Abstract
Iron regulatory protein 2 (IRP2) is an RNA-binding protein that regulates the posttranscriptional expression of proteins required for iron homeostasis such as ferritin and transferrin receptor 1. IRP2 RNA-binding activity is primarily regulated by iron-mediated proteasomal degradation, but studies have suggested that IRP2 RNA binding is also regulated by thiol oxidation. We generated a model of IRP2 bound to RNA and found that two cysteines (C512 and C516) are predicted to lie in the RNA-binding cleft. Site-directed mutagenesis and thiol modification show that, while IRP2 C512 and C516 do not directly interact with RNA, both cysteines are located within the RNA-binding cleft and must be unmodified/reduced for IRP2-RNA interactions. Oxidative stress induced by cellular glucose deprivation reduces the RNA-binding activity of IRP2 but not IRP2-C512S or IRP2-C516S, consistent with the formation of a disulfide bond between IRP2 C512 and C516 during oxidative stress. Decreased IRP2 RNA binding is correlated with reduced transferrin receptor 1 mRNA abundance. These studies provide insight into the structural basis for IRP2-RNA interactions and reveal an iron-independent mechanism for regulating iron homeostasis through the redox regulation of IRP2 cysteines.
Iron is an essential nutrient required for a variety of cellular processes, including DNA synthesis, respiration, and heme biosynthesis. However, ferrous iron readily reacts with hydroperoxides to produce hydroxyl radicals that can cause cellular damage. As both iron excess and deficiency are deleterious, cells have developed mechanisms to ensure that iron levels are sufficient for cellular need but at the same time limit iron toxicity.
Iron regulatory proteins 1 and 2 (IRP1 and IRP2) are the key iron sensors in mammalian cells (32, 48). IRPs are cytosolic proteins that bind RNA stem-loops known as iron-responsive elements (IREs) located in the 5′ or 3′ untranslated regions of mRNAs encoding proteins involved in iron homeostasis. IRP binding to a 5′ IRE present in ferritin (iron storage) or ferroportin (iron exporter) mRNAs represses protein translation. IRP binding to 3′ IREs, such as those in transferrin receptor 1 (TfR-1) and divalent metal transporter 1 (DMT1) (iron importers), stabilizes the mRNA and increases protein expression. While both IRPs function as RNA-binding proteins when iron content is low, increased cellular iron regulates IRP1 and IRP2 by different mechanisms. Increased cellular iron results in the assembly of an [4Fe-4S] cluster in the RNA-binding cleft of IRP1, which allows IRP1 to function as a cytosolic aconitase. Unlike IRP1, IRP2 does not coordinate an [4Fe-4S] cluster or function as an aconitase. Instead, the RNA-binding activity of IRP2 is reduced by iron-dependent polyubiquitylation and proteasomal degradation. The role of IRP2 as the predominant RNA-binding protein in vivo has been established in murine knockout models in which Irp1−/− mice display no overt phenotype, whereas Irp2−/− mice develop microcytic anemia and locomotor deficits (8, 13, 14, 28).
Several studies have shown that IRP2 RNA-binding activity is sensitive to perturbations in cellular redox status in animal models and cultured cells. Rats injected with the glutathione synthesis inhibitor phorone showed a decrease in IRP2 RNA-binding activity within 3 h of treatment that was partially reversed after treating extracts with a reductant (5). Another study showed decreased IRP2 RNA-binding activity within 1 h in an ischemia/reperfusion model in rats (46). Loss of IRP2 RNA-binding activity in this model could be blocked with the thiol antioxidant N-acetylcysteine, indicating that oxidative modification of thiols was the likely cause of reduced IRP2 RNA binding. In addition, studies carried out with cultured cells have reported the presence of an inactive RNA-binding form of IRP2 that could be activated by treating extracts with a reductant (19, 22, 42). Taken together, these studies indicate that IRP2 contains critical cysteines that regulate IRP2 RNA-binding activity during oxidative stress in vivo. The identity of these cysteines and the mechanism(s) by which IRP2 RNA binding is regulated have not been determined.
In this study, we explore the role of cysteines in regulating IRP2 iron-mediated degradation and RNA-binding activity. While we find that cysteines do not regulate the iron-dependent degradation of IRP2, our data indicate that two cysteines, C512 and C516, are located in the terminal-loop binding pocket and lie in close proximity to the terminal loop of the IRE. We also show that IRP2 RNA-binding activity can be regulated by changes in cellular redox through the oxidation and reduction of IRP2 C512 and C516. These data reveal structural elements that may regulate the selectivity of IRP2-IRE interactions. In addition, our studies reveal an iron-independent mechanism for regulating IRP2 RNA-binding activity during oxidative stress.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis.
Coding sequences for wild-type human IRP2 and mouse/human IRP1 (composed of the first 256 bp of the IRP1 mouse coding sequence) containing two FLAG (FLAG2 or FLAG) sequences (GACTACAAGGATGACGACGATAAG) at the 5′ end were cloned into pcDNA5/FRT/TO (Invitrogen) using PmeI restriction sites and screened for directionality. Cysteine mutants were generated from wild-type FLAG2-IRP2 or FLAG2-IRP1 vectors by following the QuikChange II site-directed mutagenesis kit (Stratagene) protocol. Primers used for mutagenesis are listed in Table S1 in the supplemental material. Sequencing confirmed the presence of the desired mutations, and each construct was subcloned into an unmutagenized vector. Redox-sensitive green fluorescent protein (roGFP2) cDNA was obtained from S. James Remington (University of Oregon).
Cell culture and nutrient deprivation assay.
All cell culture reagents (unless otherwise indicated) were obtained from Invitrogen. HEK293 cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 4% heat-inactivated fetal bovine serum (FBS), 100 μg/ml penicillin, and 100 μg/ml streptomycin at 37°C in ambient air with 5% CO2. For transient transfections, one 10-cm plate of HEK293 cells was transfected with 1 μg DNA for 4 h in Opti-MEM using Lipofectamine 2000. Stable cell lines were generated by following the manufacturer's guidelines using the Flp-In TREx HEK293 system (Invitrogen) and cultured in DMEM containing 9% FBS, 100 μg/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml hygromycin, and 15 μg/ml blasticidin.
For nutrient deprivation and add back experiments, HEK293 cells were plated on poly-d-lysine hydrobromide (Sigma)-treated plates to obtain cells at 50 to 60% confluence the following day and pretreated overnight with or without 100 μM desferrioxamine mesylate (DFO) (Sigma). Pretreated cells were washed twice with phosphate-buffered saline (PBS) and then treated with serum- and glucose-free medium (Invitrogen catalog no. 11996-025) for 0 to 8 h. In some experiments, serum- and glucose-free medium was supplemented with either 4.5 mg/ml (∼25 mM) d-glucose (Sigma), 4% FBS, or 4% dialyzed FBS (1,500-molecular-weight cutoff) with or without 100 μM DFO. Cells were harvested in Triton buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, and a cocktail of protease inhibitors [Roche]), and whole-cell lysates were cleared by centrifugation at 12,000 × g for 10 min.
Immunoblot analysis.
For immunoblotting, whole-cell lysate was boiled in lithium dodecyl sulfate sample buffer (Invitrogen) and then analyzed by 8% or 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to a Hybond-ECL nitrocellulose membrane (Amersham) and probed with the following antibodies: FLAG M2 monoclonal antibody (MAb; Sigma), transferrin receptor 1 MAb (Zymed), actin MAb (Calbiochem), β-tubulin MAb (Zymed), chicken anti-rat IRP1 polyclonal antibody (52), or rabbit anti-rat IRP2 polyclonal antibody (16). Horseradish peroxidase-conjugated secondary antibodies were bound and proteins were visualized using Western Lighting Chemiluminescence Reagent Plus (PerkinElmer Life Sciences). Membranes were stripped for 10 min at ∼65°C in stripping buffer (62.5 mM Tris-HCl [pH 6.8], 100 mM β-mercaptoethanol [β-ME], and 2% SDS).
Protein degradation assays.
