Abstract
Pax6 is important in the development of the pancreas and was previously shown to regulate pancreatic endocrine differentiation, as well as the insulin, glucagon, and somatostatin genes. Prohormone convertase 2 (PC2) is the main processing enzyme in pancreatic α cells, where it processes proglucagon to produce glucagon under the spatial and temporal control of 7B2, which functions as a molecular chaperone. To investigate the role of Pax6 in glucagon biosynthesis, we studied potential target genes in InR1G9 α cells transfected with Pax6 small interfering RNA and in InR1G9 clones expressing a dominant-negative form of Pax6. We now report that Pax6 controls the expression of the PC2 and 7B2 genes. By binding and transactivation studies, we found that Pax6 indirectly regulates PC2 gene transcription through cMaf and Beta2/NeuroD1 while it activates the 7B2 gene both directly and indirectly through the same transcription factors, cMaf and Beta2/NeuroD1. We conclude that Pax6 is critical for glucagon biosynthesis and processing by directly and indirectly activating the glucagon gene through cMaf and Beta2/NeuroD1, as well as the PC2 and 7B2 genes.
The transcription factor Pax6, which is important in the development of the pancreas, regulates glucagon, insulin, and somatostatin gene transcription (42, 52). Pax6 is expressed in all cells of the endocrine pancreas, as well as in the nose, eye, and the central nervous system (25, 63, 68). Mutations in Pax6 are related to diabetes mellitus, aniridia, brain malformation, and other anomalies (37, 43). Pax6 homozygous mutant mice fail to form islets; show a decreased number of β, δ, and pancreatic polypeptide-producing cells; a lack of α cells; and an increase in ghrelin-positive ɛ cells without any change in endocrine pancreatic mass (26, 59). Pax6 has been previously proposed to control proglucagon gene expression through binding to the G1 and G3 elements on its promoter (42, 47).
Proglucagon is a multifunctional precursor expressed in α cells of the pancreatic islets, L cells of the small and large intestines, and selected neurons of the brain (29). Proglucagon contains multiple cleavage sites recognized with various degrees of efficiency by prohormone convertase 1/3 (PC1/3) and PC2. In this way, proglucagon can be processed in different cells to yield a unique mixture of products with differing functions (reviewed in reference 3). Thus, in pancreatic α cells, abundant expression of PC2 is associated with nearly exclusive production of glucagon (49, 50, 72), a 29-amino-acid hormone which, together with insulin, intricately controls gluconeogenesis and glycogenolysis (reviewed in references 18, 23, and 58). Conversely, in intestinal L cells, processing, mediated mainly by PC1/3, leads to the production of glucagon-like peptides 1 and 2 and glicentin (10, 14, 45, 72). Following cleavage by PC2 or PC1/3, carboxypeptidase E removes the C-terminal basic residues exposed after endoproteolytic processing of the prohormone precursor by the PCs (65).
The PC2 enzyme processes a variety of hormones and is crucial for proglucagon processing in α cells (11, 16, 23, 49, 50). The islets of PC2-null mice contain a reduced number of β cells and a markedly increased number of α and δ cells, reflecting the defective processing of proglucagon to glucagon and of somatostatin 28 to somatostatin 14. Another level in the regulation of PC2 activity is that PC2 is synthesized as a proconvertase (pro-PC2) that undergoes proteolysis. In the endoplasmic reticulum early secretory pathway, a complex of pro-PC2 and pro-7B2 is formed in a Ca2+-dependent manner (2, 5). The 7B2 chaperone contains two binding sites for the PC2 enzyme; one at the C terminus acts as a potent inhibitor of 7B2, while the other is a polyproline segment (64). PC2 and 7B2 are transported together to the trans-Golgi network, where furin cleaves both precursors, followed by dissociation of the two proteins; consequently, mature PC2 is able to cleave the prohormone (reviewed in reference 33). Therefore, 7B2 couples PC2 activity in a temporal and spatial manner to activate hormone production (4, 7, 55, 56). Reduced expression of 7B2 leads to less active PC2 with potential consequences for glucose tolerance with age (5, 35, 55). Interestingly, 7B2-null mice have multiple metabolic hormonal abnormalities and die of Cushing's disease (14, 53, 54, 71).
Several studies have been conducted so far in order to better understand the transcriptional regulation and gene organization of PC2 and 7B2. PC2 was previously reported to be regulated by Egr1, repressor element 1/neuron restrictive silencer element, thyroid hormone receptor α1, retinoid X receptor, and Beta2/NeuroD1 (28, 32, 57, 73). Additionally, several elements were found on the 7B2 promoter, namely, a cyclic AMP-responsive element, an AP-1-binding site, a POU protein-binding element (Pit1/GHF1), and thermal stress response and heat shock elements. Additionally, protein kinases A and C were demonstrated to activate the 7B2 promoter (6, 19, 30, 33). Even though it is widely agreed that PC2 and 7B2 are coexpressed and regulated in the same cells, the mechanism behind this coregulation remains unknown.
To better understand the role of Pax6 in α-cell function, proglucagon gene regulation, and glucagon biosynthesis, we used two systems: α cells transfected with Pax6 small interfering RNA (siRNA) and α-cell clones expressing a dominant-negative (DN) form of Pax6 composed of the first 306 amino acids of the human Pax6 protein lacking the transactivation domain but able to bind its target promoters and thus preventing wild-type (WT) Pax6 from binding and activating transcription. We then screened for target genes of Pax6 by quantifying the expression levels of genes known to be critical for α-cell development and function. We now report a decrease of about 50% in proglucagon after inhibition of Pax6 in α cells, in agreement with previous observations indicating that Pax6 can bind to two control elements of the glucagon promoter, G1 and G3, and activate transcription (42), as well as a marked decrease in PC2 and 7B2 mRNA and protein levels. We further demonstrate, by binding and transcriptional assays, that Pax6 indirectly regulates the PC2 and 7B2 genes through cMaf and Beta2/NeuroD1 and has a direct positive transcriptional effect on the 7B2 gene promoter.
We conclude that Pax6 is critical for glucagon biosynthesis in α cells, acting directly and indirectly on the transactivation of the glucagon and 7B2 genes through cMaf and Beta2/NeuroD1 and indirectly on the PC2 gene.
MATERIALS AND METHODS
Cell culture.
InR1G9 (hamster glucagon-producing cells) (60), A5 (monoclonal InR19G cells stably transfected with the pRC-CMV vector), C4 (monoclonal InR19G cells stably transfected with the pRC-CMV-Pax6-DN306 vector) (Y. Gosmain, E. Marthinet, A. Guerardel, A. Mamin, L. S. Katz, V. M. Schwitzgebel, and J. Philippe, submitted for publication; 21), αTC.1 (mouse glucagon-producing cells) (46), BHK21 (non-islet Syrian baby hamster kidney cells), and HEK293 (human embryonic kidney cells) cells were grown in RPMI 1640 medium supplemented with 5% fetal calf serum, 5% newborn calf serum, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. A5 and C4 clones were grown with G418 in the medium at 400 μg/ml.
RNA preparation and RT-PCR analysis.
Total RNA was isolated from αTC.1 and InR1G9 cells as described in reference 20. cDNA obtained was analyzed by quantitative real-time reverse transcription (RT)-PCR. Each assay was performed in duplicate, and validation of the real-time PCR runs was done by evaluation of the melting temperature of the products and by the slope and error obtained with the standard curve. The analyses were performed with the LightCycler software (Roche Diagnostics, Rotkreuz, Switzerland). Results were corrected for TATA-binding protein (TBP) or actin mRNA levels.
Transient transfection assays.
BHK21 and HEK293 cells were transfected by the calcium phosphate technique (22), while InR1G9 cells were transfected by Lipofectamine 2000 (Invitrogen) in six-well plates with 5 μl of Lipofectamine, 1 μg of the reporter, and 0.1 μg of the effector (unless mentioned otherwise) for each transfection. Cells were cotransfected with either pSV2A-PAP or RSV-Luc in order to monitor transfection efficiency.
Promoter analysis.
Cell extracts were collected 48 h after transient transfection, and promoter activity was assessed by measurement of chloramphenicol acetyltransferase (CAT) activity as previously described (41). Quantification of the acetylation forms was done with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Placental alkaline phosphatase (PAP) and luciferase (Luc) activities were measured by spectrophotometer and luminometer, respectively. A minimum of three independent transfections were carried out, each of them in duplicate. The total amount of DNA used for transfection was kept constant by adding the pSG5 or pCDNA3 vector.
EMSA.
Ten micrograms of nuclear extract from HEK293 cells overexpressing Pax6, cMaf, MafB, Beta2/NeuroD1, or E47 was added to electrophoretic mobility shift assay (EMSA) reaction buffer [20 mmol/liter HEPES (pH 7.9), 5 mmol/liter MgCl2, 0.5 mmol/liter EDTA, 50 mmol/liter KCl, 1 mmol/liter dithiothreitol, 6.25% glycerol, 1 μg bovine serum albumin, 2 μg salmon sperm DNA, 2 μg poly(dI-dC)]. EMSAs were performed as described previously (40); for the oligonucleotides used, see Table S1 in the supplemental material.
Probes were annealed with the corresponding antisense probes and labeled with [γ-32P]dATP by the T4 polynucleotide kinase (NEB, Frankfurt, Germany). Fifty thousand counts per minute of each annealed probe was added to each reaction mixture. For supershift experiments, nuclear extracts (10 μg) were preincubated with the respective antibody for 10 min at room temperature before the addition of 32P-labeled DNA probe and subsequent incubation at room temperature for 10 min. Competition was carried out with a 200-fold excess of either the annealed unlabeled probe or a mutated probe (see Table S1 in the supplemental material) annealed with the corresponding reverse complement probe.
