Abstract
Calcium signaling is a central regulator of cardiomyocyte growth and function. Calmodulin is a critical mediator of calcium signals. Because the amount of calmodulin within cardiomyocytes is limiting, the precise control of calmodulin expression is important for the regulation of calcium signaling. In this study, we show for the first time that calmodulin levels are regulated posttranscriptionally in heart failure. The cardiomyocyte-restricted microRNA miR-1 inhibited the translation of calmodulin-encoding mRNAs via highly conserved target sites within their 3′ untranslated regions. In keeping with its effect on calmodulin expression, miR-1 downregulated calcium-calmodulin signaling through calcineurin to NFAT. miR-1 also negatively regulated the expression of Mef2a and Gata4, key transcription factors that mediate calcium-dependent changes in gene expression. Consistent with the downregulation of these hypertrophy-associated genes, miR-1 attenuated cardiomyocyte hypertrophy in cultured neonatal rat cardiomyocytes and in the intact adult heart. Our data indicate that miR-1 regulates cardiomyocyte growth responses by negatively regulating the calcium signaling components calmodulin, Mef2a, and Gata4.
In the face of biomechanical stress, the heart develops compensatory hypertrophy, the hallmark of which is an increase in the size of cardiomyocytes. Over time, these hypertrophic changes become maladaptive, resulting in decreased contractile performance, cardiomyocyte loss, and, ultimately, heart failure (16). These changes in cardiomyocyte phenotype are linked to pathological changes in cardiomyocyte gene expression (24). Calcium, in conjunction with the calcium-binding protein calmodulin (CaM), is a critical mediator of hypertrophy signaling. Ca/CaM activates the phosphatase calcineurin (CN), leading to nuclear accumulation and the activation of the transcription factor nuclear factor of activated T cells (NFAT). The activation of the CN-NFAT pathway is necessary and sufficient for cardiomyocyte hypertrophy (25, 40). The transcription factor myocyte enhancer factor 2 (Mef2) is a second major target of Ca/CaM signaling (29), and Mef2 is a crucial regulator of cardiomyocyte growth (42, 44). Both NFAT and Mef2 regulate cardiac gene expression in collaboration with Gata4, another key regulator of cardiac hypertrophy (4, 21, 28).
miRNAs are a recently discovered class of small (∼22-nucleotide) RNAs that regulate gene expression through effects on the stability and translation of mRNA transcripts (1). miRNAs negatively regulate target mRNAs through sequence-specific interactions between a “seed sequence” at the 5′ end of the miRNA and a “seed match” sequence in the 3′ untranslated region (UTR) of the target mRNA (20). miRNA expression is altered in human heart failure (12, 35, 39), and specific miRNAs have been linked to cardiac development and function (38). Among these are microRNA-1 (miR-1), a microRNA selectively expressed in striated muscle, including the heart (2, 19).
Here, we show that miR-1 is downregulated in a murine heart failure model. Reduced levels of miR-1 expression were associated with broad alterations in the expression of predicted target genes. miR-1 negatively regulated the expression of the key calcium-signaling components CaM and Mef2a. miR-1 attenuated calcium-dependent signaling through the CN-NFAT pathway. Consistent with these effects on gene expression and calcium signaling, miR-1 reduced cardiomyocyte hypertrophy in both cultured neonatal rat cardiomyocytes and the intact adult heart.
MATERIALS AND METHODS
Mice.
MHCα-CN mice were a gift from Jeffery Molkentin (25). The mice were maintained by crossing transgenic males with B6C3F1 females (Charles River Labs). Samples were collected from males at 2 months of age unless otherwise noted. Intramyocardial adenoviral injections were performed in FVB/N mice (Charles River Labs) under ultrasound guidance (Visualsonic Vevo 770). Mice were anesthetized with 2% isoflurane and injected via a 30-gauge needle with adenovirus (2 × 109 infectious units/40 μl, divided equally between four independent locations). One week after injection, osmotic minipumps (catalog number 2002; Alzet) containing either isoproterenol (ISO) (30 mg/kg of body weight/day) or vehicle were implanted subcutaneously. Two weeks after implantation, mice were sacrificed, and cardiomyocytes were dissociated by retrograde collagenase perfusion (5). Cells were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The area of X-Gal-stained cardiomyocytes was measured by a blinded observer from digital images using a StereoInvestigator apparatus (MBF BioSciences). All procedures with animals were performed under protocols approved by the Institutional Animal Care and Use Committee.
Tissue culture.
P19CL6 cells were cultured and induced to undergo cardiac differentiation as described previously (10). Neonatal rat ventricular cardiomyocytes (NRVMs) were prepared as described previously (30) and transferred into serum-free medium on the day after isolation (day 1). Adenovirus was added on day 1. Agonist stimulation (100 nM endothelin-1 [ET-1] or 1 μM ISO) was performed on day 3. NFAT-luciferase and NFAT nuclear localization assays were performed on day 4, and cell size measurements were performed on day 5. NFAT-luciferase was delivered by adenovirus and normalized to total protein in cell lysates using a previously described NFAT-luciferase reporter adenovirus (40). This construct contains 9 NFAT sites upstream of a minimal promoter, followed by luciferase. For measurements of cell size, NRVMs were stained with antiactinin antibody (Sigma) and DAPI (4′,6′-diamidino-2-phenylindole). Over 200 cells from randomly acquired images were examined by a blinded observer. Experiments were reproduced two to three times.
Gene expression measurement.
miRNA expression profiles were obtained using a liquid bead array as previously described (23). We applied a global normalization strategy using locally weighted smooth-spline curve fitting for all miRNAs detected (33). In brief, we evenly divided the distribution of bead intensities into three groups. Sample normalization was performed by curve fitting within each intensity range. To identify the overall similarity of miRNA profiles between groups, we used unsupervised hierarchical clustering with the complete linkage algorithm for both samples and features, using the 59 expressed miRNAs (see Table S1 in the supplemental material) and the Pearson correlation coefficient as a similarity measure. miRNA target genes were predicted by miRANDA or TargetScanS, version 2.1 (14, 20). mRNA expression profiling was performed using Affymetrix GeneChip 430 v2.0 microarrays as described previously (4).
Quantitative reverse transcription-PCR (qRT-PCR) was performed as described previously (4). Primer sequences or sources for qRT-PCR assays are listed in Table S1 in the supplemental material. Gene expression was normalized to U6 or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) for miRNAs and mRNAs, respectively. The miR-133a/b qRT-PCR assay did not distinguish between miR-133a and miR-133b, and the miR-30b/c assay did not distinguish between miR-30b and -30c and did not detect miR-30a, -30d, and -30e (data not shown). Northern blotting was performed as described previously (11), with ExpressHyb (Clontech) as the hybridization buffer.
Western blotting was performed using antibodies for CaM (1:1,000; Upstate), Gata4 (SC-1237; Santa Cruz), Mef2a (a gift from M. Greenberg), and GAPDH (1:5,000; Research Diagnostics). Protein expression was normalized to GAPDH expression.
miRNA expression, knockdown, and sensing.
