Abstract
Human DNA ligase I (hLigI) participates in DNA replication and excision repair via an interaction with proliferating cell nuclear antigen (PCNA), a DNA sliding clamp. In addition, hLigI interacts with and is inhibited by replication factor C (RFC), the clamp loader complex that loads PCNA onto DNA. Here we show that a mutant version of hLigI, which mimics the hyperphosphorylated M-phase form of hLigI, does not interact with and is not inhibited by RFC, demonstrating that inhibition of ligation is dependent upon the interaction between hLigI and RFC. To examine the biological relevance of hLigI phosphorylation, we isolated derivatives of the hLigI-deficient cell line 46BR.1G1 that stably express mutant versions of hLigI in which four serine residues phosphorylated in vivo were replaced with either alanine or aspartic acid. The cell lines expressing the phosphorylation site mutants of hLigI exhibited a dramatic reduction in proliferation and DNA synthesis and were also hypersensitive to DNA damage. The dominant-negative effects of the hLigI phosphomutants on replication and repair are due to the activation of cellular senescence, presumably because of DNA damage arising from replication abnormalities. Thus, appropriate phosphorylation of hLigI is critical for its participation in DNA replication and repair.
In mammalian cells, DNA replication occurs at discrete sites known as replication foci that contain DNA replication proteins. Proliferating cell nuclear antigen (PCNA) is a homotrimeric DNA sliding clamp that plays a key role in the synthesis of both leading and lagging strands by tethering DNA polymerases and other replication proteins to duplex DNA (18, 19). In addition, it also makes important contributions to the assembly of replication foci by recruiting various replication factors to these locations. For example, a conserved amino acid sequence identified in human DNA ligase I (hLigI) and other replication proteins as being required for localization to replication foci was subsequently shown to be a PCNA binding motif, also known as a PIP box (25, 37). Thus, PCNA binding is one mechanism by which DNA replication proteins are recruited to replication foci (25).
hLigI also interacts with replication factor C (RFC), a heteropentameric clamp loader that loads PCNA onto DNA during DNA replication (17). RFC inhibits joining by hLigI, but this inhibition is alleviated by PCNA in a reaction that requires the physical interaction between hLigI and PCNA (17). Such pairwise physical and functional interactions also occur among the homologous Saccharomyces cerevisiae proteins (34). More recently, it has been shown that hLigI also interacts with the hRad9-hRad1-hHus1 heterotrimeric clamp and its cognate loader, hRad17-RFC, which mediate cell cycle checkpoints activated by either DNA replication blockage or DNA damage (19, 30, 32, 35). In contrast to the inhibitory effect of RFC, hRad17-RFC weakly stimulates DNA joining by hLigI (30). While there is compelling evidence that the interaction between the hLigI PIP box and PCNA plays a critical role in cellular DNA replication and repair (16, 25), the biological significance of the interactions between hLigI and RFC and between hLigI and cell cycle checkpoint clamp and clamp loader complexes has not been established.
hLigI becomes increasingly phosphorylated during S phase, resulting in a hyperphosphorylated form in the G2 and M phases (11). Three serine residues, Ser 51, Ser 76, and Ser 91, are phosphorylated by Cdk2/cyclin A in a cell cycle-dependent manner (11, 14). Phosphorylation of Ser 91 at the G1/S transition is required for the phosphorylation of Ser 76 and the appearance of the hyperphosphorylated form of hLigI in M phase. In contrast, Ser 66, which is constitutively phosphorylated by casein kinase II, is dephosphorylated in a cell cycle-dependent manner (29). Replacement of these four phosphorylated serine residues with aspartic acid residues that mimic the charge of phosphorylated serine residues abolished the association of hLigI with replication foci in transient-transfection assays, whereas replacement of the same residues with alanine residues did not (11). Surprisingly, neither of these amino acid changes had any significant effect on PCNA binding in vitro (11). Interestingly, hRad17-RFC preferentially interacts with and stimulates a nonphosphorylated form of hLigI (30). Following DNA damage, hLigI is dephosphorylated (24, 30) and there is an increased association between hLigI and hRad17 (30), suggesting that posttranslational modification of hLigI regulates its interaction with the checkpoint clamp loader.
In this study, we show that the interaction between RFC and hLigI is regulated by hLigI phosphorylation. Specifically, a mutant version of hLigI that mimics the hyperphosphorylated M-phase form of hLigI interacts with PCNA but not RFC. Notably, this mutant version is not inhibited by RFC, demonstrating that the inhibition of DNA joining is dependent upon the physical interaction between RFC and hLigI. Furthermore, we show that the interaction between RFC and hLigI is required for efficient Okazaki fragment joining and long-patch base excision repair (BER) and that expression of phosphorylation site mutants of hLigI not only fails to complement the DNA damage sensitivity of hLigI-deficient cells but also induces cellular senescence.
MATERIALS AND METHODS
Proteins.
Wild-type hLigI and a mutant version (FA) in which the adjacent phenylalanine residues within the N-terminal PIP box were replaced by alanine residues (16, 25) were purified from baculovirus-infected insect cells as described previously (16, 36). To determine the effect of phosphorylation on protein-protein interactions, hLigI purified from baculovirus-infected insect cells was treated with λ protein phosphatase as recommended by the supplier (New England Biolabs). Aliquots of hLigI protein were also subjected to mock phosphatase treatment in the absence of λ protein phosphatase and incubation with λ protein phosphatase in the presence of phosphatase inhibitors. Unmodified wild-type hLigI was purified following overexpression in Escherichia coli (7).
A pVP-Flag5 plasmid encoding an N-terminal Flag-tagged version of hLigI (16) was mutated by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The mutations altered the coding sequence such that serine residues 51, 66, 76, and 91 were all replaced by either alanine or aspartic acid residues. After verification of the nucleotide sequence by DNA sequencing, N-terminal Flag-tagged mutant hLigI cDNAs were subcloned into the mammalian expression vector pRC/RSV. In addition, cDNAs encoding Flag-tagged versions of wild-type and mutant hLigI cDNAs were subcloned into the bacterial expression vector pRSF-Duet1, resulting in the presence of an additional His tag at the N terminus. Tagged versions of wild-type and mutant hLigI were purified from E. coli extracts by SP Sepharose Fastflow, Source Q, and Superdex 200 column chromatography.
Recombinant RFC was purified from baculovirus-infected insect cells as described previously (33).
Pull-down assays.
Glutathione S-transferase (GST) fusion proteins that contain PCNA (GST-PCNA), the N-terminal 320 residues of hRad17 (GST-N-hRad17), and the N-terminal 584 residues of RFC p140 (GST-N-p140) were expressed and purified as described previously (15, 17, 30). To prepare beads for pull-down assays, GST and GST fusion proteins (5 μg of each) were incubated with a 20-μl slurry of glutathione-Sepharose beads (Amersham Biosciences). For assays with GST and GST-N-hRad17, the beads were equilibrated and washed with 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol (DTT), 0.1% Igepal CA-630 (Sigma-Aldrich), 20 μg/ml bovine serum albumin prior to incubation with purified hLigI for 1 h at 4°C. For assays with GST, GST-PCNA, and GST-N-p140 beads, purified hLigI was adenylated by incubation with [α-32P]ATP (10 mCi/mmol; GE Healthcare) in 60 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM DTT, and 50 μg/ml bovine serum albumin for 15 min at 25°C. Equal amounts of adenylated protein were incubated with the beads liganded by GST, GST-PCNA, or GST-N-p140 that had been equilibrated in 50 mM HEPES-KOH, pH 7.5, 100 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 0.1% Igepal CA-630, 10% glycerol at 25°C for 30 min. Glutathione beads were collected by centrifugation and washed extensively with their equilibration buffer prior to resuspension in 20 μl sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. After separation by SDS-PAGE, purified hLigI was detected by immunoblotting while labeled, adenylated hLigI was visualized by PhosphorImager analysis.