All reaction mixtures were loaded onto a 6% nondenaturing polyacrylamide gel, and after electrophoresis, the gel was exposed to X-ray film for 24 h.
Chromatin immunoprecipitation (ChIP) assay.
InR1G9 and αTC1 cells were formaldehyde (1%) cross-linked for 10 min at room temperature; the reaction was stopped by the addition of 0.125 M glycine, followed by washing with ice-cold phosphate-buffered saline. Cells were lysed, and chromatin extracts were prepared and fragmented by sonication on ice. Protein A-Sepharose beads (for the Beta2/NeuroD1 ChIP assay, protein G beads) were used for cMaf, MaB, and Pax6 ChIP assays (Amersham). Beads equilibrated with radioimmunoprecipitation assay buffer were added to 50 μg of chromatin extracts for preclearance for 1 h. After centrifugation, supernatants were incubated overnight at 4°C with 10 μg of anti-Pax-6 (from Simon Saule, Curie Institute), anti-Beta2/NeuroD1 N19 (Santa Cruz), anti-cMaf or anti-MafB (Bethyl), and rabbit or goat immunoglobulin G (IgG; Santa Cruz). DNA-protein-antibody complexes were then incubated for 3 h with protein A- or protein G-Sepharose beads and subsequently rinsed five times with radioimmunoprecipitation assay buffer at 4°C, once with LiCl buffer, and finally with Tris-EDTA buffer as described previously (24). RNA was eliminated by incubation with 25 μg/ml RNase A for 3 h at 65°C. Proteins were eliminated by overnight incubation with proteinase K (200 μg; Applichem) at 45°C. After phenol precipitation of the chromatin, DNA was resuspended in water and used as a template for PCR. For the PCR primer sets used for analysis of the different transcription factors, see Table S2 in the supplemental material. Products were amplified for 32 cycles at an annealing temperature of 56 to 59°C with Invitrogen Taq polymerase and analyzed on a 3% agarose gel stained with ethidium bromide.
siRNAs.
Two different specific sequences of stealth siRNA (Invitrogen) were designed for the Pax6, cMaf, MafB, and Beta2/NeuroD1 hamster mRNA sequences. A control scrambled sequence was designed to have the same GC content. InR1G9 cells grown in six-well plates were transfected sequentially with 100 nM siRNA on the first day and 200 nM on the second day with 5 μl of Lipofectamine 2000 (Invitrogen). Total RNA and nuclear extract were isolated 48 h after the first transfection.
Total cell extract and Western blot assays.
Cells were grown in 10-cm plates to 80% confluence, washed with phosphate-buffered saline, trypsinized, and resuspended in prechilled lysis buffer (50 mM Tris, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 2 mM EGTA, protease inhibitor cocktail [Roche]). Cells were then sonicated three times for 10 s on ice and centrifuged at 12,000 × g for 10 min at 4°C, and the protein concentration of the supernatant phase was measured. Fifteen micrograms of total extracts was boiled in Laemmli buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with 12% acrylamide for Western blot analyses. Blots were probed with a 1:2,000 dilution of the PC2 antibody, a generous gift from Nabil Seidah, Clinical Research Institute of Montreal, or a 1:25 dilution of 7B2 antibodies MON-102 and MON-144, generous gifts from Wim J. M. van de Ven, Nijmegen University. Membranes were stripped and blotted with a 1:500 dilution of TFIIE-α antibody (Santa Cruz) as a loading control.
Plasmids.
The mouse PC2 promoter (positions −730 to +439) was cloned into pGL2-Basic. All human PC2 promoter constructs cloned into pCAT-Basic were kindly provided by Donald F. Steiner, University of Chicago (39), and pCAT-Basic and Enhancer-CMV-Basic were a generous gift from Joseph Orly, Hebrew University. The serial deletions of the mouse 7B2 promoter were cloned into the pGL2-Basic vector. The mouse Egr1 cDNA-encoding plasmid was obtained from Gerald Thiel, Saarland University (40). The mouse cMaf, MafB, NeuroD1, and E47 cDNAs cloned into the pSG5-ATG expression vector and the DN-Maf cDNA cloned into the pcDNA3 plasmid were provided by Roland Stein, Vanderbilt University. The mouse Pax6 (p46) cDNA was subcloned into pSG5 in our laboratory. The DN-Beta2/NeuroD1 cDNA cloned in pcDNA3 was obtained from Haeyoung Suh-Kim, Catholic University (9).
Site-directed mutagenesis.
Site-directed mutagenesis was performed with the mutated oligonucleotides also used in EMSAs for competition with mutated probes. PCR was performed with Taq Turbo (Stratagene) for 18 cycles following digestion with DpnI (Stratgen), products were transformed into bacteria, and extracted plasmids were sequenced for the presence of the mutation.
RESULTS
Both PC2 and 7B2 are decreased in cells deficient in Pax6 activity.
We demonstrate elsewhere (Gosmain et al., submitted; 21) that in hamster α cells (InR1G9 cells used, as opposed to mouse αTC cells, for their high transfection efficiency) a specific knockdown of cellular Pax6 with siRNA results in decreased expression of the glucagon gene, as well as of certain transcription factors important for glucagon gene transcription, such as cMaf, Beta2/NeuroD1, and Isl1, compared to the same cells transfected with a scrambled stealth RNA.
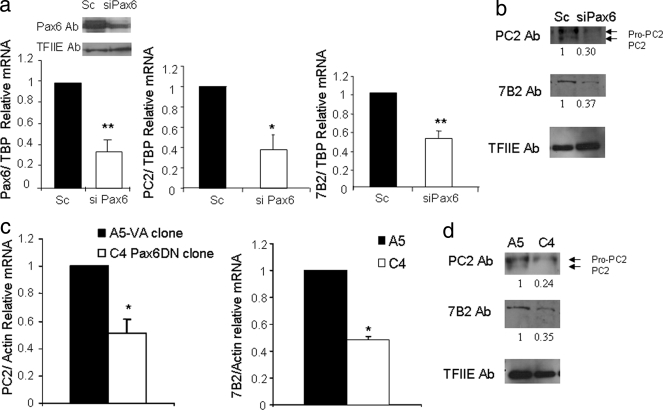
To more fully characterize the role of Pax6 in glucagon biosynthesis, we measured the mRNA levels of the PC2 enzyme, implicated in proglucagon processing, upon transfection of Pax6 siRNA into InR1G9 cells. With a knockdown of 65% of Pax6 mRNA, significant 62.3 and 70% decreases in PC2 mRNA and protein levels, respectively, were observed (Fig. 1a and b). Interestingly, the ratio of pro-PC2 to PC2 was markedly higher (>90%) in cells transfected with Pax6 siRNA than in scrambled control transfections. This high proportion of premature PC2, leading to impaired processing of the PC2 protein, was previously described in cells deficient in 7B2 (5, 34, 35). We thus measured 7B2 protein and mRNA levels and found decreases of 63 and 49%, respectively (Fig. 1 and b).
FIG. 1.
Pax6 regulates the PC2 and 7B2 genes. (a) Real-time quantitative RT-PCR from RNA obtained from InR1G9 cells transfected with a scrambled (Sc) or Pax6 siRNA (siPax6) for 48 h. The analysis was performed with the LightCycler software (Roche Applied Science). The results were corrected for TBP mRNA levels. The relative mRNA levels are presented as the means ± standard errors of the means (SEM) of at least three independent experiments. *, P < 0.05; **, P < 0.01. Left corner, Western blot analyses of Pax6 and TFIIE from nuclear extracts from InR1G9 cells transfected with scrambled or Pax6 siRNA for 48 h. Ab, antibody. (b) Western blot analyses of PC2, 7B2, and TFIIE from total extracts from InR1G9 cells transfected with scrambled or Pax6 siRNA for 48 h. (c) Real-time quantitative RT-PCR analysis of RNA obtained from the A5 (pRC-CMV-expressing) and C4 (Pax6-DN306-expressing) clones. The analysis was performed with the LightCycler software (Roche Applied Science). Left panel, measurement of PC2 mRNA, right panel, measurement of 7B2 mRNA. The results were corrected for actin mRNA levels. The relative mRNA levels are presented as the means ± SEM for at least three independent experiments. *, P < 0.05; **, P < 0.01. (d) Western blot analyses of PC2, 7B2, and TFIIE from total cell extracts from A5 and C4 clones.
We then confirmed the importance of Pax6 for PC2 and 7B2 gene expression with InR1G9 glucagon-producing clones stably expressing a DN form of Pax6, the C4 clone, and a control clone expressing the empty vector (pRN-CMV), the A5 clone (as characterized elsewhere [Gosmain et al., submitted; 21]). PC2 mRNA levels were decreased by 49% in the C4 clone compared to those in the A5 clone (Fig. 1c). An even greater decrease of 76% in PC2 protein levels was observed in the C4 clone (Fig. 1b). We observed 65 and 52% decreases in the 7B2 protein and mRNA levels, respectively, in the Pax6-DN306, compared to those in the control clone (Fig. 1c and d).
By contrast, the mRNA levels of two other processing enzymes, furin and PC1/3, measured in the A5 and C4 clones, as well as in cells transfected with siPax6 or a scrambled control, showed no significant changes. Of note, only trace amounts of PC1/3 were detected in both the A5 and C4 clones or in WT InR1G9 cells (data not shown).
