Abstract
Genetic and experimental evidence points to a critical involvement of the atypical mammalian orphan receptor DAX-1 in reproductive development and steroidogenesis. Unlike conventional nuclear receptors, DAX-1 appears not to function as a DNA-bound transcription factor. Instead, it has acquired the capability to act as a transcriptional corepressor of steroidogenic factor 1 (SF-1). The interplay of DAX-1 and SF-1 is considered a central, presumably ligand-independent element of adrenogonadal development and function that requires tight regulation. This raises a substantial interest in identifying its modulators and the regulatory signals involved. Here, we uncover molecular mechanisms that link DAX-1 to the ubiquitin modification system via functional interaction with the E3 ubiquitin ligase RNF31. We demonstrate that RNF31 is coexpressed with DAX-1 in steroidogenic tissues and participates in repressing steroidogenic gene expression. We provide evidence for the in vivo existence of a corepressor complex containing RNF31 and DAX-1 at the promoters of the StAR and CYP19 genes. Our data suggest that RNF31 functions to stabilize DAX-1, which might be linked to DAX-1 monoubiquitination. In conclusion, RNF31 appears to be required for DAX-1 to repress transcription, provides means to regulate DAX-1 in ligand-independent ways, and emerges as a relevant coregulator of steroidogenic pathways governing physiology and disease.
DAX-1 (dosage-sensitive sex reversal, adrenal hypoplasia congenita critical region on the X chromosome, gene 1; NR0B1) is an atypical member of the nuclear receptor (NR) family. It has key roles in the development and maintenance of reproductive functions and steroid hormone biosynthesis in mammals. The human DAX-1 gene was originally identified on the basis of duplications of an X-linked locus, DSS, involved in sex determination (3). Furthermore, mutations in DAX-1 cause the X-linked form of adrenal hypoplasia congenita, an inherited disorder of adrenal gland development that is commonly associated with hypogonadotropic hypogonadism during pubertal maturation (35, 51). Genetic and experimental evidence has revealed an antagonistic relationship between DAX-1 and another NR family member, steroidogenic factor 1 (SF-1; NR5A1), which is coexpressed with DAX-1 throughout the hypothalamic-pituitary-adrenogonadal axis (26, 32). Multiple studies, including the characterization of transcriptional functions and the phenotypic analysis of knockout mouse models (1, 37), suggest that SF-1 primarily acts as a transcriptional activator while DAX-1 appears to act as a repressor of gene transcription. Major steroidogenic targets include cytochrome P450s (e.g., CYP19 aromatase), cholesterol transporters (e.g., steroidogenic acute regulator protein StAR), and hydroxysteroid dehydrogenases. The interplay between DAX-1 and SF-1 is considered a central element in adrenogonadal function that requires tight regulation, raising a substantial interest in identifying its modulators and the underlying molecular mechanisms.
Our current understanding of DAX-1 action is crucially linked to the unique position of this protein within the NR family. Although DAX-1 has a putative ligand-binding domain (LBD), recent structural studies show that the ligand-binding pocket is absent and thus support a model in which DAX-1 relies entirely on ligand-independent regulatory mechanisms (40). Furthermore, DAX-1 lacks the characteristic NR zinc-finger DNA-binding domain but has instead a unique N-terminal repeat domain. This multifunctional domain mediates direct interactions with NRs via LXXLL motifs (53), carries RNA binding ability, and binds to single-stranded promoter regions (26, 30). While the biological relevance of these intriguing functions remains to be clarified, it is the specific interaction with SF-1 in conjunction with the intrinsic repressor function that classifies DAX-1 as a true corepressor of gene transcription. The demonstration that naturally occurring mutations related to adrenal hypoplasia congenita abolish DAX-1 repressor activity (18) by causing misfolding and cytoplasmic accumulation of DAX-1 (27) emphasizes that DAX-1 repression is critical for appropriate reproductive development and steroidogenesis. Peculiarities of DAX-1, in comparison to other repressing NRs, include a requirement of LBD helix 12 for the recruitment of corepressors such as N-CoR and Alien (2, 9). However, it remains unclear whether these corepressors are involved in SF-1 antagonism in vivo, thus specifying a need for further investigations to characterize components of the DAX-1 corepressor complex.
Considering the importance of posttranslational modifications in regulating NR function and the possible lack of ligand regulation in the case of DAX-1, surprisingly little is known as to what extent posttranslational modifications impact DAX-1 function. Covalent and reversible conjugation of ubiquitin or ubiquitin-like proteins, such as SUMO, has emerged as a common topic in discussions of transcriptional pathways (13, 21). A specific conjugation event is usually a three-step process involving an E1 activating enzyme, one of several E2 conjugating enzymes, and one of hundreds of E3 ligases, which confer substrate specificity. Recent studies in the NR field point to a requirement of polyubiquitination-dependent proteasomal degradation for efficient ligand-dependent transcription and coregulator exchange, while SUMOylation appears linked to transcriptional repression (11, 39). These studies collectively emphasize that coregulators are crucial and probably primary targets for the recruitment of E3 ligases to NRs. In an effort to identify novel regulatory components of DAX-1 action, we describe here specific connections to the ubiquitin modification system via RNF31, a member of the ring-between-ring (RBR) family of E3 ubiquitin ligases. Our study provides evidence that RNF31 associates with DAX-1, is expressed in steroidogenic tissues, triggers DAX-1 ubiquitination and stabilization, and participates in the repression of endogenous steroidogenic target genes by participating in a chromatin-bound corepressor complex. Thus, RNF31 appears to be required for DAX-1 to repress transcription and provides means to regulate DAX-1 function via ubiquitination. This highlights RNF31 as a physiologically relevant coregulator of steroidogenic pathways.
MATERIALS AND METHODS
Cell culture, stable, and transient transfections.
The NCI-H295R (H295R) adrenocortical carcinoma cell line (ATCC CRL-2128) was maintained in Dulbecco's modified Eagle's medium-F12 medium (1:1; Gibco, Invitrogen) supplemented with 2.5% Nu-Serum (BD Bioscience) and 1% ITS+ culture supplement (BD Bioscience). COS-7 and HeLa cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 2 mM l-glutamine (Invitrogen). Cells were transfected using Lipofectamine 2000 (Invitrogen) as instructed by the manufacturer. The green fluorescent protein (GFP)-DAX-1-expressing Flp-In 293 cell line was generated using the Flp-In system (Invitrogen) in accordance with the manufacturer's instructions.
Plasmids.
All derivatives expressing RNF31 (pGAD-hRNF31, pEYFP-RNF31, pRFP-RNF31, pFlag-RNF31, and pMyc-RNF31) or DAX-1 (pGBT9-DAX-1, pFlag-DAX-1, pFlag-DAX-1-ubiquitin, and pEGFP-DAX-1) were made according to standard PCR-based cloning procedures using human RNF31 (ATCC 6611620) and human DAX-1 (53) cDNAs as templates. PCR fragments were inserted into pGAD (Clontech), pEYFP-C2 (modified from C1; Clontech), pFlag or pMyc (modified from pcDNA3; Invitrogen), pGBT9 (Clontech), or pEGFP-C2 by using EcoRI sites. Ubiquitin wild-type (WT)- and K0R mutant-expressing plasmids were generated by PCR-based cloning and inserted into either pcDNA3-His or pcDNA3-3xHA expression vectors. Identities of all plasmids were verified by sequencing.
Other plasmids used were pSG5-hDAX-1, pGEX4T-1-hDAX-1 N, pGEX4T-1-hDAX-1 C, and pGEX4T-1-hDAX-1 R3 (53); pcDNA-SF-1 (kindly provided by K. L. Parker); pCMV-HA-ubiquitin (kindly provided by D. Bohmann); pcDNA-HA ubiquitin WT (kindly provided by A. M. Weissman); pcDNA-His6-ubiquitin (kindly provided by D. Xirodimas [50]); UbcH1, UbcH5, UbcH7, and UbcH8-GFP (kindly provided by H. Ardley [33]); and pGL2 (Promega)-derived luciferase reporter plasmids pStAR-LUC (kindly provided by D. M. Stocco) and pCYP19-LUC (kindly provided by C. D. Clyne and E. R. Simpson).
Antibodies.
An anti-human DAX-1 antibody was raised in a rabbit (AgriSera) toward an N-terminal peptide (amino acids [aa] 163 to 193) and affinity purified on an peptide column. An anti-RNF31 antibody was raised in a rabbit (AgriSera) against glutathione S-transferase (GST)-human RNF31 C terminus (aa 670 to 1072) and validated using RNF31 overexpression and small interfering RNAs (siRNAs). Other antibodies used were anti-DAX-1 2F4 (mouse monoclonal [51); anti-RNF31 (rabbit polyclonal; Abcam); anti-SF-1 (rabbit polyclonal; Upstate); anti-StAR (rabbit polyclonal; Affinity Bioreagents); anti-Flag F7425 (rabbit polyclonal), anti-Flag F4042 (mouse monoclonal), and anti-β-actin A5316 (mouse monoclonal; all from Sigma-Aldrich); anti-Myc 9E10 (mouse monoclonal; Santa Cruz); anti-GFP antibody (rabbit polyclonal; Invitrogen); and anti-hemagglutinin (anti-HA, mouse monoclonal; Covance).