Stable Flp-In TREx HEK293 cell lines expressing FLAG-recombinant IRPs were induced overnight with 1 μg/ml tetracycline. Induced cells were then washed twice in PBS and chased in complete medium supplemented with 100 μg/ml ferric ammonium citrate (FAC) or 100 μM DFO. For dot blot assays, whole-cell lysate (5 μg) was dot blotted onto a Hybond-ECL nitrocellulose membrane in triplicate. Immunoblotting was performed with FLAG M2 MAb and immunofluorescent secondary mouse antibody (Rockland), and then quantification was performed with an Odyssey infrared imaging system (Licor). For analysis by SDS-PAGE, whole-cell lysate was analyzed by 8% SDS-PAGE and immunoblotted as described above. Protein half-life (t1/2) was determined by linear regression.
RNA EMSA.
Electrophoretic mobility shift assay (EMSA) analysis was performed as previously described (16) with minor modifications. Briefly, whole-cell lysate (10 μg) was incubated with a 32P-labeled ferritin-L IRE probe for 10 min at room temperature. Heparin (Sigma) (50 μg/μl) and RNase T1 (Roche) (1 U/μl) were added simultaneously to the lysates for 10 min, followed by the addition of FLAG or IRP2 antibody for an additional 10 min to supershift FLAG-tagged recombinant IRP or endogenous IRP2 protein, respectively. Samples were analyzed on 5% nondenaturing polyacrylamide gels. The gels were dried and exposed to a PhosphorImager screen for quantification. Samples were incubated with diamide (Sigma), N-ethylmaleimide (NEM; Sigma), or iodoacetamide (IAM; Sigma) for 10 min at 4°C prior to or after the addition of the 32P-labeled IRE probe. β-ME (Sigma) was added to lysates at a final concentration of 1% or 0.5% to activate latent IRP1 or IRP2, respectively, prior to the addition of the 32P-labeled IRE (4).
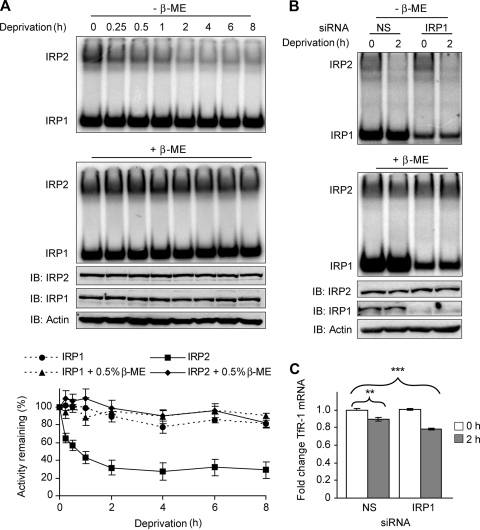
IRP1 siRNA and transferrin receptor 1 qRT-PCR.
HEK293 cells (3 × 105) were plated overnight on a 35-mm plate in DMEM containing 10% FBS without penicillin or streptomycin. The following day cells were transfected with 100 nM of either nonspecific (Dharmacon D-001206-13) or IRP1 (Dharmacon L-010037-01) small interfering RNA (siRNA) with 5 μl DharmaFECT 1 for 24 h by following the manufacturer's protocol. Cells were then trypsinized and plated in complete medium containing 4% FBS for ∼60 h. Cells were again trypsinized and plated at 50 to 60% confluence overnight in medium containing 10% FBS. Nutrient deprivation was performed as described above, and then cells were harvested in Triton buffer for EMSA and immunoblotting analyses or Trizol for quantitative reverse transcription-PCR (qRT-PCR) analysis of TfR-1 mRNA as described previously (49).
Fluorescence-activated cell sorter (FACS) analysis.
HEK293 cells transiently transfected with roGFP2 and treated with nutrient deprivation as described above were trypsinized and washed twice with PBS prior to analysis. Cells were excited with laser lines at 405 nm and 488 nm on a FACSCanto II (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star, Inc.).
Modeling of IRP1 and IRP2 structure.
Models for IRP1 and IRP2 were generated using the first approach mode in SWISS-MODEL (44). Sequences for human IRP1 and human IRP2 without the 73-amino-acid (aa) region were submitted for modeling using the Protein Data Bank file 2ipy (47) as a template. Graphic manipulation was performed with USCF Chimera (36).
Statistics.
A two-tailed Student t test was performed to determine statistical significance.
RESULTS
Location of critical IRP2 cysteine residues and predicted model of IRP2 in complex with ferritin-H RNA.
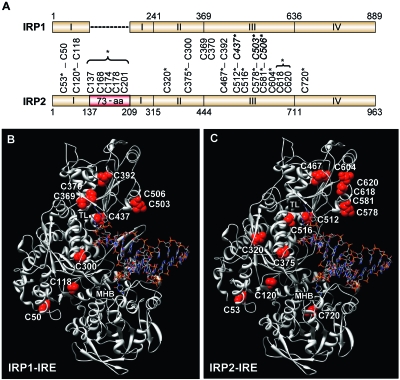
Human IRP1 contains nine cysteine residues, three of which (C437, C503, and C506) coordinate [4Fe-4S] cluster binding (Fig. 1A). Thiol mutagenesis and modification analyses have revealed that IRP1 C437 is critical for not only [4Fe-4S] coordination but also IRP1 RNA binding and protein stability (7, 23, 38, 51). Sequence alignment between IRP1 and IRP2 shows that seven of the nine cysteine residues in human IRP1 are conserved in human IRP2, including the three cysteines required for IRP1 [4Fe-4S] cluster binding (IRP2 C512, C578, and C581) (Fig. 1A). Five of the 18 cysteines in IRP2 are located in a 73-aa region that is not required for RNA binding or iron degradation (3, 17, 50, 53). Whether the other 13 cysteine residues have a role in IRP2 RNA-binding activity or iron degradation has not been determined.
FIG. 1.
Location and structural comparison of cysteine residues in IRP1 and IRP2. (A) Schematic diagram comparing cysteine residues in human IRP1 and IRP2. Conserved cysteines are connected by vertical lines. IRP1 cysteines required for [4Fe-4S] cluster binding are italicized and in boldface (C437, C503, and C506). The 73-aa region is in red, and the asterisk indicates that the five cysteines were mutated in one construct. Asterisks denote mutated cysteine residues. (B and C) Structure for human IRP1 (B) and predicted structure of human IRP2 (without the 73-aa region) (C) bound to the ferritin-H IRE. Both IRPs show IRE contacts at the terminal loop (TL) and midhelix bulge (MHB). Cysteine residues are indicated. The five cysteines located in the 73-aa region of IRP2 (C137, C168, C174, C178, and C201) are not included in the model.
To characterize the role of cysteines in regulating IRP2 functions, all 18 cysteines were mutated to serine either individually or in combination (Fig. 1A). As controls, we generated IRP1 [4Fe-4S] cluster binding mutants (IRP1-C437S, -C503S, -C506S, -C503/506S, and -C437/503/506S). To predict how specific cysteine residues may contribute to IRP2 RNA binding, we used the crystal structure of IRP1 in complex with the ferritin-H IRE (Fig. 1B) (47) to model an IRP2-RNA complex (Fig. 1C). Our model predicts that IRP2 C512 (the counterpart of IRP1 C437) and C516, which is unique to IRP2, lie within the terminal-loop binding cleft and suggests that these two cysteines may have a role in regulating IRP2 RNA binding.
All IRP2 cysteine mutants are sensitive to iron-dependent degradation.
We first determined whether cysteine residues regulate the iron-dependent degradation of IRP2. HEK293 cells stably expressing FLAG-tagged wild-type IRP1 (IRP1-wt), IRP2-wt, or IRP cysteine mutants were induced with tetracycline overnight and then chased in medium containing the iron chelator DFO or FAC (see Fig. S1 in the supplemental material). As expected, the t1/2 of IRP2-wt decreased in the presence of FAC (t1/2 = 7.4 ± 0.2 h) whereas the t1/2 of IRP1-wt was >24 h in the presence of both DFO and FAC (Table 1). Mutation of cysteines required for IRP1 [4Fe-4S] binding (IRP1-C437S and IRP1-C437/503/506S) sensitized IRP1 to iron-induced degradation as previously reported (7, 51). Mutation of IRP2 C320 or C618/620 caused a slight increase in protein turnover in the absence of excess iron (t1/2 = 15.7 ± 0.3 and 16.6 ± 0.7 h, respectively), suggesting that these residues may be important for overall IRP2 protein stability. Importantly, all IRP2 cysteine mutants were sensitive to iron-dependent protein turnover at a rate similar to that for IRP2-wt. These data indicate that IRP2 cysteines do not have a role in iron-mediated degradation.