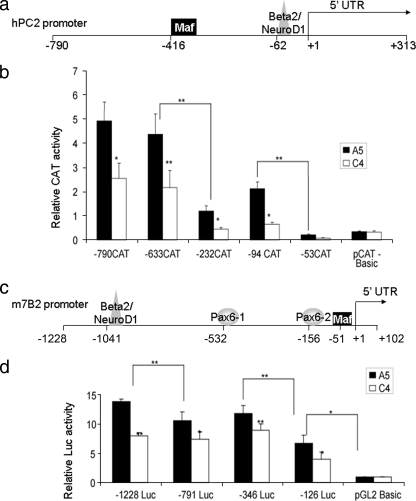
To assess whether Pax6 affects PC2 and 7B2 gene expression at the level of transcription, we transfected A5 and C4 cells with plasmids containing several deletions of either the human PC2 promoter linked to the CAT reporter gene (Fig. 2b) or the mouse 7B2 promoter linked to the Luc reporter gene (Fig. 2d). With the PC2 promoter, we observed two major decreases in activity in the control A5 clone, between bp −633 and −233 and between bp −94 and −53, indicating that these promoter segments contain important DNA control elements. When comparing the A5 and C4 clones, a significant decrease in activity was found for −790 CAT, −633 CAT, −232 CAT, and −94 CAT of the PC2 promoter in C4, suggesting that Pax6 may directly or indirectly regulate PC2 gene transcription down to bp −94. For the 7B2 promoter, we also identified three important regions in the A5 clone, between bp −1228 and −791, between bp −346 and −126, and within bp −126. In addition, all of the promoter deletions tested resulted in a significant decrease in activity between the A5 and C4 clones, indicating that Pax6 may regulate 7B2 promoter activity even further downstream of position −126 (Fig. 2d).
FIG. 2.
PC2 and 7B2 promoter structures, activities, and deletions. (a) Putative transcription factors that bind to the human PC2 promoter according to the Genomatix database. UTR, untranslated region. (b) Transcription from the PC2 promoter in A5 and C4 clones. Clones were transfected with various deletions of the human PC2 promoter linked to the CAT reporter gene; nucleotide numbering corresponds to the transcription initiation site. Results represent relative CAT and PAP activities over basal activity. Values are the means ± the standard errors of the means (SEM) from at least three separate experiments, each performed twice. *, P < 0.05; **, P < 0.01. (c) Putative transcription factors that bind to the mouse 7B2 promoter according to the Genomatix database. (d) Transcription from the 7B2 promoter in A5 and C4 clones. Clones were transfected with various deletions of the mouse 7B2 promoter linked to the Luc reporter; nucleotide numbering corresponds to the transcription initiation site. Results represent relative Luc and PAP activities over basal activity. Values are the means ± SEM mean from at least three separate experiments, each performed twice. *, P < 0.05; **, P < 0.01.
Transcription factor binding to the PC2 and 7B2 promoters.
The PC2 promoter sequence is highly conserved between rodents and humans (69.4% identity), while there is more divergence in the 7B2 promoter (56.6%). Some transcription factor binding sites on the 7B2 promoter are, however, strongly conserved (such as the thermal stress response and heat shock element binding elements) (8).
In order to determine whether Pax6 directly interacts with the PC2 or 7B2 gene promoter, we used the Genomatix (http://www.genomatix.de) database to find putative pancreatic transcription factor binding sites. For both promoters, we looked 3 kb upstream of the transcription initiation site and at the 5′ untranslated region. No Pax6 binding sites were found in the mouse and human PC2 promoters, whereas two conserved putative binding sites, one large-Maf and one Beta2/NeuroD1, were identified (Fig. 2a). These factors are of particular interest since they have previously been reported to regulate the transcription of the insulin and glucagon genes (12, 20, 38). Moreover, both the cMaf and Beta2/NeuroD1 transcription factors have been demonstrated to be decreased in the Pax6-DN306 InR1G9 clones and upon transfection with Pax6 siRNA, indicating that Pax6 positively regulates these genes (Gosmain et al., submitted; 21). A first indication of the possible relevance of Maf and Beta2/NeuroD1 for PC2 gene transcription derives from experiments shown in Fig. 2b. Indeed, deletion of the putative Maf binding site (corresponding to positions −416 to −404 in the human PC2 promoter) led to a threefold decrease in basal promoter activity in the A5 clone. Similarly, deletion of the Beta2/NeuroD1 binding site (positions −62 to −51 in the hPC2 promoter) resulted in an even stronger decrease in activity.
For the mouse 7B2 promoter, the Genomatix database search revealed two Pax6 putative binding sites, one large-Maf putative binding site, and one Beta2/NeuroD1 putative binding site (Fig. 2c), while for the human promoter, only the two Pax6 (−175 to −164 and +319 to +330) putative binding sites and the Maf (+289 to +299 in the 5′ untranslated region) putative binding site were identified. Of note, deletion of bp −1228 to −791 of the mouse 7B2 promoter, containing the Beta2/NeuroD1 site, resulted in decreased transcriptional activity (Fig. 2d). Further deletion of bp −791 to −346, containing the putative distal Pax6 site, did not change the transcriptional activity, while a nearly 50% drop was observed upon the deletion of bp −346 to −126, containing the more proximal binding site (Fig. 2d). The first 126 bp of the 7B2 promoter, containing the putative Maf site, displayed a basal activity 2.25-fold higher than the activity of the promoterless vector (Fig. 2d), suggesting that the Maf site may be functional. Our data thus suggest that the Beta2/NeuroD1, proximal Pax6, and Maf sites may be functional, although other transcription factors are likely to play additional roles in the activation of the 7B2 promoter.
Pax6, cMaf, MafB, and Beta2/NeuroD1 activate the PC2 and 7B2 gene promoters.
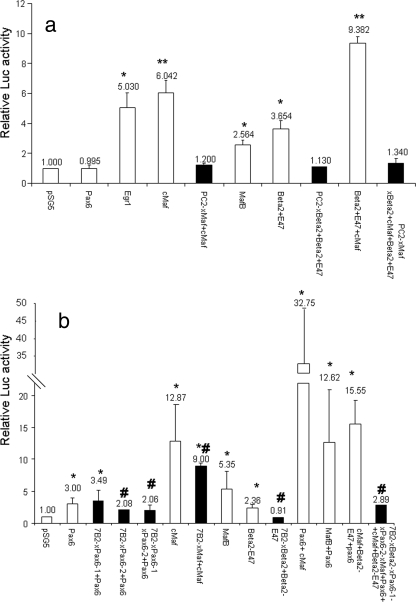
In order to determine the functionality of the identified transcription factor binding sites, we transiently transfected the −730 PC2 promoter linked to a CAT reporter into the heterologous BHK21 cell line together with cMaf, MafB, Pax6, Beta2/NeuroD1, E47, or a combination of these factors.
Egr1 was used as a positive control for the activation of the PC2 promoter as previously reported (28, 30). cMaf, MafB, and Beta2/NeuroD1-E47 (Beta2/NeuroD1 functions as a heterodimer with the ubiquitous basic helix-loop-helix transcription factor E47) (12, 39), but not Pax6, were all able to activate the PC2 promoter (Fig. 3a). An additive effect was observed when cMaf was overexpressed with Beta2/NeuroD1-E47 (Fig. 3a). The activation was specific inasmuch as mutations of the large-Maf and Beta2 sites reduced activity (solid bars). These data indicate that cMaf, MafB, and Beta2/NeuroD1-E47, but not Pax6, can interact with and transactivate the PC2 gene promoter. By contrast, the 7B2 gene promoter (−1228 bp) was activated not only by cMaf, MafB, and Beta2/NeuroD1-E47 in BHK21 cells but also by Pax6 (Fig. 3b), although the decrease in activation by Pax6 and Beta2 was small. A marked synergistic effect of cMaf or MafB with Pax6 was observed, similar to that previously reported for the glucagon gene promoter (20, 44). Specific mutations of the proximal Pax6, Maf, and Beta2 sites significantly reduced activity (solid bars) (Fig. 3a and b).
FIG. 3.
Activation of the PC2 and 7B2 promoters by ectopic expression of Pax6, cMaf, MafB, and Beta2/NeuroD1 in a heterologous cell line. (a) BHK21 cells were transfected with the −730 WT mouse PC2 promoter (open bars), the empty reporter vector (pGL2-Basic), a site-directed mutation-containing form of the large-Maf (xMaf) or Beta2/NeuroD1 (xBeta2) site, or both sites (solid bars) fused to the Luc reporter gene along with the Pax6, cMaf, MafB, Beta2/NeuroD1 (Beta2), and E47 cDNAs and the Egr1 cDNA used as a positive control. Results represent relative Luc and PAP activities over the basal level for each construct. *, P < 0.05; **, P < 0.01 compared to the transfection of the promoter with pSG5. (b) BHK21 cells were transfected with the −1228 WT mouse 7B2 promoter (open bars), the empty reporter vector (pGL2-Basic), or a site-directed mutation-containing form of the large-Maf (xMaf), Beta2/NeuroD1 (xBeta2), Pax6 (xPax6-1, or xPax6-2) site, alone or combination (solid bars), fused to the Luc reporter together with the Pax6, cMaf, MafB, Beta2/NeuroD1, and E47 cDNAs. Results represent relative Luc and PAP activities over the basal level for each construct. Values are the means ± the standard errors of the means from at least three separate experiments, each performed twice. *, P < 0.05; **, P < 0.01 (compared to the transfection of the promoter with pSG5); #, P < 0.05 (compared to the WT 7B2 plasmid together with the respective transcription factor).
