Abstract
PR-Set7/Set8/KMT5A is the sole enzyme known to catalyze monomethylation of histone H4 lysine 20 (H4K20) and is present only in multicellular organisms that compact a large fraction of their DNA. We found that mouse embryos that are homozygous null mutants for the gene PR-Set7 display early embryonic lethality prior to the eight-cell stage. Death was due to the absence of PR-Set7 catalytic activity, since microinjection of the wild type, but not a catalytically inactive version, into two-cell embryos rescued the phenotype. A lack of PR-Set7 activity resulted not only in depletion of H4K20me1 but also in reduced levels of the H4K20me2/3 marks catalyzed by the Suv4-20h1/h2 enzymes, implying that H4K20me1 may be essential for the function of these enzymes to ensure the dimethylated and trimethylated states. Embryonic stem cells that were inducibly deleted for PR-Set7 passed through an initial G2/M phase, but the progeny were defective at the subsequent S and G2/M phases, exhibiting a delay in their cell cycle, accumulation at G2/M, massive DNA damage, and improper mitotic chromosome condensation. Cell cycle analysis after synchronization indicated that the defects were a consequence of decreased H4K20me1 due to the absence of PR-Set7. Most importantly, the lack of H4K20me1 also resulted in defects in chromosome condensation in interphase nuclei. These results demonstrate the critical role of H4K20 monomethylation in mammals in a developmental context.
Posttranslational modifications (PTMs) on histones influence intra- and internucleosomal interactions and thereby contribute to the diversity in nucleosome and chromatin structure that impacts distinct genomic processes. Among these modifications, histone methylation had been considered to be relatively stable, but recent studies demonstrated that, similar to the other PTMs, it too is subject to regulation. Many methylating and demethylating enzymes that target different lysine and arginine residues have been identified. These enzymes have distinct specificities with respect to the methylation status (monomethyl, dimethyl, and trimethyl) of each residue. Furthermore, increasing numbers of proteins harboring the motifs that specifically recognize various methylated residues have been identified. These proteins, or effectors, mediate/regulate elaborate chromatin-based processes, such as gene expression, which are dictated by the presence of the PTMs. Thus, the catalysis/removal of PTMs and their recognition by effectors constitute an intricately designed system that is key to genomic integrity and function.
One of the residues of histone H4 that can be monomethylated, dimethylated, or trimethylated is lysine 20. Recent comprehensive analysis of H4 modifications with top-down mass spectrometry in human and in Drosophila melanogaster cells revealed that the dimethyl group is deposited on the majority of total H4K20, indicative of the wide distribution of H4K20me2 on chromatin (18, 34). On the other hand, H4K20me1 and H4K20me3 are relatively few in abundance. The study also showed that H4K20 methylation status changes dynamically during the cell cycle and, notably, 98% of newly synthesized histone H4 becomes dimethylated within a few cell cycles (18). Each H4K20 methylation state is known to exhibit distinct functions. H4K20me2 serves as a binding site for 53BP1 and is involved in the DNA repair pathway (2, 24). H4K20me1 is located on active genes, suggesting a positive role in transcription (1, 32). However, this modification is recognized by the malignant brain tumor domain of L3MBTL1, which results in local compaction of the chromatin and repression of transcription (12, 30, 31). Indeed, the H4K20me1 mark has been associated with the inactive X chromosome during X inactivation, providing a further link with silencing (14). The trimethylated form of H4K20 is enriched in pericentromeric heterochromatin (25), and global changes in H4K20me3 levels have been implicated in tumorigenesis (4). Thus, the dynamic changes in H4K20 methylation status, the means by which the different states of H4K20me become associated with distinct genomic regions, and how H4K20me status contributes to the particular functions associated with these genomic regions in vivo are of great importance.
In higher eukaryotes, several enzymes have been reported to methylate H4K20 in vitro. PR-Set7/Set8/KMT5A is an exclusive monomethylase (33) present only in multicellular organisms, while Suv4-20h1/KMT5B and Suv4-20h2/KMT5C were shown to have both dimethyltransferase and trimethyltransferase activities (25), and a homologue, Set9/KMT5, is present in Schizosaccharomyces pombe (24). Loss-of-function studies in Drosophila cells and in mammalian cells demonstrated that these enzymes are responsible for H4K20 methylation in vivo (2, 13, 23, 28, 34).
Analysis of a PR-Set7 null mutant demonstrated that PR-Set7 performs a critical function(s) during embryonic development and in gene silencing in Drosophila cells (13, 16). Moreover, the expression level of PR-Set7 changes dynamically during the cell cycle of human transformed cells (HeLa cells), which suggests a role in regulating cell cycle-related chromosome function, especially during mitosis (20). In accordance with this, neuroblasts from the Drosophila null mutant exhibited abnormal chromosomes during mitosis when PR-Set7 was absent. The ATR-mediated checkpoint was also activated (23), yet mutant cells did not display increased levels of γ-H2AX, which normally occurs during the onset of DNA repair.
In the case of mammalian cells, extensive studies using a knockdown approach in transformed cells have reported the impact that the loss of PR-Set7 has on cell cycle progression. However, the conclusions of these studies differ somewhat (8-11, 18, 27, 28). These discrepancies may be due to inefficient knockdown of the PR-Set7 levels or to differences in susceptibility to the absence of PR-Set7 between cell types. For example, in recent reports the knockdown of PR-Set7 in human U2OS cells led to impaired cell proliferation and DNA damage that was accompanied by an increase in γ-H2AX (9, 10, 28), but the same was not true in the case of HeLa cells, even though a significant decrease in PR-Set7 and H4K20me1 was achieved (28). The cell cycle delay in U2OS cells was deemed a consequence of intra-S-phase checkpoint activation mediated by the ATR and Chk1 pathway (10, 28). The accumulation of DNA double-strand breaks and γ-H2AX was dependent on the initiation of DNA replication and the homologous recombination pathway since the effect was suppressed by the simultaneous knockdown of Cdc45 and Rad51 (10). Another group observed that PR-Set7 knockdown in HEK293 cells caused cells to arrest at G2 with DNA damage, chromosome decondensation, and aberrant centrosome amplification (8). Certain studies have also shown that PR-Set7 and PCNA, a component of the DNA replication machinery, interact directly via the PCNA-interacting protein box of PR-Set7 and colocalize during S phase, although this was seen only in the presence of proteasome inhibitor (28). The authors proposed that PR-Set7 functions during S phase by interacting with PCNA and thereby facilitating DNA replication and efficient DNA repair. However, another group has reported opposite findings, concluding that PR-Set7 is excluded from the nucleus during S phase and degraded at the G1/S border through its ubiquitination by SCF/Skp2 (35). Taken together, these rather discordant findings suggested that the various effects observed following PR-Set7 depletion might be cell type specific and due to genetic differences in the transformed cell lines studied. Indeed, the genetic instability and deregulation of many cell cycle control genes in transformed cell lines potentially limit their use in defining the roles of factors such as PR-Set7. Thus, in order to address the role of PR-Set7 in normal mammalian cells, we generated PR-Set7-deficient mice and analyzed their phenotype during development in vivo. We also generated embryonic stem (ES) cells containing a conditional knockout of PR-Set7. We demonstrate that the monomethyltransferase activity of PR-Set7 is essential for progression through cleavage stages during early embryonic development. We also show that its absence in ES cells resulted in massive DNA damage, decondensed chromosomes, and accumulation of cells at the G2/M phase of the cell cycle. Our results demonstrate that PR-Set7 is critical for early embryonic development and chromosome structure and stability in mice.
MATERIALS AND METHODS
Targeting vector and generation of PR-Set7 conditional and null mice.
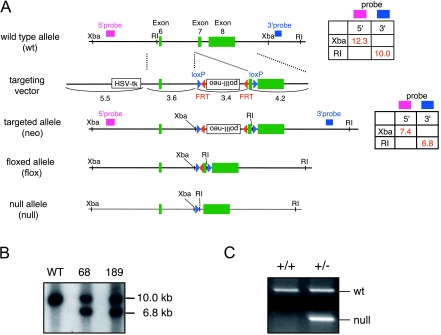
A mouse strain 129/Sv female RPCI-22 bacterial artificial chromosome (BAC) library was screened with mouse PR-Set7 probes (RPCI DNA microarray facility) designed based on the PR-Set7 cDNA sequence. The targeting vector was constructed by first amplifying a 3.6-kb left homologous arm and a 4.2-kb right homologous arm by PCR from the 129/Sv BAC clone 219K14 and then using pBluescript II KS(+) as a shuttle vector to place these arms into the targeting vector. The targeting vector was a derivative of the ploxPNT vector in which the PGKneo cassette was replaced with an FLP recombination target-flanked polII-neo cassette derived from pBluescript-polII-neo, via appropriate linkers. The targeting vector was designed to place the first loxP site into a region upstream of exon 7 that encodes the beginning of the SET domain of PR-Set7. Another loxP site was cloned downstream of exon 7 such that the two loxP sites were in the same orientation. A thymidine kinase gene cassette was located outside of the homologous region (Fig. 1A). 129S1/Sv-derived W9.5 ES cells were electroporated with the linearized targeting construct and maintained on subconfluent embryonic fibroblasts. G418 and ganciclovir-resistant ES cell colonies were selected and expanded. The ES cells were scored for homologous recombination by Southern blotting (Fig. 1B). Four homologous recombinants were identified with 5′- and 3′-flanking probes out of around 600 clones. Two targeted ES cells were injected into C57BL/6 blastocysts, and highly chimeric male mice were bred with C57BL/6 females to generate heterozygous PR-Set7neo/+ mice. Deletion of the neomycin cassette and exon 7 in vivo was achieved by crossing the mice with an ACT-FLPe (22) and Sox2-cre (5) transgenic line (Jackson Laboratory) to produce PR-Set7flox/+ and PR-Set7+/−, respectively. Mice from both lines were bred with C57BL/6 mice to remove ACT-FLPe and the Sox2-cre transgene, respectively. The PR-Set7flox/+ mice were intercrossed to generate homozygous PR-Set7flox/flox mice. The PR-Set7+/− mice were intercrossed to generate homozygous null mice. Mice were genotyped with the primer sets PRS7-7386U (5′-GTAAGAGAACTTTGAATGG-3′) and PRS7-7617L (5′-AGGCAGGGGGAGGAT-3′). For the wild-type (wt) allele, both primers amplified a 585-bp fragment, while the insertion of two loxP sites increased the size of the PCR fragment to 794 bp for the floxed allele (flox). The same primer sets amplified a 250-bp fragment for the null allele.