For miRNA expression, pcDNA6.2-GW/EmGFP-miR (Invitrogen) was modified by replacing emerald green fluorescent protein (GFP) with LacZ. A genomic fragment containing the miR-1 precursor was PCR amplified and cloned downstream of LacZ using primers described previously (7). Two nucleotide substitutions were introduced by PCR mutagenesis to yield miR-1mut. Adenoviral DNA was generated by recombinase-mediated transfer into pAd/CMV/V5-Dest (Invitrogen) using the Gateway cloning system. Adenovirus was produced by transfection into QBI293 cells (HEK293 subline; MP Biomedicals) and purified on cesium chloride gradients. Adenovirus was used at a multiplicity of infection of 10 to 100. At this dose, virtually all cardiomyocytes (>95%) were transduced. Antagomirs were purchased from Dharmacon. Primer sequences are shown in Table S1 in the supplemental material.
miRNA sensors were constructed from pMIR-Report (Ambion). 3′ UTRs of Calm1, Calm2, and Mef2a or the perfect-match sequence of miR-1 (miR-1 pm) or miR-208 pm were cloned using the PCR primers indicated in Table S1 in the supplemental material. Luciferase assays were performed by cotransfection of miRNA expression, miRNA sensor, and pRL-TK (Promega) internal control plasmid into QBI293 cells. Dual-luciferase assays (Promega) were performed. At least four biological replicates were performed, and results were independently reproduced two to three times.
Statistics.
Unless otherwise indicated, two group comparisons were performed by Welch's t test using JMP software v.5.1 (Cary, NC). The false-discovery rate of each miRNA was calculated using the Significance Analysis of Microarrays package (34). For Affymetrix transcriptome data, we use the MAS5 summary algorithm and linear scaling to median intensity. Probe sets below the detection threshold across all the samples were excluded from further analysis. Remaining probe sets representing the same RefSeq transcript ID were averaged. Error bars indicate standard errors of the means.
Microarray data accession number.
Gene expression and miRNA expression data were submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE13874.
RESULTS
Downregulation of cardiomyocyte-enriched miRNAs.
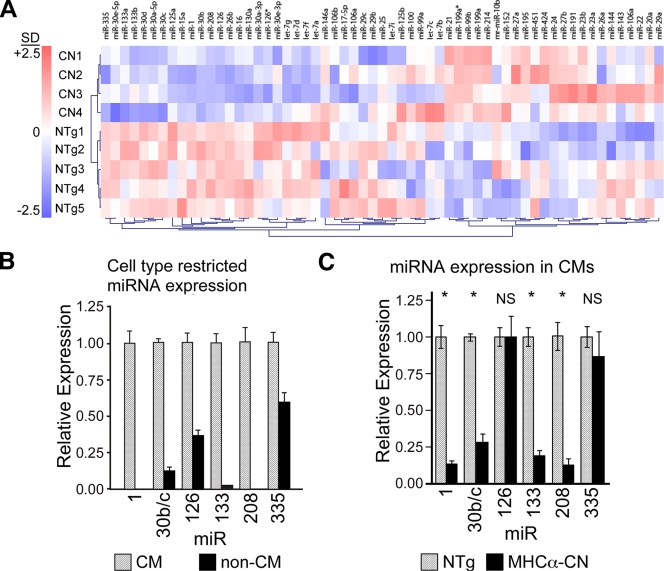
To measure miRNA expression changes in murine heart failure, we profiled miRNA expression in 2-month-old MHCα-CN and nontransgenic (NTg) control hearts. MHCα-CN mice develop marked cardiac hypertrophy by 1 month of age and severe systolic dysfunction by 2 months (22). Out of 261 miRNAs represented in the array, 59 were detectably expressed (Fig. 1A; see Table S2 in the supplemental material). Unsupervised clustering based on expression profiles segregated mice by genotype (Fig. 1A), indicating a systematic alteration of miRNA expression. Although some miRNAs tended toward increased expression, these did not reach statistical significance (see Table S2 in the supplemental material). Seven miRNAs belonging to six miRNA families were significantly downregulated (P < 0.05 by uncorrected Welch's t test; false-discovery rate of < 0.001) (Table 1). The cardiac-enriched miRNAs miR-1, miR-208, and miR-133b were downregulated. The other miR-133 family member, miR-133a, tended to have significant downregulation (P = 0.051). Within the miR-30-5p family, all five members were either significantly downregulated (30b, 30e-5p, and 30d) (P < 0.05) or tended to have significant downregulation (30c and 30a-5p) (P < 0.07). These results were validated by qRT-PCR (Table 1).
FIG. 1.
Altered miRNA expression levels in the MHCα-CN murine heart failure model. (A) Unsupervised clustering of miRNA expression profile in murine heart failure induced by transgenic expression of constitutively activated CN, compared with NTg. The color scale displays relative expression levels compared to the mean over all nine samples, where 2.5 in the color bar indicates 2.5 standard deviations (SD) from the nine-sample mean. (B) miRNA expression in wild-type cardiomyocytes (CMs) and adherent nonmyocytes (non-CMs) dissociated from nontransgenic hearts from 2-month-old animals. miRNA expression was normalized to U6 expression and displayed relative to the miRNA level in the CM fraction. (C) Expression of miRNAs in dissociated cardiomyocytes from MHCα-CN and NTg 2-month-old hearts normalized to U6 and displayed relative to expression in NTg hearts. qRT-PCR was used to measure miRNA expression (n = 3 for NTg, and n = 7 for CN). *, P < 0.002. NS, not significant.
TABLE 1.
Differentially expressed miRNAs in the MHCα-CN heart failure modela
| miRNA | Bead array
|
qRT-PCR
|
||
|---|---|---|---|---|
| Relative expression level | P value | Relative expression level | P value | |
| miR-1 | 0.53 | 0.018 | 0.63 | 0.028 |
| miR-15a | 0.53 | 0.052 | ||
| miR-16 | 0.63 | 0.051 | 0.77 | NS |
| miR-30b | 0.45 | 0.017 | 0.48b | 0.009 |
| miR-30d | 0.59 | 0.048 | ||
| miR-30e-5p | 0.45 | 0.022 | ||
| miR-133a | 0.63 | 0.051 | 0.48c | 0.075 |
| miR-133b | 0.63 | 0.033 | 0.48c | 0.075 |
| miR-126 | 0.53 | 0.052 | 0.67 | 0.028 |
| miR-208 | 0.5 | 0.032 | 0.67 | 0.036 |
| miR-335 | 0.29 | 0.008 | 0.43 | 0.009 |
The level of miRNA expression was measured in MHCα-CN and NTg myocardium using a liquid bead array. The mean expression level was compared by Welch's t test. In each case, the false-discovery rate was less than 5%. In selected cases, we independently measured gene expression levels in the same samples by qRT-PCR. Seven miRNAs in six families were differentially expressed. Relative expression indicates the ratio of expression in mutant compared to NTg myocardia. NS, not significant.
The qRT-PCR assay measured both miR-30b and -30c and did not detect miR-30a, -30d, or -30e.
The qRT-PCR assay measured both miR-133a and miR-133b.