TABLE 1.
Degradation of FLAG-IRP1 and FLAG-IRP2 cysteine mutants
| Construct (n) | Mean t1/2 (h) ± SEMa for treatment with:
|
|
|---|---|---|
| DFO | FAC | |
| IRP1-wt (3) | >24 | >24* |
| IRP1-C437S (3) | >24 | 9.3 ± 0.7* |
| IRP1-C503S | NDb | ND |
| IRP1-C506S | ND | ND |
| IRP1-C503/506S | ND | ND |
| IRP1-C437/503/506S (3) | >24 | 7.8 ± 0.2 |
| IRP2-wt (7) | >24 | 7.4 ± 0.2 |
| IRP2-C53S (3) | >24 | 7.6 ± 0.3 |
| IRP2-C120S (3) | >24 | 8.0 ± 0.4 |
| IRP2-Δ73 (3) | >24 | 8.3 ± 0.6 |
| IRP2-C320S (6) | 15.7 ± 0.3* | 7.8 ± 0.3 |
| IRP2-C375S (3) | >24 | 8.1 ± 0.2 |
| IRP2-C467S (3) | >24 | 7.3 ± 0.3 |
| IRP2-C512/516S (3) | >24 | 7.3 ± 0.1 |
| IRP2-C512/578/581S (3) | >24 | 6.2 ± 0.1* |
| IRP2-C512/516/578/581S (3) | >24 | 7.1 ± 0.4 |
| IRP2-C516S (3) | >24 | 7.0 ± 0.1 |
| IRP2-C604S (3) | >24 | 7.4 ± 0.3 |
| IRP2-C618/620S (6) | 16.6 ± 0.7* | 6.4 ± 0.3* |
| IRP2-C720S (3) | >24 | 7.2 ± 0.2 |
Significance was determined on the basis of comparison to IRP2-wt for DFO or FAC treatments; *, P < 0.02.
ND, not determined.
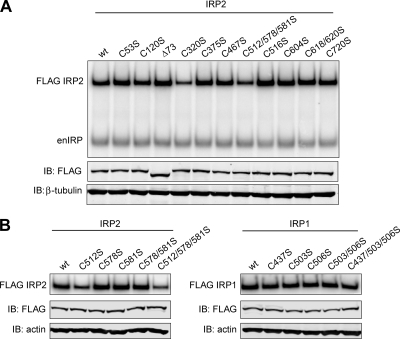
IRP2 C320 and C512 are required for optimal RNA binding.
IRP2 RNA-binding activity can be maximally activated in vitro with the addition of 0.5% β-ME, indicating a role for cysteines in regulating IRP2-IRE interactions (4). To identify critical cysteines, EMSA analysis was performed on IRP2 cysteine mutants using a 32P-labeled ferritin-L IRE probe. Of the 11 IRP2 cysteine mutants tested, IRP2-C320S and IRP2-C512/578/581S showed ∼50% and ∼40% reduction, respectively, in RNA-binding activity compared to IRP2-wt even in the presence of 0.5% β-ME (Fig. 2A). The reduction in RNA-binding activity of IRP2-C320S is likely due to its inherent instability (Table 1). The reduction in IRP2-C512/578/581S RNA-binding activity is interesting given that IRP2 C512 is predicted to lie within the IRE terminal-loop binding pocket (Fig. 1C). EMSA analysis of single mutants from this region revealed that mutation of IRP2 C512 alone reduced the RNA-binding activity of IRP2, similar to results for the IRP2-C512/578/581S mutant (Fig. 2B, left). In contrast, mutation of one or all of these conserved cysteines residues in IRP1 (C437, C503, and/or C506) did not affect RNA binding (Fig. 2B, right), as previously shown (23, 38). These data suggest that the thiol group of IRP2 C512 is necessary for optimal IRP2-IRE interactions and indicate that the terminal-loop binding pocket of IRP2 is more sensitive to thiol mutagenesis than that of IRP1.
FIG. 2.
Relative RNA-binding activities of IRP1 and IRP2 cysteine mutants. (A and B) Flp-In TREx HEK293 cells stably expressing FLAG-tagged proteins were treated with tetracycline overnight to induce protein expression. RNA-binding activity was measured by EMSA by incubating cell lysates (10 μg) with a 32P-labeled ferritin-L IRE probe. Lysates from cells expressing IRP2 and IRP1 recombinant proteins were treated with 0.5% and 1% β-ME, respectively. (A) Cell lysates (12 μg) were immunoblotted (IB) and then probed simultaneously with FLAG and β-tubulin or actin antibodies. FLAG antibody was used to supershift recombinant IRP2-IRE complexes (FLAG IRP2) away from endogenous IRP-IRE complexes (enIRP). (B) Only FLAG-supershifted IRP-IRE complexes are shown (FLAG IRP2; FLAG IRP1).
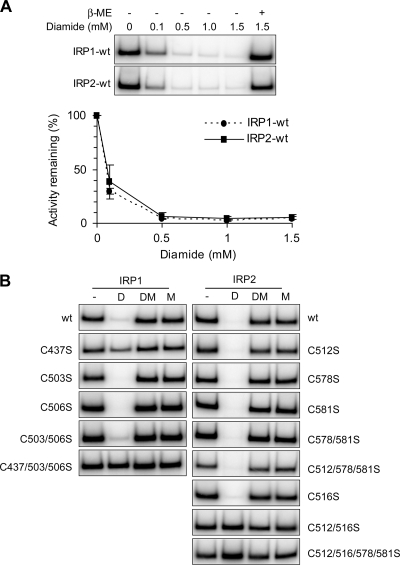
In vitro modification shows that IRP2 C512 and C516 must be reduced and unmodified to bind RNA.
In vitro modification of IRP1 C437 by diamide or NEM inhibits IRP1 RNA binding (23, 38). IRP2 RNA-binding activity is also decreased with diamide and NEM, but the residue(s) sensitive to modification has not been identified (19, 37). To determine if the conserved IRP1 C437 residue in IRP2 (C512) and/or other cysteine residues are sensitive to thiol-modifying reagents, lysates from cells expressing IRP1-wt, IRP2-wt, or IRP cysteine mutants were treated with the reversible thiol-oxidizing reagent diamide and RNA-binding activity was determined by EMSA. A diamide titration revealed that the RNA-binding activity was maximally reduced for both IRP1-wt and IRP2-wt by 0.5 mM diamide and could be reversed by β-ME treatment (Fig. 3A). Extracts from cells expressing IRP cysteine mutants were treated with 0.5 mM diamide alone, 0.5 mM diamide followed by β-ME, or β-ME alone prior to EMSA analysis (Fig. 3B). Consistent with previous studies (23, 38), mutation of IRP1 C437 alone was sufficient to eliminate IRP1 diamide sensitivity (Fig. 3B, left). In contrast, the respective IRP2-C512S mutant was sensitive to diamide treatment (Fig. 3B, right). Analysis of other IRP2 cysteine mutants showed that mutation of both IRP2 C512 and C516 was required to eliminate IRP2 diamide sensitivity (Fig. 3B, right; see Fig. S2 in the supplemental material). These data indicate that both IRP2 C512 and C516 must be reduced for IRP2 to interact with RNA.
FIG. 3.
Sensitivity of IRP1 and IRP2 cysteine mutants to diamide. Flp-In TREx HEK293 cells stably expressing FLAG-tagged IRPs were treated with tetracycline overnight to induce protein expression. (A) Cell lysates (10 μg) were treated with increasing concentrations of diamide, with or without β-ME (IRP1, 1%; IRP2, 0.5%) prior to EMSA. Quantification of three independent experiments is shown. Error bars represent the standard errors of the means. (B) Cell lysates (10 μg) were untreated (−) or treated with 0.5 mM diamide (D), 0.5 mM diamide followed by β-ME (DM), or β-ME alone (M) prior to EMSA. Only FLAG-supershifted IRP-IRE complexes are shown.