FIG. 1.
Targeting strategy of PR-Set7 knockout mice. (A) Schematic representation of the targeting strategy to generate PR-Set7 knockout mice showing the C-terminal region of wt and genetically engineered alleles of PR-Set7. Exons are shown as green boxes, and loxP and FLP recombination target sites are shown as blue and red triangles, respectively. The exons encoding the SET domain, the relevant restriction sites (XbaI and EcoRI), the sizes of the restriction fragments used for genotyping, and the position of the probes are indicated. The floxed allele functions as the wt and contains two loxP sites flanking exon 7 that include the beginning of the SET domain. Cre-mediated and FLPe-mediated recombination of targeted alleles results in a null allele and a floxed allele, respectively. HSV-tk, herpes simplex virus thymidine kinase. (B) Southern blotting to confirm homologous recombination in two ES clones (68 and 189) used for chimera production. Genomic DNA was digested with EcoRI and hybridized with 3′ probe as indicated in panel A. (C) PCR genotyping of PR-Set7 wt (+/+) and heterozygous (+/−) mice.
All animals used in the studies were handled with care, and all experiments were carried out in accordance with federal guidelines and the guidelines from French legislation and institutional policies.
In vitro embryo culture and apoptosis assay.
Preimplantation embryos were collected at 2.5 days postcoitum (dpc) by oviduct flushing from intercrosses with PR-Set7+/− males and females. Embryos were cultured in KSOM droplets under mineral oil under a 5% CO2 atmosphere at 37°C for 24 h for the apoptosis assay and genotyping. Terminal deoxynucleotidyltransferase-mediated dUTP-fluorescein nick end labeling (TUNEL) staining was performed with the in situ cell death detection kit (Roche). A CaspaTag caspase-3 and caspase-7 in situ assay kit with sulforhodamine (Millipore) was used for in situ detection of activated caspase-3 and caspase-7.
Embryo collection and mRNA microinjections for rescue experiments.
Embryos were collected from PR-Set7+/− superovulated females (∼6 weeks old) that were crossed with PR-Set7+/− males as described previously (7). The two-cell-stage embryos were collected at 44 h after human chorionic gonadotropin injection. A single blastomere of the two-cell-stage embryos was microinjected with 1 to 2 pl of 500 ng/μl of capped mRNA encoding hemagglutinin (HA)-tagged wt PR-Set7 or its catalytic mutant (R265G) that was transcribed in vitro in combination with 100 ng/μl of mRNA for green fluorescent protein (GFP). After injection embryos were cultured in KSOM under a 5% CO2 atmosphere at 37°C until they were fixed for immunostaining.
Immunofluorescence and confocal analysis for embryos.
After removal of the zona pellucida with acid Tyrode's solution (Sigma), embryos were washed three times in phosphate-buffered saline (PBS) and fixed as described previously (29). After permeabilization with 0.5% Triton X-100 in PBS, embryos were washed three times in PBS-T (0.1% Tween 20 in PBS), blocked in 3% bovine serum albumin in PBS-T, and incubated with the primary antibodies H4K20me1 (1:250 dilution; Abcam) and H3S10P (1:250 dilution; Millipore) for ∼12 h at 4°C. Embryos were then washed, blocked, and incubated for 2 h at 25°C with the corresponding secondary antibodies (Cy3-conjugated goat anti-rabbit; Jackson ImmunoResearch). DNA was stained with 4′-6-diamidino-2-phenylindole (DAPI), and embryos were mounted in Vectashield (Vector Laboratories). Some embryos were fixed without removing the zona pellucida but were immunostained in a similar way, and nuclei were stained with YOYO-1 iodide (Invitrogen). Confocal microscopy was performed using an HC Plan Apochromat ×20.0 objective/0.70 numerical aperture [NA] immersion/correction ring in an inverted Leica SP2 acoustico-optical beam splitter confocal microscope.
Isolation and culture of inducibly targeted ES cell lines.
Homozygous CAGGS-CreER (CreERT) transgenic female mice (6) (Jackson Laboratory) were crossed with heterozygous male PR-Set7+/− mice to obtain hemizygous offspring with the PR-Set7+/−; CreERT genotype. Heterozygous female PR-Set7+/−; CreERT mice were mated with heterozygous floxed male PR-Set7flox/+ mice. ES cell lines were established from 3.5-dpc blastocysts (vaginal plug detection on day 0.5) according to the methods described previously (21). These ES cells were genotyped as PR-Set7flox/−; CreERT (IKE5-2 cells) or PR-Set7flox/+; CreERT (IKE5-1 cells). These lines were found to be karyotypically XO and XY, respectively. Another cell line, IKX5 with no CreERT transgene (PR-Set7flox/+), was also used as a control for some experiments. Finally, the wt PGK12.1 ES cell line (a gift from N. Brockdorff) was also used as a control. ES cell lines were cultured, grown, and maintained on monolayers of mitomycin C-treated male primary feeder cells or on gelatin-coated flasks when maintained feeder free. ES cells were maintained in an undifferentiated state in Dulbecco's modified Eagle's medium (Sigma), 15% fetal calf serum (Gibco), 0.1 μM 2-mercaptoethanol (Sigma), and 1,000 U/ml leukemia inhibitory factor (Millipore). To induce deletion of the gene PR-Set7, cells were cultured in medium containing 1 μM 4-hydroxytamoxifen (4-OHT; Sigma). To determine cell growth, 5 × 104 ES cells were plated on 3.5-cm dishes. The cells were cultured for 4 days with or without 4-OHT, and the number of viable cells was counted using Vi-CELL XR 2.03 (Beckman Coulter).
Cell culture and synchronization.
HeLa and F9 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. To arrest F9 cells in prometaphase (PM), cells were treated with 0.4 μg/ml nocodazole (NCZ; Sigma) for 16 h. To synchronize HeLa cells at the G1/S border with a double-thymidine block, cells were treated with 2 mM thymidine (Sigma) for 16 h, released into fresh medium for 9 h, and blocked again with 2 mM thymidine for 16 h. Cells were then released into fresh medium, and time points were taken every 2 h. ES cells were synchronized by growing them in the presence of 0.4 μg/ml NCZ for 12 h, followed by growth in 2 mM thymidine for 24 h (in the presence or absence of 4-OHT), and were released into fresh medium for 14 h.
Flow cytometry.
Cells were trypsinized, washed once in cold PBS, and fixed in 5 ml 70% ethanol on ice for 30 min. Cell aliquots were immunostained with 100 μl of a 1:200 dilution of anti-H4K20me1 antibody (Abcam) for 1 h. Cells were then washed twice with PBS and incubated with anti-rabbit Alexa Fluor 488 (1:200; Invitrogen) for 30 min. For cell cycle analyses, cells were washed and resuspended in PBS containing RNase A (100 μg/ml) and propidium iodide (40 μg/ml). At each time point, ∼5 × 105 cells were used for fluorescence-activated cell sorting (FACS) analysis. Samples were run on a FACScan (Becton Dickinson), and data analysis was performed using CellQuest (Becton Dickinson) and ModFit LT (Verity Software House).
Protein extracts and immunoblotting.
Cells were lysed with buffer A (10 mM HEPES, pH 7.9, 0.1% NP-40, 5 mM MgCl2, 250 mM sucrose) containing protease inhibitors (Roche) and centrifuged. Nuclear pellets were lysed with buffer B (25 mM HEPES, pH 7.9, 20% glycerol, 0.8 M NaCl, 1.5 mM MgCl2, 0.1 mM EDTA), sonicated, and centrifuged. Protein was separated on sodium dodecyl sulfate-polyacrylamide gels, and Western blots were incubated with primary antibodies against the following proteins: H4K20me1 (Abcam), H4K20me2 (Millipore), H4K20me3 (Abcam), H4K16Ac (Millipore), H4 (Millipore), H3S10P (Sigma), H3K4me1 (Millipore), H3K4me2 (Millipore), H3K4me3 (in house), H3K27me3 (Millipore), H3K9me3 (Millipore), PR-Set7 (in house), H3 (Cell Signaling), GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Ambion), and β-tubulin (Abcam).
Immunofluorescence in ES cells.
ES cells cultured on gelatin-coated coverslips were fixed in 3% paraformaldehyde for 10 min at room temperature. Permeabilization of cells was performed on ice in PBS containing 0.5% Triton X-100 for 4 min. After three washes in 1× PBS, cells were blocked in PBS-1.0% bovine serum albumin (Gibco) for 15 min at room temperature and then incubated with the primary antibody for 40 min at room temperature in a humid chamber. Antibodies were used at the following dilutions: 1:300 for anti-γ-H2AX monoclonal antibody (JBW103; Upstate), 1:250 for anti-ATMS1981ph rabbit antibody (Rockland) 1:200 for anti-H4K20me1 rabbit antibody (07-440; Upstate), 1:300 for anti-H3S10P monoclonal antibody (3H10; Upstate), and 1:200 for anti-H4K20me3 rabbit antibody (ab9053; Abcam). After three 5-min washes in 1× PBS, cells were incubated with secondary antibodies (568 immunoglobulin G highly cross-adsorbed; Alexa Fluor 488) for 30 min at room temperature in a humid chamber. After three 5-min washes in 1× PBS, DNA was counterstained for 2 min in 0.2 mg/ml DAPI, followed by a final wash in 1× PBS. Coverslips were mounted in 90% glycerol, 0.1× PBS, 0.1% p-phenylenediamine, pH 9 (Sigma). Chromosome spreads for immunofluorescent labeling were prepared using the protocol previously described for spermatocyte metaphase spreads (19), as adapted for mouse ES cells. Mitotic cells were collected by the shake-off method and were resuspended in 100 mM sucrose (pH 8.2). A small volume of the cell suspension was transferred into a drop of fixative (1% paraformaldehyde, 5 mM sodium borate [pH 9.2], and 0.15% Triton X-100) on a Denhardt's solution-coated coverslip and dispersed on the coverslip for 20 min at room temperature. After being washed with PBS, lysed nuclei were dried and used for immunofluorescence. A 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay was performed with the Click-iT EdU assay kit according to the manufacturer's instructions (Invitrogen). For the preparation of chromosome spreads for DAPI staining to assess chromosomal structure, floating mitotically arrested cells were removed by the shake-off method. Adherent mitotically arrested cells were also isolated with trypsin. The pooled mitotic cells were then incubated in hypotonic solution (75 mM KCl) for 10 min, followed by fixation in Carnoy's fixative (3:1 ratio of methanol/acetic acid). Fixed cells were dropped onto a glass microscope slide and air dried; DNA was counterstained with 100 ng/μl DAPI and mounted in 90% glycerol, 0.1× PBS, 0.1% p-phenylenediamine at pH 9 (Sigma).