Heart failure is accompanied by significant myocardial fibrosis (see Fig. S1a and S1b in the supplemental material) and a decreased proportion of cardiomyocytes to nonmyocytes. In principle, decreased myocardial miRNA expression levels could be due to decreased levels of expression in cardiomyocytes and/or to the dilution of cardiomyocytes by nonmyocytes. To distinguish these possibilities, we fractionated the adult heart into nonmyocyte and cardiomyocyte populations (greater than 90% pure) (see Fig. S1c in the supplemental material) by collagenase perfusion and differential centrifugation. Of the six miRNAs differentially expressed in myocardial tissue, four were substantially enriched in cardiomyocytes (miR-1, miR-133a/b, miR-30b/c, and miR-208), and two were not (miR-126 and miR-335) (Fig. 1B). The four cardiomyocyte-enriched miRNAs showed significantly decreased levels of expression in MHCα-CN compared to NTg cardiomyocytes (Fig. 1C) (P < 0.05), while the two miRNAs with less enrichment in cardiomyocytes were downregulated in noncardiomyocytes and unchanged in cardiomyocytes (data not shown).
In failing cardiomyocytes, mRNA expression becomes more similar to that of fetal cardiomyocytes (13). To determine if this generalization also applies to miRNAs, we measured the level of cardiomyocyte-enriched miRNAs at several developmental time points (embryonic days 12.5 and 16.5, postnatal days 0 and 14, and 2 months). In each case, miRNA expression levels increased through fetal and perinatal development and into adulthood (see Fig. S2 in the supplemental material) and decreased in heart failure. Thus, miRNA expression in the failing hearts indeed changed to become more similar to the fetal miRNA expression pattern.
Global changes in mRNA abundance reflect altered miRNA expression.
miRNAs influence both mRNA stability and translation. An altered expression of miRNAs during developmental processes was reflected in corresponding changes in the transcriptome (9). Consequently, we hypothesized that the downregulation of cardiomyocyte-enriched miRNAs would be associated with an upregulation of predicted mRNA targets at a frequency greater than expected by chance. To test this hypothesis, we obtained genome-wide measurements of mRNA levels in MHCα-CN and NTg hearts. Target genes of miR-1, miR-30, and miR-133 were predicted by conservation of their target regulatory sequences in the 3′ UTRs of four to five vertebrate species (TargetScanS algorithm) (20). miR-208 predictions were not available for this algorithm. In the whole transcriptome, out of 12,902 detectable genes, 2,101 (16%) were upregulated at a significance threshold of a P value of <0.005 (Welch's t test). In comparison, out of 208 predicted miR-1 targets with detectable expression, 62 (30%) were upregulated (see Tables S3 and S4 in the supplemental material). The likelihood that this or a larger proportion would occur in a random sampling of all detectable genes is 9.3 × 10−7 (Fisher's exact test) (Fig. 2A). miR-30 and miR-133 targets were also upregulated at frequencies unlikely to occur by chance (Fig. 2A; see Tables S3 and S4 in the supplemental material). These results were not sensitive to the specific significance threshold used to identify upregulated genes (see Table S4 in the supplemental material).
FIG. 2.
miRNAs broadly influence gene expression. (A) mRNA abundance in NTg and MHCα-CN hearts was measured by Affymetrix microarrays (n = 4). Genes were grouped into four sets: all genes with detectable expression levels, miR-1 targets, miR-30 targets, and miR-133 targets. Target genes were predicted by TargetScanS. For each set, the percentage of upregulated (P < 0.005) genes was calculated. The likelihood that a randomly selected subset of all genes would yield a fraction of upregulated genes equal to or greater than the upregulated fraction in miRNA target sets was calculated by Fisher's exact test. This value is displayed within each bar. (B) Cardiomyocyte differentiation in P19CL6 cells was associated with a marked upregulation of miR-1, -133, and -208 expression. miR-30b/c showed a less-dynamic range of expression. Expression was displayed relative to day 10, which was defined as 1. (C) Affymetrix microarrays (n = 3) were used to measure mRNA levels at days 6 and 10 of P19CL6 differentiation. Downregulated genes (P < 0.05) were identified by Welch's t test. Predicted targets of miR-1 and -133 were disproportionately downregulated at a frequency unlikely to occur by chance (numbers within bars, Fisher's exact test).
To examine the relationship between miR-1, -30, -133, and transcript abundance and the expression of target mRNAs in an independent system, we studied the multipotent embryonal carcinoma cell line P19CL6. This cell line inducibly differentiates into beating cardiomyocytes (10) following a reproducible time course that includes the upregulation of the cardiac transcription factors Gata4 and Nkx2-5 (Fig. 2B). miR-1 and -133 were highly upregulated between days 6 and 10 of differentiation (Fig. 2B). The upregulation of miR-1 and -133 was associated with a disproportionate downregulation of predicted target genes between days 6 and 10 (Fig. 2C). The expression of miR-30b/c was not as dynamic during P19CL6 differentiation (twofold upregulation between days 6 and 10) (Fig. 2B). This change in miR-30b/c was associated with a trend toward a disproportionate downregulation of predicted miR-30 targets (P = 0.058) (Fig. 2C). Taken together, the negative relationship between miRNA level and target gene abundance in two independent systems indicates that these miRNAs broadly influence transcript abundance. These analyses do not address translational regulation, and thus, the effect of altered miRNA levels on gene expression in heart failure is likely to be even more pervasive.
miR-1 attenuates cardiomyocyte hypertrophy in vitro and in vivo.
To investigate the function of miRNAs in the adult heart, we focused on miR-1, the most highly expressed cardiac-enriched miRNA (2, 12, 19). We generated a plasmid (pcDNA-miR-1) and an adenovirus (Ad:miR-1) that express miR-1 cocistronic with LacZ (Fig. 3A). We also generated two parallel negative control plasmids (pcDNA-empty and pcDNA-miR-1mut) and adenoviruses (Ad:empty and Ad:miR-1mut). Ad:empty is the same as Ad:miR-1 except for the omission of miR-1 sequences. Ad:miR-1mut is identical to Ad:miR-1 except for two nucleotides within the critical seed sequence. The expression constructs and corresponding adenoviruses were characterized in HEK293 cells and NRVMs, respectively (Fig. 3A). Northern blotting demonstrated that pcDNA-miR-1 and Ad:miR-1 appropriately expressed mature microRNA, with Ad:miR-1 increasing miR-1 levels in NRVMs to approximately 500% of endogenous levels. On the other hand, the miR-1 mutant construct did not efficiently express miR-1mut (Fig. 3A), which is likely due to alterations of the stem-loop structure that impaired miRNA biogenesis. Nevertheless, we retained this construct as a second negative control.