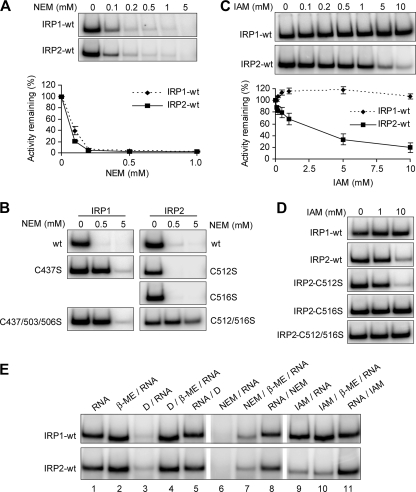
We next determined the sensitivity of IRP2 to the nonreversible alkylating reagent NEM. Similar to results for diamide, we observed that IRP1-wt and IRP2-wt were equally sensitive to increasing concentrations of NEM (Fig. 4A). To identify IRP2 cysteines responsible for NEM sensitivity, we treated lysates from cells expressing IRP1 or IRP2 cysteine mutants with low (0.5 mM) or high (5 mM) concentrations of NEM. Mutation of only IRP1 C437 eliminated IRP1 sensitivity to 0.5 mM NEM, while all IRP1 cysteine mutants remained sensitive at 5 mM NEM (Fig. 4B, left). Similar to what was found for diamide treatment, mutation of both IRP2 C512 and C516 was required to eliminate IRP2 NEM sensitivity (Fig. 4B, right). In contrast to IRP1 cysteine mutants, IRP2-C512/C516S showed reduced sensitivity to both low and high concentrations of NEM (Fig. 4B, right). These data are consistent with our diamide studies and demonstrate that both IRP2 and C516 must be unmodified to interact with RNA.
FIG. 4.
Sensitivity of IRP1 and IRP2 cysteine mutants to NEM and IAM. Flp-In TREx HEK293 cells stably expressing FLAG-tagged IRPs were treated with tetracycline overnight to induce protein expression. Cell lysates (10 μg) were treated with the indicated concentrations of NEM (A and B) or IAM (C and D) prior to EMSA. Quantification of three independent experiments is shown for panels A and C. Error bars represent the standard errors of the means. (E) EMSA was performed on cell lysates (10 μg) containing either IRP1-wt or IRP2-wt treated with β-ME (IRP1, 1%; IRP2, 0.5%), 0.5 mM diamide (D), 5 mM NEM, and/or 10 mM IAM in the order indicated by each label. Only FLAG-supershifted recombinant IRP-IRE complexes are shown.
Diamide and NEM both contain bulky side groups that may inhibit RNA-protein interactions via steric interference or distortion of the protein backbone. We therefore treated lysates containing IRP1-wt or IRP2-wt with the nonreversible alkylating reagent IAM, which contains a smaller side group than diamide or NEM. Consistent with previous studies, IRP1-wt RNA-binding activity was not sensitive to IAM treatment (Fig. 4C, top) (23, 38). In contrast, IRP2-wt RNA-binding activity was significantly decreased with increasing concentrations of IAM (Fig. 4C, bottom). Analysis of IRP2 cysteine mutants revealed that mutation of only IRP2 C516 was required to eliminate IAM sensitivity, indicating that IAM modification of IRP2 C516 but not IRP2 C512 disrupts IRP2 RNA binding (Fig. 4D). We attempted to confer IAM sensitivity to IRP1 by generating an IRP1-S441C (the counterpart to IRP2 C516) mutant, but this protein was RNA binding and aconitase deficient (see Fig. S3 in the supplemental material). These data indicate that the presence of an additional cysteine (IRP2 C516) within the RNA-binding pocket of IRP2 renders IRP2 sensitive to IAM modification and suggest significant structural differences in the pockets of IRP2 and IRP1.
Our data suggest that IRP2 C512 and C516 are located in close proximity to bases in the terminal loop of the IRE. We predict that binding RNA prior to treatment with oxidizing or alkylating reagents should protect IRP2 C512 and C516 from thiol modification. Extracts containing recombinant IRP1-wt or IRP2-wt were therefore prebound to RNA prior to treatment with diamide (0.5 mM), NEM (5 mM), or IAM (10 mM). In all cases, prebinding of IRP1-wt or IRP2-wt to RNA prevented sensitivity to these reagents (Fig. 4E, lanes 5, 8, and 11). Collectively, our in vitro analyses indicate that IRP2 C512 and C516 are located within the terminal-loop binding pocket and lie in close proximity to bases of the terminal loop but that neither cysteine directly interacts with the RNA since IAM modification of IRP2 C512 or mutation of IRP2 C516 does not disrupt RNA binding.
IRP2 RNA-binding activity is decreased during nutrient deprivation.
Some thiol-containing proteins use cysteine residues to rapidly and reversibly modify their activities in response to changes in cellular oxidative status (34). To characterize the role of cysteines in regulating IRP2 RNA binding in cells, we used glucose and serum deprivation (nutrient deprivation) to induce oxidative stress in cells. Serum and/or glucose deprivation has been shown to generate oxidative stress within 15 min after nutrient removal in cultured cells (24, 43). HEK293 cells were treated with glucose- and serum-free medium containing DFO (to prevent reactive oxygen species [ROS] production via Fenton chemistry) for up to 8 h. IRP2 RNA-binding activity was reduced by ∼40% after 15 min of nutrient deprivation and maximally decreased by ∼70% at 2 h (Fig. 5A, top and graph). Similar results were observed in COS-1 and HeLa cells (data not shown) and in HEK293 cells without DFO treatment (see Fig. S4 in the supplemental material). The reduction in IRP2 RNA binding was due to protein oxidation and not changes in IRP2 protein levels, as β-ME treatment restored IRP2 RNA-binding activity and immunoblotting showed no change in IRP2 abundance (Fig. 5A, middle and graph). IRP1 RNA-binding activity and protein levels remained relatively constant throughout the time course (Fig. 5A, top and graph). These data indicate that IRP2-RNA interactions are inhibited during nutrient deprivation due to thiol oxidation.
FIG. 5.
IRP2 RNA-binding activity and TfR-1 mRNA abundance are reduced during nutrient deprivation. (A) HEK293 cells were treated overnight with 100 μM DFO and then treated with DMEM lacking both serum and glucose (deprivation) that was supplemented with 100 μM DFO for the indicated times. Cell lysates (10 μg) were used for EMSA in the presence or absence of 0.5% β-ME. IRP2 protein was supershifted with IRP2 antibody. Cell lysates (20 μg) were immunoblotted (IB) and probed sequentially with IRP2, IRP1, and actin antibodies. Quantification of EMSA activity for three independent experiments is shown. (B) HEK293 cells were treated with nonspecific (NS) or IRP1-targeted siRNA duplexes, cultured overnight in DMEM containing 10% FBS, and then treated with DMEM lacking both serum and glucose (deprivation) for 2 h. EMSA and protein analyses were performed as for panel A. (C) HEK293 cells treated as for panel B were harvested in Trizol, and TfR-1 mRNA was quantified by qRT-PCR. Quantification of four independent experiments is shown. Significance was determined on the basis of comparison to NS-siRNA-treated cells without nutrient deprivation; **, P < 0.01; ***, P < 0.0001. Error bars in panels A and C represent the standard errors of the means.
