DOI] [PubMed] [Google Scholar]
- 52.Yang, Y., J. Bai, R. Shen, S. A. Brown, E. Komissarova, Y. Huang, N. Jiang, G. F. Alberts, M. Costa, L. Lu, J. A. Winkles, and W. Dai. 2008. Polo-like kinase 3 functions as a tumor suppressor and is a negative regulator of hypoxia-inducible factor-1 alpha under hypoxic conditions. Cancer Res. 684077-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, T., V. Kruys, G. Huez, and C. Gueydan. 2002. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem. Soc. Trans. 30952-958. [DOI] [PubMed] [Google Scholar]
- 54.Zimmerman, W. C., and R. L. Erikson. 2007. Polo-like kinase 3 is required for entry into S phase. Proc. Natl. Acad. Sci. USA 1041847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.