Microscopy.
All three-dimensional (3D) acquisitions were performed using a Delta Vision system (Applied Precision) with ×100 objective/1.35 NA and ×60 objective/1.40 NA (Olympus) and filters set for DAPI, fluorescein isothiocyanate, and rhodamine provided by Applied Precision. Acquired z planes were separated by 0.2 μm, and an average of 75 planes was taken for each nucleus. The 3D stacks were deconvolved using the Softworx software algorithm (conservative ratio method, 10 iterations; Applied Precision).
Chromosome painting.
PR-Set7flox/−; CreERT ES cells treated with 4-OHT for 0, 1, and 2 days were trypsinized, washed twice with PBS, and adhered to poly-l-lysine (Sigma-Aldrich)-coated coverslips for 10 min. The coverslips were fixed and permeabilized as described previously and then dehydrated by serial 3-min washes in 70%, 90%, and 100% ethanol, aged for 5 min at room temperature, denatured in 70% formamide with 2× SSC (1× SSC is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.0]) for 10 min at 80°C, quenched for 5 min in ice-cold 70% ethanol, dehydrated again, and then hybridized with mouse X chromosome Cy3 paint (Cambio) for 48 h at 37°C. After hybridization, the coverslips were washed three times in 50% formamide with 2× SSC at 42.5°C for 8 min and twice in 2× SSC at 42.5°C for 5 min before being stained with DAPI and mounted as described previously.
Measurement of chromosome areas.
For each experimental time point, 50 nuclei were analyzed following 3D acquisition. The images were projected in 2D, the resulting area was occupied by the chromosomal domain, and the total area of the nucleus was traced using ImageJ software. Chromosome areas were then normalized to the nuclear area.
DNA FISH.
ES cells cultured on gelatin-coated coverslips were fixed in 3% paraformaldehyde (pH 7.2) for 10 min at room temperature. Permeabilization of cells was performed on ice in PBS containing 0.5% Triton X-100 for 5 min. Coverslips were rinsed twice and stored in 70% ethanol. The coverslips were then dehydrated through an ethanol series (70%, 85%, 90%, and 100%), air-dried, and then rehydrated in 2× SSC. DNA was then denatured in 50% formamide, 2× SSC for 38 min at 80°C in an oven. The coverslips were then placed in ice-cold 2× SSC and rinsed once, and DNA fluorescence in situ hybridization (FISH) was performed. BAC-derived probes were fluorescently labeled by nick translation (Vysis) using Spectrum Green and Spectrum Red dUTP (Vysis) or Cy5 dUTP (Amersham). Hybridization involved 0.1 μg of the BAC probe (per 22- by 22-mm coverslip), which was precipitated with 10 μg salmon sperm and 5 μg Cot-1 DNA, resuspended in 6 μl formamide, denatured at 75°C for 7 min, and competed at 37°C for 30 min. A further 6 μl of hybridization buffer (Cambio) was added to the probe, and hybridization with the coverslips was performed overnight at 37°C. After three washes in 50% formamide with 2× SSC and three washes in 2× SSC at 42°C, DNA was counterstained for 2 min in 0.2 mg/ml DAPI, followed by a final wash in 2× SSC. Samples were mounted in 90% glycerol, 0.1× PBS, 0.1% p-phenylenediamine at pH 9 (Sigma).
DNA FISH signal distance measurements.
Only nuclei with a normal shape in which the three DNA FISH signals could be detected were chosen for analysis. For each experimental time point, a minimum of 30 nuclei were analyzed, following 3D acquisition and image deconvolution. Manual selection of the DNA FISH signals was performed, and distances were measured between the two centers of mass from 3D image stacks using an ImageJ plug-in. In the case of sister chromatids, the median of the line drawn between the centers of mass of the sister chromatid signals was used as the point from which to measure the distance. The nuclear volume was calculated using an ImageJ plug-in to detect 3D objects using the DAPI signal. An average PGK12.1 ES cell nuclear volume was obtained, and the intergenic distances of the PR-Set7flox/−; CreERT and PR-Set7flox/+; CreERT cells were then normalized to this value.
RESULTS
PR-Set7 null embryos exhibit embryonic lethality prior to implantation.
To investigate the in vivo role of PR-Set7 during mouse development, we used gene targeting by homologous recombination in ES cells to generate both a conditional allele and a null allele of PR-Set7 (Fig. 1A). The conditional allele contains loxP sites flanking exon 7 of PR-Set7, such that Cre-mediated recombination would result in deletion of the SET domain (encoding methyltransferase activity). Two independent mouse lines carrying the conditional allele were generated, and both were phenotypically wt. No differences between these lines were observed in any of the experiments performed.
To generate a null allele of PR-Set7, we crossed male mice carrying the conditional PR-Set7 allele with female mice containing a Sox2-Cre transgene, which expresses Cre recombinase in cleavage-stage embryos (5). Following recovery and intercrossing of double heterozygotes we recovered PR-Set7 null heterozygotes, and after a further cross with wt C57BL/6 mice we successfully obtained PR-Set7 null heterozygotes that lacked the Sox2-Cre transgene. Heterozygotes for the PR-Set7 null allele displayed a normal phenotype and were indistinguishable from their wt littermates (data not shown).
For analysis of the null phenotype of PR-Set7, we performed heterozygote intercrosses using the null allele. Among 58 newborn pups tested, no PR-Set7 null homozygotes were identified, while the wt and heterozygotes were present in a 1:2 ratio, as expected if the homozygotes displayed embryonic lethality (Table 1). To determine the stage at which PR-Set7 homozygosity resulted in lethality, we analyzed embryos arising from heterozygote intercrosses at 6.5, 7.5, and 11.5 dpc (Table 1). PCR genotyping of these embryos demonstrated that PR-Set7 homozygotes could not be recovered at these stages, indicating lethality prior to 6.5 dpc. Further examination of preimplantation embryos at 2.5 and 3.5 dpc showed that PR-Set7 homozygotes could be recovered only as cleavage-stage embryos at 2.5 dpc, but no null blastocysts could be recovered at 3.5 dpc (Table 1).
TABLE 1.
Genotypes of progeny from PR-Set7 null heterozygous intercrosses
| No. of litters | Age of mice (dpc) | No. (%) of mice with indicated genotype
|
Total no. of mice | |||
|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | NDa | |||
| 11 | Neonate | 20 (34) | 38 (66) | 0 | 58 | |
| 3 | 11.5 | 6 (46) | 7 (54) | 0 | 13 | |
| 2 | 7.5 | 6 (43) | 8 (57) | 0 | 14 | |
| 2 | 6.5 | 5 (42) | 7 (58) | 0 | 12 | |
| 8 | 3.5 | 22 (46) | 22 (46) | 0 | 4 | 48 |
| 9 | 2.5 | 22 (23) | 47 (50) | 20 (21) | 5 | 94 |
ND, genotype not determined due to failure of PCR.
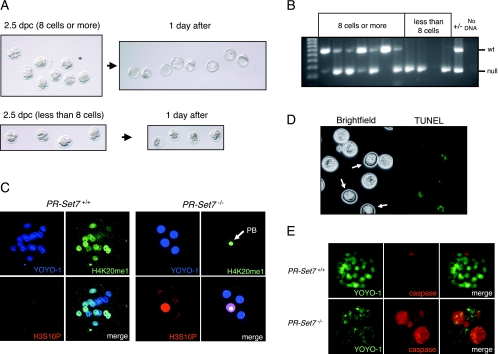
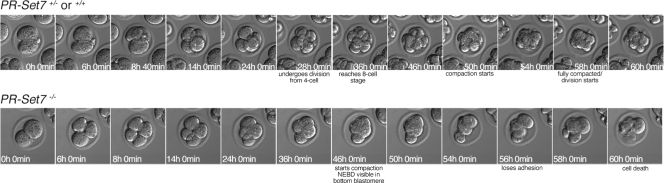
Genotyping demonstrated that 2.5-dpc embryos with eight or more cells were wt or heterozygous, while those with fewer than eight cells were homozygous mutant (Fig. 2B); genotyping of some embryos was unsuccessful due to the loss or degeneration of their DNA. To examine the fate of these embryos, we cultured embryos recovered at 2.5 dpc in vitro for 1 day. Embryos that had eight or more cells developed into blastocysts following 1 day of culture, while those with fewer than eight cells degenerated (Fig. 2A). Time-lapse analysis of embryos derived from PR-Set7+/− intercrosses confirmed that PR-Set7−/− embryos become arrested without reaching the eight-cell stage (Fig. 3). Therefore, we conclude that PR-Set7 is required for preimplantation development, most likely during the transition from the four-cell stage to the eight-cell stage.
FIG. 2.