RFC p140 was labeled with [35S]methionine by coupled in vitro transcription and translation and then partially purified by ammonium sulfate precipitation as described previously (6). Flag-tagged derivatives of hLigI purified from E. coli (1 μg of each) were conjugated to 15 μl of anti-Flag M2 affinity gel beads (Sigma) in 50 mM HEPES-KOH, pH 7.5, 100 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 0.1% Igepal CA-630, and 10% glycerol for 60 min at 4°C. The beads were washed extensively and then incubated with in vitro-translated RFC p140 in the same buffer for 60 min at 4°C. After collection by centrifugation and washing, the beads were resuspended in 20 μl SDS-PAGE sample buffer. Labeled RFC p140 eluted from the beads was detected by PhosphorImager analysis after separation by SDS-PAGE.
DNA-joining assays.
DNA-joining assays with purified wild-type and mutant versions of hLigI and a biotinylated nicked DNA substrate were carried out in the presence or absence of RFC as described previously (17).
Generation of 46BR.1G1 cell lines that stably express Flag-tagged versions of hLigI phosphorylation site mutants.
46BR.1G1 cells were transfected with pRC/RSV plasmid encoding the hLigI expression constructs using the Lipofectamine transfection reagent (Invitrogen) according to the manufacturer's directions. After selection for resistance to G418, single colonies were isolated. The level of hLigI protein in these clones was determined by immunoblotting with antibodies against hLigI (15) and the Flag epitope (Sigma). Clones that stably expressed tagged hLigI at levels similar to that of endogenous hLigI in wild-type simian virus 40-immortalized human fibroblasts were chosen for further analysis. Derivatives of 46BR.1G1 cells that stably express tagged versions of wild-type hLigI and a PCNA-binding-defective mutant have been described elsewhere (16).
Proliferation of 46BR.1G1 cell lines that stably express Flag-tagged versions of hLigI phosphorylation site mutants.
To measure cell proliferation, 105 cells were seeded in 60-mm dishes and then cultured at 37°C in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum containing 0.5 mg/ml of G418. After 24, 48, 72, 96, and 120 h, cells were washed with phosphate-buffered saline, trypsinized, and then counted using a particle counter (Beckman Instruments). Cell cycle distributions within asynchronous cultures were determined by fluorescence-activated cell sorter analysis in the Flow Cytometry Core of the University of Maryland Marlene and Stewart Greenebaum Cancer Center. To measure DNA synthesis, 105 cells were seeded in 60-mm dishes in DMEM containing 10% fetal bovine serum and 0.5 mg/ml G418. After 3 days, [methyl-3H]thymidine (40 to 60 Ci/mmol; GE Healthcare, Piscataway, NJ) was added (final concentration, 1 μCi/ml) and incubation was continued for 30 min. Cells were then washed with phosphate-buffered saline and resuspended in 0.5 M trichloroacetic acid. After extensive washing with 0.4 M trichloroacetic acid, acid-insoluble radioactivity was measured by liquid scintillation counting in an LS6500 multipurpose scintillation counter (Beckman, Fullerton, CA).
Expression of proteins involved in cell cycle checkpoints and senescence.
To detect expression of senescence-associated β-galactosidase (10), cells were fixed and stained using the senescence β-galactosidase staining kit (Cell Signaling Technology, Beverly, MA) according to the manufacturer's instructions. Cell staining was visualized using a Nikon Eclipse TE200 microscope, and images were processed using Adobe Photoshop Elements (Adobe, San Jose, CA). To determine the level of cell cycle checkpoint proteins, a whole-cell extract was prepared from ∼107 cells as described previously (13). Protein concentration was determined using the method of Bradford (4). Proteins in the whole-cell extracts (40 μg) were detected by immunoblotting after separation by SDS-PAGE using the following antibodies: anti-hLigI (rabbit polyclonal, 1:2,500), anti-Flag (mouse monoclonal, M-2; Sigma; 1:1,000), anti-β-actin (mouse monoclonal, AC-15; Abcam; 1:15,000), anti-RFC1 (rabbit polyclonal; Genetex; 1:2,000), anti-hRad17 (rabbit polyclonal, H-300; Santa Cruz Biotechnology; 1:1,000), anti-PCNA (mouse monoclonal, PC-10; Abcam; 1:1,000), anti-ATM (rabbit polyclonal, ab91; Abcam; 1:1,000), anti-ATR (goat polyclonal, N-19:sc-1887; Santa Cruz Biotechnology; 1:1,000), anti-Chk1 (mouse monoclonal, G-4; Santa Cruz Biotechnology; 1:1,000), anti-Chk2 (mouse monoclonal, B-4; Santa Cruz Biotechnology; 1:1,000), anti-p53 (mouse monoclonal DO1; Oncogene; 1:1,000), anti-p53 phospho-Ser15 (mouse monoclonal, 16G8; Cell Signaling; 1:1,000), anti-RB (mouse monoclonal, 1F8; NeoMarkers; 1:500), and anti-p16 (rabbit polyclonal, C-20; Delta BioLabs; 1:500).
Cell survival assay.
Derivatives of the 46BR.1G1 cell line (105 cells) were plated in six-well plates in DMEM supplemented with 10% fetal bovine serum and G418 (500 μg/ml). Various concentrations of methyl methanesulfonate (MMS) (0, 3, 10, and 30 μM) were added to the medium, and cells were cultured in drug-containing media for 5 days. Surviving cells were counted using an improved Neubauer chamber.
Extract assays of gap-filling synthesis-dependent ligation.
Nuclear extracts were prepared from derivatives of the 46BR.1G1 cell line (∼108 cells) according to the published protocol (8) with minor modifications. Briefly, cells were incubated in hypotonic buffer (20 mM HEPES-KOH, pH 7.8, 1 mM MgCl2, 5 mM KCl, 1 mM DTT) containing a cocktail of protease inhibitors for 1 h on ice and then lysed by Dounce homogenization (15). Nuclei were collected by centrifugation and then resuspended in 20 mM HEPES-KOH, pH 7.9, 20% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT plus a cocktail of protease inhibitors and then homogenized. After centrifugation, the supernatant was dialyzed against 20 mM HEPES-KOH, pH 7.9, 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 1 mM DTT plus a cocktail of protease inhibitors. The dialyzed nuclear extract was aliquoted and then flash-frozen at −70°C.
To measure gap-filling synthesis-dependent ligation by nuclear extracts (39), a linear duplex DNA with a 25-nucleotide gap was constructed by annealing two oligonucleotides to a complementary 58-mer oligonucleotide. A single labeled nucleotide was added to the 3′ end of the 18-mer that contributes the 5′ terminus to the gap by incubation with E. coli Klenow fragment (New England Biolabs, Ipswich, MA) and [α-32P]dGTP (50 μCi, 3,000 Ci/mmol; Perkin-Elmer, Waltham, MA). The labeled DNA substrate (7.5 pmol) was incubated with nuclear extract (7.5 μg) in reaction buffer containing 20 mM HEPES-KOH, pH 7.5, 70 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 mM ATP, and 200 μM deoxynucleoside triphosphates at 37°C. Aliquots (10 μl) were removed at various time intervals and added to an equal volume of formamide to stop the reaction. After separation by denaturing gel electrophoresis, labeled oligonucleotides were detected and quantitated by PhosphorImager analysis.