Protein-protein interaction assays. (i) Yeast two-hybrid assays.
Yeast two-hybrid screenings were performed using pGBT9-human DAX-1 WT (aa 1 to 470) and pGBT9-DAX-1 N (aa 2 to 199) as bait and an activation domain (pGAD)-tagged mouse embryo day 16.5 library in the Saccharomyces cerevisiae AH109 strain (BD Biosciences/Clontech). For the yeast two-hybrid quantitative assays, the Y187 strain was cotransformed with pGAD-RNF31 and pGBT9-DAX-1 variants, and liquid β-galactosidase assays were performed according to the manufacturer's instructions.
(ii) Coimmunoprecipitations.
COS-7 cells were transfected with the indicated constructs. Whole-cell extract preparation and coimmunoprecipitation were performed as previously described (17).
(iii) GST pull-down assays.
Bacterially expressed GST-DAX-1 N (aa 1 to 199), C (aa 200 to 470), and R3 (aa 115 to 199) fusion proteins were batch purified using glutathione Sepharose 4B (GE Healthcare). Beads were washed, and in vitro-translated [35S]methionine-labeled RNF31 was added. After being incubated and washed, proteins were eluted by boiling in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading buffer and separated by SDS-polyacrylamide gel electrophoresis. Radiolabeled RNF31 was detected using autoradiography film (GE Healthcare).
Expression analysis. (i) ISH.
Five oligonucleotide probes homologous to mouse and rat RNF1 mRNAs (nucleotides 319 to 350, 663 to 695, 1070 to 1103, 2220 to 2253, and 2459 to 2492; GenBank accession number AB385175) and three probes homologous to mouse and rat Dax-1 mRNAs (nucleotides 20 to 52, 107 to 140, and 243 to 276; GenBank accession number NM_007430) were used for in situ hybridization (ISH). Several probes against nonrelated mRNAs with known expression patterns and with similar lengths and GC contents were used as controls. The addition of a 100-fold excess of nonlabeled probes quenched all signal. The ISH was carried out as described in detail previously (42).
(ii) Immunohistochemistry.
Immunohistochemistry was performed as described previously (38). Briefly, paraffinized sections of mouse adrenals were subjected to antigen retrieval by boiling in 10 mM citric acid, pH 6.0, and then left undisturbed for 30 min. Sections were incubated in 3% hydrogen peroxide for 15 min and then blocked with 3% bovine serum albumin in TBST (0.1 M Tris-HCl, pH 7.6, 154 mM NaCl, 0.01% Tween 20). Both anti-RNF31 (rabbit polyclonal) and anti-DAX-1 (mouse monoclonal) were diluted 1:100 in TBST and incubated overnight at 4°C. Sections were then incubated with either biotinylated goat anti-rabbit (1:200; Dako) or goat anti-mouse (1:200; Dako) secondary antibodies for 1 h at room temperature.
(iii) Immunocytochemistry.
Cells were fixed in 1.5% formaldehyde, permeabilized with phosphate-buffered saline-Triton X-100 (0.2%) and blocked with 1% goat serum (Jackson ImmunoResearch). Anti-Flag (1:1,000) was detected with Alexa Fluor 488 goat anti-rabbit immunoglobulin G (Molecular Probes) after incubation for 1 h, and nuclear DNA was stained using DAPI (4′,6-diamidino-2-phenylindole; 1:500). Images in Fig. 2A to C were visualized using a TCS SP multiband confocal imaging system (Leica), and images in Fig. 2D and E were visualized on a Leica ASMDW instrument.
FIG. 2.
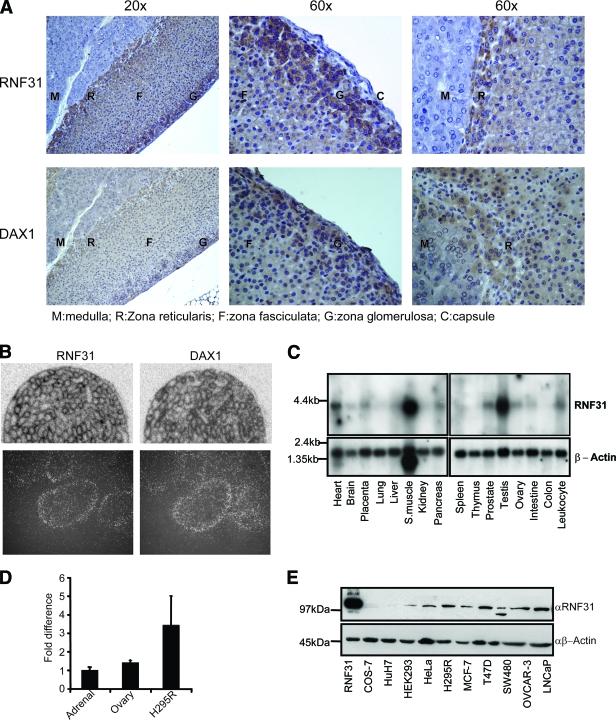
Expression of RNF31 in tissues and cell lines. (A) Paraffin-embedded mouse adrenal glands were stained with RNF31 rabbit polyclonal or anti-DAX-1 mouse monoclonal antibody. (B) ISH of specific RNF31 or DAX-1 oligonucleotide probe on sections of mouse testes. The lower panels show dipped sections at higher magnifications. (C) Northern blots containing mRNA from a panel of human tissues were probed with 32P-labeled RNF31 and β-actin cDNAs. S. muscle, skeletal muscle. (D) RNF31 mRNA expression levels in commercially available human RNA from adrenals and ovaries as well as in the H295R human adrenocortical carcinoma cell line were quantified. Tissues and cell line samples were analyzed in quadruplicate. Mean relative expression levels (± standard errors of the means) in ovaries and the H295R cell line were compared to the expression level in adrenals, which was set to 1.0. (E) Whole-cell extracts from a panel of cell lines were analyzed by Western blots using anti-RNF31 (αRNF31) and anti-β-actin (αβ-Actin) antibodies. Lanes: 1, overexpressed human RNF31 in COS-7 cells (RNF31); 2, monkey kidney (COS-7); 3, human hepatoma (Huh7); 4, human kidney (HEK293); 5, human cervix (HeLa); 6, human adrenal cortex (H295R); 7, human breast cancer (MCF-7); 8, human breast cancer (T47D); 9, human colon cancer (SW-480); 10, human ovary (OVCAR-3); 11, human prostate cancer (LNCaP).
Ubiquitination assays. (i) His-ubiquitin pull-down assays.
COS-7 cells in 10-cm dishes were transfected with 1 μg pcDNA3-His-ubiquitin, 1 to 2 μg pcDNA3-Flag-DAX-1, and 3 μg pcDNA3-Myc-RNF31 or pcDNA3-Myc-RNF31 deletion constructs in the indicated combinations. siRNA-treated HeLa cells were transfected with 2 μg Flag-DAX-1 and 1 μg His-ubiquitin 48 h after siRNA treatment. Twenty-four hours after transfection, cells were lysed, and His-ubiquitin-conjugated proteins were purified as described in reference 50, with a few modifications. Briefly, cells were lysed in a 6 M guanidine-HCl-containing buffer, 25 μl HIS-select magnetic Ni-nitrilotriacetic acid beads (Sigma Aldrich) were added, and samples were incubated 4 h at room temperature. Beads were then washed four times for 5 min each in 1-ml wash buffers and eluted in 50 μl of elution buffer (50). Eluates were analyzed by Western blotting using an anti-Flag (DAX-1) rabbit polyclonal antibody.
(ii) Ubiquitin immunoprecipitation assays.
Myc-RNF31 and Flag-DAX-1 were coexpressed with HA-tagged ubiquitin, K0R mutant ubiquitin, or HA alone in COS-7 cells. Cell extracts were prepared under denaturing conditions, diluted to lower detergent concentrations, and subjected to immunoprecipitation using anti-Flag rabbit polyclonal antibody (Sigma-Aldrich) and protein A-Sepharose (GE Healthcare) for 4 h at 4°C. After being washed, precipitates were eluted by boiling in SDS sample buffer and analyzed by Western blotting using indicated antibodies.
Protein stability assays.
H295R cells were seeded in six-well plates 24 h before transfection. Cells were transfected with 0.5 μg pCDNA3-Flag-RNF31 or pCDNA3-Flag. Cycloheximide (100 μM) was added to all samples 24 h posttransfection. Samples were harvested in radioimmunoprecipitation assay buffer at designated time points. Equal amounts of protein were analyzed by Western blotting.
siRNA and qPCR.