We next questioned whether the loss of IRP2 RNA-binding activity was sufficient to alter the levels of an IRE-regulated mRNA such as TfR-1 mRNA. HEK293 cells contain high levels of IRP1 RNA-binding activity when grown under standard conditions, but in the low oxygen tension found in most tissues IRP1 is an aconitase (30, 31). Because the contribution of IRP2 RNA-binding activity in regulating TfR-1 mRNA stability may be masked by high levels of IRP1 RNA-binding activity, we treated cells with siRNA targeted to endogenous IRP1 and then quantified TfR-1 mRNA levels after 2 h of nutrient deprivation. EMSA showed an ∼70% reduction in total IRP1 RNA-binding activity in IRP1 siRNA-treated cells (Fig. 5B, top and middle). qRT-PCR analysis showed a 10% reduction in TfR-1 mRNA after nutrient deprivation in nonspecific-siRNA-treated cells and a 20% reduction in TfR-1 mRNA in IRP1 siRNA-treated cells (Fig. 5C). The reduction in mRNA levels was specific for TfR-1 mRNA, as ferritin-L mRNA levels were not decreased during nutrient deprivation (data not shown). These data suggest that loss of IRP2 RNA-binding activity during nutrient deprivation is associated with reduced TfR-1 mRNA abundance.
IRP2 C512 and C516 are oxidized during nutrient deprivation.
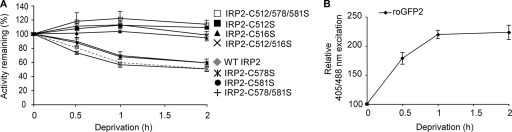
Our data indicate that the oxidation of a critical cysteine residue(s) during nutrient deprivation decreases IRP2 RNA binding. From our in vitro studies, we hypothesized that IRP2 C512 and/or C516 may be the critical cysteine(s). HEK293 cells were therefore transiently transfected with IRP2-wt or IRP2 cysteine mutants and the recombinant proteins were tested for sensitivity to nutrient deprivation. The RNA binding of IRP2-wt, IRP2-C578S, IRP2-C581S, and IRP2-C578/581S decreased during nutrient deprivation (Fig. 6A) and was restored by β-ME treatment (see Fig. S5 in the supplemental material). In contrast, the RNA-binding activities of IRP2-C512S, IRP2-C516S, IRP2-C512/516S, and IRP2-C512/578/581S did not decrease with nutrient deprivation (Fig. 6A; see Fig. S5 in the supplemental material). These data indicate that oxidation of both IRP2 C512 and C516 is required to inhibit the formation of an IRP2-RNA complex during nutrient deprivation. While we cannot exclude the possibility of individual thiol modification, our data suggest that a disulfide bond that inhibits IRP2-IRE interactions is formed between IRP2 C512 and C516.
FIG. 6.
IRP2 C512 and C516 are oxidized during nutrient deprivation. (A and B) HEK293 cells were transiently transfected with FLAG-tagged IRP2-wt or cysteine mutants (A) or roGFP2 (B), cultured overnight in DMEM containing 100 μM DFO, and then treated with DMEM lacking both serum and glucose (deprivation) that was supplemented with 100 μM DFO for the indicated times. (A) Cell lysates (10 μg) were used for EMSA. Quantification of three independent experiments for each recombinant protein is shown. (B) Cells were analyzed by FACS. The relative 405/488 nm excitation ratios for three independent experiments are shown. Error bars represent the standard errors of the means.
To confirm the presence of oxidative stress during nutrient deprivation and to determine if a disulfide bond can be formed under these conditions, we assayed the effect of nutrient deprivation on a redox-sensitive green fluorescent protein (roGFP2) (10, 18). roGFP2 is a cytosolic protein that contains two cysteine residues located on the surface of the protein near the chromophore. Oxidative stress induces the formation of a reversible disulfide bond between these cysteines that causes protonation of the chromophore, resulting in an increased excitation spectrum at 405 nm with a compensatory loss at 488 nm. HEK293 cells were transiently transfected with roGFP2 and treated with nutrient deprivation, and the 405/488-nm excitation ratio was determined by FACS analysis. We observed a significant increase in roGFP2 oxidation after 30 min of nutrient deprivation, with a maximal increase of greater than twofold occurring at ∼2 h (Fig. 6B). These data indicate that nutrient deprivation generates oxidative stress that can induce the formation of a disulfide bond in a redox-sensitive protein.
IRP2 RNA-binding activity is sensitive to changes in intracellular glucose.
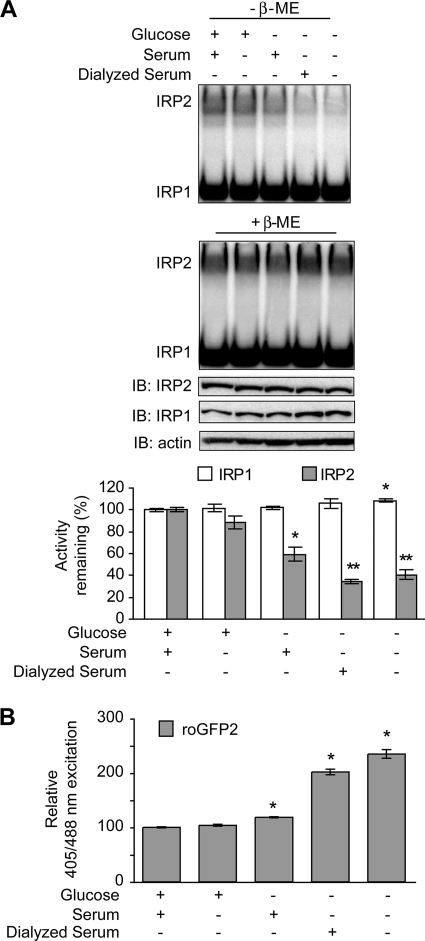
We next determined whether either serum or glucose withdrawal alone could reduce IRP2 RNA-binding activity. HEK293 cells were treated for 2 h in complete medium (containing both glucose and serum) or medium lacking either glucose or serum or lacking both glucose and serum. Serum deprivation alone did not significantly alter IRP2 RNA-binding activity (Fig. 7A, top and graph). In contrast, glucose deprivation resulted in an ∼40% decrease in IRP2 RNA binding, while removal of both glucose and serum reduced IRP2 RNA-binding activity by ∼70%. The modest reduction in IRP2 RNA binding observed with glucose deprivation alone compared to glucose and serum deprivation may be due to the presence of residual glucose found in serum (∼1 μg/ml). When cells were treated with dialyzed serum, IRP2 RNA binding was reduced and the reduction was similar to that caused by glucose and serum deprivation. Oxidative stress, as assayed by determining roGFP2 oxidation, was found to be increased under conditions of glucose, but not serum, deprivation (Fig. 7B). These data indicate that glucose withdrawal alone causes oxidative stress in HEK293 cells and the subsequent loss of IRP2 RNA-binding activity.
FIG. 7.
Glucose deprivation reduces IRP2 RNA binding. (A) HEK293 cells were cultured overnight in DMEM containing 100 μM DFO and then treated with DMEM lacking both serum and glucose that was supplemented with 100 μM DFO and d-glucose (4.5 mg/ml), serum (4%), or dialyzed serum (4%) for 2 h, as indicated. Cell lysates (10 μg) were used for EMSA in the presence or absence of 0.5% β-ME. IRP2 was supershifted with IRP2 antibody. Cell lysates (20 μg) were immunoblotted (IB) and probed sequentially with IRP2, IRP1, and actin antibodies. Quantification of EMSA activity for three independent experiments is shown. Significance was determined by comparing the RNA-binding activities of IRP1 or IRP2 in medium containing both glucose and serum to those of IRP1 or IRP2 in nutrient-depleted samples, respectively. *, P < 0.05; **, P < 0.005. (B) HEK293 cells were transiently transfected with roGFP2 and treated with nutrient deprivation as described for panel A. Cells were analyzed by FACS. The relative 405/488 nm excitation ratios for six independent experiments are shown. Significance was determined on the basis of comparison to cells treated with medium containing both glucose and serum; *, P < 0.0001. Error bars in panels B and C represent the standard errors of the means.