PR-Set7 knockout embryos arrest at late G2 or M phase prior to the eight-cell stage and exhibit decreased H4K20me1, increased H3S10P, and apoptosis. (A) Microscopic analysis of embryos at 2.5 dpc that were collected and transferred to in vitro culture for a day. (Top) Embryos having eight or more cells developed into blastocysts after 1 day of culture. (Bottom) Those having less than eight cells did not develop into blastocysts and degenerated. (B) Genotyping results of in vitro-cultured embryos shown in panel A. Embryos that developed into blastocysts were wt (+/+) or heterozygous (+/−). All the embryos that had less than eight cells at 2.5 dpc were homozygous (−/−). (C) Immunofluorescence analysis using anti-H4K20me1 and anti-H3S10P antibodies. (Left) A 2.5-dpc embryo wt for PR-Set7 exhibits variable levels of H4K20me1 in each blastomere, and the H3S10P signal is negative. (Right) PR-Set7 KO embryos arrest with nearly undetectable levels of H4K20me1 in the blastomeres but with clear staining in the polar body (PB; arrow). H3S10P shows speckled or diffuse staining in all blastomeres. (D) TUNEL assay performed on 2.5-dpc embryos after in vitro culture for a day. Presumptive PR-Set7 KO embryos showed positive signals (white arrows) as opposed to other embryos that showed few, weak signals and developed into blastocysts. (E) Comparison of homozygous null (PR-Set7−/−) versus wt (PR-Set7+/+) 2.5-dpc embryos in an in situ caspase assay after in vitro culture for a day. The PR-Set7−/− embryo shows strong caspase-3 and caspase-7 activity.
FIG. 3.
Time-lapse analysis of embryos derived from PR-Set7+/− crosses. Embryos were obtained 44 h post-human chorionic gonadotropin injection from 6-week-old females. Two-cell-stage embryos were cultured in KSOM covered with paraffin oil in a 37°C chamber under a 5% CO2 atmosphere. Embryos were imaged under differential interference contrast optics with the ×20 objective of an inverted Leica microscope. Images were captured every 20 min in different z planes (covering 60 μm) and in four different fields using an automated stage controller.
PR-Set7 null embryos arrest during the cleavage stage and exhibit decreased H4K20me1, increased H3S10P, and apoptosis.
To determine the effects of PR-Set7 depletion on its catalytic product, monomethylated H4K20, we collected embryos from PR-Set7 heterozygous intercrosses at 2.5 dpc and scored them for H4K20me1 levels using immunofluorescence, followed by single-embryo genotyping. In the wt embryos, blastomeres showed variable levels of H4K20me1, likely reflective of their different cell cycle stages (Fig. 2C, left). This is in line with PR-Set7 normally being expressed during G2/M (20). In contrast, the H4K20me1 signal was undetectable in the nuclei of blastomeres of null embryos. Only residual H4K20me1 signal was observed in the second polar body of null embryos (Fig. 2C, right). This result strongly suggests that the production of H4K20me1 is dependent on zygotic PR-Set7 expression and that PR-Set7 is the major enzyme catalyzing H4K20me1 at this stage of development. Moreover, this result indicates that the maternal pool of PR-Set7 from the oocyte cytoplasm is insufficient to support development of the embryo beyond two rounds of cleavage. By using immunofluorescence and scoring for H3S10P levels, which are normally associated with condensed chromatin during mitosis, we examined the stage in the cell cycle during which PR-Set7−/− embryos become arrested. Most of the blastomeres in PR-Set7−/− embryos showed a sustained speckled or diffuse pattern for H3S10P (Fig. 2C, right), which suggests a cell cycle arrest at late G2 or early M phase, while PR-Set7+/+ embryos showed positive staining as expected only when the blastomeres undergo mitosis.
Given the early and rapid lethality of PR-Set7−/− embryos, we next tested if mutant cells underwent apoptosis. Embryos at 2.5 dpc were cultured for 1 day in vitro and then analyzed by TUNEL assay. Embryos that developed into blastocysts showed very few positive TUNEL signals, as expected (Fig. 2D). However, the embryos that did not progress past the eight-cell stage exhibited a widespread and strong TUNEL signal (Fig. 2D). To confirm that PR-Set7 null embryos were apoptotic, we tested them for caspase-3 and caspase-7 levels in situ using the fluorescent-labeled DEVD peptide. While wt blastocysts exhibited a positive signal only in the polar body (Fig. 2E, top), the PR-Set7 null blastomeres exhibited strong positive signals reflecting caspase-3 and caspase-7 activation accompanying apoptosis (Fig. 2E, bottom).
PR-Set-7 methyltransferase activity rescues the developmental arrest in null embryos and restores H4K20me1 levels.
We next investigated whether the early lethality displayed by the PR-Set7 null embryos was specifically due to the absence of PR-Set7 methyltransferase activity. To this end, we injected one cell of two-cell embryos derived from PR-Set7+/− intercrosses with in vitro-transcribed mRNA encoding either wt HA-tagged PR-Set7 or the HA-tagged PR-Set7 mutant in its catalytic domain (R265G). In each case, GFP mRNA was coinjected and served both as a lineage tracer to identify the injected cells and as a positive marker for mRNA expression (Fig. 4A). In total, we injected 11 embryos with GFP mRNA alone, 20 with wt PR-Set7 and GFP mRNAs, and another 20 with mutant PR-Set7 and GFP mRNAs. Based on Mendelian ratios, approximately 25% of the injected embryos in each case were expected to be PR-Set7−/−. As expected, approximately 75% of the embryos reached the blastocyst stage and cavitated normally in all three cases. Furthermore, approximately one-fourth of the embryos arrested or fragmented after the four- to eight-cell stage in the case of GFP alone (n = 3/11), as expected, and this was also the case when we injected mutant PR-Set7/GFP (n = 6/20) (Fig. 4B). Single-embryo genotyping confirmed that these arrested embryos were indeed PR-Set7−/− (data not shown). In contrast, we did not observe such fragmentation in the presumably null embryos (n = 7/20) that had been injected with wt PR-Set7/GFP mRNAs, although these embryos were not completely normal in appearance (likely because only one of the two cells in the two-cell embryo was microinjected) (Fig. 4B, bottom right). We also observed formation of a blastocoel cavity in three out of these seven embryos (Fig. 4C). Thus, the catalytic activity of PR-Set7 was specifically required to rescue the lethal phenotype associated with PR-Set7−/− embryos.
FIG. 4.
The methyltransferase activity of PR-Set7 is required to rescue the knockout embryo. (A) Schematic representation of HA-tagged PR-Set7, either wt or mutant, in the catalytic SET domain. The experimental strategy for microinjection of mRNAs encoding GFP- and HA-tagged PR-Set7, either wt or mutant (mut), into one cell of a two-cell embryo is shown. The embryos were derived from intercrossing mice heterozygous for the PR-Set7 null allele. (B) Bright-field microscopy and fluorescence analyses for GFP expression of embryos microinjected with the mRNAs are indicated to the left. Injection of wt but not mutant PR-Set7 mRNA into one cell of a two-cell-stage embryo rescues the injected cells in PR-Set7 null embryos. Note that in these rescued embryos, cell proliferation and in some instances avitation are observed by day 3 (bottom, black arrows). (C) As described for panel B, a blastomere corresponding to the injected side of a two-cell-stage embryo that was presumably PR-Set7−/− developed into a blastocyst-like structure and exhibited formation of a blastocoelic cavity. The white broken line shows an uninjected blastomere(s) (e.g., GFP negative), which is presumably degenerated by day 3. (D) Analysis of an equivalent to an approximately eight-cell-stage embryo after injection of one cell of a two-cell embryo with wt PR-Set7 and GFP mRNAs. The panels show DAPI staining, indirect immunofluorescence using anti-H4K20me1 antibodies, and direct fluorescence from GFP expression. The last indicates the progeny of the injected blastomere (dashed white line). H4K20me1 appears distributed throughout the nuclei of the injected cells (dashed line; the arrow points to the polar body [PB]) but not in the other half of the embryo. The reduced H4K20me1 levels and DNA fragmentation observed in this other half indicate that the embryo is PR-Set7−/−. In contrast to that of noninjected cells, the nuclear morphology of the injected cells seemed normal. (E) Single-embryo genotyping of the embryo shown in panel D confirmed it to be PR-Set7−/−. PR-Set7+/+ and PR-Set7+/− genotypes are shown as controls on the right.
We similarly coinjected wt PR-Set7 and GFP mRNAs into PR-Set7 null embryos at the two-cell stage and then examined the H4K20me1 levels at what should correspond to the approximately eight-cell stage using GFP fluorescence to identify positive cells (Fig. 4D, right). H4K20me1 appeared distributed throughout these nuclei, but not in the other half of such embryos, suggesting that injection of PR-Set7 mRNA restored H4K20me1 levels (Fig. 4D, middle). Indeed, the genotype confirmed that these were null mutant embryos (Fig. 4E, left). Of note, the nuclear morphology of these injected cells appeared normal (Fig. 4D, left) compared to that of cells within the same embryo that did not exhibit PR-Set7 or GFP expression and which were likely to be apoptotic given the fragmented appearance of their DNA.
Sharp increase in H4K20me1 and PR-Set7 expression at G2/M.
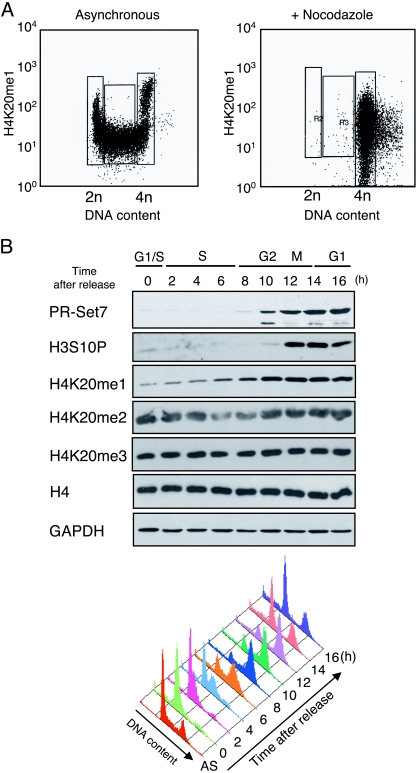
As a prelude to the studies described below for ES cells, we characterized the levels of the PR-Set7 product, H4K20me1, during the cell cycle in asynchronous mouse embryonic carcinoma F9 and HeLa cells using FACS analyses. The levels of H4K20me1 were increased ∼10-fold during the course of a cell cycle in F9 cells (Fig. 5A, top). When F9 cells were arrested at PM by treatment with NCZ, the level of H4K20me1 was approximately half that of the peak mean intensity during the cell cycle (Fig. 5A, bottom). The peak intensities at G2/M were considerably greater than would be expected based upon the increase in DNA content, demonstrating that histone H4 is highly monomethylated at lysine 20 during this stage of the cell cycle. These maximal levels were maintained until anaphase, after which the levels of H4K20me1 decreased concomitantly with the reduced copy number of the genome after cytokinesis, that being half of the peak M-phase intensity at early G1. The level was reduced during G1 and maintained as such throughout S phase. Similar results were observed when PR-Set7 expression and H4K20me were analyzed in HeLa cells (Fig. 5B). The observed increase in PR-Set7 and H4K20me1 during G2 and M phase suggests a possible function in M-phase chromatin, and this is in agreement with previous results showing that PR-Set7 associates with mitotic chromosomes (20).