Cells were transfected with Dharmafect I (Dharmacon) and 50 to 100 nM siRNAs as instructed by the manufacturer. Oligonucleotides used were siGENOME SMARTpool RNF31 D-0021419 (siRNF31), nontargeting control D-001206-14 (siLUC), nontargeting control D-001210-01 (siCONTROL), siGENOME NR0B1 D-003409-03, and siGENOME SMARTpool NR5A1 M-003429 (siSF-1), all from Dharmacon. Samples were harvested 72 h after transfection and analyzed for protein levels using Western blot analysis with indicated antibodies or for mRNA levels using quantitative PCR (qPCR). For qPCR, total RNA was prepared using the RNeasy minikit (Qiagen) according to the manufacturer's instructions. RNA concentrations were determined on a Nanodrop (ND-1000) machine. cDNA was synthesized using Superscript III (Invitrogen) reverse transcriptase and random hexamer primers. All qPCR experiments were run on an ABI 7500 instrument with SYBR green technology. qPCR data were analyzed with the comparative ΔΔCT method using 18S rRNA to normalize the expression data. Values are presented as changes relative to the negative control siRNA values, which were set to 1. Results from representative experiments with standard deviations are shown. Primer pairs were designed in Primer Express (ABI) and were as follows: for human RNF31, forward primer ACCCCCTATTGAGAGAGATTGCT and reverse primer TGGAGCCTGGGACAGAGG; for mouse RNF31, forward primer TGCAGTGGCTGCTACAACG and reverse primer GTTAGGGTCTGGACATTTATTCTTGG; for human StAR, forward primer TGGGCATCCTTAGCAACCA and reverse primer GGGACCACTTTACTCATCACTTTGT; for mouse StAR, forward primer TTTAAGGTGAGCGAGGGCA and reverse primer CCGGGATGGTTCTATCAGAAAC; for human CYP19, forward primer TGAGGATCCCTTTGGACGAA and reverse primer AATAACCTTGGATTTTAACCACGATAG; for mouse CYP19, forward primer GCCCAATGAATTTACCCTTGAA and reverse primer TACCTGTAGGGAACATTCTTCTCAAA; for human SF-1, forward primer TTTGTCTGCCTCAAGTTCATCATC and reverse primer CAGGAACTTCAAATCCAGGCTG; and for 18S rRNA, forward primer GGGAGCCTGAGAAAC and reverse primer GGGTCGGGAGTGGGT.
For luciferase assays, siRNA-treated cells were transfected 48 h after siRNA treatment with 250 ng promoter construct and 150 ng β-galactosidase reporter (internal control). Forty-eight hours later, cells were analyzed for luciferase and β-galactosidase activity. Luciferase values were normalized against β-galactosidase values and compared to the negative control siRNA value, which was set to 1. Representative values with standard deviations are shown.
ChIP assay.
siRNA-treated H295R cells in 10-cm dishes were cross-linked in 1% formaldehyde for 15 min at room temperature. Cross-linking was quenched in 125 mM glycine for 5 min, and cells were washed and harvested in phosphate-buffered saline. Cell pellets were resuspended in lysis buffer (50 mM HEPES, pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, complete protease inhibitors [Roche]) and sonicated to generate DNA fragments with sizes between 0.2 and 0.8 kb. Lysates were then subjected to chromatin immunoprecipitation (ChIP) as described in reference 41. Isolated DNA and inputs were analyzed by PCR and qPCR. Antibodies used were anti-DAX-1(rabbit polyclonal, raised in-house); anti-SF-1 (Upstate); anti-RNF31 (rabbit polyclonal; Abcam); anti-RNA polymerase II (Pol II) H-224, anti-CREB/CREM H-74 X-12, anti-C/EBPβ H7, and anti-CBP A-22 (all from Santa Cruz); and SRC2 (T73620; Transduction Laboratories). Primers for PCR analysis were as follows: for pStAR, forward primer CCCCACAAACGGCCAAA and reverse primer CCATCACTCACTGTGCAAAGGA; for pCYP19, forward primer ACTAGAGATGGCCTGAGTGAGTCA and reverse primer TTGGGCATCGTTGAGGTCTT; and StAR exon 7 (negative control), forward primer TTGCTTTATGGGCTCAAGAATG and reverse primer GGAGACCCTCTGAGATTCTGCTT.
RESULTS
Identification of RNF31 as a DAX-1-associated protein.
To identify DAX-1-associated proteins, we performed yeast two-hybrid screenings using either WT human DAX-1 or the N terminus as bait and repeatedly identified cDNAs encoding RNF31 (aliases, ZIBRA, FLJ10111, PAUL, and HOIP). Domain analysis revealed that RNF31 contains a C-terminal RBR motif characteristic of E3 ubiquitin ligases of the RBR family (12), various putative ubiquitin-binding motifs including a central ubiquitin-associated (UBA) domain and three Zn_RBZ domains (zinc finger domains, Ran-binding protein type), and an N-terminal PUB (putative AAA ATPase-binding) domain (Fig. 1A).
FIG. 1.
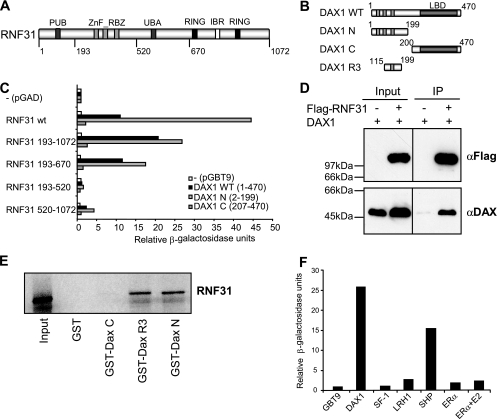
RNF31 associates with DAX-1. (A) Domain structure of human RNF31 (aa 1 to 1072). PUB, putative AAA ATPase-binding domain; ZnF_RBZ, putative zinc-finger ubiquitin-binding domain; RING-IBR-RING, E3 ubiquitin ligase domain. (B) Domain structures of human DAX-1 (WT, aa 1 to 470) and derivatives used for interaction assays. N, N-terminal domain consisting of three repeats (R1 to R3); C, C-terminal repressor domain homologous to the NR LBD; R3, third repeat region. (C) Gal4-DAX-1 WT, N terminus, or C terminus constructs were tested for interaction with GAD-RNF31 variants in a yeast two-hybrid liquid β-galactosidase assay. Relative β-galactosidase units represent absolute units relative to the value for the GAD negative control (Gal4 activation domain only). The expression of RNF31 constructs was verified by Western blotting (see Fig. S1 in the supplemental material). (D) COS-7 cells were cotransfected with DAX-1, Flag-RNF31, or Flag as indicated. Whole-cell extracts were subjected to immunoprecipitation (IP) using a rabbit polyclonal anti-Flag (αFlag) antibody and analyzed by Western blotting with anti-DAX-1 (αDAX) and anti-Flag mouse monoclonal antibodies. (E) Glutathione-Sepharose-bound GST-DAX-1 constructs and GST alone were probed for interaction with in vitro-translated 35S-labeled RNF31 in a GST pull-down assay. (F) pGAD-RNF31 was tested for interaction with various NRs by using a yeast two-hybrid liquid β-galactosidase assay. Relative β-galactosidase units represent absolute units relative to the value for the GBT9 negative control (empty vector). ERα, estrogen receptor α.
The interaction between DAX-1 constructs (Fig. 1B) and RNF31 was verified and further characterized by yeast two-hybrid analysis (Fig. 1C), coimmunoprecipitations from mammalian cell extracts (Fig. 1D), and GST pull-down assay (Fig. 1E). The RNF31-binding domain of DAX-1 was mapped to the N-terminal repeat domain (Fig. 1C and E), with the third repeat being sufficient for interaction. This repeat region is not present in conventional NRs but shares limited homology to the N terminus of the orphan receptor SHP (NR0B2). In agreement with this, RNF31 was shown to interact with SHP but failed to interact with the DAX-1 target receptors SF-1, LRH-1, and estrogen receptor α (Fig. 1F). Finally, the DAX-1-binding region of RNF31 was mapped to a central region (aa 193 to 670) (Fig. 1C), not overlapping with the RBR domain or conserved in other RBR E3s (12).
Analysis of tissue distribution and coexpression.
To investigate whether RNF31 is coexpressed with DAX-1 in steroidogenic target tissues, we determined its mRNA and protein expression in mouse, rat, and human tissues (Fig. 2; see Fig. S2 in the supplemental material). Immunohistochemistry revealed that RNF31 protein is expressed and coexpressed with DAX-1 in the rodent adrenal cortex (Fig. 2A; see Fig. S2A and B in the supplemental material). Coexpression is particularly evident in the outer layer of the cortex (zona glomerulosa), a known site of DAX-1 expression and function (34), while no expression of RNF31 is evident in the medulla or capsule. At the subcellular level, RNF31 seems to be expressed in both the nucleus and cytoplasm, consistent with the intracellular dynamics of overexpressed RNF31 (see below).