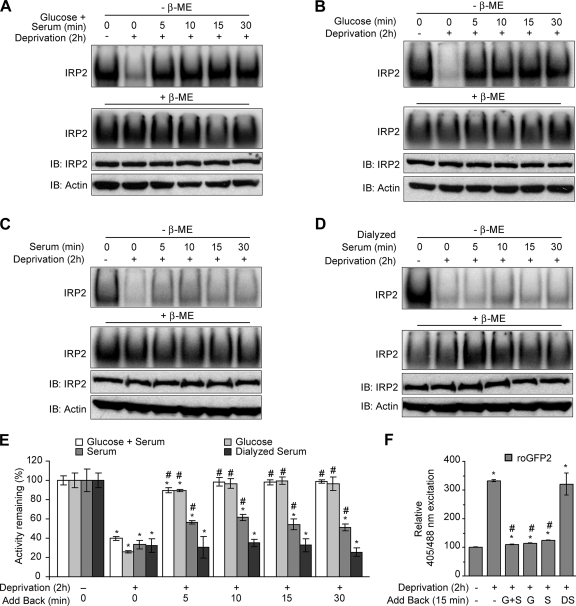
To determine if IRP2 RNA-binding activity could be restored in cells after nutrient deprivation, HEK293 cells were starved of both glucose and serum for 2 h and then glucose and serum, glucose alone, serum alone, or dialyzed serum alone was restored to cells for 5, 10, 15, or 30 min. EMSA analysis showed that addition of both glucose and serum or glucose alone completely recovered IRP2 RNA-binding activity by 10 min (Fig. 8A, B, and E). In contrast, the addition of serum alone only partially recovered IRP2 RNA-binding activity while the addition of dialyzed serum had no effect (Fig. 8C to E). Analysis of cellular redox using roGFP2 revealed that oxidative stress, induced by nutrient deprivation, was decreased after restoration of glucose and/or serum (which contains glucose) but not dialyzed serum (Fig. 8F). Taken together, these data indicate that IRP2 RNA binding is regulated in HEK293 cells by oxidative stress caused by glucose deprivation.
FIG. 8.
Glucose supplementation restores IRP2 RNA-binding activity after nutrient deprivation. (A to D) HEK293 cells were cultured overnight in DMEM containing 100 μM DFO and then treated with DMEM lacking both serum and glucose (deprivation) that was supplemented with 100 μM DFO for 2 h. (A) d-Glucose (4.5 mg/ml) and serum (4%), (B) d-glucose (4.5 mg/ml), (C) serum (4%), or (D) dialyzed serum (4%) was then restored to the medium for the indicated times. Cell lysates (10 μg) were used for EMSA in the presence or absence of 0.5% β-ME. Only IRP2 supershifted complexes are shown. Cell lysates (20 μg) were immunoblotted (IB) and probed sequentially with IRP2 and actin antibodies. (E) Quantification of EMSA activity for three independent experiments for panels A to D. Significance was determined within each supplementation group. *, P < 0.05 compared to no treatment; #, P < 0.05 compared to 2-h deprivation. (F) HEK293 cells were transiently transfected with roGFP2 and treated with nutrient deprivation for 2 h, and then glucose and serum (G+S), glucose (G), serum (S), or dialyzed serum (DS) was added back for 15 min at the concentrations indicated in panels A to D. Cells were analyzed by FACS. The relative 405/488 nm excitation ratios for three independent experiments are shown. Significance was determined on the basis of comparison to no treatment (*, P < 0.005) or 2-h deprivation (#, P < 0.001). Error bars in panels E and F represent the standard errors of the means.
DISCUSSION
Structural role for IRP2 C512 and C516 in IRE binding.
Our model of IRP2 in complex with the ferritin-H IRE predicts that IRP2 C512 and C516 are located in close proximity to the bases of the terminal loop within the binding pocket of IRP2. The data we report here verify this model. First, we show that modification of either IRP2 C512 or C516 with diamide or NEM eliminates IRP2 RNA binding. This is likely a result of steric interference with the IRE terminal loop or altered structure of the IRP2 terminal-loop binding pocket. Second, we show that mutation of IRP2 C512, but not IRP2 C516, to serine significantly reduces IRP2 RNA binding compared to that of IRP2-wt. However, modification of IRP2 C512 with IAM, which contains a smaller side group than diamide or NEM, does not affect RNA binding. These data indicate that neither IRP2 C512 nor IRP2 C516 directly interacts with the RNA but suggest that the substitution of a serine residue at IRP2 C512 may alter the chemical environment of the terminal-loop binding pocket. Finally, we show that modification of IRP2 C516 with IAM inhibits IRP2 RNA binding. These data suggest that bases of the terminal loop may be positioned closer to IRP2 C516 than to IRP2 C512 such that modification of IRP2 C516 with the small IAM group sterically inhibits formation of the IRP2-IRE complex. These studies also suggest that the structures of the terminal-loop binding pockets of IRP1 and IRP2 are different since the RNA-binding activity of IRP2 but not that of IRP1 is affected by thiol mutagenesis and IAM modification. Taking these results together, we propose that the presence of two cysteine residues within the terminal-loop binding pocket of IRP2 results in pocket that is more constricted than that in IRP1. We also predict that this constrained environment requires precise positioning of the IRE terminal loop within the binding pocket of IRP2 and therefore contributes to the selectivity of IRP2-IRE interactions (see below).
IRP1 and IRP2 IRE-binding selectivity.
Several studies have shown that IRE-containing mRNAs are not equally regulated by iron. For example, dietary iron supplementation in rats results in an ∼100-fold increase in ferritin protein in liver, whereas mitochondrial aconitase is increased only by ∼2-fold (6, 27, 41). In addition, ferritin protein and TfR-1 mRNA levels were significantly altered in IRP2−/− mice while ferroportin protein and DMT1 mRNA levels showed little change (13, 30). The differential regulation of IRE-containing mRNAs by IRP1 and IRP2 is likely due to differences in both IRE affinity and tissue-specific expression (12, 26, 29, 30).
IRP1 and IRP2 display different affinities to natural and synthetic IRE-containing mRNAs in vitro (21, 26). The differences in IRP1 and IRP2 RNA-binding affinity have been attributed to structural variations in the midhelix bulge, which is located in the stem of the IRE (Fig. 1B and C) (20, 21, 26). The midhelix bulge is composed of either a complex internal loop/bulge (IL/B) (present only in ferritin IREs) or a more simple C bulge (found in all other IREs) (39). IRP1 has been shown to promiscuously bind to both C bulge- and IL/B-containing IREs while IRP2 binds selectively to IL/B-containing IREs, indicating that an IL/B is critical for high-affinity IRP2 RNA binding (26). Nuclear magnetic resonance spectroscopy has shown that an IL/B is more flexible than a C bulge and that mutation of the IL/B is reflected in an altered conformation of the terminal loop (1, 15, 25). These studies suggest that the flexibility of the IL/B allows for an “induced fit” of the terminal loop within the binding pocket of the IRPs.
We hypothesize that the orientation of the IRE terminal loop within the binding pocket of the IRPs is also a critical determinant in IRE selectivity. The promiscuous binding of IRP1 to both IL/B and C bulge IREs is consistent with a more open terminal-loop binding pocket that is insensitive to thiol mutagenesis or IAM modification and tolerant to variances in the positioning of the terminal loop within the pocket (20, 21, 23, 38). Unlike IRP1, IRP2 shows increased sensitivity to thiol mutagenesis and IAM modification, suggesting that the binding pocket of IRP2 is more constricted and requires more precise orientation of the terminal loop within the pocket for high-affinity IRP2-IRE binding. This model is supported by studies showing that IRP2 is less tolerant to sequence and structural variations in the terminal loop of the IRE than IRP1 (20, 21, 25). Taking these results together, we suggest that IRP2 binds with high affinity to IL/B-containing IREs because the IL/B is required to efficiently position the terminal loop within the constricted environment of the terminal-loop binding pocket of IRP2.
Regulation of IRP2 RNA-binding activity during oxidative stress.
Our data suggest that during oxidative stress induced by glucose deprivation a disulfide bond that inhibits IRP2 RNA binding is formed between IRP2 C512 and C516. Dupuy et al. inferred from the crystal structure of IRP1 that IRP2 C512 and C516 may be close enough to form a disulfide bond (11). Due to technical difficulties with mass spectrometry, we were unable to confirm the presence of a disulfide bond in IRP2, but we hypothesize that a disulfide bridge between IRP2 C512 and C516 would close off the terminal-loop binding pocket and prevent insertion of the IRE terminal loop.