Long-patch BER.
Assays for long-patch BER of a synthetic abasic (AP) site analog were conducted as previously described (21). Briefly, 10 ng of a circular double-stranded DNA carrying a synthetic AP site (tetrahydrofuran) within a unique SacI site was incubated with 5 μg of the nuclear extract from 46BR.1G1 cells and an indicated amount of hLigI purified from E. coli in a 40-μl reaction mixture. After 60 min of incubation at 37°C, the DNA substrate was recovered by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation and then digested with AP endonuclease 1 followed by SacI. Subsequently, the digested samples were subjected to electrophoresis in a 1% agarose gel in Tris-borate-EDTA buffer and stained with SYBR green. The ratio of nicked circular DNA (unrepaired DNA) and the SacI-linearized DNA (repaired DNA) was quantitated from gel images taken with a digital camera.
RESULTS
Phosphorylation of hLigI modulates its interaction with RFC.
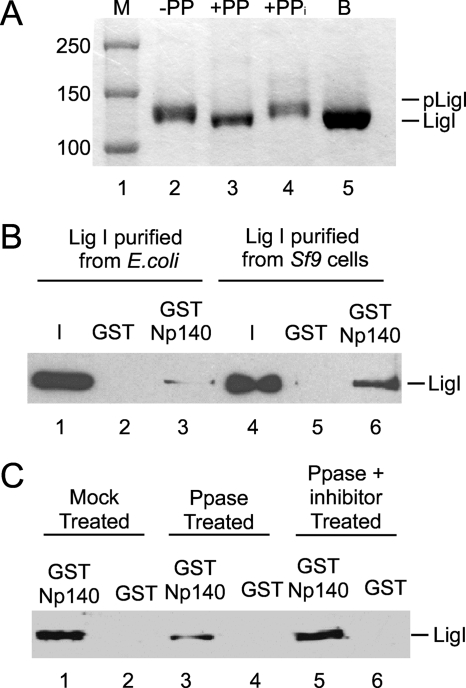
Previously we demonstrated that hLigI interacts with the two subunits, hRad17 and RFC p140, that are unique to the hRad17-RFC and RFC clamp loader complexes, respectively (17, 30). Since hLigI is phosphorylated during cell cycle progression (11) and hRad17 preferentially binds to unmodified hLigI (30), we asked whether the interaction with RFC p140 is also modulated by phosphorylation of hLigI. As expected, the mobility of phosphorylated hLigI purified from insect cells was lower than that of unmodified hLigI purified from E. coli unless the hLigI from insect cells was preincubated with λ phosphatase (Fig. 1A). In initial studies, we compared the levels of binding of phosphorylated and unmodified hLigI to the N-terminal half of RFC p140 (Fig. 1B). Approximately three- to fourfold-more phosphorylated hLigI than unmodified hLigI was retained by glutathione beads liganded by the N terminus of RFC p140 fused to GST (Fig. 1B, compare lanes 3 and 6). To demonstrate that this difference in binding was due to phosphorylation, hLigI purified from insect cells was incubated with λ phosphatase. As expected, this treatment significantly reduced the binding of hLigI to the GST-p140 beads (Fig. 1C, compare lanes 1 and 3) whereas incubation with λ phosphatase in the presence of phosphatase inhibitors did not (Fig. 1C, compare lanes 1, 3, and 5).
FIG. 1.
RFC preferentially interacts with phosphorylated hLigI. (A) hLigI (0.8 μg) purified from insect cells was incubated without λ protein phosphatase (−PP, lane 2) and with λ protein phosphatase either in the absence (PP, lane 3) or in the presence of phosphatase inhibitors (PPi, lane 4). hLigI (1 μg) purified from E. coli was also used (B, lane 5). Proteins were separated by SDS-PAGE with Coomassie blue. The positions of molecular mass standards (M, lane 1) and phosphorylated (pLigI) and unmodified (LigI) hLigI are indicated on the left and right, respectively. Numbers at left are molecular masses in kilodaltons. (B) The binding of hLigI purified from E. coli (lanes 1 to 3) or insect cells (lanes 4 to 6) to GST (lanes 2 and 5) or GST-N-p140 beads (lanes 3 and 6) was detected by immunoblotting. Lanes 1 and 4 contain 10% of the hLigI input (0.1 μg hLigI). (C) hLigI (1 μg) from insect cells was mock treated (lanes 1 and 2), λ phosphatase treated (lanes 3 and 4), or coincubated with λ phosphatase and λ phosphatase inhibitor (lanes 5 and 6). hLigI in the eluates was detected by immunoblotting.
Replacement of serine residues 51, 66, 76, and 91 with aspartic acid abolishes the interaction of hLigI with RFC but not with PCNA.
Using electron spray mass spectrometry analysis, we identified 10 phosphorylated amino acids in hLigI purified from insect cells. These included residues characterized by the Montecucco lab (11, 29), residues identified in recent analyses of the human phosphoproteome (3, 26), and several novel phosphorylation sites (A. Yang and A. E. Tomkinson, unpublished results). Given the large number of phosphorylation sites, several of which have not been characterized in human cells, we decided against a systematic site-directed mutagenesis approach to identify the combinations of phosphorylation sites that enhance binding to RFC p140. Instead we chose to focus on the four serine residues, Ser 51, 66, 76, and 91, because phosphorylation of at least two of these residues is required for the generation of the M-phase hyperphosphorylated form of hLigI that does not associate with PCNA (11). Furthermore, replacement of the four serine residues with aspartic acid to mimic phosphorylation was shown to abrogate targeting of hLigI to replication foci but had no effect on PCNA binding in vitro (11).
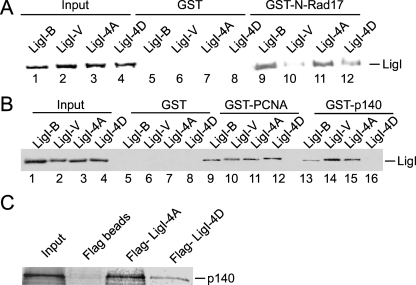
Wild-type hLigI and mutant versions in which all four serine residues were replaced with either alanine (4A) or aspartic acid (4D) were expressed in E. coli and then purified. Previously, we showed that hRad17 preferentially interacts with nonphosphorylated hLigI (30). In accord with these results, wild-type hLigI and the 4A version of hLigI exhibited similar levels of binding to the N-terminal region of hRad17 (Fig. 2A, lanes 9 and 11) and this binding was much greater than that of phosphorylated hLigI purified from insect cells (Fig. 2A, lane 10). Notably, the binding of the 4D version of hLigI (Fig. 2A, lane 12) was reduced compared with that of the 4A version and wild-type hLigI purified from E. coli, albeit not to the same extent as that with phosphorylated hLigI from insect cells (Fig. 2A, lane 10). Thus, the aspartic acid substitutions appear to mimic, at least in part, phosphorylation at the four serine residues.