Direct binding of transcription factors to the PC2 and 7B2 promoters and their relative contributions to promoter activation.
We thus sought to determine whether the Maf and Beta2/NeuroD1 sites are playing a role in PC2 gene expression in α cells.
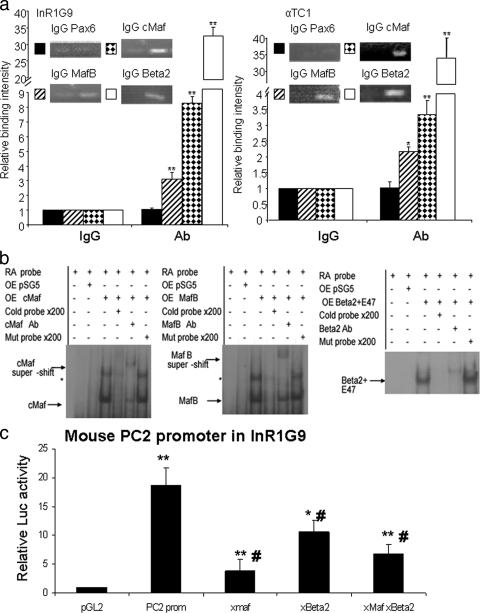
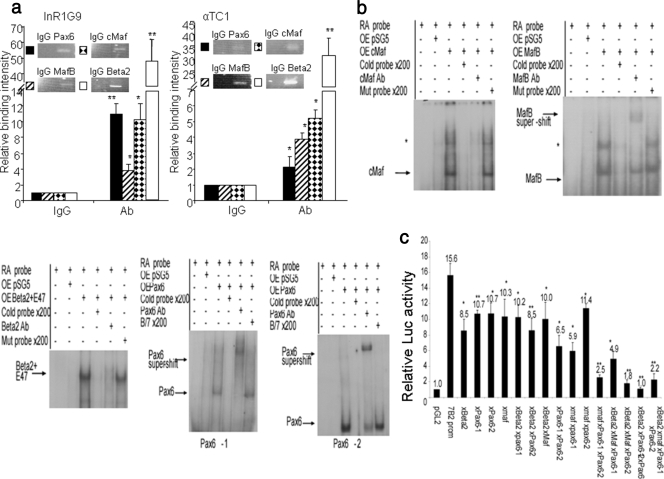
We first carried out ChIP experiments to confirm the binding of the two Mafs expressed in α cells, cMaf and MafB, as well as Beta2/NeuroD1. MafB, cMaf, and Beta2/NeuroD1 were all able to bind to the PC2 promoter both in InR1G9 and in αTC1, but not Pax6 (Fig. 4a).
FIG. 4.
Direct binding of cMaf, MafB, and Beta2/NeuroD1 to the PC2 promoter. (a) In vivo binding of large-Mafs and Beta2/NeuroD1 to the PC2 promoter by ChIP analysis. Histograms represent the relative binding of the Pax6, cMaf, MafB, and Beta2/NeuroD1 proteins to the PC2 gene promoter in InR1G9 and αTC1 cells. Binding intensity data are expressed relative to IgG immunoprecipitation (nonspecific binding) and are presented as the means ± the standard errors of the means (SEM) for at least three independent experiments. *, P < 0.05; **, P < 0.01. (b) EMSAs were performed with labeled oligonucleotides containing large-Maf and Beta2/NeuroD1 putative binding sites on the PC2 gene promoter in the presence of nuclear extracts from HEK cells overexpressing (OE) the empty vector (pSG5), cMaf, MafB, or Beta2/NeuroD1/E47. Probes correspond to the positions of the Maf (−659 to −629) and Beta2/NeuroD1 (−72 to −42) binding sites on the mouse PC2 promoter. *, possible homodimer or heterodimer of Maf. Mutated oligonucleotides correspond to mutated sites on the PC2 promoter. Ab, antibody. (c) Site-directed mutation-containing forms of the large-Maf (xMaf) or Beta2/NeuroD1 (xBeta2) site or both sites on the mouse PC2 promoter. InR1G9 cells were transfected with the mutated mouse PC2 promoter plasmids linked to the Luc reporter. Results represent relative Luc and PAP activities over basal activity. Values are the means ± (SEM) from at least three separate experiments, each performed twice. *, P < 0.05; **, P < 0.01 (compared to the pGL2Basic empty vector) #, P < 0.05 (compared to the WT mouse PC2 promoter).
We further confirmed the specific binding of the cMaf, MafB, and Beta2/NeuroD1 transcription factors on oligonucleotides containing the respective binding sites of the PC2 promoter by EMSA (positions −659 to −628 for Mafs and −72 to −41 for Beta2/NeuroD1 in the mouse PC2 gene promoter, Fig. 4b).
To further establish the relevance of these sites, we transfected the mutated promoters in InR1G9 cells; their activities were significantly lower compared to that of the WT promoter, underlining the functional importance of these sites for PC2 gene expression (Fig. 4c).
We then investigated whether cMaf, MafB, Beta2/NeuroD1, and Pax6 bind to the 7B2 promoter in vivo with the ChIP assay in both the InR1G9 and αTC1 cell lines (Fig. 5a). All four transcription factors were found to interact with the 7B2 promoter in both cell lines. We also demonstrated specific binding of these factors to the 7B2 promoter with oligonucleotides containing each of the four binding sites, −1050 to −1016 for Beta2/NeuroD1, −539 to −505 and −163 to −129 for Pax6, and −57 to −21 for the Mafs, by EMSA (Fig. 5b). Of note, the distal Pax6-binding site also interacted specifically with Pax6, although its deletion or mutation did not change the transcriptional activity (Fig. 2d and 5b).
FIG. 5.
Direct binding of Pax6, cMaf, MafB, and Beta2/NeuroD1 on the 7B2 promoter. (a) In vivo binding of Pax6, cMaf, MafB, and Beta2/NeuroD1 (Beta2) on the PC2 promoter by ChIP analysis. Histograms represent the relative binding of the Pax6, cMaf, MafB, and Beta2/NeuroD1 proteins on the 7B2 gene promoter in InR1G9 and αTC1 cells. Binding intensity data are expressed relative to IgG immunoprecipitation (nonspecific binding) and are presented as the means ± the standard errors of the means (SEM) for at least three independent experiments. *, P < 0.05. (b) EMSAs were performed with labeled oligonucleotides containing the large-Maf, Beta2/NeuroD1, and Pax6 putative binding sites on the 7B2 gene promoter in the presence of nuclear extracts from HEK cells overexpressing (OE) the empty vector (pSG5), cMaf, MafB, Beta2/NeuroD1/E47, or Pax6. Binding experiments were performed with probes corresponding to the positions of the Beta2/NeuroD1 (−1050 to −1016), Pax6-1 (−539 to −505), pax6-2 (−163 to −129), and large-Maf (−57 to −21) binding sites on the mouse 7B2 promoter. *, Possible heterodimer or homodimer of Maf. (c) Site-directed mutation-containing forms of large-Maf (xMaf), Beta2/NeuroD1 (xBeta2), Pax6 (xPax6-1, and xPax6-2) alone or combined with the mouse 7B2 promoter. InR1G9 cells were transfected with the mutated plasmids of the mouse 7B2 promoter linked to the Luc reporter. Results represent relative Luc and PAP activities over basal activity. Values are the means ± SEM mean from at least three separate experiments, each performed twice. *, P < 0.05; **, P < 0.01 (compared to the WT mouse 7B2 promoter).
Transfection of the 7B2 promoter mutated at one or two transcription factor binding sites in InR1G9 cells resulted in only minor decreases in activity (Fig. 5c), whereas all combinations of three, as well as four, mutations resulted in significantly decreased activity compared to that observed for the single- and double-mutated promoters, suggesting possible cooperative interactions between binding sites (Fig. 5c). Of note, mutation of the distal Pax6 site also resulted in decreased activity, suggesting a potential functional role for this site.
Specific silencing of cMaf and Beta2/NeuroD1 results in decreased transcription of the PC2 and 7B2 genes.
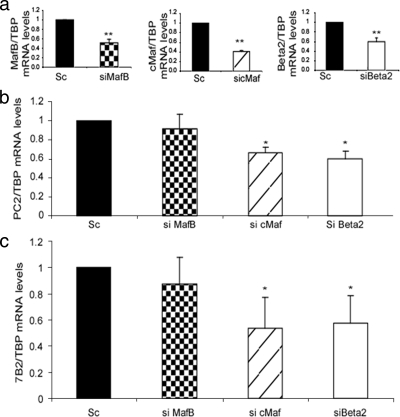
To further investigate the role of cMaf, MafB, and Beta2/NeuroD1 in PC2 and 7B2 gene transcription in α cells, we used specific siRNAs. InR1G9 cells were transfected with siRNA specific for MafB, cMaf, or Beta2/NeuroD1, and PC2 and 7B2 mRNA levels were assessed by real-time RT-PCR. The knockdown efficiency of each siRNA was 40 to 60% for the siRNA combinations used (Fig. 6a). PC2 mRNA levels were decreased by 34 and 42% upon silencing of cMaf or Beta2/NeuroD1 (Fig. 6b), whereas 7B2 mRNA levels were inhibited by 49 and 40% (Fig. 6c). No significant effect on PC2 and 7B2 levels was found upon transfection of the MafB siRNA (Fig. 6b and c). These results suggest that cMaf and Beta2/NeuroD1 are implicated in the transcriptional regulation of PC2 and 7B2.