FIG. 3.
miR-1 inhibited cardiomyocyte hypertrophy in cultured neonatal cardiomyocytes. (A) microRNA expression constructs. Constructs drove the expression of LacZ followed by no insert (empty [emp]), the miR-1 stem-loop (miR-1), or an miR-1 stem-loop containing two point mutations in the seed sequence (miR-1mut). The plasmid constructs were transfected into 293 cells, or their adenoviral derivatives were transduced into NRVMs. Northern blotting showed the appropriate expression of miR-1, increasing miR-1 levels in NRVMs by fivefold. The miR-1mut constructs did not efficiently express the mutated sequence. (B) Measurement of microRNA activity with a luciferase reporter. An assay sequence of interest was positioned downstream of luciferase. miRNA activity at the assay sequence reduced luciferase activity. To measure miR-1 activity, we used the reverse complement of miR-1 (1pm). The reverse complement of miR-208 was the negative control (208pm). Luciferase activity was normalized to activity for the cotransfected internal control (pRL-TK). For each luciferase sensor, normalized luciferase activity in the presence of the empty expression plasmid was assigned a value of 1. (C) Overexpression of miR-1, but not miR-1mut, reduced NRVM size at baseline and in response to ET-1 or ISO stimulation. Representative images of NRVMs stained with α-actinin (green) and DAPI (blue) are shown. *, P < 0.001 versus empty vector; †, P < 0.02. (D) Antagomir against miR-1 specifically inhibited miR-1 activity in NRVMs. NRVMs were transfected with miR-1 pm or miR-208 pm luciferase reporters and an internal control (pRL-TK) and then treated with the indicated antagomir. Luciferase activity was normalized to the internal control. For each luciferase sensor, normalized luciferase activity in the presence of GFP antagomir was assigned a value of 1. *, P < 0.001. (E) miR-1 knockdown with a specific antagomir (Antag) increased NRVM size at baseline and in response to ET-1 compared to the control antagomir directed against an unrelated sequence (GFP). Representative images of NRVMs stained with anti-actinin (green) and DAPI (blue) are shown. PBS, phosphate-buffered saline. *, P < 0.05; †, P < 0.001.
To validate the biological effectiveness of the miR-1 expression construct, we measured miR-1 activity with a “miR-1 pm sensor” composed of a luciferase reporter containing the reverse complement of miR-1 in its 3′ UTR (Fig. 3B). As anticipated, miR-1, but not miR-1mut or empty, constructs strongly suppressed the activity of the miR-1 pm sensor (Fig. 3B). The effect was specific to the miR-1 sensor, as there was no effect on the activity of a sensor containing a perfect match for an unrelated miRNA (miR-208; “miR-208 pm sensor”) (Fig. 3B).
We then asked whether the overexpression of miR-1 alters the hypertrophy of NRVMs in response to stimulation with the hypertrophic agonists ET-1 and ISO. miR-1 overexpression reduced the basal size of NRVMs compared to controls (Fig. 3C). Stimulation with either ET-1 or ISO increased NRVM size in the control groups, but this response was abolished by miR-1 overexpression. This result indicated that miR-1 is a negative regulator of cardiomyocyte hypertrophy.
Cardiomyocyte hypertrophy is associated with the activation of a fetal gene program, including Nppa, Nppb, and Acta1 (skeletal α-actin) (13). To determine the effect of miR-1 overexpression on this process, we measured the levels of expression of these genes in ET-1-stimulated NRVMs treated with Ad:miR-1 or control adenoviruses. Ad:miR-1 attenuated the expression of Nppa, Nppb, and Acta1 in NRVM stimulated with ET-1 (see Fig. S3 in the supplemental material), consistent with negative regulation of cardiomyocyte hypertrophy by miR-1.
To further establish a role for miR-1 in the regulation of cardiomyocyte growth, we used a loss-of-function approach. Cholesterol-linked RNA oligonucleotide (“antagomir”) (18) directed against miR-1 (Antag:miR-1) was effective in antagonizing miR-1 activity in NRVMs, as shown by the markedly diminished inhibition (upregulation) of the miR-1 pm reporter (Fig. 3D). The inhibitory activity of Antag:miR-1 was sequence specific, as it did not inhibit miR-208 activity (Fig. 3D).
We then asked if miR-1 loss of function altered NRVM cell size at baseline or after treatment with a hypertrophic agonist. NRVMs were cultured in the presence of Antag:miR-1 or Antag:GFP, and cell size was measured after treatment with vehicle (phosphate-buffered saline; baseline condition) or with ET-1. At both baseline and after ET-1 stimulation, NRVM size was significantly increased by an miR-1 loss of function (Fig. 3E). These data further indicate that miR-1 is an important regulator of cardiomyocyte hypertrophy.
To address the effect of miR-1 on the hypertrophy of adult cardiomyocytes in vivo, we used adenoviral gene transfer to overexpress miR-1 in adult hearts. Under ultrasound guidance, we injected Ad:miR-1 or Ad:miR-1mut into the left ventricular wall (Fig. 4A). The injected mice were then treated with ISO or vehicle. After 2 weeks, cardiomyocytes were dissociated from hearts by collagenase perfusion. About 10% of dissociated cardiomyocytes were infected by Ad:miR-1 or Ad:miR-1mut, as determined by staining for LacZ (Fig. 4B). At baseline, LacZ+ cardiomyocytes did not differ in size between the Ad:miR-1 and Ad:miR-1mut groups (Fig. 4C). However, Ad:miR-1 significantly blocked cardiomyocyte hypertrophy in response to chronic ISO infusion (Fig. 4C). Thus, miR-1 antagonizes ISO-induced cardiomyocyte hypertrophy in intact adult hearts as well as in NRVMs.
FIG. 4.
miR-1 inhibited adult cardiomyocyte hypertrophy. (A) Transthoracic intramyocardial injection of adenovirus under ultrasound guidance. A schematic of the ultrasound image is shown, with a needle penetrating the left ventricular myocardium (LV). (B) Dissociated cardiomyocytes. Cardiomyocytes with adenoviral miR-1 or miR-1mut overexpression were identified by the expression of LacZ, which stains blue with X-Gal. (C) Quantitation of cell size after expression of miR-1 or miR-1mut. miR-1 blocked cardiomyocyte hypertrophy in response to ISO. *, P < 0.001. NS, no significant difference (n = 4 per group).
miR-1 negatively regulates CaM and Ca/CaM-CN-NFAT signaling.
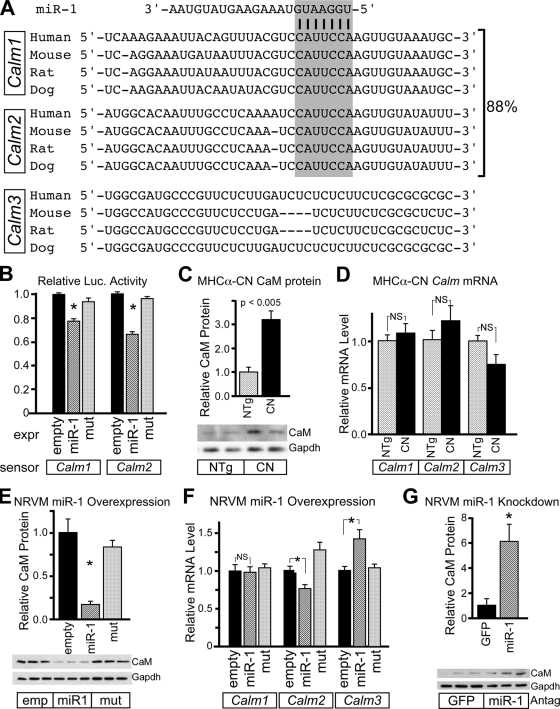
To identify mRNA targets of miR-1 that mediate the effect of miR-1 on cardiomyocyte hypertrophy, we used computational algorithms to find genes containing evolutionarily conserved miR-1 seed match sequences in their 3′ UTRs (3, 17, 20). Two interesting candidates were the CaM-encoding genes Calm1 and Calm2. In mammals, three separate genes (Calm1 to Calm3) encode identical CaM proteins and differ in their UTRs (36). In heart, Calm1 and Calm2 comprise 88% of CaM-encoding transcripts, as determined by sequence-based counting of transcript tag sequences (15). Both contain highly conserved miR-1 seed match sequences in their 3′ UTRs (Fig. 5A). When cloned downstream of luciferase, these sequences mediated the downregulation of luciferase activity by miR-1 (Fig. 5B).