FIG. 5.
Marked changes in H4K20me1 and PR-Set7 expression at G2/M. (A) FACS analysis of asynchronous or NCZ-arrested F9 cells as indicated. The G1, S, and G2/M populations are indicated. Each panel plots the fluorescent signal intensity (log scale) of the H4K20me1 antibody in arbitrary units against DNA content in arbitrary units. (B) (Top) Western analysis of histone H3 and H4 modifications and PR-Set7 protein in synchronized HeLa cells using the antibodies indicated to the left. GAPDH provides a loading control. Cells released from G1 arrest after a double-thymidine block were analyzed every 2 h as indicated at the top. (Bottom) Cell cycle phases were confirmed by FACS analyses, and the data are represented by a histogram.
The levels of all H4K20-methylated states decrease in PR-Set7 conditional knockout ES cells.
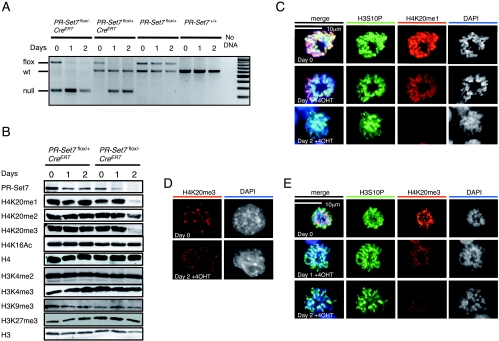
Given the very early lethality associated with the PR-Set7 knockout, we decided to examine the repercussions of PR-Set7 depletion at the cellular level. To this end, we established ES cells carrying a conditional allele of PR-Set7, together with a tamoxifen-inducible Cre recombinase. PR-Set7flox/+ mice were mated with PR-Set7+/−; CreERT mice that harbor the PR-Set7 heterozygous null allele and a transgenic allele for expression of a fusion protein containing the Cre recombinase and the ligand binding domain of the human estrogen receptor (CreERT) driven by the CAGGS promoter (6). Elimination of the SET domain of PR-Set7 in ES cells can be achieved after activating the CreERT recombinase by the addition of 4-OHT to the culture medium. Two ES cell lines containing the CreERT allele were derived from blastocysts, one of which had the PR-Set7flox/− genotype and the other a PR-Set7flox/+ genotype. Activation of the CreERT recombinase in each case resulted in the successful isolation of derivatives lacking the floxed allele (Fig. 6A). Two cell lines that did not contain the CreERT allele provided negative controls and showed no change in the genotype of PR-Set7 upon the addition of 4-OHT (Fig. 6A).
FIG. 6.
The levels of all three methylation states of H4K20 decrease in PR-Set7 conditional knockout ES cells. (A) Genotype analysis of PR-Set7 conditional knockout ES cells (see Materials and Methods) as a function of time of CreERT activation with 4-OHT as indicated at the top (see text). Both the XEN cell line that has PR-Set7flox/+ but lacks CreERT and PGK12.1 ES cells were used as negative controls for 4-OHT treatment. The expected migration of DNA fragments containing floxed, wt, or null PR-Set7 alleles is shown to the left. PR-Set7+/− and PR-Set7−/− ES cells were derived from IKE5-1 (PR-Set7flox/+; CreERT) and IKE5-2 (PR-Set7flox/−; CreERT) ES cells, respectively, after conditional knockout of PR-Set7 during 2 days of 4-OHT treatment. (B) Western blot analyses using the antibodies are shown to the left, and nuclear protein isolated from the cells indicated as a function of time of 4-OHT treatment is also indicated at the top. The levels of H4K20me1, H4K20me2, and H4K20me3 declined appreciably in the case of PR-Set7−/− ES cells, derived by treating PR-Set7flox/−; CreERT ES cells with 4-OHT for 2 days. (C, D, and E) Immunofluorescence analysis using anti-H4K20me1 antibody (C) or anti-H4K20me3 antibody (D and E) and PR-Set7flox/−; CreERT cells either untreated or treated with 4-OHT for 1 or 2 days as indicated to the left.
We first examined the levels of PR-Set7 before and after 4-OHT treatment in the PR-Set7flox/+; CreERT and PR-Set7flox/−; CreERT cells (Fig. 6B). In accordance with the genotyping results, the levels of PR-Set7 decreased to almost 50% after treatment with 4-OHT in the case of PR-Set7flox/+; CreERT and to nearly undetectable levels after 4-OHT treatment in the case of the PR-Set7flox/−; CreERT. We next compared the levels of the different H4K20-methylated states and other methylated residues of histone H3 as a function of 4-OHT treatment. We did not detect changes in the levels of H4K20me1, H4K20me2, or H4K20me3 nor in any of the histone H3 and H4 modifications tested in the case of PR-Set7flox/+; CreERT cells. This suggests that an approximately 50% drop in PR-Set7 levels in ES cells has no effect on global levels of these histone modifications. However, in the case of PR-Set7flox/−; CreERT cells, treatment with 4-OHT gave rise to ES cells depleted of -Set7 and of H4K20me1. These cells were also severely reduced in their levels of H4K20me3 and to a lesser extent in H4K20me2 after 2 days of treatment. The levels of histone H4K16Ac were not affected nor were the levels of any of the H3 modifications tested, and global H3 and H4 levels also remained unchanged (Fig. 6B). While the reduced levels of H4K20me1 were expected, given that PR-Set7 is the only enzyme responsible for this modification, the decline in H4K20me2 and H4K20me3 was unexpected but suggests that H4K20me1 may be the substrate for further methylation by the Suv4-20h1/h2 methylases, a hypothesis confirmed by recent studies (26, 34). An alternate possibility is that PR-Set7 functions as a dimethylase and trimethylase in vivo. However, this is not supported by structural studies demonstrating that PR-Set7 functions exclusively as a monomethyltransferase (33).
To further investigate the observed change in H4K20 methylation status, immunofluorescence studies were performed following 4-OHT treatment using H4K20me1- and H4K20me3-specific antibodies (Fig. 6C to E). Both H4K20me forms show a strong enrichment on mitotic chromosomes of ES cells. After 1 day of 4-OHT treatment the levels of H4K20me1 on PM chromosomes showed a visible decrease, and by 2 days of 4-OHT treatment H4K20me1 had almost disappeared (Fig. 6C). H4K20me3 accumulation on metaphase chromosomes, particularly at pericentromeric regions, was evident prior to 4-OHT treatment (Fig. 6E). However, upon treatment this mark was also reduced, although less so than that observed for H4K20me1, in agreement with the results discussed above (Fig. 6B). In interphase nuclei, H4K20me3 is known to localize at pericentromeric heterochromatin (25). Although the DAPI-dense chromocenter was detectable before 4-OHT treatment, after 2 days of 4-OHT treatment the signal no longer colocalized to chromocenters or with DAPI-dense regions (Fig. 6D). Thus, the loss of PR-Set7 resulted in rapid reduction of H4K20me1 and also impacted on H4K20me2/3 levels, implying that H4K20me1 is the substrate for further methylation by the Suv4-20h1/h2 methyltransferases. This underlines the importance of the PR-Set7 enzyme in the generation of all H4K20 methylation states.
Conditional PR-Set7 knockout cells accumulate at G2/M phase and exhibit mitotic chromosome decondensation.
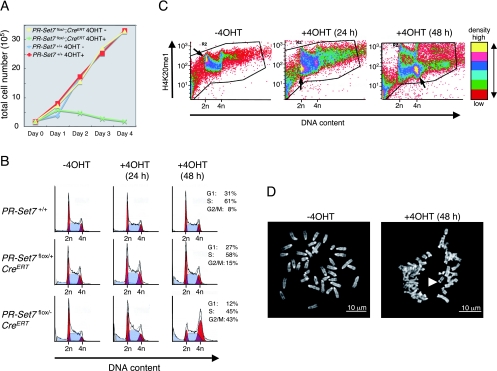
In order to gain insight into the cellular defects induced by a lack of PR-Set7, we examined the effect of PR-Set7 depletion on cell proliferation using the PR-Set7flox/−; CreERT and control wt cells, with or without 4-OHT treatment. A defect in cell growth was apparent only in the case of 4-OHT-treated PR-Set7flox/−; CreERT cells (Fig. 7A). Given that H4K20me1 levels change as a function of the cell cycle (20) (Fig. 5), we set out to determine at what cell cycle stage this defect occurred. FACS analysis was performed on IKE5-2 (PR-Set7flox/−; CreERT), IKE5-1 (PR-Set7flox/+; CreERT), and control PGK12.1 (PR-Set7+/+) cells with and without 4-OHT treatment. Once again, only in the case of 4-OHT-treated PR-Set7flox/−; CreERT cells was there a detectable change in cell cycle distribution with the conditional knockout of PR-Set7 leading to a significant accumulation at G2/M by day 2 of 4-OHT treatment (Fig. 7B).
FIG. 7.