Glucose deprivation is a known inducer of oxidative stress (45). In several glucose-deprived cell lines, H2O2 and superoxide anion were shown to be elevated and reduced glutathione levels decreased (2, 45). These alterations are thought to be due in part to reduced production of NADPH via the pentose-phosphate pathway, which provides electrons for the reduction of glutathione and thioredoxin (45). These data suggest that H2O2 and/or superoxide anion may be responsible for IRP2 oxidation during glucose deprivation and that NADPH may be required to maintain IRP2 in a reduced state via reduction of glutathione or thioredoxin. However, we did not detect increased H2O2 levels in HEK293 cells during glucose deprivation using 2′,7′-dichlorofluorescein (data not shown). In addition, treatment with N-acetylcysteine (glutathione precursor) or buthionine sulfoximine (glutathione synthesis inhibitor) did not alter the effect of glucose deprivation or restoration on IRP2 RNA binding, respectively (data not shown). These data suggest that H2O2 and glutathione are not involved in regulating IRP2 RNA binding in nutrient-deprived HEK293 cells. This idea is consistent with in vitro studies showing that IRP2 RNA binding was not affected by H2O2 treatment, and that inactivation of IRP2 RNA binding by nitric oxide could be reversed with thioredoxin but not glutathione (33, 35). Future studies will focus on identifying ROS that oxidize IRP2 C512 and C516 during oxidative stress.
Oxidative stress induced by doxorubicin, phorone, or ischemia/reperfusion in rodents led to decreased IRP2 RNA-binding activity concomitant with increased ferritin and decreased TfR-1 protein abundance (5, 9, 46). Similarly, we find that the abundance of TfR-1 mRNA is decreased during oxidative stress induced by glucose deprivation. Taken together, these studies indicate that, during oxidative stress, IRP2 RNA-binding activity may be decreased to reduce iron uptake by TfR-1 and increase iron sequestration by ferritin. This would lead to reduced free iron and limit the further production of ROS by Fenton chemistry. This model is supported by studies with Irp2−/− primary cortical cells showing that increased ferritin expression attenuated oxidant stress-induced cytotoxicity, presumably by increased iron chelation (40). A reduction in IRP2 RNA-binding activity during glucose deprivation could therefore function as a protective mechanism to limit iron toxicity during conditions of acute pro-oxidant stress.
Supplementary Material
Acknowledgments
We thank S. James Remington for the roGFP2 cDNA. We are grateful to Dennis Winge, Don Ayer, Steve Lessnick, and Janet Lindsley for comments on both the experiments and manuscript. Special thanks go to Matthew Williams for FACS assistance and William Walden for advice regarding the IRP structures.
This work was supported by NIH grants R01GMS45201 (to E.A.L.). K.B.Z. and M.L.W. were supported by NIH Hematology training grant T32DK00715.
Footnotes
Published ahead of print on 17 February 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Addess, K. J., J. P. Basilion, R. D. Klausner, T. A. Rouault, and A. Pardi. 1997. Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins. J. Mol. Biol. 27472-83. [DOI] [PubMed] [Google Scholar]
- 2.Anykin-Burns, N., I. M. Ahmad, Y. Zhu, L. W. Oberley, and D. R. Spitz. 2009. Increased levels of superoxide and hydrogen peroxide mediate the differential susceptibility of cancer cells vs. Normal cells to glucose deprivation. Biochem J. 41829-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourdon, E., D.-K. Kang, M. C. Ghosh, S. K. Drake, J. Wey, R. L. Levine, and T. A. Rouault. 2003. The role of endogenous heme synthesis and degradation domain cysteines in cellular iron-dependent degradation of IRP2. Blood Cells Mol. Dis. 31247-255. [DOI] [PubMed] [Google Scholar]
- 4.Bouton, C., H. Hirling, and J.-C. Drapier. 1997. Redox modulation of iron regulatory proteins by peroxynitrite. J. Biol. Chem. 27219969-19975. [DOI] [PubMed] [Google Scholar]
- 5.Cairo, G., L. Tacchini, G. Pogliaghi, E. Anzon, A. Tomasi, and A. Bernelli-Zazzera. 1995. Induction of ferritin synthesis by oxidative stress. J. Biol. Chem. 270700-703. [DOI] [PubMed] [Google Scholar]
- 6.Chen, O. S., K. L. Schalinske, and R. S. Eisenstein. 1997. Dietary iron intake modulates the activity of iron regulatory proteins and the abundance of ferritin and mitochondrial aconitase in rat liver. J. Nutr. 127238-248. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, S. L., A. Vasanthakuman, S. A. Anderson, C. Pondarre, C. M. Koh, K. M. Deck, J. S. Pitula, C. J. Epstein, M. D. Fleming, and R. S. Eisenstein. 2006. Iron-responsive degradation of iron-regulatory protein 1 does not require the Fe-S cluster. EMBO J. 25544-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooperman, S. S., E. G. Meyron-Holtz, H. Olivierre-Wilson, M. C. Ghosh, J. P. McConnell, and T. A. Rouault. 2005. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood 1061084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corna, G., B. Galy, M. Hentze, and G. Cairo. 2006. IRP1-independent alterations of cardiac iron metabolism in doxorubicin-treated mice. J. Mol. Med. 84551-560. [DOI] [PubMed] [Google Scholar]
- 10.Dooley, C. T., T. M. Dore, G. T. Hanson, W. C. Jackson, S. J. Remington, and R. Y. Tsien. 2004. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 27922284-22293. [DOI] [PubMed] [Google Scholar]
- 11.Dupuy, J., A. Volbeda, P. Carpentier, C. Darnault, J.-M. Moulis, and J. C. Fontecilla-Camps. 2006. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure 14129-139. [DOI] [PubMed] [Google Scholar]
- 12.Erlitzki, R., J. C. Long, and E. C. Theil. 2002. Multiple, conserved iron-responsive elements in the 3′-untranslated region of transferrin receptor mRNA enhance binding of iron regulatory protein 2. J. Biol. Chem. 27742579-42587. [DOI] [PubMed] [Google Scholar]
- 13.Galy, B., D. Ferring, B. Minana, O. Bell, H. G. Janser, M. Muckenthaler, K. Schumann, and M. W. Hentze. 2005. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2). Blood 1062580-2589. [DOI] [PubMed] [Google Scholar]
- 14.Galy, B., S. M. Holter, T. Klopstock, D. Ferring, L. Becker, S. Kaden, W. Wurst, H.-J. Grone, and M. W. Hentze. 2006. Iron homeostasis in the brain: complete iron regulatory protein 2 deficiency without symptomatic neurodegeneration in the mouse. Nat. Genet. 38967-969. [DOI] [PubMed] [Google Scholar]
- 15.Gdaniec, Z., H. Sierzputowska-Gracz, and E. C. Theil. 1998. Iron regulatory element and internal loop/bulge structure for ferritin mRNA studied by cobalt(iii) hexammine binding, molecular modeling, and NMR spectroscopy. Biochemistry 371505-1512. [DOI] [PubMed] [Google Scholar]
- 16.Guo, B., Y. Yu, and E. A. Leibold. 1994. Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J. Biol. Chem. 26924252-24260. [PubMed] [Google Scholar]
- 17.Hanson, E. S., M. L. Rawlins, and E. A. Leibold. 2003. Oxygen and iron regulation of iron regulatory protein 2. J. Biol. Chem. 27840337-40342. [DOI] [PubMed] [Google Scholar]
- 18.Hanson, G. T., R. Aggeler, D. Oglesbee, M. Cannon, R. A. Capaldi, R. Y. Tsien, and S. J. Remington. 2004. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 27913044-13053. [DOI] [PubMed] [Google Scholar]
- 19.Henderson, B. R., and L. C. Kühn. 1995. Differential modulation of the RNA-binding proteins IRP-1 and IRP-2 in response to iron. J. Biol. Chem. 27020509-20515. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, B. R., E. Menotti, C. Bonnard, and L. C. Kuhn. 1994. Optimal sequence and structure of iron-responsive elements. Selection of RNA stem-loops with high affinity for iron regulatory factor. J. Biol. Chem. 26917481-17489. [PubMed] [Google Scholar]