FIG. 5.
miR-1 negatively regulates CaM and CaM-dependent NFAT signaling. (A) Conservation of miR-1 seed match sequence (gray box) in mammalian Calm1 and Calm2, which account for 88% of cardiac CaM-encoding transcripts (15). (B) Calm1 and Calm2 3′ UTRs contain miR-1-repressible sequences. When 3′ UTRs from Calm1 and Calm2 were positioned downstream of luciferase, luciferase activity was reduced by a cotransfected miR-1 expression plasmid. QBI293 cells were transfected with the indicated miR-1 expression (expr), luciferase sensor, and pRL-TK internal control plasmids. (C) The CaM protein was significantly upregulated in 2-month-old MHCα-CN hearts compared to nonlittermate controls. A representative Western blot is shown (n = 4). (D) CaM upregulation in MHCα-CN hearts was posttranscriptional. Calm1 to Calm3 mRNAs were measured by qRT-PCR. CN overexpression did not significantly alter mRNA levels of Calm1 to Calm3. (E) Adenoviral expression of miR-1 in NRVMs significantly reduced levels of the CaM protein compared to miR-1mut and empty (emp) negative controls. A representative Western blot is shown (n = 3). (F) Adenoviral miR-1 overexpression in NRVMs did not significantly alter Calm1 and Calm2 mRNA levels. There was a compensatory upregulation of Calm3 (n = 3). *, P < 0.05. (G) Antagomir (Antag)-mediated miR-1 knockdown increased CaM protein levels in NRVMs compared to unrelated antagomir against GFP (n = 3 per group). A representative Western blot is shown. *, P < 0.05. NS, not significant.
miR-1 was downregulated in MHCα-CN mice. Therefore, we asked if Calm1 and Calm2 were upregulated, consistent with a role of miR-1 in regulating these genes in vivo. By immunoblotting, CaM expression was upregulated threefold in 2-month-old MHCα-CN mice compared to NTg littermates (Fig. 5C). The regulation of CaM occurred posttranscriptionally, as Calm1, Calm2, and Calm3 mRNA levels were not significantly changed in MHCα-CN mice (Fig. 5D).
To further test the hypothesis that miR-1 directly downregulates CaM expression, we measured CaM levels in NRVMs after a gain or loss of function of miR-1. Adenoviral overexpression of miR-1 significantly downregulated the CaM protein (Fig. 5E). miR-1 overexpression did not affect Calm1 mRNA abundance and weakly reduced Calm2 mRNA levels, suggesting that the effect was largely at the level of translation (Fig. 5F). Calm3 mRNA was upregulated with miR-1 overexpression, potentially as a compensatory response to CaM downregulation. Conversely, antagomir knockdown of miR-1 upregulated CaM (Fig. 5G). Collectively, these data indicate that miR-1 negatively regulates CaM.
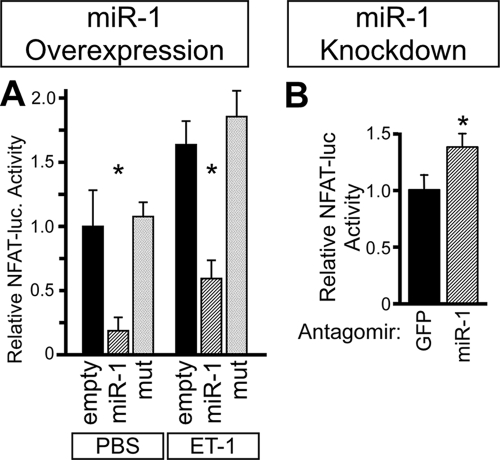
Given the crucial role of Ca/CaM signaling in cardiomyocyte hypertrophy, we hypothesized that miR-1 antagonizes cardiomyocyte hypertrophy by inhibiting the signaling of Ca/CaM-dependent signaling pathways. Ca/CaM binds to and activates the phosphatase CN, which dephosphorylates the transcription factor NFAT and thereby promotes its nuclear translocation. NFAT activation by Ca/CaM-CN is essential for cardiomyocyte hypertrophy (25). Therefore, we asked if miR-1 negatively regulates NFAT signaling. Adenoviral miR-1 overexpression inhibited an NFAT-dependent luciferase reporter in NRVMs both at baseline and after stimulation with ET-1 (Fig. 6A). On the other hand, the reduction of miR-1 activity by antagomir treatment increased NFAT-dependent reporter activity (Fig. 6B).
FIG. 6.
miR-1 negatively regulates NFAT signaling. (A) miR-1 overexpression inhibited NFAT-luciferase activity. NRVMs were treated with adenovirus containing an NFAT-dependent luciferase reporter and with the indicated miRNA-expressing adenoviruses. Levels of NFAT activity were increased by ET-1 and were decreased by miR-1 overexpression. *, P < 0.01 versus empty vector. (B) miR-1 knockdown increased NFAT-luciferase activity. NRVMs were treated NFAT-luciferase adenovirus and miR-1 or unrelated antagomir (GFP). *, P < 0.05 (n = 3). PBS, phosphate-buffered saline.
miR-1 negatively regulates Mef2a and Gata4.