FIG. 6.
Silencing of cMaf and Beta2/NeuroD1 results in decreased PC2 and 7B2 promoter mRNA levels. InR1G9 cells were transfected for 48 h with siRNA directed against MafB, cMaf, or Beta2/NeuroD1 (Beta2) or with a scrambled (Sc) control stealth siRNA. RNA was then isolated after 48 h, and a real-time RT-PCR was performed. (a) Quantitative real-time RT-PCR of MafB, cMaf, or Beta2/NeuroD1 over TBP to evaluate the silencing efficiency of the respective siRNA. (b) PC2 mRNA levels as measured by real-time RT-PCR. InR1G9 cells were transfected with an siRNA against cMaf, MafB, or Beta2/NeuroD1 or with a scrambled siRNA. Results were corrected for TBP mRNA levels. (c) 7B2 mRNA levels as measured by real-time RT-PCR. InR1G9 cells were transfected with an siRNA against cMaf, MafB, or Beta2/NeuroD1 or with a scrambled siRNA. Results were corrected for TBP mRNA levels. Values are the means ± the standard errors of the means from at least three separate experiments, each performed twice. *, P < 0.05; **, P < 0.01.
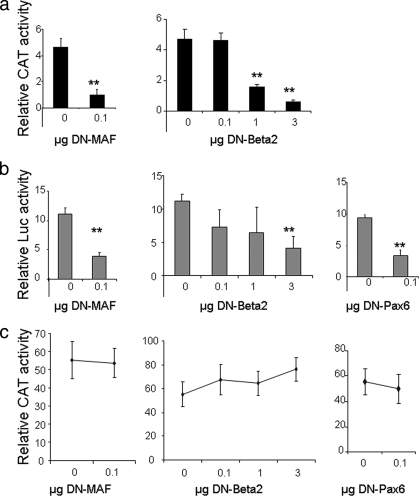
To assess the relevance of these factors in situ, we also used DN forms of Maf and Beta2/NeuroD1. A DN form of MafA which binds large-Maf consensus sequences and prevents activation of transcription by competing with the endogenous Mafs was overexpressed in InR1G9 cells (data not shown). This resulted in a significant fourfold decrease in PC2 promoter activity (Fig. 7a, left panel), while a DN form of Beta2/NeuroD1 previously shown to dominantly inhibit WT Beta2/NeuroD1 activity (9) resulted in dose-dependent inhibition (Fig. 7a, right panel). The high levels of the DN-Beta2/NeuroD1 plasmid required to inhibit PC2 promoter activity were similar to those previously reported by Cho et al. to decrease insulin gene transcription (9), suggesting that the DN form of Beta2/NeuroD1 is a weak inhibitor of WT Beta2/NeuroD1 or that Beta2/NeuroD1 is a weak activator of gene transcription.
FIG. 7.
The DN forms of Beta2/NeuroD1 and Maf inhibit transcription from the PC2 promoter. (a) InR1G9 cells were transiently transfected with the −633-bp PC2 promoter together with the cDNA that codes for DN-Beta2/NeuroD1 (DN-Beta2) or DN-Maf. Results represent relative CAT and Luc activities over the basal levels for each construct. (b) InR1G9 cells were transiently transfected with the −1,228-bp 7B2 promoter together with the cDNA that codes for DN-Beta2/NeuroD1, DN-Maf, or DN-Pax6. Results represent relative Luc and PAP activities over the basal levels for each construct. (c) InR1G9 cells were transiently transfected with the Enhancer-CMV-Basic reporter used as a control, together with increasing concentrations of the cDNA encoding the DN form of the transcription factor. Results represent relative CAT and Luc activities over the basal levels for each construct. The total DNA transfected was kept constant at 3 μg. Values are the means ± the standard errors of the means from at least three separate experiments, each performed twice. **, P < 0.01 (compared with the control).
Overexpression of the DN forms of MafA and Beta2/NeuroD1 led to quantitatively comparable results for the 7B2 promoter (Fig. 7b). In addition, transient overexpression of DN-Pax6 also resulted in inhibition of 7B2 promoter activity (Fig. 7b).
In order to verify that these transcriptional effects are specific to the respective promoters and not due to a general decrease in transcription, we transfected cells with the Enhancer-CMV-Basic reporter together with the DN form of Beta2/NeuroD1, MafA, or Pax6. No significant change in the transcription level of the CAT reporter under the control of the CMV promoter was observed (Fig. 7c).
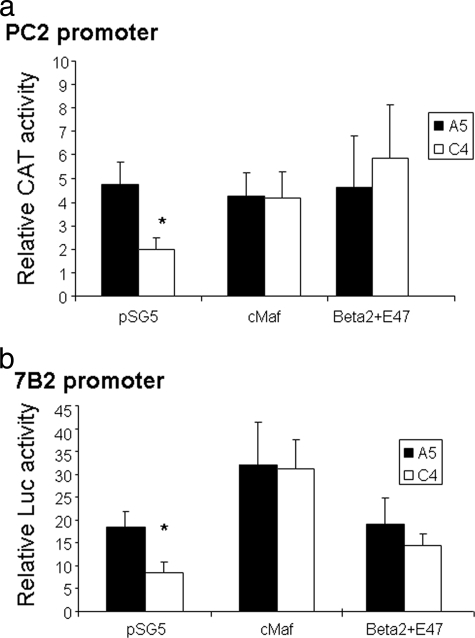
The cMaf and Beta2/NeuroD1 transcription factors are able to rescue the low transcription levels of PC2 and 7B2 in Pax6-DN306 clones.
To investigate whether the observed decrease in PC2 and 7B2 gene expression in Pax6-DN306 clones was indeed secondary to a decrease in cMaf or Beta2/NeuroD1 gene expression, we overexpressed cMaf and Beta2/NeuroD1 in the Pax6 DN clone, C4. The cMaf and Beta2/NeuroD1 proteins were both able to correct the low PC2 and 7B2 transcription levels in cells expressing the DN-Pax6 protein to that observed in the control A5 clone (Fig. 8a and b). Each factor compensated for the other and was able to rescue PC2 and 7B2 transcription in the Pax6-DN306 clone in this experiment, probably because of the high expression levels reached in the nucleus.
FIG. 8.
Overexpression of cMaf or Beta2/NeuroD1+E47 can rescue the decreased PC2 and 7B2 promoter activities in Pax6-DN306 clones. PC2 and 7B2 promoter activities in Pax6-DN306 clones overexpressing cMaf and Beta2/NeuroD1. The A5 and C4 clones were transfected with either the −633-bp PC2 human promoter linked to the CAT reporter or the empty reporter vector (pCAT-Basic) (a) or to the −1,228-bp mouse 7B2 promoter or the empty reporter (pGL2-Basic) (b) together with the annotated transcription factors. Results represent relative CAT and Luc or Luc and PAP activities over the basal levels for each construct. Values are the means ± the standard errors of the means from at least three separate experiments, each performed twice. *, P < 0.05 (compared with the control).
DISCUSSION
The present and previous results demonstrate the importance of Pax6 as a major regulator of glucagon biosynthesis through the activation of glucagon gene transcription by direct binding to the G1 and G3 elements on the glucagon promoter (42, 47), as well as by the activation of genes implicated in glucagon gene transcription such as cMaf, Beta2/NeuroD1, and Isl1, as demonstrated elsewhere (Gosmain et al., submitted; 21). We also show that Pax6 is important for proglucagon processing through the activation of the PC2 and 7B2 genes directly and indirectly.
Our data suggest that Pax6 indirectly regulates PC2 gene transcription through cMaf and Beta2/NeuroD1 activation in an additive manner, indicating that both of them play independent activating roles. Both cMaf and Beta2/NeuroD1 are decreased in DN-Pax6 clones and in cells transfected with Pax6 siRNA (Gosmain et al., submitted). Yatoh et al. have previously demonstrated the implication of Beta2/NeuroD1 in PC2 transcription regulation in hepatocytes by transfection studies. However, the mechanism of this activation was not investigated (73). The 7B2 gene is regulated by Pax6, cMaf, and Beta2/NeuroD1. A small decrease in transcriptional activity was observed upon the deletion of one or two of these sites, while a marked effect was observed upon the mutation of three sites in InR1G9 cells, suggesting that in our system the combination of all of these transcription factors is needed for the optimal control of 7B2 promoter activity. Indeed, deletion of all of the transcription factor binding sites in the 7B2 promoter results in complete silencing of the promoter, underlining the importance of these sites for 7B2 gene transcription. A synergistic effect between cMaf and Pax6 is observed, similar to the observed effect on the glucagon gene promoter (20). We previously demonstrated that cMaf can directly interact with Pax6 and form a heterodimeric complex on the G1 element of the glucagon gene promoter. However, the cMaf and Pax6 binding sites in G1 are next to each other while in the 7B2 promoter, the proximal Pax6 site and the Maf site are separated by approximately 100 bp. Determination of whether DNA looping allowing these two sites to be in close proximity in vivo, thus facilitating protein-protein interactions, or whether the interaction of additional cofactors bridging cMaf and Pax6 is involved requires further experiments. Overexpression of Beta2/NeuroD1 inhibited the transcriptional effects observed with cMaf alone and with cMaf and Pax6 (data not shown). A possible explanation for this is that Beta2/NeuroD1 might act both as an activator and as a repressor of transcription, depending on its cellular localization (27). Indeed, overexpression of Beta2/NeuroD1 together with other transcription factors might cause its shuttling outside of the nucleus and hence inhibit transcription. Alternatively, Beta2/NeuroD1 may activate or inhibit transcription, depending on its nuclear levels. The fact that its binding site is not conserved in the human promoter raises doubts as to its relevance in humans. Further work with human islets is needed to address this question. The role of Beta2/NeuroD1 in the activation of the mouse 7B2 gene is, however, clearly supported by our data. It is obvious that 7B2 is not regulated only by these three transcription factors; a more precise picture of the regulation of the 7B2 gene promoter should emerge from the identification of additional factors.
siRNAs directed to silence specifically the expression of Pax6, cMaf, or Beta2/NeuroD1 are capable of decreasing the levels of PC2 and 7B2 in InR1G9 cells, while MafB siRNA had no effect. Combined with the results obtained from the transactivation experiments, where cMaf is a more potent activator of the PC2 and 7B2 promoters than MafB is, we propose that cMaf and MafB are not interchangeable and redundant but functionally different even though they can bind and activate the same sites. It is still possible, however, since MafB is more abundant than cMaf in InR1G9 cells (20), that the 60% decrease in MafB obtained with its specific siRNA is not sufficient to affect PC2 or 7B2 gene expression while the same decrease in c-Maf results in clear effects. However, our results indicate, at least in InR1G9 cells, that MafB cannot compensate for the decrease in cMaf and strongly suggest the specificity of cMaf for PC2 and 7B2 activity.