Conditional PR-Set7 KO ES cells accumulate at G2/M phase. (A) Growth curve for PR-Set7flox/−; CreERT and PR-Set7+/+ as a function of 4-OHT treatment as indicated. PR-Set7flox/−; CreERT cells showed a slow growth in the presence of 4-OHT, whereas the cells continued to proliferate in its absence. (B) FACS analyses of the cells are indicated to the left as a function of the number of days of 4-OHT treatment as indicated at the top. Conditional knockout of PR-Set7 generated by 2 days of 4-OHT treatment of PR-Set7flox/−; CreERT cells led to significant accumulation at the G2/M phase (bottom right). (C) The fluorescent signal intensity (log scale) of the antibody against H4K20me1 in arbitrary units is plotted against the DNA content in arbitrary units for PR-Set7flox/−; CreERT cells treated with 4-OHT for the number of days indicated above each plot. Six color divisions represent the plot density from highest to lowest as follows: yellow, magenta, blue, blue-green, green, and red. The highest density area in each panel is indicated by an arrow. (D) Morphology of chromosome in PR-Set7flox/−; CreERT cells without 4-OHT treatment (left) and with 4-OHT treatment (right) for 2 days. (Right) The arrowhead shows separated sister centromeres. −4OHT, without 4-OHT treatment; +40HT, with 4-OHT treatment.
In wt ES cells, FACS analyses demonstrated that the cell cycle dynamics of H4K20me1 were similar to those found in F9 cells (Fig. 5A), with G1-phase cells displaying relatively high levels of H4K20me1 (Fig. 7C, left). After 24 h of 4-OHT treatment of the PR-Set7flox/−; CreERT cells, the levels of H4K20me1 in the G1 phase were decreased (Fig. 7C, middle), although some cells accumulated at the G2/M phase without an obvious decrease in H4K20me1. By 48 h, most of the cells had accumulated at G2/M, exhibiting low levels of H4K20me1 (Fig. 7C, right). We also noticed an accumulation of floating, rounded cells (possibly in mitosis) and an increase in cells with an apoptotic appearance after 24 h of 4-OHT treatment. To clarify the composition of the G2/M population, we evaluated the mitotic index (MI) of cells before and after 4-OHT treatment (see Materials and Methods). In untreated ES cells, we observed an MI of ∼5%; following 1 day of 4-OHT treatment, the MI increased to 12%, as expected from the G2/M accumulation described above. However, after 2 days the MI fell to less than 5%, implying that the arrested mitotic population probably died by 48 h of treatment. Chromosome spreads were prepared from cells following 2 days of 4-OHT treatment. Essentially all mitoses appeared normal on days 0 and 1. At day 2, the centromeres and sister chromatids appeared separated in 44% of the cells (Fig. 7D, right) compared to those at day 0 (Fig. 7D, left). In fact, 85% of mitoses appeared abnormal in some way at day 2 (data not shown). This is consistent with precocious sister chromatid separation and abnormal chromosomes observed during mitosis in PR-Set7 knockout cells.
The analyses described above demonstrate that there are clearly defects in PR-Set7 null ES cells at G2/M phase. We wished to address whether there are also any S-phase defects in these cells. Previous reports have implicated the methyltransferase activity of PR-Set7 as being pivotal for the interaction of PR-Set7 with PCNA during S phase (9, 10, 28). However, it was not clear whether it was the presence of PR-Set7 protein during S phase or the methylated histone that was required for cell cycle progression. We addressed this question using conditional knockout ES cells synchronized by treatment with NCZ and thymidine (Fig. 8A). First, asynchronous (AS) PR-Set7flox/−; CreERT cells were treated with NCZ for 12 h to arrest them at PM (Fig. 8A). The arrest was confirmed by FACS analysis (Fig. 8C). PR-Set7 protein was abundant in PM cells compared to asynchronous cells following NCZ treatment (Fig. 8B). Following release from NCZ, the cells were then treated with excess thymidine in order to block DNA synthesis. The gene PR-Set7 was then deleted using 4-OHT during this 24-h thymidine treatment (Fig. 8A). The cells were then analyzed every 2 hours following this 24 h treatment. The majority of cells were found to be in mid- to late S phase at 0 h (Fig. 8C). No difference in H4K20me1 levels could be detected in cells with or without 4-OHT at this time, although PR-Set7 was completely absent in 4-OHT-treated cells (Fig. 8B). PR-Set7 protein has been reported to be degraded at the G1/S border and kept low during S phase by proteasome-mediated degradation (35). As expected, PR-Set7 protein could not be detected after release, demonstrating that newly synthesized PR-Set7 was also undetectable (Fig. 8B). Control cells were treated with thymidine alone for 24 h, but expression of PR-Set7 was not affected (Fig. 8B).
FIG. 8.
Cell cycle analysis after PR-Set7 depletion. (A) Experimental design. PR-Set7flox/−; CreERT cells were synchronized with NCZ for 12 h followed by treatment with thymidine in the presence or absence of 4-OHT for 24 h and released into fresh ES medium. Samples were collected for FACS analysis and protein extraction at the time points shown at the top (black triangles). (B) Changes of the level of PR-Set7 protein and H4K20me1. Shown are Western blot analyses using the antibodies shown to the left and nuclear protein isolated from the cells at the time points indicated at the top. PR-Set7 protein was undetectable at 0 h after 24 h of treatment with 4-OHT (bottom). Both β-tubulin and histone H4 are shown as loading controls. (C) FACS analyses of the cells at each time point as indicated at the top. Note that cells treated with 4-OHT (bottom) show slower cell cycle progression between 8 h and 12 h than the cells untreated with 4-OHT (top) and accumulate at early S phase at 12 h (bottom right). AS, asynchronous.
The PR-Set7-depleted cells did not exhibit an obvious delay in S phase and subsequent G2/M phase after release from the thymidine block compared with control cells between 0 h and 8 h (Fig. 8C). This was surprising because we expected the cells to arrest during S or G2/M as a consequence of PR-Set7 protein being absent after 0 h. The difference in H4K20me1 levels became obvious after 4 h when the cells went through their first mitotic phase without PR-Set7 protein (Fig. 8B). When cells were analyzed by FACS following release from the thymidine block, the PR-Set7-depleted cells showed an obvious accumulation at the beginning of the following S phase between 8 h and 12 h (Fig. 8C).
Thus, the PR-Set7 protein itself did not appear to be essential for progression through the first S phase and G2/M following its depletion. Rather, cell cycle progression appeared to become disturbed in the following S phase, in other words, in the cells that had already passed through M phase once without PR-Set7 protein. This suggests that the S-phase phenotype reported in PR-Set7-depleted cells may not be due to the lack of PR-Set7 protein per se but may instead be due to alterations in H4K20 methylation that occurred during the previous cell cycle. These include both an increase in unmethylated H4K20 (H4K20me0) and a decrease in H4K20me1, H4K20me2, and H4K20me3. We favor the idea that it is increased H4K20me0 and/or decreased H4K20me1 levels rather than decreased H4K20me2 and H4K20me3 levels that cause this S-phase delay (see Discussion).
Conditional knockout of PR-Set7 leads to increased γ-H2AX foci in ES cells.
We next assessed whether PR-Set7 depletion results in DNA damage induction such as formation of double-strand breaks (15), which could result in activation of a G2/M checkpoint. We checked for the presence of DNA damage foci in PR-Set7 knockout cells using γ-H2AX as a marker after pulse labeling with EdU to identify S-phase cells. A substantial increase in the number of γ-H2AX foci was already detectable by day 1 of 4-OHT treatment, and this increased further by day 2 (Fig. 9A). This increase was seen in both EdU-positive and EdU-negative cells (data not shown), implying that it is not S-phase specific. Furthermore, in S-phase cells, although some of the γ-H2AX foci colocalized with sites of EdU incorporation, most did not. We tested whether the γ-H2AX foci were related to DNA damage by costaining with antibodies against another marker, phospho-ATM(S1981). The majority of the γ-H2AX foci costained for phospho-ATM(S1981) in PR-Set7flox/−; CreERT cells after tamoxifen-induced deletion of PR-Set7 (Fig. 9B). The extent of DNA damage induction following 4-OHT treatment was assessed by counting the number of γ-H2AX/phospho-ATM colocalizing foci (Fig. 9C). The percentage of cells that had more than 20 foci increased substantially by day 1 of 4-OHT treatment, and by day 2 more than 50% of the cells displayed colocalization of γ-H2AX and phospho-ATM. We conclude that ES cells undergo massive DNA damage after depletion of PR-Set7. Although this may be partially due to S-phase defects, the high levels of γ-H2AX foci that are also present in non-S-phase cells suggests that damage may also result from other defects during G2/M phase (data not shown).
FIG. 9.
DNA synthesis decreases during S phase, and γ-H2AX foci increase after conditional knockout of PR-Set7 in ES cells. (A and B) Immunofluorescence analyses using untreated PR-Set7flox/−; CreERT cells (−4OHT) or those exposed to 4-OHT for 48 h [+4OHT (48 h)] as indicated at the top. Colocalization of antibody specific to γ-H2AX with either EdU or antibody specific to phospho-ATM is shown (merge). Phosphorylation of γ-H2AX at serine 139 and of the ATM kinase at serine 1981 occurs in response to DNA damage. (C) The percentage of PR-Set7flox/−; CreERT cells exhibiting the indicated number of foci of colocalized γ-H2AX and phospho-ATM as a function of 4-OHT treatment for the number of hours shown at the bottom. The number of colocalized foci increased dramatically after knockout of PR-Set7.
Unlike the situation in ES cells, we were unable to detect a major change in γ-H2AX in PR-Set7−/− embryos, nor were significant differences seen for the 53BP1 DNA repair protein (Fig. 10). Thus, the embryonic phenotype differs from the ES cell phenotype at this level and implies that the lethality in the PR-Set7 null embryo is not necessarily due to increased DNA damage.
FIG. 10.
Analysis of 53BP1 and γ-H2AX in embryos from PR-Set7+/− intercrosses. (A) Embryos from PR-Set7+/− intercrosses were recovered between the four- and the eight-cell stage and immediately processed for immunostaining with antibodies against 53BP1 and γ-H2AX. Shown are full projections of stack sections taken every 1 μm or merged single-section images as indicated. Note that 53BP1 displays both cytoplasmic and nuclear localization. After imaging, embryos were genotyped individually as indicated. (B) Single sections at a higher magnification of two representative nuclei from either PR-Set7 null, wt, or heterozygous embryos are shown. No major differences in levels of either 53BP1 or γ-H2AX were detected between PR-Set7 heterozygous embryos, wt embryos, and null embryos.
Conditional knockout of PR-Set7 leads to chromosomal decondensation in ES cells.