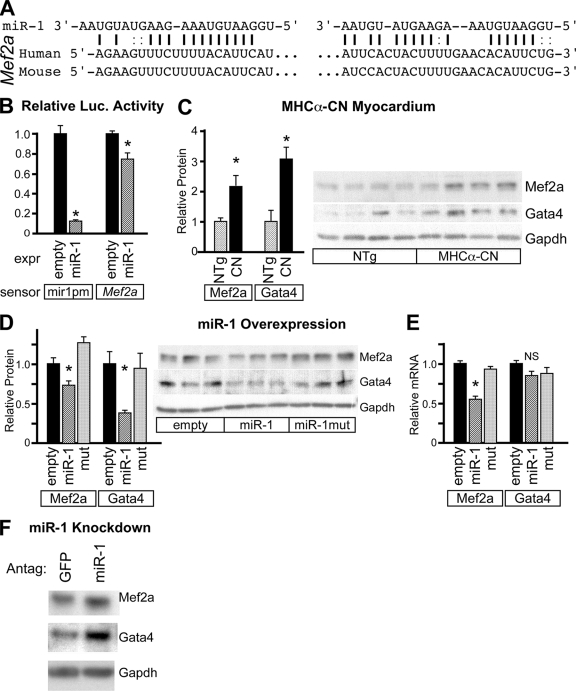
The transcription factor Mef2 is an important regulator of cardiomyocyte hypertrophy downstream of Ca/CaM (29). Mef2a has two conserved miR-1 recognition sequences in its 3′ UTR (Fig. 7A). These sequences conferred miR-1 responsiveness to a luciferase reporter (Fig. 7B). The Mef2a protein was upregulated in MHCα-CN mice, consistent with miR-1 downregulation in this model (Fig. 7C). miR-1 overexpression in NRVMs decreased levels of the Mef2a protein (Fig. 7D). Mef2a mRNA was downregulated by miR-1 overexpression, suggesting that miR-1 decreased Mef2a transcript stability (Fig. 7E). On the other hand, miR-1 knockdown in NRVMs increased Mef2a protein levels (Fig. 7F). Collectively, these data indicate that miR-1 negatively regulates Mef2a expression.
FIG. 7.
miR-1 negatively regulates Mef2a. (A) The 3′ UTR of Mef2a contains two conserved miR-1 recognition sequences. (B) The Mef2a 3′ UTR contains miR-1-repressible sequences. 3′ UTR sequences from Mef2a containing predicted miR-1 recognition sequences were cloned downstream of luciferase. Luciferase activity was reduced by a cotransfected miR-1 expression (expr) plasmid. QBI293 cells were transfected with the indicated miR expression, luciferase sensor, and pRL-TK internal control plasmids. (C) Mef2a and Gata4 proteins were upregulated in MHCα-CN myocardium compared to littermate controls. (D) Adenoviral expression of miR-1 in NRVMs significantly reduced levels of the Mef2a and Gata4 proteins compared to miR-1mut and the empty negative control. A representative Western blot is shown. (E) Adenoviral expression of miR-1 significantly downregulated Mef2a but not Gata4 mRNA levels. mRNA levels were determined by qRT-PCR. (F) Antagomir-mediated miR-1 knockdown increased levels of the Mef2a and Gata4 proteins in NRVMs compared to levels of unrelated antagomir against GFP. *, P < 0.05. NS, not significant.
In cardiomyocytes, NFAT and Mef2a cooperate with the transcription factor Gata4 to activate hypertrophic gene expression (25). The level of the Gata4 protein was increased in MHCα-CN mice (Fig. 7C). miR-1 overexpression in NRVMs decreased levels of the Gata4 protein compared to controls (Fig. 7D), and miR-1 knockdown increased Gata4 protein levels (Fig. 7F). Interestingly, the Gata4 mRNA level was unchanged (Fig. 7E), suggesting that Gata4 is regulated posttranscriptionally downstream of miR-1. Gata4 lacks predicted miR-1 recognition sequences, leading us to hypothesize that the miR-1 regulation of Gata4 is indirect.
DISCUSSION
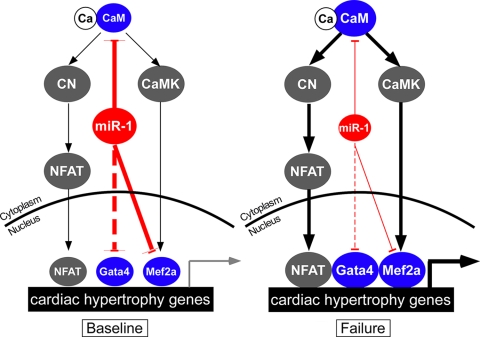
microRNAs are emerging as powerful regulators of gene expression and cellular phenotype. We found that the expression of multiple cardiac-enriched miRNAs was downregulated in a murine heart failure model. Altered expression of these miRNAs had a pervasive effect on the cardiac transcriptome and likely has a greater effect on the proteome. The cardiac-enriched microRNA miR-1 negatively regulated cardiac hypertrophy by downregulating the expression of CaM, Gata4, and Mef2a, key mediators of calcium-dependent signaling (Fig. 8).
FIG. 8.
miR-1 inhibits calcium-dependent signaling at multiple levels. Cardiac hypertrophy is regulated by calcium, which complexes with CaM to activate CN-NFAT and calcium-CaMK-Mef2 transcriptional pathways. Both of these pathways are stimulated by protein-protein interactions with Gata4. At baseline, miR-1 attenuates cardiomyocyte hypertrophy by downregulating the expression of CaM, Mef2, and Gata4. In heart failure, miR-1 expression is downregulated. This favors increased levels of expression of Gata4, Mef2a, and CaM. Increased levels of CaM augment Ca/CaM signaling through CN-NFAT (Fig. 6) and potentially also through CaMK. This results in an increased level of transcription of genes downstream of NFAT, Gata4, and Mef2a, promoting cardiac hypertrophy. The dotted line indicates indirect regulation.
Altered expression of miRNAs in heart failure.
In a heart failure model induced by the transgenic expression of activated CN, we observed the downregulation of the cardiac-enriched miRNAs miR-1, miR-133, miR-30b, and miR-208. Altered expression of these miRNAs has been observed in some, but not all, studies of miRNA expression in murine and human heart failure (see Table S5 in the supplemental material). miR-1 expression has been found to be upregulated, downregulated, and unchanged in heart disease (6, 8, 12, 31, 35, 39, 43). The divergent results likely reflect differences in disease models, myocardial regions sampled, assay platforms, normalization methods, control samples, and statistical analyses. In this study, we validated high-throughput data with qRT-PCR from both myocardium and purified cardiomyocyte samples, where we found that expression differences were more robust for cardiomyocyte-enriched miRNAs. We also demonstrated for the first time that miR-1, miR-133, miR-30b, and miR-208 are highly enriched in the cardiomyocyte fraction of the myocardium.
miR-1, miR-133, miR-30b, and miR-208 are each highly upregulated from fetal to newborn stages and again in early postnatal life between 2 and 14 days of life (see Fig. S2 in the supplemental material). In our heart failure model, each of these miRNAs was downregulated, suggesting a return to a more-fetal expression profile. This is reminiscent of the activation of the mRNA “fetal gene program” in the failing heart and is perhaps driven by an overlapping transcriptional program.
miR-1 attenuates cardiomyocyte hypertrophy.