The regulation of PC2 and 7B2 by Pax6 and Beta2/NeuroD1 might occur not only in pancreatic α cells but also in insulin- and somatostatin-producing cells. In insulin-producing cells, PC2 and 7B2 are coexpressed along with Pax6, Beta2/NeuroD1, MafA, and cMaf, suggesting that the same mechanisms of PC2 and 7B2 gene regulation might operate in β cells (20, 25, 45). Similarly, in somatostatin-producing cells, Pax6 is important for the control of hormone gene expression (59) and PC2 is probably also expressed in δ cells. Although this is disputed (45, 61, 62), PC2 knockout studies have revealed impaired processing of proinsulin, proglucagon, and prosomatostatin (13-15). It is therefore possible that Pax6 regulates the biosynthesis and processing not only of proglucagon but also of the two other major hormones of the pancreas, insulin and somatostatin.
Immunohistochemistry studies of the human pancreas indicate that PC2 and 7B2 are colocalized in α and β cells (45, 62), strengthening the close relationship of PC2 and 7B2. These observations are coherent with the function of 7B2 as a molecular chaperone of PC2.
Islet cells expressing 7B2 and PC2 are also immunoreactive for furin (45), which is implicated in PC2 activation. Our results indicate, however, that the furin gene is not a target of Pax6 but likely the target of more ubiquitous transcription factors as furin is widely expressed in neuroendocrine and nonneuroendocrine tissues (31, 51, 56, 67). Furthermore, during development, furin is expressed earlier, on embryonic day 7.5 (48, 74), than Pax6, which is expressed on embryonic day 9.0 (52, 63). Since furin is implicated in the activation of not only PC2 and 7B2 but also a variety of growth factors and hormones, receptors, plasma proteins, etc. (reviewed in reference 36), there are good reasons to think that it might be regulated differently from PC2. Similar reasons might be evoked for PC1/3, although our model system, α cells, may not be adequate to study the regulation of PC1/3. Indeed, PC1/3 mRNA was hardly detected and not altered in the Pax6-DN306 clone in agreement with data obtained in α cells of PC2 knockout mice (15, 66). The reported induction of PC1/3 in an α-cell line not expressing PC2 (αTCΔPC2 cells) is thus likely to be specific for this cell line (70, 72).
Since Pax6 has been previously demonstrated to be a highly important gene in the development of the pancreas, it seems likely that it will also directly or indirectly control genes implicated in endocrine cell function. We show here that, in α cells, Pax6 not only controls glucagon gene transcription but also regulates the transcription of other factors important for α-cell function, glucagon gene transcription, and proglucagon processing. In addition, our data indicate that 7B2 is also directly regulated by Pax6 while PC2 is regulated only by targets of Pax6, thus suggesting a regulatory mechanism in which 7B2 prevents PC2 from being misfolded or acting prematurely on its target hormones by Pax6 regulation.
Metabolic labeling analyses of the C4 clone revealed markedly decreased intracellular proglucagon and glucagon levels compared to those in the control clone (data not shown). This is probably a consequence of decreased glucagon gene transcription secondary to decreased Pax6, cMaf, Beta2/NeuroD1, and Isl1 (Gosmain et al., submitted; 21), which is proposed to directly control glucagon gene expression (1, 12, 17, 20, 69). This prevented us from quantifying the processed proglucagon peptides in the A5 and C4 clones. However, the decrease in 7B2 activity in the Pax6-DN306 clone resulted in altered pro-PC2 processing, indicating the functional relevance of our results.
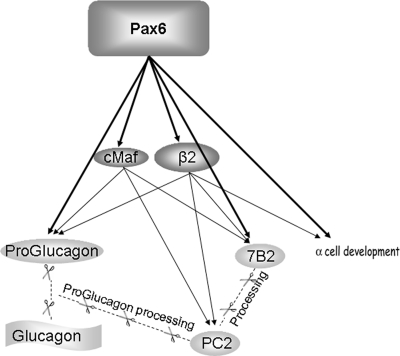
In conclusion, we describe a transcriptional network in which Pax6 is an important gene for glucagon biosynthesis. Pax6 controls glucagon gene transcription by directly binding to its promoter, as well as activating other factors implicated in glucagon gene regulation. Among these factors are cMaf and Beta2/NeuroD1, which also control PC2 and 7B2 gene transcription, the 7B2 gene also being controlled directly by Pax6 (Fig. 9).
FIG. 9.
Schematic representation of the role of Pax6 in glucagon biosynthesis through glucagon gene transcription and proglucagon processing.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation; Novo-Nordisk, Switzerland; and the Biology and Cancer Research Foundation.
We thank members of the J. Philippe laboratory and V. M. Schwitzgebel for discussions, R. W. James (University of Geneva) for critical reading of the manuscript, and M. J. Tsai (University of Texas M. D. Anderson Cancer Center) for helpful advice regarding the Beta2/NeuroD1 antibody. We also thank S. Saule (CNRS/UMR, Institute Curie), N. G. Seidah (Clinical Research Institute of Montreal), and W. J. van de Ven (University of Nijmegen) for generously providing Pax6, PC2, and 7B2 antibodies, respectively, and S. Singh (University of Texas M. D. Anderson Cancer Center), D. F. Steiner (University of Chicago), J. Orly (Hebrew University), G. Thiel (Saarland University), H. Suh-Kim (Catholic University, Seoul), and R. Stein (Vanderbilt University) for generously providing the DN form of human Pax6 (Pax6-DN306), PC2 promoter constructs, pCAT-Basic vector, Egr1, DN-Beta2 and cMaf, MafB, NeuroD1, E47, and DN-Maf expression vectors, respectively.