FIG. 2.
Interaction of hLigI phosphomutants with RFC. (A) Wild-type hLigI purified from insect cells (LigI-V) and E. coli (LigI-B) and hLigI 4A (LigI-4A) and 4D (LigI-4D) purified from E. coli (1 μg of each) were incubated with GST (lanes 5 to 8) or GST-N-Rad17 (lanes 9 to 12) beads as indicated. hLigI was detected in the eluates by immunoblotting. The input lanes (1 to 4) contain 10% of the hLigI input (0.1 μg hLigI). (B) Labeled adenylated wild-type hLigI purified from insect cells (LigI-V) and E. coli (LigI-B) and hLigI 4A (LigI-4A) and 4D (LigI-4D) purified from E. coli (1 μg of each) were incubated with GST (lanes 5 to 8), GST-PCNA (lanes 9 to 12), or GST-p140 (lanes 13 to 16) beads as indicated. Radiolabeled hLigI was detected in the eluates by PhosphorImager analysis. The input lanes (1 to 4) contain 10% of the hLigI input (0.1 μg hLigI). (C) Labeled in vitro-translated RFC p140 was incubated with anti-Flag beads liganded by no protein, Flag-LigI 4A, and Flag-LigI 4D as described in Materials and Methods. RFC p140 in the eluates from the beads was detected by PhosphorImager analysis. The left lane contains 10% of the input RFC p140.
In agreement with the evidence that hLigI phosphorylation does not regulate its interaction with PCNA (11), replacement of the four serine residues with aspartic acid did not significantly alter PCNA binding (Fig. 2B, lane 12). However, the 4D version was markedly defective in binding to the N terminus of RFC p140 compared with the 4A version (Fig. 2B, compare lanes 15 and 16) and wild-type hLig I purified from E. coli (Fig. 2B, compare lanes 13 and 16). To confirm that the aspartic acid substitutions do indeed reduce binding to RFC p140, we performed a reciprocal experiment with in vitro-translated RFC p140 and beads liganded by Flag-tagged derivatives of hLigI purified from E. coli. As expected, significantly less RFC p140 was retained on the beads liganded by the 4D version of hLigI (Fig. 2C, lanes 3 and 4).
Interaction with RFC inhibits hLigI catalytic activity.
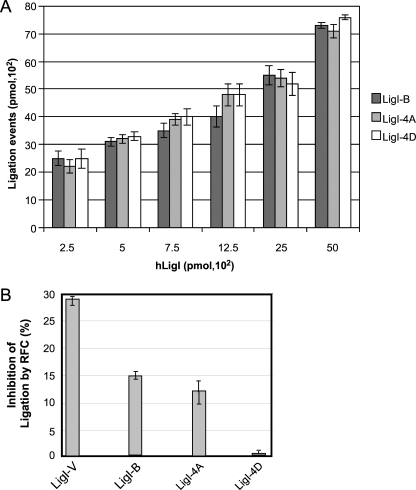
The DNA-joining activities of the 4A and 4D versions of hLigI were indistinguishable from that of wild-type hLigI purified from E. coli (Fig. 3A). This was not unexpected, because the four serine residues being replaced reside within the noncatalytic N-terminal region of hLigI. In previous studies with yeast and human proteins, we have shown that RFC inhibits joining by the replicative DNA ligase (17, 34). Since both RFC and hLigI interact with 3′ termini at interruptions in duplex DNA, it is possible that RFC inhibits hLigI by binding to DNA nicks, thereby excluding hLigI from its substrate. The defect in RFC binding exhibited by the 4D version of hLigI allowed us to delineate the role of the physical interaction between RFC and hLigI in the inhibition of DNA joining. Specifically, we compared the abilities of RFC to inhibit versions of hLigI that differ in their RFC binding properties. Phosphorylated wild-type hLigI from insect cells, which binds most effectively to RFC (Fig. 1), was inhibited about 30% by RFC, while bacterially expressed unmodified wild-type and 4A versions of hLigI, which bind less well to RFC, were inhibited by only 15% and 12%, respectively (Fig. 3B). Strikingly, the bacterially expressed 4D version of hLigI, which is defective in RFC binding, is not inhibited by RFC (Fig. 3B). Together these results demonstrate that RFC does not inhibit joining by simply occluding the DNA nick and that the inhibition of DNA joining by RFC is dependent upon the physical interaction between RFC and hLigI.
FIG. 3.
DNA-joining activity of hLigI phosphomutants; inhibition of hLigI phosphomutants by RFC. (A) The joining of a nicked DNA substrate (1 pmol) by wild-type hLigI (LigI-B) and hLigI phosphomutants (LigI-4A and LigI-4D) purified from E. coli was measured as described in Materials and Methods. The results shown graphically were compiled from three independent experiments with the error bars indicating standard deviations. (B) The joining of a nicked DNA substrate (1 pmol) by the indicated versions of hLigI (1 pmol) was measured in the absence or presence of RFC (1 pmol) as described in Materials and Methods. Results were from two independent experiments and are expressed as the percent inhibition of ligation by RFC. The error bars indicate standard deviations.
Effect of phosphorylation site mutants of hLigI on cell proliferation.
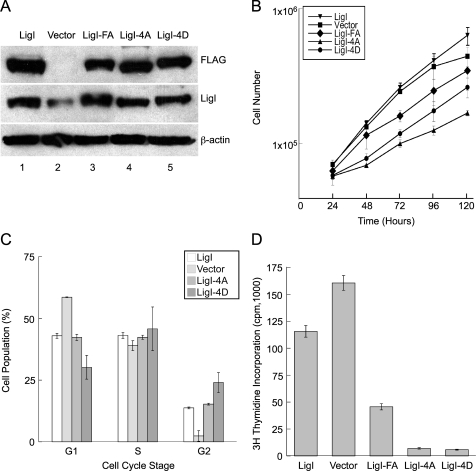
Previously we have shown that replacement of key adjacent phenylalanine residues within the hLigI PIP box with alanine residues disrupts PCNA binding but has no effect on catalytic activity (16). In addition, we showed that expression of the PCNA-binding-defective version of hLigI (FA) in hLigI-deficient 46BR.1G1 cells failed to correct replication and repair defects (1, 16, 28). To provide evidence that phosphorylation of hLigI serine residues 51, 66, 76, and 91 is biologically significant, we isolated stable derivatives of 46BR.1G1 cells expressing similar levels of Flag-tagged versions of wild-type hLigI and the 4A and 4D mutants (Fig. 4A). In agreement with published studies (16), expression of wild-type hLigI did not significantly enhance the initial rate of proliferation of 46BR.1G1 cells (Fig. 4B). In contrast, expression of the mutant versions of hLigI reduced the initial rate of cell proliferation, with the phosphorylation site mutants (4D and 4A) having a greater effect than the PCNA-interaction-defective mutant (FA) (Fig. 4B). Fluorescence-activated cell sorting analysis of asynchronous cell populations revealed that, although the 4D cell line exhibited a small increase in the fraction of cells in S phase, the cell cycle distributions of the 4A cell line and the complemented derivative expressing wild-type hLigI were essentially the same (Fig. 4C). In contrast, the parental 46BR.1G1 cell population contained a higher fraction of G0/G1 cells.
FIG. 4.