ISH revealed coexpression of RNF31 and DAX-1 mRNA in the seminiferous epithelium of rodent testes (Fig. 2B, upper panel). Coexpression was made further evident by analyzing dipped sections, showing seminiferous tubuli at a higher magnification (Fig. 2B, lower panel) and during postnatal testis development toward puberty (see Fig. S2C in the supplemental material).
Multiple tissue Northern blots revealed the presence of a ∼4-kb mRNA in various human tissues, including testes (high levels), ovaries, and prostates (Fig. 2C). qPCR analysis confirmed that RNF31 mRNA is expressed in human steroidogenic tissues and in the adrenocortical cell line H295R, the only human steroidogenic cell line known to express DAX-1 (Fig. 2D). Western blot analysis confirmed expression of RNF31 protein in adrenal H295R cells but also revealed its presence in a variety of DAX-1-negative human cell lines (Fig. 2E).
A variety of experimental methods were utilized to investigate the intracellular localization of RNF31 and its possible colocalization with DAX-1 (Fig. 3). We observed that RNF31 upon overexpression seemed to be primarily (>90% of transfected cells) localized in the cytoplasm (Fig. 3A). As both immunohistochemistry (Fig. 2A; see Fig. S2A and B in the supplemental material) and subcellular fractionations (data not shown) suggest that endogenous RNF31 is found in both nuclear and cytoplasmic compartments, we considered that RNF31 could be subject to active nuclear export mechanisms. This was confirmed as yellow fluorescent protein-tagged RNF31 relocalized from the cytoplasm to the nucleus following incubation with the export inhibitor leptomycin B (LMB) (Fig. 3A). In agreement with previous observations, DAX-1 localization was not affected by LMB. Consistent with RNF31 being actively exported, we identified a high-probability nuclear export signal (aa 151-LEVLLLRTELSLLL; underlined residues represent matches to the accepted nuclear export signal consensus [24]).
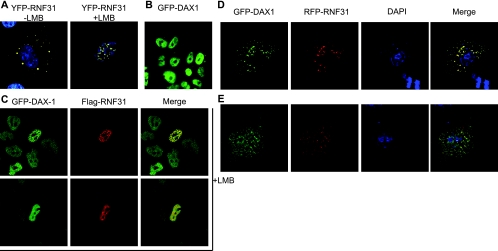
FIG. 3.
Intracellular localization analysis of RNF31 and DAX-1. (A) Yellow fluorescent protein (YFP)-RNF31 (yellow) was expressed in COS-7 cells in the absence (left) or presence (right) of 5 nM of the nuclear export inhibitor LMB. DAPI (blue) shows staining of nuclear DNA. (B) Nuclear localization of GFP-DAX-1 upon stable integration in Flp-In 293 cells. (C) Flag-RNF31 (red) was transfected to GFP-DAX-1-containing Flp-In 293 cells. Cells were treated with 5 nM LMB and analyzed for colocalization (yellow) of DAX-1 and RNF31. Representative images show colocalization in the nucleus in distinct loci (upper panels) or uniformly distributed (lower panels). (D and E) COS-7 cells were transfected with red fluorescent protein (RFP)-RNF31 (red) and GFP-DAX-1 (green). Cells were fixed, and nuclear DNA was stained with DAPI (blue). (D) Mainly cytoplasmic colocalization (yellow) is representative of cells with high expression levels of RFP-RNF31 and GFP-DAX-1. (E) A more even distribution is representative of cells with low expression levels.
To monitor the assumed dynamic interplay of RNF31 with DAX-1, we generated a Flp-In 293 cell line stably expressing GFP-DAX-1. Due to single-copy integration via Flp-In recombination, DAX-1 levels and localization appear to be uniform (Fig. 3B) and thus mimic the endogenous situation (see Fig. S3C in the supplemental material). We then overexpressed RNF31 in this cell line and observed colocalization, with GFP-DAX-1 primarily in the cytoplasm (data not shown). However, in LMB-treated GFP-DAX-1-expressing cells, colocalization of RNF31 was observed in the nucleus (Fig. 3C). We then essentially confirmed the results mentioned above by using transient expression in COS-7 cells (Fig. 3D and E) combined with quantitative single-cell analysis (see Fig. S3A and B in the supplemental material). In the majority of these cells, DAX-1 and RNF31 colocalized in the cytoplasm (Fig. 3D); however, we also observed nuclear colocalization in the absence of LMB in approximately 10% of cells. These cells expressed only small amounts of both RNF31 and DAX-1, indicating that the level of overexpression affects the observed localization pattern. In light of this, the focus-like structures present in some cells (Fig. 3C and D) may be a consequence of overexpression, especially since similar foci are not observed with endogenous DAX-1, which is uniformly distributed in H295R cells (Fig. 3C).
Taken together, the data above indicate that RNF31 and DAX-1 are coexpressed in steroidogenic cells of the adrenal and testis and that RNF31 localization and colocalization with DAX-1 are dynamically regulated at the intracellular level.
Characterization of DAX-1 ubiquitination.
RNF31 is an enigmatic member of the RBR family of putative E3 ubiquitin ligases, and the physiological substrates and mechanism of RNF31 ligase activity have not yet been described (12). As our interaction and expression studies suggest DAX-1 to be a physiologically relevant RNF31 substrate, we next addressed the role of RNF31 in ubiquitination of DAX-1 (Fig. 4; see Fig. S4 in the supplemental material).
FIG. 4.
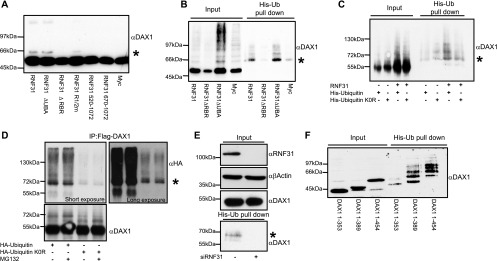
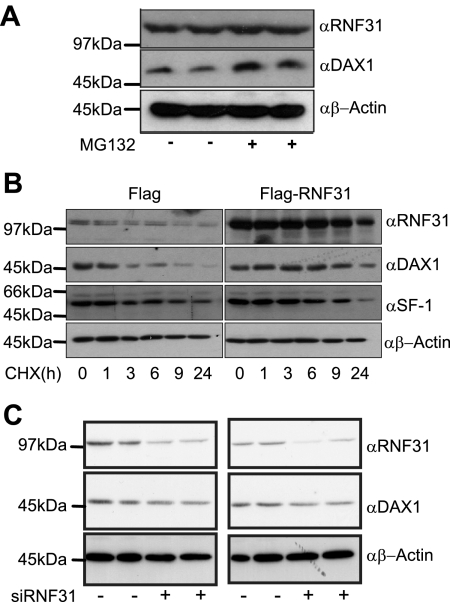
RNF31 promotes monoubiquitination of DAX-1 in an RBR domain-dependent manner. α, anti; *, monoubiquitinated DAX-1. (A) Flag-DAX-1 was expressed with the indicated RNF31 derivatives in COS-7 cells. Whole-cell extracts were analyzed by Western blotting. The presence of an extra signal ∼10 kDa (*) above WT Flag-DAX-1 indicates possible monoubiquitination of DAX-1. (B) COS-7 cells were transfected with His-ubiquitin, Flag-DAX-1 and indicated Myc-RNF31 constructs. His-ubiquitinated (His-Ub) proteins were isolated from denatured whole-cell extracts. Isolated proteins and input samples (whole-cell extracts) were analyzed by Western blotting with anti-Flag (DAX-1) antibody. Signal in His-Ub pull-down lanes corresponds to ubiquitinated DAX-1. (C) Flag-DAX-1 and Myc or Myc-RNF31 were expressed with either His-ubiquitin or a lysine-less ubiquitin mutant incapable of forming ubiquitin chains (His-ubiquitin K0R). His-tagged proteins were isolated and analyzed together with whole-cell extracts by Western blot analysis to detect the presence of DAX-1. (D) Flag-DAX-1, Myc-RNF31, and either HA-ubiquitin or HA-ubiquitin K0R were expressed in COS-7 cells. Whole-cell extracts were prepared and Flag-DAX-1 was immunoprecipitated (IP) using an anti-Flag rabbit polyclonal antibody. Precipitates were analyzed by Western blotting with an anti-HA mouse monoclonal antibody to detect ubiquitinated DAX-1. Both a short exposure and a long exposure of the Western blot are shown. The DAX-1 blot represents input samples. (E) HeLa cells were treated with either control or RNF31-specific siRNA for 48 h before transfection for 24 h with Flag-DAX-1 and His-ubiquitin. DAX-1 expression and knockdown of RNF31 were analyzed by Western blotting using β-actin as loading control. Ubiquitinated proteins were isolated and analyzed by Western blotting with anti-Flag (DAX-1) antibody. (F) His-ubiquitinated proteins were isolated from cells transfected with His-ubiquitin, Myc-RNF31, and indicated Flag-DAX-1 C-terminal deletion constructs. Results were analyzed by Western blotting using anti-Flag (DAX-1) antibody.