Aside from the S-phase perturbation, increase in DNA damage, and mitotic chromosomal defects described above, we also assessed whether the interphase chromatin organization of ES cells might be perturbed after knockout of PR-Set7. An increase in nuclear volume and sensitivity to micrococcal nuclease has previously been reported in cells knocked down for PR-Set7 (8). We observed no general increase in nuclear volume following 1 or 2 days of 4-OHT treatment in PR-Set7flox/−; CreERT ES cells (data not shown). To address whether there is any chromosomal decondensation, we took a 3D DNA FISH approach using a chromosome paint probe for the X chromosome. The area occupied by the single X chromosome in PR-Set7flox/−; CreERT ES cells following 0, 1, or 2 days of 4-OHT treatment was measured in z projections of 3D stacks for 50 nuclei and normalized to the nuclear area (Fig. 11A). We observed a significant increase (63%; P < 0.001; unpaired t test) in the size of the mean area occupied by the X chromosome in the PR-Set7flox/−; CreERT cells between day 0 and day 2 of 4-OHT treatment (Fig. 11B). To confirm this global chromosomal decondensation in 3D, the interphase distances of DNA FISH signals for genes across the single X chromosome (G6pdx, Fgd1, and Ureb1) were measured in 3D stacks (Fig. 11C and D). We observed a progressive increase in intergenic distances between G6pdx and Fgd1 loci (Fig. 11D, top left) as well as between G6pdx and Ureb1 loci (Fig. 11D, top right) in the PR-Set7flox/−; CreERT cells at day 1 and day 2 of 4-OHT treatment. There was no observed increase between Fgd1 and Ureb1 loci in the PR-Set7flox/−; CreERT cells (Fig. 11D, top left) or the G6pdx and Ureb1 loci in the PR-Set7flox/+; CreERT control cells (Fig. 11D, top right). We conclude that loss of PR-Set7 causes significant, global chromosomal decondensation at interphase that might contribute to the perturbations observed during mitosis described above.
FIG. 11.
Increased chromosomal territory area and intergenic distances indicative of global chromosomal decondensation following conditional knockout of PR-Set7 in ES cells. (A) DNA FISH using an X chromosome paint probe on PR-Set7flox/−; CreERT cells after 0, 1, and 2 days of 4-OHT treatment. Bar, 10 μm. (B) Quantification of relative chromosome area for each time point. The area of the X chromosome in z projections of 3D stacks was measured and normalized to the nuclear area for 50 nuclei at each point of 4-OHT treatment. Significance values are shown at the top. (C) (Left) Triple-probe DNA FISH using BAC probes for the X-linked genes Ureb1 (Cy5), G6pdx (Spectrum Red [SR]), and Fgd1 (Spectrum Green [SG]) following 0, 1, and 2 days of 4-OHT treatment of PR-Set7flox/−; CreERT ES cells. DAPI results are shown in blue. Maximum intensity projections of 3D image stacks (five to seven planes) are shown. (Right) Map of the X chromosome showing the positions of the genes tested. (D) PR-Set7flox/−; CreERT cells show an increase in intergenic distances after 4-OHT treatment. Intergenic distances for each DNA FISH probe are shown following day 0, 1, and 2 for 4-OHT-treated PR-Set7flox/−; CreERT cells and for the G6pdx-Ureb1 probes for day 0 and 2 for 4-OHT-treated PR-Set7flox/+; CreERT cells. Distances were measured between the two centers of mass of the DNA FISH signals from 3D stacks of images and then normalized to the nuclear volume using an ImageJ plug-in (n = 30 per day of treatment). +4OHT, 4-OHT treated.
DISCUSSION
The deficiency in PR-Set7/Set8/KMT5A reported here had a profound effect on mouse development, leading to early embryonic lethality. The methyltransferase activity of PR-Set7 was indispensable for embryonic development at cleavage stages up to the eight-cell stage. The conditional gene inactivation of PR-Set7 in ES cells resulted in reduction of all three methylation states for H4K20, chromosome decondensation, massive DNA damage with subsequent delay in S-phase progression, and cell cycle arrest at G2/M. All of these are likely due to the decrease in H4K20me1. These findings underscore the pivotal role of the PR-Set7 methyltransferase activity in maintaining chromosome stability during normal cell cycle progression and demonstrate that it is essential during the earliest stages of mammalian development.
Some of the previously reported studies using small interfering RNA to knock down PR-Set7 in cultured, transformed cell lines revealed no detectable effects on the cell cycle (11, 18, 27), although others showed impaired cell proliferation and/or DNA damage, resulting in an increase in γ-H2AX (8-10, 28). These discrepancies are likely due to the different levels of PR-Set7 knockdown achieved and perhaps to the different types of tumor cell lines used. The phenotype that we observed in a PR-Set7 null mutant (knockout) context, which results in clear cell cycle perturbation and rapid cell death following full depletion of the PR-Set7 protein, was consistent both in ES cells and in embryos.
H4K20 methylation and its modifying enzymes during cell cycle.
PR-Set7 protein is a highly specific nucleosomal monomethyltransferase that targets lysine 20 of histone H4 as previously shown biochemically (16) and structurally (33). In accordance with this, the global levels of H4K20me1 were markedly reduced after tamoxifen-induced deletion of PR-Set7 in the ES cells studied here; thus, PR-Set7 is indeed the major monomethyltransferase of H4K20 in vivo. Yet the global levels of H4K20me3 and H4K20me2, marks set by the Suv4-20h1/h2 enzymes (25), were also either markedly reduced or substantially decreased, respectively. This points to a direct relationship between the establishment of H4K20me1, H4K20me2, and H4K20me3, with H4K20me1 being catalyzed by PR-Set7 and then serving as the primary substrate for Suv4-20h1/h2 in vivo. This is substantiated by the increased levels of H4K20me1 that accompany the complete loss of H4K20me2 and H4K20me3 in the double knockout of the genes suv4-20h1 and suv4-20h2 (26). Importantly, however, monomethylation and dimethylation/trimethylation of H4K20 have different functions. H4K20me1 is required for appropriate chromatin/chromosome condensation (our studies) and for efficient cell cycle progression, whereas H4K20me2/H4K20me3 function in response to the induction of DNA damage in mice (26), although the difference might be due to cell type differences used in these two studies.
We found that the changes in H4K20me1 levels detected in our FACS analysis reflected the changing levels of detectable PR-Set7 protein during the cell cycle with an increase at late S/G2 and a peak at M phase. This was consistent between ES, F9, and HeLa cells. Similar cell cycle dynamics of H4K20 methylation have been reported using top-down mass spectrometry (17, 18). Taken together, our data and those of previous reports (using top-down mass spectrometry) suggest that newly synthesized histones are deposited on chromatin during S phase and become monomethylated at H4K20 during the period from late S/G2 through M phase and that this persists into early G1 phase. H4K20me1 levels subsequently drop during G1 phase, likely due to H4K20me1 conversion to H4K20me2 and H4K20me3.
Embryonic lethality of PR-Set7 knockout mice.
In the context of embryonic development, null mutation of PR-Set7 results in lethality before implantation. Embryonic development was arrested at late G2 or M phase prior to the eight-cell stage, and apoptosis was induced. The methyltransferase activity of PR-Set7 was found to be essential for further embryonic development at least up to the blastocyst stage. Interestingly, the D. melanogaster PR-Set7 mutant survived to a later stage in development, with lethality occurring at a late larval stage (13). These differences are likely due to the stores of mRNA and protein that are supplied maternally. The mouse oocyte probably contains maternal PR-Set7 since H4K20me1 increased on the male pronucleus in the zygote after fertilization (our unpublished data). Our data suggest that zygotic expression of PR-Set7 is required by the four-cell stage in most mouse embryos. Given that the majority of PR-Set7 protein is thought to degrade at the G1/S border at least in cultured cells (35), if a similar turnover occurs in the embryo the maternal store would be degraded after the first cell division, at or before S phase of the two-cell embryo. Yet the PR-Set7 knockout embryos proceeded through at least one more cleavage before arresting at late G2 or M phase of the four-cell stage. This finding supports our hypothesis based on the experiments performed with ES cells, which is that the cell cycle can progress through the first S and M phases without PR-Set7, but catastrophic events occur in the following cell cycle.
DNA damage and cell cycle delay caused by the absence of H4K20 methylation.
That the ES cells proceeded through the first S phase when PR-Set7 was depleted is perhaps not unexpected given that PR-Set7 protein is normally absent from the nucleus during most of S phase (35). ES cells depleted of PR-Set7 passed through the second S phase slowly, at which point a population of cells accumulated at the second G2/M phase. We propose that the chromosomal anomalies occurred as a direct consequence of the cells having passed through the initial G2/M phase in the absence of PR-Set7 such that H4K20 methylation was reduced at that time. The massive DNA damage that we observed in chromatin with reduced methylation at H4K20 occurred spontaneously and likely gave rise to activation of cellular checkpoints at the subsequent S and G2 phases. In fact, the phenotype of PR-Set7 knockdown was previously rescued by small interfering RNA against cdc45 and rad51 activities that are essential for the initiation of DNA replication and homologous recombination, respectively (10). This suggests that they are likely involved in the null phenotype as well. This also implies that chromatin with reduced levels of methylation at H4K20 somehow impairs the DNA replication and homologous recombination processes, leading to DNA double-strand breaks.
Since PR-Set7 catalyzes H4K20me1, the substrate for H4K20me2 and H4K20me3 (see above), this raises the issue of which methylation state is actually responsible for the phenotypes. The absence of H4K20me2 and H4K20me3 has recently been shown to cause a delay in S-phase entry in suv4-20h1 and suv4-20h2 double knockout mouse embryonic fibroblast cells (26). However, based on the range of defects we observed in the PR-Set7 mutant cells, we hypothesize that it is either the decrease in H4K20me1 or the increase in unmethylated H4 (H4K20me0) that is pivotal to the PR-Set7 null phenotype. In particular, the increase in γ-H2AX foci observed in the PR-Set7 null mutant ES cells occurs in the absence of treatment with DNA-damaging agents. This contrasts with the phenotype in Suv4-20h double-knockout cells where an increase in DNA damage is seen only after treatment with UV and agents such as etoposide and hydroxyurea (26). Therefore, the decrease in H4K20me2 and H4K20me3 alone cannot explain the massive increase in spontaneous γ-H2AX foci observed in PR-Set7 knockout ES cells.