While the developmental functions of miR-1 were previously highlighted (45, 46), the upregulation of miR-1 to substantial levels in the postnatal heart suggested an important function in the regulation of postnatal heart function. Consistent with this hypothesis, miR-1 attenuated cardiomyocyte hypertrophy. Our data are consistent with and significantly expand on prior work describing an antihypertrophic activity of miR-1 (6, 31). First, we use both gain- and loss-of-function approaches to support this conclusion, avoiding limitations inherent to either approach alone. Second, we show that miR-1 attenuates cardiomyocyte hypertrophy in adult cardiomyocytes in the intact heart in addition to cultured neonatal cardiomyocytes. Third, we have identified key genes downstream of miR-1 that contribute to its regulation of cardiomyocyte growth.
miR-1 overexpression decreased the size of cultured NRVMs both at baseline and after hypertrophic stimulation. In adult cardiomyocytes, miR-1 overexpression in vivo attenuated agonist-induced cardiomyocyte hypertrophy but did not significantly decrease cardiomyocyte size at baseline. This difference might be due to the approximately threefold increase in levels of miR-1 expression between neonatal and adult hearts. As a result, Ad:miR-1 drove a relatively smaller increase in miR-1 expression levels in adult compared to neonatal cardiomyocytes. Along the same lines, it is possible that the baseline effect of miR-1 is saturated in adult cardiomyocytes so that forced expression has little additional effect. Nevertheless, the observed difference in adult cardiomyocyte size under conditions of hypertrophic stimulation indicates that miR-1 continues to regulate cardiomyocyte hypertrophy in the adult heart.
miR-1 negatively regulates the calcium signaling pathway components CaM and Mef2a.
Calcium is a central regulator of cardiomyocyte function and growth. CaM is a crucial mediator of calcium signaling. Importantly, nearly all CaM in the cardiomyocyte is protein bound, indicating that CaM levels are limiting (41). Consistent with this, CaM transgenic mice with three- to fivefold-increased cardiac CaM levels develop severe cardiomyocyte hypertrophy and heart failure (27). This level of CaM upregulation is in the same range as what we observed for MHCα-CN hearts, suggesting that the posttranscriptional upregulation of CaM in MHCα-CN hearts likely contributes to heart failure pathogenesis in this model.
Because of the critical, dosage-sensitive role of CaM, cells have likely evolved rich regulatory networks to regulate CaM expression. We show for the first time that CaM expression is regulated posttranscriptionally, suggesting a novel mechanism for modulating calcium signaling. Remarkably, three separate genes (Calm1 to Calm3) encode the identical protein. Each Calm isoform has distinct untranslated regions, which may allow the isoforms to be differentially regulated by posttranscriptional mechanisms (36). Calm1 and Calm2, the two isoforms that comprise the great majority of CaM-encoding transcripts in the heart, have highly conserved miR-1 recognition sequences in their 3′ UTRs. Both gain- and loss-of-function approaches demonstrated that miR-1 negatively regulates CaM levels. Collectively, our data indicate that CaM is posttranscriptionally regulated and that this is mediated by sequence-specific interactions between miR-1 and target sequences within the Calm1 and Calm2 3′ UTRs. Other miRNAs may also contribute to the posttranscriptional regulation of Calm1 or Calm2. For instance, Calm1 contains a conserved binding site for miR-133.
The CN-NFAT signaling pathway is a key signaling pathway downstream of Ca/CaM. NFAT acts in collaboration with Gata4 to initiate transcriptional changes that are essential for cardiomyocyte hypertrophy (25). Consistent with the decreased availability of CaM with miR-1 overexpression, the activity of this pathway was attenuated by miR-1 (Fig. 8). In addition, miR-1 indirectly downregulated Gata4, which collaborates with NFATs to regulate cardiomyocyte hypertrophy (25). Thus, the miR-1 downregulation of Gata4 provides a second means by which miR-1 inhibits the activation of this prohypertrophic calcium signaling pathway.
The transcription factor Mef2 is another important mediator of the prohypertrophic effects of increased Ca/CaM signaling. Ca/CaM signaling activated Mef2 transcriptional activity, largely through Ca/CaM-dependent protein kinase (CaMK) (29). Ca/CaM-dependent signaling through CN also likely regulates Mef2 transcriptional activity (29), and dominant negative Mef2 also blocked ventricular dilatation and systolic dysfunction in MHCα-CN mice (37). Mef2A overexpression is sufficient to induce cardiac hypertrophy (42), and dominant negative inhibition of Mef2 signaling blocked cardiomyocyte hypertrophy (44). Collectively, these data indicate that Ca/CaM signaling regulates Mef2 activity and that Mef2 is a key regulator of cardiomyocyte hypertrophy. Consistent with a recent report for Caenorhabditis elegans Mef2 (32), we found that miR-1 negatively regulates Mef2a expression. Thus, our data suggest that miR-1 antagonizes Mef2a activity at three levels (Fig. 8). First, miR-1 decreases the level of expression of Mef2a. Second, miR-1 may reduce the Ca/CaM-dependent activation of Mef2a. Third, miR-1 downregulates Gata4, which physically interacts with Mef2 to regulate a subset of cardiac genes (26).
In summary, this study provides insights into the mechanisms by which miR-1 attenuates cardiomyocyte hypertrophy. miR-1 negatively regulates CaM, Mef2a, and Gata4, key components of calcium signaling pathways that are necessary for agonist-induced cardiomyocyte hypertrophy.
Supplementary Material
Acknowledgments
S.I. was supported by an AHA fellowship. This work was supported by NIH grant PO1 HL074734 and by a charitable donation from Edward P. Marram and Karen K. Carpenter.
Footnotes
Published ahead of print on 2 February 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 2.Baskerville, S., and D. P. Bartel. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betel, D., M. Wilson, A. Gabow, D. S. Marks, and C. Sander. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res. 36D149-D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisping, E., S. Ikeda, S. W. Kong, O. Tarnavski, N. Bodyak, J. R. McMullen, S. Rajagopal, J. K. Son, Q. Ma, Z. Springer, P. M. Kang, S. Izumo, and W. T. Pu. 2006. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc. Natl. Acad. Sci. USA 10314471-14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodyak, N., P. M. Kang, M. Hiromura, I. Sulijoadikusumo, N. Horikoshi, K. Khrapko, and A. Usheva. 2002. Gene expression profiling of the aging mouse cardiac myocytes. Nucleic Acids Res. 303788-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caré, A., D. Catalucci, F. Felicetti, D. Bonci, A. Addario, P. Gallo, M. L. Bang, P. Segnalini, Y. Gu, N. D. Dalton, L. Elia, M. V. Latronico, M. Høydal, C. Autore, M. A. Russo, G. W. Dorn II, O. Ellingsen, P. Ruiz-Lozano, K. L. Peterson, C. M. Croce, C. Peschle, and G. Condorelli. 2007. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13613-618. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. F., E. M. Mandel, J. M. Thomson, Q. Wu, T. E. Callis, S. M. Hammond, F. L. Conlon, and D. Z. Wang. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38228-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Y., R. Ji, J. Yue, J. Yang, X. Liu, H. Chen, D. B. Dean, and C. Zhang. 2007. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am. J. Pathol. 1701831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farh, K. K., A. Grimson, C. Jan, B. P. Lewis, W. K. Johnston, L. P. Lim, C. B. Burge, and D. P. Bartel. 2005. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 3101817-1821. [DOI] [PubMed] [Google Scholar]