Initially, we observed that coexpression of RNF31 with DAX-1, followed by direct SDS lysis (to prevent deubiquitination and degradation), revealed the presence of potentially monoubiquitinated DAX-1 species (Fig. 4A). Independent experiments confirmed that RNF31 increased the appearance of higher-molecular-weight DAX-1 species in a concentration-dependent manner (see Fig. S4A in the supplemental material).
To provide direct evidence that the modified DAX-1 species corresponds to ubiquitin conjugation, we coexpressed His-tagged ubiquitin and Flag-tagged DAX-1 with or without Myc-tagged RNF31 and isolated His-ubiquitin-conjugated proteins under denaturing conditions. After extensively washing, we eluted and analyzed the ubiquitinated proteins by Western blot analysis using a Flag (DAX-1)-specific antibody (Fig. 4B). Ubiquitin conjugation was detected in the RNF31-containing samples. Interestingly, we observed a dominant band at the expected size of monoubiquitinated DAX-1, suggesting that this might be the preferential type of modification (Fig. 4B). In these and other assays, the mobility shift of the dominant band was consistent with that observed with an artificial DAX-1-ubiquitin fusion protein (see Fig. S4D in the supplemental material).
Addressing the question whether endogenous levels of RNF31 are sufficient to ubiquitinate DAX-1, we transfected Flag-DAX-1 and His-ubiquitin to HeLa cells, which contain detectable levels of RNF31 endogenously (Fig. 2E) and have been previously utilized to identify RNF31 biochemically (22). The HeLa cells were pretreated with control or RNF31-specific siRNA oligonucleotides, the latter treatment resulting in efficient knockdown of RNF31 protein. After isolating His-ubiquitinated proteins, we could detect ubiquitinated DAX-1 in control cells but not in RNF31-deficient cells, demonstrating that endogenous RNF31 is sufficient and necessary to ubiquitinate DAX-1 (Fig. 4E).
To determine which of the RNF31 domains are involved in DAX-1 ubiquitination, we generated RNF31 derivatives lacking the C-terminal RBR domain (implicated in ubiquitin ligation), the UBA domain (potentially implicated in interactions with ubiquitin or ubiquitin-like domains in proteins), or parts of the central DAX-1-binding region (see Fig. S4C in the supplemental material). We found that both the RBR and the substrate-binding domains are required, while the UBA domain appears dispensable or even inhibitory, for efficient DAX-1 ubiquitination (Fig. 4A and B). Control Western blots and coimmunoprecipitation experiments showed that all RNF31 derivatives are expressed and that the ubiquitination-deficient RNF31 derivative lacking the RBR ubiquitin ligase domain interacts with DAX-1 in a manner comparable to that of full-length RNF31 (see Fig. S4B in the supplemental material).
The involvement of the RBR domain pointed at yet another aspect that needed to be investigated to support RNF31 function as E3 ubiquitin ligase. Usually, E3s transfer ubiquitin from E2 ubiquitin-conjugating enzymes (Ubc proteins) to the substrate in a process where direct binding between the E3 catalytic domains (i.e., the RBR domain) and E2 Ubc is necessary. To identify the E2 cooperating with RNF31, Flag-RNF31 was coexpressed with GFP-tagged E2s, and coimmunoprecipitations were performed. Interaction was observed with UbcH8 but not with Ubc7, UbcH7, or UbcH1 (see Fig. S4E in the supplemental material). RNF31 selectivity toward UbcH8 was further confirmed by intracellular localization analysis. In the absence of Flag-RNF31, both UbcH7 and UbcH8 were predominantly nuclear (see Fig. S4F in the supplemental material). Coexpression with RNF31 changed the pattern of UbcH8, resulting in cytoplasmic colocalization, while UbcH7 was unchanged (see Fig. S4G in the supplemental material). We conclude that UbcH8 is likely to cooperate with RNF31 in executing substrate ubiquitination, consistent with this E2 being implicated in binding to other RBR-type E3 ligases (33).
To provide further evidence for DAX-1 monoubiquitination, we compared the conjugation of WT ubiquitin to that of a lysine-to-arginine ubiquitin mutant (the K0R mutant). The two ubiquitin variants were cloned into the same expression vector backbone (pcDNA-His or pcDNA-3xHA) to ensure equal expression levels. Complementary pull-down assays (Fig. 4C) and immunoprecipitations (Fig. 4D) from denatured cell extracts revealed conjugation of polymerization-deficient lysine-less ubiquitin to DAX-1. While the interpretation of quantitative differences is difficult due to the possible formation of mixed chains with endogenous ubiquitin, the general patterns and mobilities of modified DAX-1 species for the two ubiquitin variants are comparable. Interestingly, the inclusion of the proteasome inhibitor MG132 did not significantly change the DAX-1 expression levels or modification pattern, arguing against DAX-1 polyubiquitination in these experiments (Fig. 4D).
In support of the specificity of RNF31 toward DAX-1 ubiquitination, we could not detect any modification of SF-1 or the estrogen receptor (data not shown). This result is in agreement with the interaction specificity of RNF31 (Fig. 1).
In initial efforts to map the DAX-1 ubiquitination sites, we used the His-ubiquitin pull-down method and analyzed DAX-1 C-terminal deletion constructs expressing aa 1 to 353, 1 to 389, and 1 to 454 (Fig. 4E). The results indicated that the region between residues 353 and 389 is preferentially ubiquitinated. This part of the DAX-1 LBD contains several lysine residues that are conserved across species (data not shown), some of which may be surface exposed and accessible to both modification and interaction with coregulators.
In conclusion, our results suggest that the major DAX-1 modification observed in different experimental settings is likely to be monoubiquitination at one or more lysine residues (multiubiquitination) possibly located within the LBD of DAX-1.
Effects of RNF31 on stability of endogenous DAX-1.
A key observation from the coexpression/colocalization studies was that DAX-1 levels were often increased but never reduced or abolished in cells expressing RNF31 (e.g., Fig. 3C and 4C; see also Fig. S4A in the supplemental material). This would be consistent with an involvement of RNF31 in DAX-1 monoubiquitination, which is not linked to proteasomal degradation. Since these experiments exclusively utilized overexpressed DAX-1, it was necessary to confirm the relevance of endogenous DAX-1. To achieve this, we used the adrenocortical carcinoma cell line H295R.
We initially observed that treatment of H295R cells with the proteasome inhibitor MG132 caused an accumulation of endogenous DAX-1 (Fig. 5A). This indicated that DAX-1 is degraded in a proteasome-dependent manner, a process triggered mainly by K48-linked polyubiquitination. To monitor the turnover of endogenous DAX-1, H295R cells containing endogenous or, by overexpression, elevated levels of RNF31 were treated with the protein synthesis inhibitor cycloheximide. The levels of DAX-1, SF-1, and actin were analyzed by Western blotting (Fig. 5B). We found that elevated levels of RNF31 seemed to stabilize DAX-1, a result incompatible with RNF31 being responsible for proteasomal degradation of DAX-1. In fact, the effect of overexpression of RNF31 was similar to that of cotreatment with cycloheximide and MG132 (data not shown). Importantly, no effect was observed for SF-1, consistent with the failure of RNF31 to interact with and to ubiquitinate SF-1 (Fig. 1F).
FIG. 5.
RNF31 stabilizes endogenous DAX-1. α, anti. (A) H295R cells were treated with 10 μM proteasome inhibitor MG132 for 6 h and analyzed by Western blotting with indicated antibodies. (B) H295R cells transfected with either Flag or Flag-RNF31 were treated with cycloheximide (CHX) 24 h posttransfection. Cells were lysed 0, 1, 3, 6, 9, and 24 h after CHX addition, and extracts were analyzed by Western blotting using indicated antibodies. (C) H295R cells were treated with either control (−) or RNF31-specific (+) siRNA. Endogenous proteins from whole-cell extracts were analyzed by Western blotting using indicated the antibodies. Results of independent transfections of each oligonucleotide are shown.
Further support for a role of RNF31 in stabilizing DAX-1 was gained using siRNA-mediated knockdown of endogenous RNF31 levels in H295R cells (Fig. 5C). Western blot analysis indicated that RNF31 depletion did not increase the steady-state protein levels of DAX-1, something that would have been expected if RNF31 was involved in DAX-1 degradation. However, in repeated independent siRNA experiments, we did observe that DAX-1 protein levels appeared reduced upon RNF31 knockdown (Fig. 5C). The expression of RNF31 was reduced but not abolished upon siRNA treatment, which may account for its effects on DAX-1 levels being more subtle than the effects caused by overexpression.
Consequences of RNF31 knockdown on DAX-1 target gene expression.