Effect of stable expression of hLigI phosphomutants in hLigI-deficient 46BR.1G1 cells on cell proliferation and DNA synthesis. (A) Whole-cell extracts (40 μg) from 46BR.1G1 cells stably transfected with the empty expression vector (Vector) and derivatives of 46BR.1G1 cells that stably express Flag-tagged versions of wild-type hLigI (LigI), the phosphomutants (LigI-4A and LigI-4D), and the PIP box mutant (LigI-FA) were separated by SDS-PAGE. Flag-tagged and endogenous hLigI were detected by immunoblotting with anti-Flag and anti-hLigI antibodies, respectively. To control for extract loading, β-actin was also detected by immunoblotting. (B) Cultures of the same derivatives of 46BR.1G1 cells were seeded in 60-mm dishes (105 cells per dish). At various time intervals, cells were washed, trypsinized, and counted using a Coulter Counter. The graph shows data compiled from three independent experiments with the error bars indicating the standard errors of the means. (C) The cell cycle distributions of asynchronous populations of the 46BR.1G1 derivatives were determined by fluorescence-activated cell sorting analysis. The graph shows data compiled from two independent experiments with the error bars indicating standard deviations. (D) The indicated 46BR.1G1 derivatives were seeded in duplicate into 60-mm dishes (105 cells per dish). After 3 days, [3H]thymidine was added for 30 min. Incorporation of [3H]thymidine into DNA was measured by liquid scintillation counting. The graph represents data from three independent experiments with the error bars indicating the standard errors of the means.
Because of the role of hLigI in DNA replication, we examined DNA synthesis as measured by incorporation of tritiated thymidine into genomic DNA. Approximately 50% more thymidine was incorporated by the parental 46BR.1G1 cells than by the stable derivative expressing wild-type hLigI (Fig. 4D). This is consistent with studies indicating that the defect in joining Okazaki fragments in 46BR.1G1 cells results in an increase in DNA synthesis because unlinked Okazaki fragments are released during DNA replication, presumably by strand displacement DNA synthesis (28). Strikingly, expression of both 4A and 4D phosphomutant versions of hLigI dramatically reduced DNA synthesis (Fig. 4D). In accord with the cell proliferation results (Fig. 4B), expression of the PCNA interaction-defective (FA) version of hLigI also reduced DNA synthesis, albeit to a lesser extent than that with the phosphomutants (Fig. 4D).
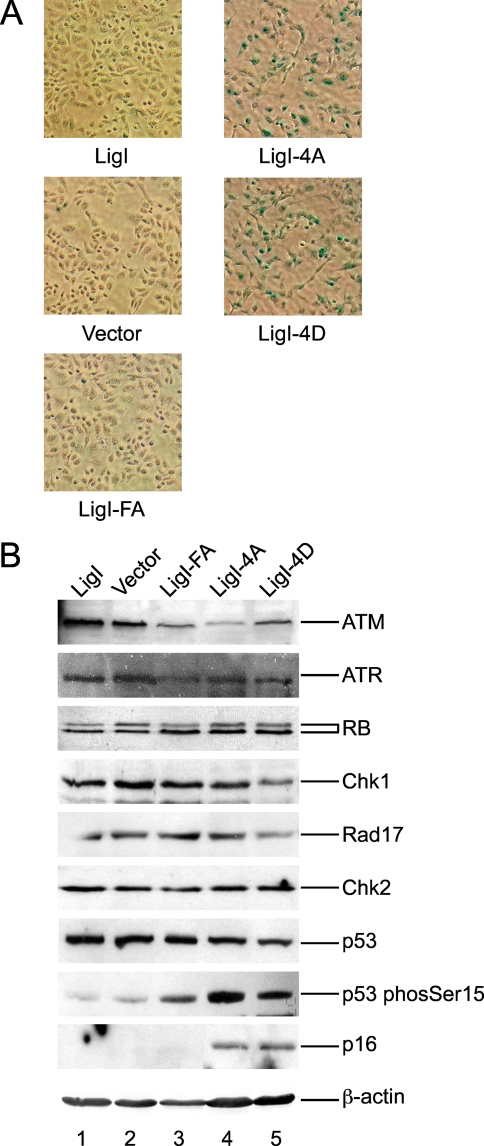
The morphology of the cells expressing the phosphomutant versions of hLigI (Fig. 5A, right panels) was clearly different from that of uncomplemented 46BR.G1 cells and the stable derivatives expressing either wild-type hLigI or the PCNA interaction mutant (Fig. 5A, left panels). A fraction of the 46BR.1G1 cells expressing the 4A and 4D versions of hLigI were increased in size and had a flattened appearance, features that are characteristic of senescent cells. In accord with this observation, a significant proportion of these cells express senescence-associated β-galactosidase (Fig. 5A) and contain elevated levels of p16, p53 phosphorylated on Ser15, and hypophosphorylated Rb (Fig. 5B), each of which is indicative of cellular senescence possibly triggered by DNA damage (2, 9). Thus, our results suggest that expression of the phosphorylation site mutant versions of hLigI results in the activation of the senescence program. Interestingly, cells expressing the 4A and 4D versions of hLigI have reduced levels of the upstream signal transduction proteins ATM, Chk1, and hRad17 (Fig. 5B). This may reflect an adaptation favoring proliferation instead of senescence.
FIG. 5.
Expression of hLigI phosphomutants in hLigI-deficient 46BR.1G1 cells induces cellular senescence. (A) Cultures of 46BR.1G1 cells transfected with the empty expression vector (Vector) and derivatives of 46BR.1G1 cells that stably express Flag-tagged versions of wild-type hLigI (LigI), the phosphomutants (LigI-4A and LigI-4D), and the PIP box mutant (LigI-FA) were seeded in six-well plates at 6 × 105 cells per well and allowed to attach overnight prior to fixation and staining for expression of β-galactosidase (magnification, ×28). (B) ATM, ATR, RB, Rad17, Chk1, Chk2, p53, p53 phosphorylated on serine 15, p16, and β-actin were detected in whole-cell extracts (40 μg) from the indicated derivatives of 46BR.1G1 cells by immunoblotting.
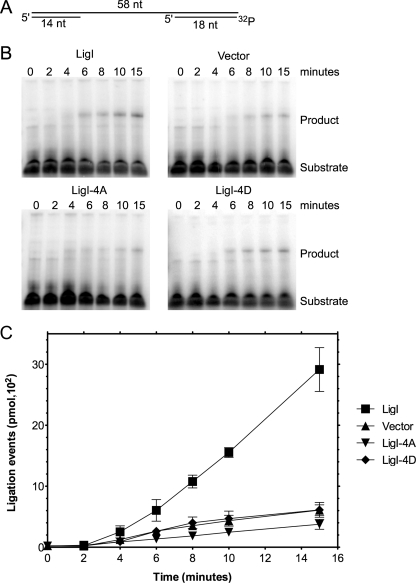
Because hLigI phosphorylation modulates its interaction with both the replicative (Fig. 1 and 2) and checkpoint (30) clamp loaders, the phenotype of the cells expressing the phosphomutant version of hLigI may reflect checkpoint activation that occurs either indirectly because of a replication defect or by a direct effect on the checkpoint protein. To more directly address the effect of hLigI phosphorylation on DNA replication, we examined the activity of extracts from 46BR.1G1 cells and derivatives of this cell line expressing wild-type and mutant versions of hLigI in assays that mimic Okazaki fragment joining. Using a gapped DNA substrate that resembles an intermediate of Okazaki fragment processing (Fig. 6A), we found that, unlike wild-type hLigI, expression of the phosphorylation site mutants in hLigI-deficient 46BR.1G1 cells failed to complement the defect in ligation-coupled gap-filling DNA synthesis in cell extract assays (Fig. 6B). Because derivatives of 46BR.1G1 cells expressing either the 4A or the 4D versions of hLigI had reduced DNA synthesis activity, we asked whether the extracts were defective in either gap-filling DNA synthesis or primer extension in assays with modified unlabeled versions of the ligation substrate. Extracts from the 4A and 4D derivatives were also defective in DNA synthesis although not to the same extent as that with the defect in ligation (see Fig. S1 in the supplemental material). These results suggest that the interaction between hLigI and RFC is critical for coordinating gap-filling synthesis and ligation.