- 10.Habara-Ohkubo, A. 1996. Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct. Funct. 21101-110. [DOI] [PubMed] [Google Scholar]
- 11.He, A., L. Zhu, N. Gupta, Y. Chang, and F. Fang. 2007. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol. Endocrinol. 212785-2794. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda, S., S. W. Kong, J. Lu, E. Bisping, H. Zhang, P. D. Allen, T. R. Golub, B. Pieske, and W. T. Pu. 2007. Altered microRNA expression in human heart disease. Physiol. Genomics 31367-373. [DOI] [PubMed] [Google Scholar]
- 13.Izumo, S., B. Nadal-Ginard, and V. Mahdavi. 1988. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc. Natl. Acad. Sci. USA 85339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.John, B., A. J. Enright, A. Aravin, T. Tuschl, C. Sander, and D. S. Marks. 2004. Human MicroRNA targets. PLoS Biol. 2e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jongeneel, C. V., M. Delorenzi, C. Iseli, D. Zhou, C. D. Haudenschild, I. Khrebtukova, D. Kuznetsov, B. J. Stevenson, R. L. Strausberg, A. J. Simpson, and T. J. Vasicek. 2005. An atlas of human gene expression from massively parallel signature sequencing (MPSS). Genome Res. 151007-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz, A. M. 1995. The cardiomyopathy of overload: an unnatural growth response. Eur. Heart J. 16(Suppl. O)110-114. [DOI] [PubMed] [Google Scholar]
- 17.Krek, A., D. Grun, M. N. Poy, R. Wolf, L. Rosenberg, E. J. Epstein, P. MacMenamin, I. da Piedade, K. C. Gunsalus, M. Stoffel, and N. Rajewsky. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37495-500. [DOI] [PubMed] [Google Scholar]
- 18.Krutzfeldt, J., N. Rajewsky, R. Braich, K. G. Rajeev, T. Tuschl, M. Manoharan, and M. Stoffel. 2005. Silencing of microRNAs in vivo with ‘antagomirs.’ Nature 438685-689. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel, and T. Tuschl. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12735-739. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 12015-20. [DOI] [PubMed] [Google Scholar]
- 21.Liang, Q., L. J. De Windt, S. A. Witt, T. R. Kimball, B. E. Markham, and J. D. Molkentin. 2001. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 27630245-30253. [DOI] [PubMed] [Google Scholar]
- 22.Lim, H. W., L. J. De Windt, J. Mante, T. R. Kimball, S. A. Witt, M. A. Sussman, and J. D. Molkentin. 2000. Reversal of cardiac hypertrophy in transgenic disease models by calcineurin inhibition. J. Mol. Cell. Cardiol. 32697-709. [DOI] [PubMed] [Google Scholar]
- 23.Lu, J., G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz, and T. R. Golub. 2005. MicroRNA expression profiles classify human cancers. Nature 435834-838. [DOI] [PubMed] [Google Scholar]
- 24.McKinsey, T. A., and E. N. Olson. 2005. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J. Clin. Investig. 115538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molkentin, J. D., J. R. Lu, C. L. Antos, B. Markham, J. Richardson, J. Robbins, S. R. Grant, and E. N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morin, S., F. Charron, L. Robitaille, and M. Nemer. 2000. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 192046-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obata, K., K. Nagata, M. Iwase, M. Odashima, T. Nagasaka, H. Izawa, T. Murohara, Y. Yamada, and M. Yokota. 2005. Overexpression of calmodulin induces cardiac hypertrophy by a calcineurin-dependent pathway. Biochem. Biophys. Res. Commun. 3381299-1305. [DOI] [PubMed] [Google Scholar]
- 28.Oka, T., M. Maillet, A. J. Watt, R. J. Schwartz, B. J. Aronow, S. A. Duncan, and J. D. Molkentin. 2006. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ. Res. 98837-845. [DOI] [PubMed] [Google Scholar]
- 29.Passier, R., H. Zeng, N. Frey, F. J. Naya, R. L. Nicol, T. A. McKinsey, P. Overbeek, J. A. Richardson, S. R. Grant, and E. N. Olson. 2000. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Investig. 1051395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pu, W. T., Q. Ma, and S. Izumo. 2003. NFAT transcription factors are critical survival factors that inhibit cardiomyocyte apoptosis during phenylephrine stimulation in vitro. Circ. Res. 92725-731. [DOI] [PubMed] [Google Scholar]
- 31.Sayed, D., C. Hong, I. Y. Chen, J. Lypowy, and M. Abdellatif. 2007. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 100416-424. [DOI] [PubMed] [Google Scholar]
- 32.Simon, D. J., J. M. Madison, A. L. Conery, K. L. Thompson-Peer, M. Soskis, G. B. Ruvkun, J. M. Kaplan, and J. K. Kim. 2008. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell 133903-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31265-273. [DOI] [PubMed] [Google Scholar]
- 34.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 1009440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thum, T., P. Galuppo, C. Wolf, J. Fiedler, S. Kneitz, L. W. van Laake, P. A. Doevendans, C. L. Mummery, J. Borlak, A. Haverich, C. Gross, S. Engelhardt, G. Ertl, and J. Bauersachs. 2007. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 116258-267. [DOI] [PubMed] [Google Scholar]
- 36.Toutenhoofd, S. L., and E. E. Strehler. 2000. The calmodulin multigene family as a unique case of genetic redundancy: multiple levels of regulation to provide spatial and temporal control of calmodulin pools? Cell Calcium 2883-96. [DOI] [PubMed] [Google Scholar]
- 37.van Oort, R. J., E. van Rooij, M. Bourajjaj, J. Schimmel, M. A. Jansen, R. van der Nagel, P. A. Doevendans, M. D. Schneider, C. J. van Echteld, and L. J. De Windt. 2006. MEF2 activates a genetic program promoting chamber dilation and contractile dysfunction in calcineurin-induced heart failure. Circulation 114298-308. [DOI] [PubMed] [Google Scholar]
- 38.van Rooij, E., N. Liu, and E. N. Olson. 2008. MicroRNAs flex their muscles. Trends Genet. 24159-166. [DOI] [PubMed] [Google Scholar]
- 39.van Rooij, E., L. B. Sutherland, N. Liu, A. H. Williams, J. McAnally, R. D. Gerard, J. A. Richardson, and E. N. Olson. 2006. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA 10318255-18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkins, B. J., Y. S. Dai, O. F. Bueno, S. A. Parsons, J. Xu, D. M. Plank, F. Jones, T. R. Kimball, and J. D. Molkentin. 2004. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ. Res. 94110-118. [DOI] [PubMed] [Google Scholar]
- 41.Wu, X., and D. M. Bers. 2007. Free and bound intracellular calmodulin measurements in cardiac myocytes. Cell Calcium 41353-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, J., N. L. Gong, I. Bodi, B. J. Aronow, P. H. Backx, and J. D. Molkentin. 2006. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J. Biol. Chem. 2819152-9162. [DOI] [PubMed] [Google Scholar]
- 43.Yang, B., H. Lin, J. Xiao, Y. Lu, X. Luo, B. Li, Y. Zhang, C. Xu, Y. Bai, H. Wang, G. Chen, and Z. Wang. 2007. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 13486-491. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, Y., J. F. Ransom, A. Li, V. Vedantham, M. von Drehle, A. N. Muth, T. Tsuchihashi, M. T. McManus, R. J. Schwartz, and D. Srivastava. 2007. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129303-317. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, Y., E. Samal, and D. Srivastava. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436214-220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.