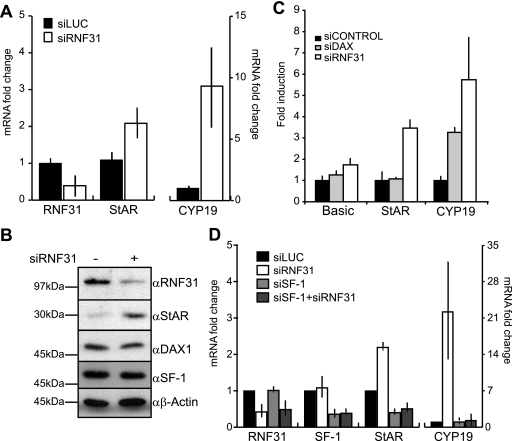
We utilized RNA interference to investigate if RNF31 would affect DAX-1-dependent target gene expression in H295R cells. siRNA-mediated knockdown of endogenous RNF31 expression resulted in maximally 70% reduction at both mRNA and protein levels (Fig. 6A, B, and D) compared to the mRNA and protein levels of cells transfected with nonsilencing control siRNAs. We chose to analyze the effects of RNF31 knockdown on transcription of the StAR and CYP19 (aromatase) genes, both classic SF-1 target genes known to be regulated by DAX-1 (14, 19, 46, 52). Intriguingly, we observed that RNF31 depletion resulted in increased StAR and CYP19 mRNA levels (Fig. 6A), while no significant change was seen for SF-1 and DAX-1 mRNA levels (data not shown). Western blot analysis further revealed that knockdown of RNF31 caused a considerable increase in StAR protein levels (Fig. 6B). As the siRNF31 reagent used was a pool of four different oligonucleotides, we also investigated the effect of the individual oligonucleotides and found that all four siRNAs knocked down RNF31 mRNA, albeit with different efficiencies (see Fig. S5A in the supplemental material).
FIG. 6.
Knockdown of RNF31 upregulates DAX-1 steroidogenic target StAR and CYP19 genes in H295R cells. (A) H295R cells treated with siRNA targeting RNF31 (siRNF31) or a nontargeting oligonucleotide (siLUC) for 72 h were assayed for relative RNF31, StAR, and CYP19 mRNA levels by using qPCR. Data are presented as values for the siRNF31-treated cells relative to the negative control (siLUC) value, which was set to 1. Error bars represent standard deviations. (B) Whole-cell extracts from H295R cells treated with siRNF31 or siLUC were analyzed by Western blotting with indicated antibodies. α, anti. (C) Luciferase reporter constructs of StAR or CYP19 promoter or luciferase reporter (Basic) alone were transfected into H295R cells 48 h after treatment with siRNF31, siGENOME NR0B1 D-003409-03 (siDAX-1), or siCONTROL (nontargeting oligonucleotide). Twenty-four hours after transfection, cell lysates were assayed for luciferase activity. Data are presented as induction values relative to the negative control (siCONTROL) value, which was set to 1 for each promoter construct. Error bars correspond to standard deviations. (D) qPCR analysis of H295R cells treated with siLUC, siRNF31, siSF-1 or siRNF31, and siSF-1. Data are presented as described for panel A.
To investigate whether the observed effect was due to transcriptional events at the established promoter regions of these genes, we next assayed the activity of StAR and CYP19 promoter-luciferase reporter plasmids in siRNA-treated H295R cells. The results confirmed that the increases in StAR and CYP19 mRNA levels were due to increased promoter activity in siRNF31-treated cells compared to that in the control-treated cells (Fig. 6C). Additionally, attempts to knock down DAX-1 (maximally 25 to 50% mRNA reduction; data not shown) revealed an increase of reporter activity in the case of CYP19, supporting the concept that RNF31 and DAX-1 act in the same transcriptional pathway.
Furthermore, we wanted to investigate if the effects of RNF31 depletion are linked to SF-1/DAX-1 and not to additional transcription factors involved in the regulation of CYP19 and StAR expression (29, 43). We hypothesized that a simultaneous knockdown of SF-1 should antagonize the positive effect of the RNF31 knockdown if RNF31 cooperates with DAX-1 repression of SF-1. In fact, we observed this effect on both StAR and CYP19 mRNA levels (Fig. 6D). siSF-1 reduced the expression levels of these genes, and the reduction was not relieved by simultaneous knockdown of RNF31. These results indicate that RNF31 is functionally dependent on the presence of SF-1 at the promoters. To provide additional evidence that RNF31 cooperates with DAX-1 in repressing SF-1 target genes, we analyzed the role of RNF31 in the mouse adrenocortical Y-1 cell line (see Fig. S5B in the supplemental material). Y-1 cells share steroidogenic pathways, including that for StAR expression, with H295R cells but do not express DAX-1 (25). We found no increase in the mRNA levels of StAR despite efficient knockdown of RNF31 (CYP19 expression could not be detected).
Collectively, these data demonstrate that RNA interference-mediated reduction of endogenous RNF31 levels causes an elevation of SF-1-dependent transcription of key steroidogenic genes that are known to be repressed by DAX-1 in vivo.
In vivo analysis of promoter recruitment and coregulator assembly.
To provide evidence that the transcriptional effects observed were a consequence of RNF31 action at the promoters and to further study the connection of RNF31 with DAX-1 repression, we established ChIP assays for the StAR and CYP19 genes in H295R cells. The combination of the ChIP assay with siRNA-mediated depletion of RNF31 or SF-1 allowed us to investigate the requirement of different factors for DAX-1 recruitment and coregulator assembly on the promoters (Fig. 7A).
FIG. 7.
RNF31 is necessary for DAX-1 corepressor complex recruitment and stability at the human StAR and CYP19 promoters. (A) H295R cells treated for 72 h with siLUC, siSF-1, or siRNF31 were subjected to ChiP using the indicated antibodies. Precipitated DNA was analyzed with conventional PCR (agarose gel) and qPCR (graph) to determine the recruitment of transcription factors and coregulators to StAR (left) and CYP19 (right) promoters. qPCR data are presented as changes relative to the siLUC (control) value, which was set to 1. IgG, immunoglobulin G. (B) Proposed model of the role of RNF31 in corepressor complex assembly (repressed promoters; left) and exchange upon activation of steroidogenic transcription (active promoters; right). Ub, ubiquitin; HDAC, histone deacetylase; GTFs, general transcription factors; SFRE, SF-1 response element.
Under control conditions (siLUC), recruitment of SF-1 can be readily detected, along with additional “primary” transcription factors such as CREM/CREB or C/EBPβ, which are implicated in regulating StAR and CYP19 transcription (29, 43). Consistent with basal expression of these genes, recruitment of coactivators such as CBP and SRC2, along with RNA Pol II, is evident. Most intriguing, however, was the recruitment of RNF31 together with DAX-1 and SMRT (but not N-CoR; data not shown), a core component of conventional NR corepressor-histone deacetylase complexes.
The depletion of RNF31 (via siRNF31) further substantiated the existence of a corepressor complex, containing RNF31, DAX-1, and SMRT, and revealed a requirement of RNF31 for complex assembly and/or recruitment. The fact that SF-1 remains promoter-bound suggests a dominant role of RNF31 in determining DAX-1 recruitment and supports a function in stabilizing DAX-1. Release of corepressors allowed enhanced recruitment of a CBP/SRC2/Pol II coactivator complex, consistent with transcriptional activation upon RNF31 depletion as indicated by the siRNA experiments (Fig. 6).
Depletion of SF-1 (via siSF-1) caused the apparent dissociation of all corepressors and coactivators despite the continuous presence of other primary transcription factors (CREB/CREM and C/EBPβ) at the promoters. This is consistent with the transcription profiling showing that the basal expressions of both StAR and CYP19 are dependent on the presence of SF-1 (Fig. 6D). Therefore, SF-1 appears essential in mediating recruitment and exchange of both corepressors, including RNF31 and DAX-1, and coactivators.
Taken together, these data confirm the essential role of RNF31 in mediating transcriptional repression in cooperation with DAX-1. They indicate that RNF31 is crucial for (i) assembling a corepressor complex containing DAX-1 and SMRT, (ii) DAX-1 recruitment and stability at promoters, and (iii) function in pathways that depend on SF-1.
DISCUSSION
The regulation of DAX-1 transcriptional activity has been studied since the discovery of the protein in 1994. Although it has been established that DAX-1 is a crucial component in the regulation of steroidogenesis and in the development of steroidogenic and reproductive tissues, the precise mechanisms behind this regulation are poorly understood, both at the transcriptional level and at the posttranslational level. Our study addresses key issues regarding DAX-1 protein function, firstly by identifying the E3 ubiquitin ligase RNF31 as a corepressor component of DAX-1 transcription and secondly by characterizing ubiquitination as the first example of a posttranslational modification of DAX-1 that may be directly linked to transcriptional repression. Our findings imply that RNF31 has significant coregulatory potential in DAX-1 pathways governing steroidogenesis.