- 21.Henderson, B. R., E. Menotti, and L. C. Kühn. 1996. Iron regulatory proteins 1 and 2 bind distinct sets of RNA target sequences. J. Biol. Chem. 2714900-4908. [DOI] [PubMed] [Google Scholar]
- 22.Henderson, B. R., C. Seiser, and L. C. Kuhn. 1993. Characterization of a second RNA-binding protein in rodents with specificity for iron-responsive elements. J. Biol. Chem. 26827327-27334. [PubMed] [Google Scholar]
- 23.Hirling, H., B. R. Henderson, and L. C. Kühn. 1994. Mutational analysis of the [4Fe-4S]-cluster converting iron regulatory factor from its RNA-binding form to cytoplasmic aconitase. EMBO J. 13453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang, S. I., H. W. Choi, and I. Y. Kim. 2008. Redox-mediated modification of PLZF by SUMO-1 and ubiquitin. Biochem. Biophys. Res. Commun. 3691209-1214. [DOI] [PubMed] [Google Scholar]
- 25.Ke, Y., H. Sierzputowska-Gracz, Z. Gdaniec, and E. C. Theil. 2000. Internal loop/bulge and hairpin loop of the iron-responsive element of ferritin mRNA contribute to maximal iron regulatory protein 2 binding and translational regulation in the iso-iron-responsive element/iso-iron regulatory protein family. Biochemistry 396235-6242. [DOI] [PubMed] [Google Scholar]
- 26.Ke, Y., J. Wu, E. A. Leibold, W. E. Walden, and E. C. Theil. 1998. Loops and bulge/loops in iron-responsive element isoforms influence iron regulatory protein binding. Fine-tuning of mRNA regulation? J. Biol. Chem. 27323637-23640. [DOI] [PubMed] [Google Scholar]
- 27.Kim, H.-Y., T. LaVaute, K. Iwai, R. D. Klausner, and T. A. Rouault. 1996. Identification of a conserved and functional iron-responsive element in the 5′-untranslated region of mammalian mitochondrial aconitase. J. Biol. Chem. 27124226-24230. [DOI] [PubMed] [Google Scholar]
- 28.LaVaute, T., S. Smith, S. Cooperman, K. Iwai, W. Land, E. Meyron-Holtz, S. K. Drake, G. Miller, M. Abu-Asab, M. Tsokos, R. Switzer, A. Grinberg, P. Love, N. Tresser, and T. A. Rouault. 2001. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 27209-214. [DOI] [PubMed] [Google Scholar]
- 29.Leibold, E. A., L. C. Gahring, and S. W. Rogers. 2001. Immunolocalization of iron regulatory protein expression in the murine central nervous system. Histochem. Cell Biol. 115195-203. [DOI] [PubMed] [Google Scholar]
- 30.Meyron-Holtz, E. G., M. C. Ghosh, K. Iwai, T. LaVaute, X. Brazzolotto, U. V. Berger, W. Land, H. Ollivierre-Wilson, A. Grinberg, P. Love, and T. A. Rouault. 2004. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 23386-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyron-Holtz, E. G., M. C. Ghosh, and T. A. Rouault. 2004. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science 3062087-2090. [DOI] [PubMed] [Google Scholar]
- 32.Muckenthaler, M. U., B. Galy, and M. W. Hentze. 2008. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 28197-213. [DOI] [PubMed] [Google Scholar]
- 33.Oliveira, L., C. Bouton, and J.-C. Drapier. 1999. Thioredoxin activation of iron regulatory proteins. Redox regulation of RNA binding after exposure to nitric oxide. J. Biol. Chem. 274516-521. [DOI] [PubMed] [Google Scholar]
- 34.Paget, M. S. B., and M. J. Buttner. 2003. Thiol-based regulatory switches. Annu. Rev. Genet. 3791-121. [DOI] [PubMed] [Google Scholar]
- 35.Pantopoulos, K., G. Weiss, and M. W. Hentze. 1996. Nitric oxide and oxidative stress (H2O2) control mammalian iron metabolism by different pathways. Mol. Cell. Biol. 163781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Counch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF chimera—a visualization system for exploratory research and analysis. J. Comp. Chem. 251605-1612. [DOI] [PubMed] [Google Scholar]
- 37.Phillips, J. D., B. Guo, Y. Yu, F. M. Brown, and E. A. Leibold. 1996. Expression and biochemical characterization of iron regulatory proteins 1 and 2 in Saccharomyces cerevisiae. Biochemistry 3515704-15714. [DOI] [PubMed] [Google Scholar]
- 38.Philpott, C. C., D. Haile, T. A. Rouault, and R. D. Klausner. 1993. Modification of a free Fe-S cluster cysteine residue in the active iron-responsive element-binding protein prevents RNA binding. J. Biol. Chem. 26817655-17658. [PubMed] [Google Scholar]
- 39.Piccinelli, P., and T. Samuelsson. 2007. Evolution of the iron-responsive element. RNA 13952-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regan, R. F., Z. Li, M. Chen, X. Zhang, and J. Chen-Roetling. 2008. Iron regulatory proteins increase neuronal vulnerability to hydrogen peroxide. Biochem. Biophys. Res. Commun. 3756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schalinske, K. L., O. S. Chen, and R. S. Eisenstein. 1998. Iron differentially stimulates translation of mitochondrial aconitase and ferritin mRNAs in mammalian cells. Implications for iron regulatory proteins as regulators of mitochondrial citrate utilization. J. Biol. Chem. 2733740-3746. [DOI] [PubMed] [Google Scholar]
- 42.Schalinske, K. L., and R. S. Eisenstein. 1996. Phosphorylation and activation of both iron regulatory proteins 1 and 2 in HL-60 cells. J. Biol. Chem. 2717168-7176. [DOI] [PubMed] [Google Scholar]
- 43.Scherz-Shouval, R., E. Shvets, E. Fass, H. Shorer, L. Gil, and Z. Elazar. 2007. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 261749-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwede, T., J. Kopp, N. Guex, and M. C. Peitch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 313381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simons, A. L., I. M. Ahmad, D. M. Mattson, K. J. Dornfeld, and D. R. Spitz. 2007. 2-Deoxy-D-glucose combined with cisplatin enhances cytotoxicity via metabolic oxidative stress in human head and neck cancer cells. Cell Res. 673364-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tacchini, L., S. Recalcati, A. Bernelli-Zazzera, and G. Cairo. 1997. Induction of ferritin synthesis in ischemic-reperfused rat liver: analysis of the molecular mechanisms. Gastroenterology 113946-953. [DOI] [PubMed] [Google Scholar]
- 47.Walden, W. E., A. I. Selezneva, J. Dupuy, A. Volbeda, J. C. Fontecilla-Camps, E. C. Theil, and K. Volz. 2006. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science 3141903-1908. [DOI] [PubMed] [Google Scholar]
- 48.Wallander, M. L., E. A. Leibold, and R. S. Eisenstein. 2006. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta 1763668-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallander, M. L., K. B. Zumbrennen, E. S. Rodansky, S. J. Romney, and E. A. Leibold. 2008. Iron-independent phosphorylation of iron regulatory protein 2 regulates ferritin during the cell cycle. J. Biol. Chem. 28323589-23598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, J., G. Chen, M. Muchenthaler, B. Galy, M. W. Hentze, and K. Pantopolous. 2004. Iron-mediated degradation of IRP2, an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol. Cell. Biol. 24954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, J., C. Fillebeen, G. Chen, A. Biederbick, R. Lill, and K. Pantopoulos. 2007. Iron-dependent degradation of apo-IRP1 by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 272423-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, Y., E. Radisky, and E. A. Leibold. 1992. The iron-responsive element binding protein. Purification, cloning, and regulation in rat liver. J. Biol. Chem. 26719005-19010. [PubMed] [Google Scholar]
- 53.Zumbrennen, K. B., E. S. Hanson, and E. A. Leibold. 2008. HOIL-1 is not required for iron-mediated IRP2 degradation in HEK293 cells. Biochim. Biophys. Acta 1783246-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.