FIG. 6.
Effect of the expression of hLigI phosphomutants on the defect in the repair of gapped DNA of hLigI-deficient 46BR.1G1 cells. (A) Schematic representation of the labeled gapped DNA substrate. The position of the labeled phosphate group is indicated by 32P. (B) Repair of the gapped DNA substrate (7.5 pmol) by nuclear extracts (7.5 μg) from 46BR.1G1 cells transfected with the empty expression vector (Vector) and derivatives of 46BR.1G1 cells that stably express Flag-tagged versions of wild-type hLigI (LigI) and the phosphomutants (LigI-4A and LigI-4D) was measured as a function of time. The positions of the labeled substrate and ligated product are indicated. (C) Graphic representation of three independent experiments that were performed in duplicate. The error bars represent the standard errors of the means.
Expression of phosphorylation site mutants of hLigI increases the sensitivity of hLigI-deficient 46BR.1G1 cells to killing by MMS.
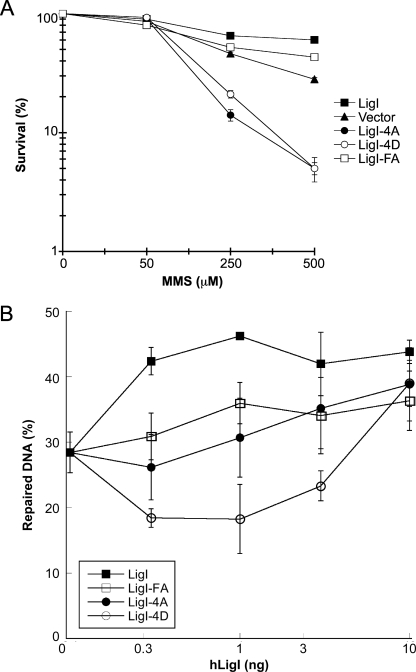
RFC, PCNA, and hLigI also participate in the long-patch BER subpathway of BER (23). Previously, we have shown that the hLigI mutant cell line 46BR.1G1 is hypersensitive to killing by MMS because of a defect in long-patch BER and that efficient long-patch BER is dependent upon the interaction between PCNA and hLigI (16). This prompted us to determine the effect of expressing the hLigI phosphorylation site mutants on the DNA damage sensitivity of 46BR.1G1 cells. As expected, expression of wild-type hLigI significantly increased resistance to MMS whereas the PIP box mutant (FA) of hLigI was less effective at restoring resistance to MMS (Fig. 7A). In contrast, expression of the phosphorylation site mutants 4A and 4D markedly increased the hypersensitivity of the 46BR.1G1 cells to killing by MMS (Fig. 7A). Thus, it appears that expression of the hLigI phosphorylation site mutants has a dominant-negative effect on both replicative DNA synthesis and the repair of DNA alkylation damage.
FIG. 7.
Effect of the expression of hLigI phosphomutants on the sensitivity of hLigI-deficient 46BR.1G1 cells to MMS; effect of hLigI phosphomutants on long-patch BER activity of nuclear extracts from hLigI-deficient 46BR.1G1 cells. (A) Cultures of 46BR.1G1 cells transfected with the empty expression vector (Vector) and derivatives of 46BR.1G1 cells that stably express Flag-tagged versions of wild-type hLigI (LigI), the phosphomutants 4A (LigI-4A) and 4D (LigI-4D), and the PIP box mutant (LigI-FA) were seeded in triplicate at 4 × 104 cells per dish. The next day, cells were incubated with MMS at the indicated concentration for 1 h and then the medium containing MMS was replaced with fresh medium. After 5 days, surviving cells were counted using an improved Neubauer chamber. The graph is a compilation of two independent experiments with the error bars representing standard deviations. (B) The repair of a circular substrate containing a single synthetic AP site (10 ng) by a nuclear extract of 46BR.1G1 cells (5 μg) supplemented with versions of hLigI purified from E. coli—wild-type hLigI (LigI), PCNA-interaction-defective version of hLigI (LigI-FA), and the hLigI phosphomutants (LigI-4A and LigI-4D)—was measured as described in Materials and Methods. The graph is a compilation of three independent experiments with the error bars representing standard deviations.
To more directly determine the influence of hLigI phosphorylation on the ligation step of long-patch BER, we examined the effect of adding purified unmodified versions of hLigI to 46BR.1G1 extracts on the repair of a circular DNA substrate with a single synthetic AP site that cannot be repaired by short-patch BER (16, 23). In accord with the results that we obtained with phosphorylated versions of hLigI purified from insect cells (16), addition of wild-type hLigI corrected the defect in AP site repair more efficiently than did the FA version that is defective in binding to PCNA. To determine whether unmodified versions of hLigI are phosphorylated by the cell extract, we performed similar assays in the presence of [γ-32P]ATP. Wild-type hLigI and the 4A and 4D mutants were labeled by incubation in the cell extract (see Fig. S2 in the supplemental material), indicating that these proteins are phosphorylated by kinases in the cell extract and that phosphorylation is not limited to Ser 51, Ser 66, Ser 76, and Ser 91. At lower concentrations, the 4D phosphomutant version of hLigI inhibited the BER activity of the cell extract whereas the activity of the 4A phosphomutant was similar to that of the FA version that is defective in binding to PCNA (Fig. 7B). Together these results demonstrate that, like the interaction with PCNA (16), hLigI phosphorylation and the interaction between hLigI and RFC are critical for efficient long-patch BER.
DISCUSSION
Previously we and others showed that hLigI physically and functionally interacts with the clamp loader and clamp complexes involved in DNA replication and cell cycle checkpoints (15-17, 30-32, 35). Although the in vitro interactions with the PCNA and hRad9-hRad1-hHus1 clamps are not sensitive to the phosphorylation status of hLigI (11, 30), we have shown that the checkpoint clamp loader hRad17-RFC preferentially binds to unphosphorylated hLigI (30). Here we demonstrate that the interaction of RFC with hLigI is also influenced by phosphorylation and that this posttranslational modification plays a critical role in regulating the in vivo functions of hLigI.
The interaction between PCNA and hLigI is required to target hLigI to replication foci and for coordinating DNA ligation with the DNA synthesis and processing reactions involved in Okazaki fragment maturation and joining, as well as during long-patch BER (16, 22, 25). In contrast, relatively little is known about the functional and biological significance of the interaction between RFC and hLigI. Here we have shown that a phosphorylation site mutant version of hLigI (4D), in which four serine residues were replaced with aspartic acid residues to mimic the hyperphosphorylated M-phase-specific form, is defective in interacting with RFC but retains PCNA binding activity. In transient-transfection assays with COS7 cells, the 4D but not the 4A version of hLigI was defective in targeting to replication foci (11). However, we did not observe a defect in the localization of either the 4D or the 4A version of hLigI to replication foci when these proteins were stably expressed in hLigI-deficient 46BR.1G1 cells. Similar results have been obtained by the Montecucco lab (30a). Thus, it appears that the stable association of hLigI with replication factories is not dependent upon its interaction with RFC.