RNF31 adds a previously unidentified component to the intricate relationship between DAX-1 and SF-1 and emerges as a physiologically relevant corepressor of SF-1 transcription in steroidogenic tissues. Our data are compatible with a regulatory model in which RNF31 cooperates with DAX-1 in antagonizing SF-1 activation (Fig. 7B) and describe for the first time the in vivo existence of an SF-1 corepressor complex consisting of DAX-1, RNF31, and SMRT at the steroidogenic promoters of the human StAR and CYP19 genes. We demonstrate that RNF31 is necessary for the stable association of the DAX-1 corepressor complex with chromatin-bound SF-1, thereby inhibiting the recruitment of coactivators and Pol II and controlling basal transcription levels of SF-1 target genes. Since DAX-1 is assumed to occupy the AF-2 coactivator interaction surface of SF-1, RNF31 presumably antagonizes SF-1 coactivators, including acetyltransferases such as SRC proteins or GCN5 (10, 48). We additionally provide evidence that the presence of SF-1 at the promoters is required for recruitment of the RNF31 corepressor complex, consistent with the observation that SF-1 knockdown abolished expression of StAR and CYP19. This suggests that the in vivo association of DAX-1 and RNF31 with chromatin is dependent on the presence of SF-1. The generality of this suggestions remains to be tested, as DAX-1 may also directly bind to DNA hairpin structures in the human and mouse StAR promoters (30, 52). As the hairpin is located in proximity to the SF-1-binding site, one could imagine that cooperative interactions of DAX-1 with SF-1 and DNA are necessary for promoter recruitment of DAX-1 in vivo.
It is surprising that posttranslational modifications of DAX-1 have not been previously described, especially as the lack of ligand binding dictates a need for alternative mechanisms. In contrast, a number of studies have highlighted the regulatory importance of such modifications in the case of SF-1, despite indications that SF-1 may bind phospholipids as natural ligands (23, 28). These studies suggest that phosphorylation and acetylation stimulate SF-1 activity (6) (15), SUMOylation inactivates SF-1 (7), and polyubiquitination triggers proteasomal degradation of SF-1 (8). Our results imply that RNF31-dependent ubiquitination may affect the transcriptional activity and the stability of DAX-1. Only lately has the covalent and reversible ubiquitination of coregulators and NRs been recognized to have regulatory functions beyond degradation. One example is the coactivator SRC3, which appears to be regulated by monoubiquitination leading to increased transcriptional coactivation (49). Ubiquitination certainly has the potential for changing the interactome of a protein both by generating new interaction surfaces and by eliminating old interaction surfaces. Thus, the conjugation of ubiquitin to DAX-1 adds an intriguing regulatory mechanism to consider when investigating DAX-1 corepressor complex function in vivo.
We have shown here that RNF31 is involved in ubiquitination of DAX-1, the first RNF31 substrate shown to be modified among the few described candidates (5, 12). The assumption that DAX-1 is monoubiquitinated (at one or several sites) is consistent with the stabilizing effect of RNF31 on DAX-1 observed in a variety of independent experimental systems. It is conceivable that even subtle changes in DAX-1 function and stability caused by RNF31 action and ubiquitination could significantly impact the assembly of coregulator complexes on SF-1-dependent promoters and thereby influence the transcriptional outcomes. Indeed, we demonstrated that RNF31 knockdown destabilizes chromatin-bound DAX-1 complexes, promoting coregulator exchange and allowing increased transcriptional activation. Related molecular events are likely to occur during activation of steroidogenesis (30, 47), highlighting an important area of future investigations regarding the role of RNF31.
The stabilizing role of RNF31 may appear provocative, given that E3 ligases are usually assumed to catalyze polyubiquitination, which is linked to proteasomal degradation. However, compared to the RING and HECT families, RBR-type E3s, including RNF31, are poorly characterized with regard to substrate range, molecular mechanisms, and consequences of ubiquitination (12). Few of the RBR members have as yet convincingly been demonstrated to function as E3 ubiquitin ligase on natural substrate proteins. Also, the structure and function of the signature RBR domain is not fully understood. In direct support of the mechanisms we propose here for RNF31, recent reports suggest that the RBR protein Parkin, known for its genetic association with Parkinson's disease, can trigger monoubiquitination (16, 20). These and additional recent studies concluded that RBR E3 function does not result in degradation of substrates but instead leads to stabilization (31, 36). Stabilization could occur via several mechanisms other than E2-dependent monoubiquitination, including direct binding of the RBR ligase to (i) free, mono- or polyubiquitinated substrates, (ii) free ubiquitin or polyubiquitin chains, or (iii) other E3 ligases. Each of these scenarios is plausible for RNF31, as it has distinct E3 ligase and ubiquitin-binding domains and potentially dimerizes with other RBR ligases. Indeed, RNF31 has been reported to associate with RNF54, which catalyzed the formation of atypical ubiquitin chains on a model substrate in vitro (22) and independently was implicated in transcriptional functions (44).
Of particular significance is the identification of CYP19 (aromatase) as a target of RNF31. A major phenotype of the DAX-1 knockout mouse model was upregulation of CYP19 gene expression in testicular Leydig cells, leading to male infertility due to elevated levels of local estrogens (46). Although StAR expression appeared unchanged in the testes of these DAX-1 knockout mice, cellular studies and human adrenal phenotypes strongly suggest StAR to be subject to DAX-1 repression (19, 52), consistent with our results derived from human adrenocortical cells. This is in agreement with assumptions that key components and mechanisms of DAX-1 pathways in steroidogenic gene expression are conserved between humans and mice (26). Considering recent indications that DAX-1 action may not be restricted to steroidogenic pathways, it is likely that the functional interplay of RNF31 with DAX-1 goes beyond modulation of SF-1-dependent steroidogenic gene expression. For example, many DAX-1-containing cell types also express LRH-1 (NR5A2), the structural and functional SF-1 homologue. This not only suggests yet another feasible target for RNF31 action, it also indicates alternative routes to regulate steroidogenic gene expression in the ovary, endometrium, and mammary gland, in which LRH-1 can fulfill roles of SF-1. Further, the expression and functional relationship of RNF31, DAX-1, and NR5A receptors are of substantial interest with regard to endocrine tumorigenesis, such as that for adrenocortical tumors (4), pituitary adenomas, epithelial ovarian carcinoma, and prostate and breast cancers. Notably, the first report describing the cloning of human RNF31 (ZIBRA) characterizes its expression in human breast cancer cells (45). Finally, DAX-1 is expressed in embryonic stem cells and appears to be required for maintenance of pluripotency (32). These possibilities raise a profound interest in the role of RNF31 during early development, cell differentiation, and cancer, implicating major directions for future research.
Supplementary Material
Acknowledgments
We are grateful to H. Ardley, D. Bohmann, C. D. Clyne, K. L. Parker, E. R. Simpson, D. M. Stocco, A. M. Weissman, and D. Xirodimas for kindly providing plasmids. Agrisera AB (Vännäs, Sweden) is acknowledged for assistance with the generation of anti-DAX-1 and anti-RNF31 antibodies.
This work was supported by grants from the Swedish Research Council (E.T.), the Swedish Cancer Society (E.T.), the European Union Network CASCADE (J.-Å.G.), the Medical Research Fund of Tampere University Hospital (M.P.H.), and the Association Recherche sur le Cancer (E.L.).