Footnotes
Published ahead of print on 17 February 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Artner, I., J. Le Lay, Y. Hang, L. Elghazi, J. C. Schisler, E. Henderson, B. Sosa-Pineda, and R. Stein. 2006. MafB—an activator of the glucagon gene expressed in developing islet α- and β-cells. Diabetes 55297-304. [DOI] [PubMed] [Google Scholar]
- 2.Ayoubi, T. A., H. L. van Duijnhoven, W. J. van de Ven, B. G. Jenks, E. W. Roubos, and G. J. Martens. 1990. The neuroendocrine polypeptide 7B2 is a precursor protein. J. Biol. Chem. 26515644-15647. [PubMed] [Google Scholar]
- 3.Bataille, D. 2007. Pro-protein convertases in intermediary metabolism: islet hormones, brain/gut hormones and integrated physiology. J. Mol. Med. 85673-684. [DOI] [PubMed] [Google Scholar]
- 4.Benjannet, S., A. M. Mamarbachi, J. Hamelin, D. Savaria, J. S. Munzer, M. Chretien, and N. G. Seidah. 1998. Residues unique to the pro-hormone convertase PC2 modulate its autoactivation, binding to 7B2 and enzymatic activity. FEBS Lett. 42837-42. [DOI] [PubMed] [Google Scholar]
- 5.Benjannet, S., D. Savaria, M. Chretien, and N. G. Seidah. 1995. 7B2 is a specific intracellular binding protein of the prohormone convertase PC2. J. Neurochem. 642303-2311. [DOI] [PubMed] [Google Scholar]
- 6.Braks, J. A., C. A. Broers, J. M. Danger, and G. J. Martens. 1996. Structural organization of the gene encoding the neuroendocrine chaperone 7B2. Eur. J. Biochem. 23660-67. [DOI] [PubMed] [Google Scholar]
- 7.Braks, J. A., and G. J. Martens. 1994. 7B2 is a neuroendocrine chaperone that transiently interacts with prohormone convertase PC2 in the secretory pathway. Cell 78263-273. [DOI] [PubMed] [Google Scholar]
- 8.Braks, J. A., A. M. Van Horssen, and G. J. Martens. 1996. Dissociation of the complex between the neuroendocrine chaperone 7B2 and prohormone convertase PC2 is not associated with proPC2 maturation. Eur. J. Biochem. 238505-510. [DOI] [PubMed] [Google Scholar]
- 9.Cho, J. H., I. S. Kwon, S. Kim, S. H. Ghil, M. J. Tsai, Y. S. Kim, Y. D. Lee, and H. Suh-Kim. 2001. Overexpression of BETA2/NeuroD induces neurite outgrowth in F11 neuroblastoma cells. J. Neurochem. 77103-109. [DOI] [PubMed] [Google Scholar]
- 10.Damholt, A. B., A. M. Buchan, J. J. Holst, and H. Kofod. 1999. Proglucagon processing profile in canine L cells expressing endogenous prohormone convertase 1/3 and prohormone convertase 2. Endocrinology 1404800-4808. [DOI] [PubMed] [Google Scholar]
- 11.Dhanvantari, S., N. G. Seidah, and P. L. Brubaker. 1996. Role of prohormone convertases in the tissue-specific processing of proglucagon. Mol. Endocrinol. 10342-355. [DOI] [PubMed] [Google Scholar]
- 12.Dumonteil, E., B. Laser, I. Constant, and J. Philippe. 1998. Differential regulation of the glucagon and insulin I gene promoters by the basic helix-loop-helix transcription factors E47 and BETA2. J. Biol. Chem. 27319945-19954. [DOI] [PubMed] [Google Scholar]
- 13.Furuta, M., R. Carroll, S. Martin, H. H. Swift, M. Ravazzola, L. Orci, and D. F. Steiner. 1998. Incomplete processing of proinsulin to insulin accompanied by elevation of Des-31,32 proinsulin intermediates in islets of mice lacking active PC2. J. Biol. Chem. 2733431-3437. [DOI] [PubMed] [Google Scholar]
- 14.Furuta, M., H. Yano, A. Zhou, Y. Rouille, J. J. Holst, R. Carroll, M. Ravazzola, L. Orci, H. Furuta, and D. F. Steiner. 1997. Defective prohormone processing and altered pancreatic islet morphology in mice lacking active SPC2. Proc. Natl. Acad. Sci. USA 946646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuta, M., A. Zhou, G. Webb, R. Carroll, M. Ravazzola, L. Orci, and D. F. Steiner. 2001. Severe defect in proglucagon processing in islet A-cells of prohormone convertase 2 null mice. J. Biol. Chem. 27627197-27202. [DOI] [PubMed] [Google Scholar]
- 16.Gabreëls, B. A., D. F. Swaab, D. P. de Kleijn, A. Dean, N. G. Seidah, J. W. Van de Loo, W. J. van de Ven, G. J. Martens, and F. W. Van Leeuwen. 1998. The vasopressin precursor is not processed in the hypothalamus of Wolfram syndrome patients with diabetes insipidus: evidence for the involvement of PC2 and 7B2. J. Clin. Endocrinol . Metab. 834026-4033. [DOI] [PubMed] [Google Scholar]
- 17.Gauthier, B. R., V. M. Schwitzgebel, M. Zaiko, A. Mamin, B. Ritz-Laser, and J. Philippe. 2002. Hepatic nuclear factor-3 (HNF-3 or Foxa2) regulates glucagon gene transcription by binding to the G1 and G2 promoter elements. Mol. Endocrinol. 16170-183. [DOI] [PubMed] [Google Scholar]
- 18.Gerich, J. E. 1993. Control of glycaemia. Bailliere's Clin. Endocrinol. Metab. 7551-586. [DOI] [PubMed] [Google Scholar]
- 19.Gherzi, R., H. C. Fehmann, R. Eissele, and B. Goke. 1994. Expression, intracellular localization, and gene transcription regulation of the secretory protein 7B2 in endocrine pancreatic cell lines and human insulinomas. Exp. Cell Res. 21320-27. [DOI] [PubMed] [Google Scholar]
- 20.Gosmain, Y., I. Avril, A. Mamin, and J. Philippe. 2007. Pax-6 and c-Maf functionally interact with the alpha-cell-specific DNA element G1 in vivo to promote glucagon gene expression. J. Biol. Chem. 28235024-35034. [DOI] [PubMed] [Google Scholar]
- 21.Gosmain, Y., E. Marthinet, A. Guerardel, L. S. Katz, A. Mamin, V. Schwitzgebel, and J. Philippe. 2007. The transcription factor Pax6 is a critical master gene for α-cell development. Diabetes 56(Suppl. 1)A443. [Google Scholar]
- 22.Graham, F. L., and A. J. van der Eb. 1973. Transformation of rat cells by DNA of human adenovirus 5. Virology 54536-539. [DOI] [PubMed] [Google Scholar]
- 23.Gromada, J., I. Franklin, and C. B. Wollheim. 2007. Alpha-cells of the endocrine pancreas: 35 years of research but the enigma remains. Endocr. Rev. 2884-116. [DOI] [PubMed] [Google Scholar]
- 24.Guo, S., and G. E. Sonenshein. 2004. Forkhead box transcription factor FOXO3a regulates estrogen receptor alpha expression and is repressed by the Her-2/neu/phosphatidylinositol 3-kinase/Akt signaling pathway. Mol. Cell. Biol. 248681-8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habener, J. F., D. M. Kemp, and M. K. Thomas. 2005. Minireview: transcriptional regulation in pancreatic development. Endocrinology 1461025-1034. [DOI] [PubMed] [Google Scholar]
- 26.Heller, R. S., M. Jenny, P. Collombat, A. Mansouri, C. Tomasetto, O. D. Madsen, G. Mellitzer, G. Gradwohl, and P. Serup. 2005. Genetic determinants of pancreatic epsilon-cell development. Dev. Biol. 286217-224. [DOI] [PubMed] [Google Scholar]
- 27.Itkin-Ansari, P., E. Marcora, I. Geron, B. Tyrberg, C. Demeterco, E. Hao, C. Padilla, C. Ratineau, A. Leiter, J. E. Lee, and F. Levine. 2005. NeuroD1 in the endocrine pancreas: localization and dual function as an activator and repressor. Dev. Dyn. 233946-953. [DOI] [PubMed] [Google Scholar]
- 28.Jansen, E., T. A. Ayoubi, S. M. Meulemans, and W. J. van De Ven. 1997. Regulation of human prohormone convertase 2 promoter activity by the transcription factor EGR-1. Biochem. J. 32869-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieffer, T. J., and J. F. Habener. 1999. The glucagon-like peptides. Endocr. Rev. 20876-913. [DOI] [PubMed] [Google Scholar]
- 30.Leung-Theung-Long, S., E. Roulet, P. Clerc, C. Escrieut, S. Marchal-Victorion, B. Ritz-Laser, J. Philippe, L. Pradayrol, C. Seva, D. Fourmy, and M. Dufresne. 2005. Essential interaction of Egr-1 at an islet-specific response element for basal and gastrin-dependent glucagon gene transactivation in pancreatic α-cells. J. Biol. Chem. 2807976-7984. [DOI] [PubMed] [Google Scholar]
- 31.Lou, H., A. M. Smith, L. C. Coates, N. X. Cawley, Y. P. Loh, and N. P. Birch. 2007. The transmembrane domain of the prohormone convertase PC3: a key motif for targeting to the regulated secretory pathway. Mol. Cell. Endocrinol. 26717-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mbikay, M., M. L. Raffin-Sanson, F. Sirois, L. Kalenga, M. Chretien, and N. G. Seidah. 2002. Characterization of a repressor element in the promoter region of proprotein convertase 2 (PC2) gene. Brain Res. Mol. Brain Res. 10235-47. [DOI] [PubMed] [Google Scholar]
- 33.Mbikay, M., N. G. Seidah, and M. Chretien. 2001. Neuroendocrine secretory protein 7B2: structure, expression and functions. Biochem. J. 357329-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller, L., P. Zhu, M. A. Juliano, L. Juliano, and I. Lindberg. 1999. A 36-residue peptide contains all of the information required for 7B2-mediated activation of prohormone convertase 2. J. Biol. Chem. 27421471-21477. [DOI] [PubMed] [Google Scholar]
- 35.Muller, L., X. Zhu, and I. Lindberg. 1997. Mechanism of the facilitation of PC2 maturation by 7B2: involvement in ProPC2 transport and activation but not folding. J. Cell Biol. 139625-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakayama, K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327(Pt. 3)625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishi, M., M. Sasahara, T. Shono, S. Saika, Y. Yamamoto, K. Ohkawa, H. Furuta, T. Nakao, H. Sasaki, and K. Nanjo. 2005. A case of novel de novo paired box gene 6 (PAX6) mutation with early-onset diabetes mellitus and aniridia. Diabetic Med. 22641-644. [DOI] [PubMed] [Google Scholar]