The interactions of hLigI with both RFC and PCNA may be involved in coordinating reactions during Okazaki fragment metabolism and long-patch BER. Previously, we have shown that the interaction between hLigI and PCNA is required to alleviate the inhibitory effect of RFC on DNA joining (17, 34). Here we show that the inhibition of DNA joining by RFC is dependent upon the physical interaction between RFC and hLigI. Thus, it appears that RFC interacts with and holds hLigI in an inactive conformation. Although it is possible that RFC delivers hLigI to the PCNA trimer that remains linked to duplex DNA in the vicinity of a DNA nick generated between adjacent Okazaki fragments, there are contradictory reports as to whether RFC remains associated with the 3′-OH primer and/or loaded PCNA during lagging-strand DNA synthesis by polymerase δ (27, 38). Indeed, in a recent study with purified recombinant proteins, Masuda et al. have provided evidence that RFC stably associates with PCNA and polymerase δ during DNA synthesis (20). This suggests that hLigI may instead interact with an RFC-PCNA complex remaining at the nick between Okazaki fragments. Since RFC has very weak PCNA unloading activity (5), it is possible that the interaction between RFC and hLigI not only targets hLigI to DNA nicks but also may stimulate PCNA unloading, thereby coupling the joining of Okazaki fragments with recycling of PCNA for lagging-strand DNA synthesis.
Although the 4D version of hLigI is more defective than the 4A version in binding to RFC in vitro, the phenotypes of hLigI-deficient cells expressing the 4A and 4D mutant versions of hLigI are essentially indistinguishable and are more severe than that of hLigI-deficient cells expressing the PCNA interaction mutant version of hLigI, which is defective in subnuclear targeting (16, 25). Strikingly, expression of the hLigI phosphorylation site mutants in hLigI-deficient cells induces cellular senescence. We suggest that a specific phosphorylated species of hLigI present in S phase functionally interacts with RFC during DNA replication (see Fig. S3 in the supplemental material) and that the phosphorylated hLigI from insect cells most closely resembles the S-phase species of hLigI. In this scenario, both the 4A version of hLigI, which mimics the hypophosphorylated G1 form of hLigI, and the 4D version of hLigI, which mimics the hyperphosphorylated M-phase form of hLigI, are defective in interacting with RFC and neither can be modified to generate the S-phase-specific form that interacts and functions with RFC. It appears that disrupting the interaction between hLigI and RFC has a more severe effect on DNA replication than does disrupting the interaction between hLigI and PCNA. For example, it may impact PCNA loading and/or unloading. In this scenario, the severe replication defect results in DNA damage that in turn activates the cellular senescence program. This model is supported by the results of studies by the Montecucco laboratory showing that expression of the 4D version of hLigI markedly increases the level of spontaneous DNA damage in hLigI-deficient 46BR.1G1 cells, which already have elevated levels of spontaneous DNA damage (30a). In accord with these observations, the derivatives of 46BR.1G1 cells expressing either the 4A or the 4D mutant have significantly higher levels of poly(ADP-ribose) and poly(ADP-ribosylated) proteins, an indicator of PARP-1 activation by DNA single-strand breaks, than do 46BR.1G1 cells and their derivatives expressing either wild-type hLigI or the FA mutant (see Fig. S4 in the supplemental material). Alternatively, dysregulating the interaction of hLigI with RFC and/or hRad17-RFC may directly activate signaling pathways leading to cellular senescence. Irrespective of the mechanism, expression of either the 4A or the 4D version of hLigI has a dominant-negative effect on replicative DNA synthesis, demonstrating the importance of the posttranslational regulation of hLigI for DNA replication.
The hLigI-deficient human cells are not only defective in joining Okazaki fragments but also hypersensitive to killing by simple DNA-alkylating agents because of a defect in long-patch BER (16). Although the phosphorylation site mutants of hLigI, like the PCNA interaction mutant (16), are less efficient than wild-type hLigI in correcting the long-patch BER defect of extracts from hLigI-deficient cells, expression of these mutant proteins in hLigI-deficient cells has dramatically different effects on cellular sensitivity to DNA alkylation. While expression of the PCNA-binding-defective mutant does not complement the DNA alkylation sensitivity of hLigI-deficient cells, it does result in a slight increase in cell survival (16). In contrast, expression of either the 4A or the 4D phosphorylation site mutant markedly decreases cell survival. Thus, similar to their effects on cell proliferation and DNA synthesis, expression of the hLigI phosphorylation site mutants exerts a dominant effect on the cellular response to DNA damage. Since activation of senescence pathways increases DNA damage sensitivity by suppressing signaling by Chk1 kinase (12) and the cell lines expressing either the 4A or the 4D version of hLigI have reduced levels of Chk1, we suggest that the DNA damage hypersensitivity of the cell lines expressing the phosphorylation site mutants of hLigI is caused by the activation of cellular senescence pathways rather than a defect in long-patch BER.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants GM57479 (to A.E.T.), CA92584 (to A.E.T.), and GM034559 (to J.H.).