We were unable to detect a major change in either γ-H2AX or 53BP1 in PR-Set7−/− embryos (Fig. 10). This could be partly due to the fact that rather high levels of γ-H2AX are present in wt embryos (36). Alternatively, this difference could be explained if the chromatin environment, which is thought to be globally rather different in the cleavage-stage embryo compared to that of an ES cell, is also a determinant in triggering the DNA damage response.
The cause of lethality in the PR-Set7 null cells could in fact be due to multiple defects during the cell cycle. A link with DNA replication could be envisaged in particular, given that PR-Set7 has been reported to interact with PCNA during S phase (9). However, the physiological relevance of this observation remains obscure as PR-Set7 is ubiquitinated via SCF/Skp2 at the G1/S border, triggering its degradation, and therefore the levels of PR-Set7 are very low during S phase (35). Moreover, live-cell imaging of cells following overexpression of the YFP-PR-Set7 fusion protein revealed that PR-Set7 remains outside the nucleus during S phase (35). Thus, the importance of PR-Set7 and PCNA interaction during S phase remains unclear, but it is possible that the interaction between these proteins is linked to processes other than DNA replication. Our studies confirmed this interaction in vitro, but its relevance to PR-Set7 and DNA replication remain elusive (data not shown).
PR-Set7 and global chromosome condensation.
The loss of PR-Set7 results in decreased H4K20me1 and increased unmethylated H4, which in turn leads to defects in chromosome compaction, not only in mitotic cells but also in interphase nuclei. These findings at the cytological level are consistent, albeit on a different scale, with structural studies of the nucleosome suggesting that the H4K20 residue is involved in chromatin compaction. In these studies it was demonstrated that residues 16 to 20 of histone H4 interact with two acidic patches on the carboxy-terminal α helices of histone H2A present on an adjacent nucleosome and therefore can promote chromatin compaction (3). The global chromosomal decondensation that we have reported here using a 3D FISH approach similarly suggests that the loss of H4K20me1 in PR-Set7 mutant ES cells also has an effect on chromatin compaction and the overall structure of chromosomes at interphase. Our studies show that the PR-Set7-mediated H4K20me1 mark is likely critical to establishing appropriate chromatin structure in normal ES cells and during embryonic development. Future studies using the conditional mouse mutant of PR-Set7 that we have developed here should provide further information into the exact role of H4K20 methylation in chromatin architecture, the cell cycle, and other biological functions such as gene regulation and X inactivation.
Acknowledgments
We are grateful to Zhixian Hu, Reelina Chatterjee, Deborah Hernandez, and Kettly Cabane for technical assistance and to Patrick Trojer and other members of the Reinberg lab for helpful discussions. We thank Gunnar Schotta and Thomas Jenuwein for showing us their work on Suv40h1/h2 knockout mice before publication. We also thank Gelo de la Cruz and Peter Lopez for assistance with FACS analysis, Ya-Ping Hu for technical help with ES cell screening, Kyriakos Economides and Mario Capecchi for providing the pBluescript-polII-neo plasmid, Jerry Hurwitz for providing recombinant PCNA protein, and Lynne Vales for comments on the manuscript.
This work was supported by grants from NIH HD042837 (to M.M.S.), NIH GM64844 (to D.R.), and the HHMI (to D.R.). Funding for E.H. was from the ANR, the Fondation pour la Recherche Medicale (Equipe FRM), and the Curie Institute PIC program. M.E.T.-P. acknowledges support from the PNNRE/INSERM.
Footnotes
Published ahead of print on 17 February 2009.
REFERENCES
- 1.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 2.Botuyan, M. V., J. Lee, I. M. Ward, J. E. Kim, J. R. Thompson, J. Chen, and G. Mer. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 1271361-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorigo, B., T. Schalch, K. Bystricky, and T. J. Richmond. 2003. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 32785-96. [DOI] [PubMed] [Google Scholar]
- 4.Fraga, M. F., E. Ballestar, A. Villar-Garea, M. Boix-Chornet, J. Espada, G. Schotta, T. Bonaldi, C. Haydon, S. Ropero, K. Petrie, N. G. Iyer, A. Perez-Rosado, E. Calvo, J. A. Lopez, A. Cano, M. J. Calasanz, D. Colomer, M. A. Piris, N. Ahn, A. Imhof, C. Caldas, T. Jenuwein, and M. Esteller. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37391-400. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi, S., P. Lewis, L. Pevny, and A. P. McMahon. 2002. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119(Suppl. 1)S97-S101. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi, S., and A. P. McMahon. 2002. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244305-318. [DOI] [PubMed] [Google Scholar]
- 7.Hogan, B. L., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 8.Houston, S. I., K. J. McManus, M. M. Adams, J. K. Sims, P. B. Carpenter, M. J. Hendzel, and J. C. Rice. 2008. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J. Biol. Chem. 28319478-19488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huen, M. S., S. M. Sy, J. M. van Deursen, and J. Chen. 2008. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J. Biol. Chem. 28311073-11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jørgensen, S., I. Elvers, M. B. Trelle, T. Menzel, M. Eskildsen, O. N. Jensen, T. Helleday, K. Helin, and C. S. Sorensen. 2007. The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 1791337-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julien, E., and W. Herr. 2004. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol. Cell 14713-725. [DOI] [PubMed] [Google Scholar]
- 12.Kalakonda, N., W. Fischle, P. Boccuni, N. Gurvich, R. Hoya-Arias, X. Zhao, Y. Miyata, D. Macgrogan, J. Zhang, J. K. Sims, J. C. Rice, and S. D. Nimer. 2008. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 274293-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karachentsev, D., K. Sarma, D. Reinberg, and R. Steward. 2005. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 19431-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohlmaier, A., F. Savarese, M. Lachner, J. Martens, T. Jenuwein, and A. Wutz. 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray, A. 1994. Cell cycle checkpoints. Curr. Opin. Cell Biol. 6872-876. [DOI] [PubMed] [Google Scholar]
- 16.Nishioka, K., J. C. Rice, K. Sarma, H. Erdjument-Bromage, J. Werner, Y. Wang, S. Chuikov, P. Valenzuela, P. Tempst, R. Steward, J. T. Lis, C. D. Allis, and D. Reinberg. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 91201-1213. [DOI] [PubMed] [Google Scholar]
- 17.Pesavento, J. J., C. R. Bullock, R. D. Leduc, C. A. Mizzen, and N. L. Kelleher. 2008. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J. Biol. Chem. 28314927-14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pesavento, J. J., H. Yang, N. L. Kelleher, and C. A. Mizzen. 2008. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell. Biol. 28468-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters, A. H., A. W. Plug, M. J. van Vugt, and P. de Boer. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 566-68. [DOI] [PubMed] [Google Scholar]
- 20.Rice, J. C., K. Nishioka, K. Sarma, R. Steward, D. Reinberg, and C. D. Allis. 2002. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 162225-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson, E. 1987. Embryo-derived stem cell lines. IRL Press, Oxford, United Kingdom.
- 22.Rodríguez, C. I., F. Buchholz, J. Galloway, R. Sequerra, J. Kasper, R. Ayala, A. F. Stewart, and S. M. Dymecki. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25139-140. [DOI] [PubMed] [Google Scholar]
- 23.Sakaguchi, A., and R. Steward. 2007. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J. Cell. Biol. 176155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders, S. L., M. Portoso, J. Mata, J. Bahler, R. C. Allshire, and T. Kouzarides. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119603-614. [DOI] [PubMed] [Google Scholar]
- 25.Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 181251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schotta, G., R. Sengupta, S. Kubicek, S. Malin, M. Kauer, E. Callen, A. Celeste, M. Pagani, S. Opravil, I. A. De La Rosa-Velazquez, A. Espejo, M. T. Bedford, A. Nussenzweig, M. Busslinger, and T. Jenuwein. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 222048-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, X., I. Kachirskaia, H. Yamaguchi, L. E. West, H. Wen, E. W. Wang, S. Dutta, E. Appella, and O. Gozani. 2007. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol. Cell 27636-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tardat, M., R. Murr, Z. Herceg, C. Sardet, and E. Julien. 2007. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J. Cell Biol. 1791413-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres-Padilla, M. E., A. J. Bannister, P. J. Hurd, T. Kouzarides, and M. Zernicka-Goetz. 2006. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50455-461. [DOI] [PubMed] [Google Scholar]
- 30.Trojer, P., G. Li, R. J. Sims III, A. Vaquero, N. Kalakonda, P. Boccuni, D. Lee, H. Erdjument-Bromage, P. Tempst, S. D. Nimer, Y. H. Wang, and D. Reinberg. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129915-928. [DOI] [PubMed] [Google Scholar]
- 31.Trojer, P., and D. Reinberg. 2008. Beyond histone methyl-lysine binding: how malignant brain tumor (MBT) protein L3MBTL1 impacts chromatin structure. Cell Cycle 7578-585. [DOI] [PubMed] [Google Scholar]
- 32.Vakoc, C. R., M. M. Sachdeva, H. Wang, and G. A. Blobel. 2006. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 269185-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao, B., C. Jing, G. Kelly, P. A. Walker, F. W. Muskett, T. A. Frenkiel, S. R. Martin, K. Sarma, D. Reinberg, S. J. Gamblin, and J. R. Wilson. 2005. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 191444-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, H., J. J. Pesavento, T. W. Starnes, D. E. Cryderman, L. L. Wallrath, N. L. Kelleher, and C. A. Mizzen. 2008. Preferential dimethylation of histone H4-lysine 20 by Suv4-20. J. Biol. Chem. 28312085-12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin, Y., V. C. Yu, G. Zhu, and D. C. Chang. 2008. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 71423-1432. [DOI] [PubMed] [Google Scholar]
- 36.Ziegler-Birling, C., A. Helmrich, L. Tora, and M. E. Torres-Padilla. Distribution of p53 binding protein 1 (53BP1) and phosphorylated H2A.X during mouse preimplantation development in the absence of DNA damage. Int. J. Dev. Biol., in press. [DOI] [PubMed]