- 38.Nishimura, W., T. Kondo, T. Salameh, I. El Khattabi, R. Dodge, S. Bonner-Weir, and A. Sharma. 2006. A switch from MafB to MafA expression accompanies differentiation to pancreatic β-cells. Dev. Biol. 293526-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohneda, K., R. G. Mirmira, J. Wang, J. D. Johnson, and M. S. German. 2000. The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol. Cell. Biol. 20900-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Philippe, J. 1991. Insulin regulation of the glucagon gene is mediated by an insulin-responsive DNA element. Proc. Natl. Acad. Sci. USA 887224-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philippe, J., and M. Missotten. 1990. Functional characterization of a cAMP-responsive element of the rat insulin I gene. J. Biol. Chem. 2651465-1469. [PubMed] [Google Scholar]
- 42.Philippe, J., C. Morel, and M. Cordier-Bussat. 1995. Islet-specific proteins interact with the insulin-response element of the glucagon gene. J. Biol. Chem. 2703039-3045. [DOI] [PubMed] [Google Scholar]
- 43.Philips, G. T., C. N. Stair, H. Young Lee, E. Wroblewski, M. A. Berberoglu, N. L. Brown, and G. S. Mastick. 2005. Precocious retinal neurons: Pax6 controls timing of differentiation and determination of cell type. Dev. Biol. 279308-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Planque, N., L. Leconte, F. M. Coquelle, S. Benkhelifa, P. Martin, M. P. Felder-Schmittbuhl, and S. Saule. 2001. Interaction of Maf transcription factors with Pax-6 results in synergistic activation of the glucagon promoter. J. Biol. Chem. 27635751-35760. [DOI] [PubMed] [Google Scholar]
- 45.Portela-Gomes, G. M., L. Grimelius, and M. Stridsberg. 2008. Prohormone convertases 1/3, 2, furin and protein 7B2 (secretogranin V) in endocrine cells of the human pancreas. Regul. Pept. 146117-124. [DOI] [PubMed] [Google Scholar]
- 46.Powers, A. C., S. Efrat, S. Mojsov, D. Spector, J. F. Habener, and D. Hanahan. 1990. Proglucagon processing similar to normal islets in pancreatic alpha-like cell line derived from transgenic mouse tumor. Diabetes 39406-414. [DOI] [PubMed] [Google Scholar]
- 47.Ritz-Laser, B., A. Estreicher, N. Klages, S. Saule, and J. Philippe. 1999. Pax-6 and Cdx-2/3 interact to activate glucagon gene expression on the G1 control element. J. Biol. Chem. 2744124-4132. [DOI] [PubMed] [Google Scholar]
- 48.Roebroek, A. J., L. Umans, I. G. Pauli, E. J. Robertson, F. van Leuven, W. J. van de Ven, and D. B. Constam. 1998. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase furin. Development 1254863-4876. [DOI] [PubMed] [Google Scholar]
- 49.Rouillé, Y., S. Martin, and D. F. Steiner. 1995. Differential processing of proglucagon by the subtilisin-like prohormone convertases PC2 and PC3 to generate either glucagon or glucagon-like peptide. J. Biol. Chem. 27026488-26496. [DOI] [PubMed] [Google Scholar]
- 50.Rouillé, Y., G. Westermark, S. K. Martin, and D. F. Steiner. 1994. Proglucagon is processed to glucagon by prohormone convertase PC2 in alpha TC1-6 cells. Proc. Natl. Acad. Sci. USA 913242-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salvas, A., S. Benjannet, T. L. Reudelhuber, M. Chretien, and N. G. Seidah. 2005. Evidence for proprotein convertase activity in the endoplasmic reticulum/early Golgi. FEBS Lett. 5795621-5625. [DOI] [PubMed] [Google Scholar]
- 52.Sander, M., A. Neubuser, J. Kalamaras, H. C. Ee, G. R. Martin, and M. S. German. 1997. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 111662-1673. [DOI] [PubMed] [Google Scholar]
- 53.Sarac, M. S., S. Windeatt, M. G. Castro, and I. Lindberg. 2002. Intrapituitary adenoviral administration of 7B2 can extend life span and reverse endocrinological deficiencies in 7B2 null mice. Endocrinology 1432314-2323. [DOI] [PubMed] [Google Scholar]
- 54.Sarac, M. S., A. W. Zieske, and I. Lindberg. 2002. The lethal form of Cushing's in 7B2 null mice is caused by multiple metabolic and hormonal abnormalities. Endocrinology 1432324-2332. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt, G., F. Sirois, Y. Anini, L. M. Kauri, C. Gyamera-Acheampong, E. Fleck, F. W. Scott, M. Chretien, and M. Mbikay. 2006. Differences of pancreatic expression of 7B2 between C57BL/6J and C3H/HeJ mice and genetic polymorphisms at its locus (Sgne1). Diabetes 55452-459. [DOI] [PubMed] [Google Scholar]
- 56.Seidah, N. G., S. Benjannet, J. Hamelin, A. M. Mamarbachi, A. Basak, J. Marcinkiewicz, M. Mbikay, M. Chretien, and M. Marcinkiewicz. 1999. The subtilisin/kexin family of precursor convertases. Emphasis on PC1, PC2/7B2, POMC and the novel enzyme SKI-1. Ann. N. Y. Acad. Sci. 88557-74. [DOI] [PubMed] [Google Scholar]
- 57.Shen, X., Q. L. Li, G. A. Brent, and T. C. Friedman. 2005. Regulation of regional expression in rat brain PC2 by thyroid hormone/characterization of novel negative thyroid hormone response elements in the PC2 promoter. Am. J. Physiol. Endocrinol Metab. 288E236-E245. [DOI] [PubMed] [Google Scholar]
- 58.Sloop, K. W., M. D. Michael, and J. S. Moyers. 2005. Glucagon as a target for the treatment of type 2 diabetes. Expert Opin. Ther. Targets 9593-600. [DOI] [PubMed] [Google Scholar]
- 59.St-Onge, L., B. SosaPineda, K. Chowdhury, A. Mansouri, and P. Gruss. 1997. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature 387406-409. [DOI] [PubMed] [Google Scholar]
- 60.Takaki, R., J. Ono, Y. Yokogawa, S. Kumae, M. Nakamura, H. Koyama, and A. Kawaoi. 1984. Establishment of glucagon-producing cells by cell hybridization. Diabetes 33879-887. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka, S., S. Kurabuchi, H. Mochida, T. Kato, S. Takahashi, T. Watanabe, and K. Nakayama. 1996. Immunocytochemical localization of prohormone convertases PC1/PC3 and PC2 in rat pancreatic islets. Arch. Histol. Cytol. 59261-271. [DOI] [PubMed] [Google Scholar]
- 62.Tomita, T. 2001. Immunocytochemical localization of prohormone convertase 1/3 and 2 in pancreatic islet cells and islet cell tumors. Pancreas 23172-176. [DOI] [PubMed] [Google Scholar]
- 63.Turque, N., S. Plaza, F. Radvanyi, C. Carriere, and S. Saule. 1994. Pax-QNR/Pax-6, a paired box- and homeobox-containing gene expressed in neurons, is also expressed in pancreatic endocrine cells. Mol. Endocrinol. 8929-938. [DOI] [PubMed] [Google Scholar]
- 64.van Horssen, A. M., W. H. van den Hurk, E. M. Bailyes, J. C. Hutton, G. J. Martens, and I. Lindberg. 1995. Identification of the region within the neuroendocrine polypeptide 7B2 responsible for the inhibition of prohormone convertase PC2. J. Biol. Chem. 27014292-14296. [DOI] [PubMed] [Google Scholar]
- 65.Varlamov, O., L. D. Fricker, H. Furukawa, D. F. Steiner, S. H. Langley, and E. H. Leiter. 1997. β-Cell lines derived from transgenic Cpefat/Cpefat mice are defective in carboxypeptidase E and proinsulin processing. Endocrinology 1384883-4892. [DOI] [PubMed] [Google Scholar]
- 66.Vincent, M., Y. Guz, M. Rozenberg, G. Webb, M. Furuta, D. Steiner, and G. Teitelman. 2003. Abrogation of protein convertase 2 activity results in delayed islet cell differentiation and maturation, increased alpha-cell proliferation, and islet neogenesis. Endocrinology 1444061-4069. [DOI] [PubMed] [Google Scholar]
- 67.von Eggelkraut-Gottanka, R., and A. G. Beck-Sickinger. 2004. Biosynthesis of peptide hormones derived from precursor sequences. Curr. Med. Chem. 112651-2665. [DOI] [PubMed] [Google Scholar]
- 68.Walther, C., and P. Gruss. 1991. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 1131435-1449. [DOI] [PubMed] [Google Scholar]
- 69.Wang, M., and D. J. Drucker. 1995. The LIM domain homeobox gene isl-1 is a positive regulator of islet cell-specific proglucagon gene transcription. J. Biol. Chem. 27012646-12652 . [DOI] [PubMed] [Google Scholar]
- 70.Webb, G. C., A. Dey, J. Wang, J. Stein, M. Milewski, and D. F. Steiner. 2004. Altered proglucagon processing in an α-cell line derived from prohormone convertase 2 null mouse islets. J. Biol. Chem. 27931068-31075. [DOI] [PubMed] [Google Scholar]
- 71.Westphal, C. H., L. Muller, A. Zhou, X. Zhu, S. Bonner-Weir, M. Schambelan, D. F. Steiner, I. Lindberg, and P. Leder. 1999. The neuroendocrine protein 7B2 is required for peptide hormone processing in vivo and provides a novel mechanism for pituitary Cushing's disease. Cell 96689-700. [DOI] [PubMed] [Google Scholar]
- 72.Wideman, R. D., S. D. Covey, G. C. Webb, D. J. Drucker, and T. J. Kieffer. 2007. A switch from prohormone convertase (PC)-2 to PC1/3 expression in transplanted α-cells is accompanied by differential processing of proglucagon and improved glucose homeostasis in mice. Diabetes 562744-2752. [DOI] [PubMed] [Google Scholar]
- 73.Yatoh, S., T. Akashi, P. P. Chan, H. Kaneto, A. Sharma, S. Bonner-Weir, and G. C. Weir. 2007. NeuroD and reaggregation induce β-cell specific gene expression in cultured hepatocytes. Diabetes Metab. Res. Rev. 23239-249. [DOI] [PubMed] [Google Scholar]
- 74.Zheng, M., R. D. Streck, R. E. Scott, N. G. Seidah, and J. E. Pintar. 1994. The developmental expression in rat of proteases furin, PC1, PC2, and carboxypeptidase E: implications for early maturation of proteolytic processing capacity. J. Neurosci. 144656-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.