Footnotes
Published ahead of print on 17 February 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Barnes, D. E., A. E. Tomkinson, A. R. Lehmann, A. D. Webster, and T. Lindahl. 1992. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell 69495-503. [DOI] [PubMed] [Google Scholar]
- 2.Bartkova, J., Z. Horejsi, M. Sehested, J. M. Nesland, E. Rajpert-De Meyts, N. E. Skakkebaek, M. Stucki, S. Jackson, J. Lukas, and J. Bartek. 2007. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene 267414-7422. [DOI] [PubMed] [Google Scholar]
- 3.Beausoleil, S. A., M. Jedrychowski, D. Schwartz, J. E. Elias, J. Villen, J. Li, M. A. Cohn, L. C. Cantley, and S. P. Gygi. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 10112130-12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 5.Bylund, G. O., and P. M. Burgers. 2005. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol. Cell. Biol. 255445-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J., A. E. Tomkinson, W. Ramos, Z. B. Mackey, S. Danehower, C. A. Walter, R. A. Schultz, J. M. Besterman, and I. Husain. 1995. Mammalian DNA ligase III: molecular cloning, chromosomal localization, and expression in spermatocytes undergoing meiotic recombination. Mol. Cell. Biol. 155412-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X., J. Pascal, S. Vijayakumar, G. M. Wilson, T. Ellenberger, and A. E. Tomkinson. 2006. Human DNA ligases I, III, and IV-purification and new specific assays for these enzymes. Methods Enzymol. 40939-52. [DOI] [PubMed] [Google Scholar]
- 8.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Micco, R., M. Fumagalli, A. Cicalese, S. Piccinin, P. Gasparini, C. Luise, C. Schurra, M. Garre, P. G. Nuciforo, A. Bensimon, R. Maestro, P. G. Pelicci, and F. d'Adda di Fagagna. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444638-642. [DOI] [PubMed] [Google Scholar]
- 10.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 929363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrari, G., R. Rossi, D. Arosio, A. Vindigni, G. Biamonti, and A. Montecucco. 2003. Cell cycle-dependent phosphorylation of human DNA ligase I at the cyclin-dependent kinase sites. J. Biol. Chem. 27837761-37767. [DOI] [PubMed] [Google Scholar]
- 12.Gabai, V. L., C. O'Callaghan-Sunol, L. Meng, M. Y. Sherman, and J. Yaglom. 2008. Triggering senescence programs suppresses Chk1 kinase and sensitizes cells to genotoxic stresses. Cancer Res. 681834-1842. [DOI] [PubMed] [Google Scholar]
- 13.Goetz, J. D., T. A. Motycka, M. Han, M. Jasin, and A. E. Tomkinson. 2005. Reduced repair of DNA double-strand breaks by homologous recombination in a DNA ligase I-deficient human cell line. DNA Repair (Amsterdam) 4649-654. [DOI] [PubMed] [Google Scholar]
- 14.Koundrioukoff, S., Z. O. Jonsson, S. Hasan, R. N. de Jong, P. C. van der Vliet, M. O. Hottiger, and U. Hubscher. 2000. A direct interaction between proliferating cell nuclear antigen (PCNA) and Cdk2 targets PCNA-interacting proteins for phosphorylation. J. Biol. Chem. 27522882-22887. [DOI] [PubMed] [Google Scholar]
- 15.Levin, D. S., W. Bai, N. Yao, M. O'Donnell, and A. E. Tomkinson. 1997. An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc. Natl. Acad. Sci. USA 9412863-12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin, D. S., A. E. McKenna, T. A. Motycka, Y. Matsumoto, and A. E. Tomkinson. 2000. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 10919-922. [DOI] [PubMed] [Google Scholar]
- 17.Levin, D. S., S. Vijayakumar, X. Liu, V. P. Bermudez, J. Hurwitz, and A. E. Tomkinson. 2004. A conserved interaction between the replicative clamp loader and DNA ligase in eukaryotes: implications for Okazaki fragment joining. J. Biol. Chem. 27955196-55201. [DOI] [PubMed] [Google Scholar]
- 18.Maga, G., and U. Hubscher. 2003. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 1163051-3060. [DOI] [PubMed] [Google Scholar]
- 19.Majka, J., and P. M. Burgers. 2004. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 78227-260. [DOI] [PubMed] [Google Scholar]
- 20.Masuda, Y., M. Suzuki, J. Piao, Y. Gu, T. Tsurimoto, and K. Kamiya. 2007. Dynamics of human replication factors in the elongation phase of DNA replication. Nucleic Acids Res. 356904-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumoto, Y. 2006. Base excision repair in mammalian cells. Methods Mol. Biol. 314365-375. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto, Y. 2001. Molecular mechanism of PCNA-dependent base excision repair. Prog. Nucleic Acid Res. Mol. Biol. 68129-138. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto, Y., K. Kim, J. Hurwitz, R. Gary, D. S. Levin, A. E. Tomkinson, and M. S. Park. 1999. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J. Biol. Chem. 27433703-33708. [DOI] [PubMed] [Google Scholar]
- 24.Montecucco, A., R. Rossi, G. Ferrari, A. I. Scovassi, E. Prosperi, and G. Biamonti. 2001. Etoposide induces the dispersal of DNA ligase I from replication factories. Mol. Biol. Cell 122109-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montecucco, A., R. Rossi, D. S. Levin, R. Gary, M. S. Park, T. A. Motycka, G. Ciarrocchi, A. Villa, G. Biamonti, and A. E. Tomkinson. 1998. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 173786-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen, J. V., B. Blagoev, F. Gnad, B. Macek, C. Kumar, P. Mortensen, and M. Mann. 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127635-648. [DOI] [PubMed] [Google Scholar]
- 27.Podust, V. N., N. Tiwari, S. Stephan, and E. Fanning. 1998. Replication factor C disengages from proliferating cell nuclear antigen (PCNA) upon sliding clamp formation, and PCNA itself tethers DNA polymerase delta to DNA. J. Biol. Chem. 27331992-31999. [DOI] [PubMed] [Google Scholar]
- 28.Prigent, C., M. S. Satoh, G. Daly, D. E. Barnes, and T. Lindahl. 1994. Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol. Cell. Biol. 14310-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi, R., A. Villa, C. Negri, I. Scovassi, G. Ciarrocchi, G. Biamonti, and A. Montecucco. 1999. The replication factory targeting sequence/PCNA-binding site is required in G(1) to control the phosphorylation status of DNA ligase I. EMBO J. 185745-5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song, W., D. S. Levin, J. Varkey, S. Post, V. P. Bermudez, J. Hurwitz, and A. E. Tomkinson. 2007. A conserved physical and functional interaction between the cell cycle checkpoint clamp loader and DNA ligase I of eukaryotes. J. Biol. Chem. 28222721-22730. [DOI] [PubMed] [Google Scholar]
- 30a.Soza, S., V. Leva, R. Vago, G. Ferrari, G. Mazzini, G. Biamonti, and A. Montecucco. 2009. DNA ligase I deficiency leads to replication-dependent DNA damage and impacts cell morphology without blocking cell cycle progression. Mol. Cell. Biol. 292032-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tom, S., L. A. Henricksen, M. S. Park, and R. A. Bambara. 2001. DNA ligase I and proliferating cell nuclear antigen form a functional complex. J. Biol. Chem. 27624817-24825. [DOI] [PubMed] [Google Scholar]
- 32.Toueille, M., N. El-Andaloussi, I. Frouin, R. Freire, D. Funk, I. Shevelev, E. Friedrich-Heineken, G. Villani, M. O. Hottiger, and U. Hubscher. 2004. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 323316-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlmann, F., E. Gibbs, J. Cai, M. O'Donnell, and J. Hurwitz. 1997. Identification of regions within the four small subunits of human replication factor C required for complex formation and DNA replication. J. Biol. Chem. 27210065-10071. [DOI] [PubMed] [Google Scholar]
- 34.Vijayakumar, S., B. R. Chapados, K. H. Schmidt, R. D. Kolodner, J. A. Tainer, and A. E. Tomkinson. 2007. The C-terminal domain of yeast PCNA is required for physical and functional interactions with Cdc9 DNA ligase. Nucleic Acids Res. 351624-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, W., L. A. Lindsey-Boltz, A. Sancar, and R. A. Bambara. 2006. Mechanism of stimulation of human DNA ligase I by the Rad9-Rad1-Hus1 checkpoint complex. J. Biol. Chem. 28120865-20872. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Y. C., W. A. Burkhart, Z. B. Mackey, M. B. Moyer, W. Ramos, I. Husain, J. Chen, J. M. Besterman, and A. E. Tomkinson. 1994. Mammalian DNA ligase II is highly homologous with vaccinia DNA ligase. Identification of the DNA ligase II active site for enzyme-adenylate formation. J. Biol. Chem. 26931923-31928. [PubMed] [Google Scholar]
- 37.Warbrick, E. 1998. PCNA binding through a conserved motif. Bioessays 20195-199. [DOI] [PubMed] [Google Scholar]
- 38.Yuzhakov, A., Z. Kelman, J. Hurwitz, and M. O'Donnell. 1999. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 186189-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, L., H. Dai, J. Qiu, Q. Huang, and B. Shen. 2007. Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol. Cell. Biol. 273176-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.