Footnotes
Published ahead of print on 23 February 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Achermann, J. C., J. J. Meeks, and J. L. Jameson. 2001. Phenotypic spectrum of mutations in DAX-1 and SF-1. Mol. Cell. Endocrinol. 18517-25. [DOI] [PubMed] [Google Scholar]
- 2.Altincicek, B., S. P. Tenbaum, U. Dressel, D. Thormeyer, R. Renkawitz, and A. Baniahmad. 2000. Interaction of the corepressor alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J. Biol. Chem. 2757662-7667. [DOI] [PubMed] [Google Scholar]
- 3.Bardoni, B., E. Zanaria, S. Guioli, G. Floridia, K. C. Worley, G. Tonini, E. Ferrante, G. Chiumello, E. R. McCabe, M. Fraccaro, et al. 1994. A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat. Genet. 7497-501. [DOI] [PubMed] [Google Scholar]
- 4.Barlaskar, F. M., and G. D. Hammer. 2007. The molecular genetics of adrenocortical carcinoma. Rev. Endocr. Metab. Disord. 8343-348. [DOI] [PubMed] [Google Scholar]
- 5.Bromann, P. A., J. A. Weiner, E. D. Apel, R. M. Lewis, and J. R. Sanes. 2004. A putative ariadne-like E3 ubiquitin ligase (PAUL) that interacts with the muscle-specific kinase (MuSK). Gene Expr. Patterns 477-84. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W. Y., L. J. Juan, and B. C. Chung. 2005. SF-1 (nuclear receptor 5A1) activity is activated by cyclic AMP via p300-mediated recruitment to active foci, acetylation, and increased DNA binding. Mol. Cell. Biol. 2510442-10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, W.-Y., W.-C. Lee, N.-C. Hsu, F. Huang, and B.-C. Chung. 2004. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 27938730-38735. [DOI] [PubMed] [Google Scholar]
- 8.Chen, W.-Y., J.-H. Weng, C.-C. Huang, and B.-C. Chung. 2007. Histone deacetylase inhibitors reduce steroidogenesis through SCF-mediated ubiquitination and degradation of steroidogenic factor 1 (NR5A1). Mol. Cell. Biol. 277284-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford, P. A., C. Dorn, Y. Sadovsky, and J. Milbrandt. 1998. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol. Cell. Biol. 182949-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dammer, E. B., A. Leon, and M. B. Sewer. 2007. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3′,5′-monophosphate-dependent cytochrome P450c17 transcription rate. Mol. Endocrinol. 21415-438. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, A. P., and B. W. O'Malley. 2005. Rush hour at the promoter: how the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J. Steroid Biochem. Mol. Biol. 93139-151. [DOI] [PubMed] [Google Scholar]
- 12.Eisenhaber, B., N. Chumak, F. Eisenhaber, and M.-T. Hauser. 2007. The ring between ring fingers (RBR) protein family. Genome Biol. 8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiss-Friedlander, R., and F. Melchior. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8947-956. [DOI] [PubMed] [Google Scholar]
- 14.Gurates, B., A. Amsterdam, M. Tamura, S. Yang, J. Zhou, Z. Fang, S. Amin, S. Sebastian, and S. E. Bulun. 2003. WT1 and DAX-1 regulate SF-1-mediated human P450arom gene expression in gonadal cells. Mol. Cell. Endocrinol. 20861-75. [DOI] [PubMed] [Google Scholar]
- 15.Hammer, G. D., I. Krylova, Y. Zhang, B. D. Darimont, K. Simpson, N. L. Weigel, and H. A. Ingraham. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3521-526. [DOI] [PubMed] [Google Scholar]
- 16.Hampe, C., H. Ardila-Osorio, M. Fournier, A. Brice, and O. Corti. 2006. Biochemical analysis of Parkinson's disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum. Mol. Genet. 152059-2075. [DOI] [PubMed] [Google Scholar]
- 17.Holter, E., N. Kotaja, S. Makela, L. Strauss, S. Kietz, O. A. Janne, J. A. Gustafsson, J. J. Palvimo, and E. Treuter. 2002. Inhibition of androgen receptor (AR) function by the reproductive orphan nuclear receptor DAX-1. Mol. Endocrinol. 16515-528. [DOI] [PubMed] [Google Scholar]
- 18.Ito, M., R. Yu, and J. Jameson. 1997. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol. Cell. Biol. 171476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jo, Y., and D. M. Stocco. 2004. Regulation of steroidogenesis and steroidogenic acute regulatory protein in R2C cells by DAX-1 (dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X chromosome, gene-1). Endocrinology 1455629-5637. [DOI] [PubMed] [Google Scholar]
- 20.Joch, M., A. R. Ase, C. X.-Q. Chen, P. A. MacDonald, M. Kontogiannea, A. T. Corera, A. Brice, P. Seguela, and E. A. Fon. 2007. Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol. Biol. Cell 183105-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerscher, O., R. Felberbaum, and M. Hochstrasser. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22159-180. [DOI] [PubMed] [Google Scholar]
- 22.Kirisako, T., K. Kamei, S. Murata, M. Kato, H. Fukumoto, M. Kanie, S. Sano, F. Tokunaga, K. Tanaka, and K. Iwai. 2006. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 254877-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krylova, I. N., E. P. Sablin, J. Moore, R. X. Xu, G. M. Waitt, J. A. MacKay, D. Juzumiene, J. M. Bynum, K. Madauss, V. Montana, L. Lebedeva, M. Suzawa, J. D. Williams, S. P. Williams, R. K. Guy, J. W. Thornton, R. J. Fletterick, T. M. Willson, and H. A. Ingraham. 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120343-355. [DOI] [PubMed] [Google Scholar]
- 24.la Cour, T., R. Gupta, K. Rapacki, K. Skriver, F. M. Poulsen, and S. Brunak. 2003. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 31393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lalli, E., M. H. Melner, D. M. Stocco, and P. Sassone-Corsi. 1998. DAX-1 blocks steroid production at multiple levels. Endocrinology 1394237-4243. [DOI] [PubMed] [Google Scholar]
- 26.Lalli, E., and P. Sassone-Corsi. 2003. DAX-1, an unusual orphan receptor at the crossroads of steroidogenic function and sexual differentiation. Mol. Endocrinol. 171445-1453. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann, S. G., J. M. Wurtz, J. P. Renaud, P. Sassone-Corsi, and E. Lalli. 2003. Structure-function analysis reveals the molecular determinants of the impaired biological function of DAX-1 mutants in AHC patients. Hum. Mol. Genet. 121063-1072. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y., M. Choi, G. Cavey, J. Daugherty, K. Suino, A. Kovach, N. C. Bingham, S. A. Kliewer, and H. E. Xu. 2005. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol. Cell 17491-502. [DOI] [PubMed] [Google Scholar]
- 29.Manna, P. R., M. T. Dyson, D. W. Eubank, B. J. Clark, E. Lalli, P. Sassone-Corsi, A. J. Zeleznik, and D. M. Stocco. 2002. Regulation of steroidogenesis and the steroidogenic acute regulatory protein by a member of the cAMP response-element binding protein family. Mol. Endocrinol. 16184-199. [DOI] [PubMed] [Google Scholar]
- 30.Manna, P. R., M. T. Dyson, Y. Jo, and D. M. Stocco. 2009. Role of dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X chromosome, gene 1 in protein kinase A- and protein kinase C-mediated regulation of the steroidogenic acute regulatory protein expression in mouse Leydig tumor cells: mechanism of action. Endocrinology 150187-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marteijn, J. A., L. T. van der Meer, L. van Emst, S. van Reijmersdal, W. Wissink, T. de Witte, J. H. Jansen, and B. A. Van der Reijden. 2007. Gfi1 ubiquitination and proteasomal degradation is inhibited by the ubiquitin ligase Triad1. Blood 1103128-3135. [DOI] [PubMed] [Google Scholar]
- 32.McCabe, E. R. B. 2007. DAX1: increasing complexity in the roles of this novel nuclear receptor. Mol. Cell. Endocrinol. 265/266179-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moynihan, T. P., H. C. Ardley, U. Nuber, S. A. Rose, P. F. Jones, A. F. Markham, M. Scheffner, and P. A. Robinson. 1999. The Ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHARI and H7-AP1. J. Biol. Chem. 27430963-30968. [DOI] [PubMed] [Google Scholar]
- 34.Mukai, T., M. Kusaka, K. Kawabe, K. Goto, H. Nawata, K. Fujieda, and K. Morohashi. 2002. Sexually dimorphic expression of Dax-1 in the adrenal cortex. Genes Cells 7717-729. [DOI] [PubMed] [Google Scholar]
- 35.Muscatelli, F., T. M. Strom, A. P. Walker, E. Zanaria, D. Recan, A. Meindl, B. Bardoni, S. Guioli, G. Zehetner, W. Rabl, et al. 1994. Mutations in the DAX-1 gene give rise to both X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature 372672-676. [DOI] [PubMed] [Google Scholar]
- 36.Nikolaev, A. Y., M. Li, N. Puskas, J. Qin, and W. Gu. 2003. Parc: a cytoplasmic anchor for p53. Cell 11229-40. [DOI] [PubMed] [Google Scholar]
- 37.Park, S. Y., J. J. Meeks, G. Raverot, L. E. Pfaff, J. Weiss, G. D. Hammer, and J. L. Jameson. 2005. Nuclear receptors Sf1 and Dax1 function cooperatively to mediate somatic cell differentiation during testis development. Development 1322415-2423. [DOI] [PubMed] [Google Scholar]
- 38.Robertson, K. M., M. Norgard, S. H. Windahl, K. Hultenby, C. Ohlsson, G. Andersson, and J. A. Gustafsson. 2006. Cholesterol-sensing receptors, liver X receptor alpha and beta, have novel and distinct roles in osteoclast differentiation and activation. J. Bone Miner. Res. 211276-1287. [DOI] [PubMed] [Google Scholar]
- 39.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 201405-1428. [DOI] [PubMed] [Google Scholar]
- 40.Sablin, E. P., A. Woods, I. N. Krylova, P. Hwang, H. A. Ingraham, and R. J. Fletterick. 2008. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc. Natl. Acad. Sci. USA 10518390-18395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanyal, S., A. Bavner, A. Haroniti, L. M. Nilsson, T. Lundasen, S. Rehnmark, M. R. Witt, C. Einarsson, I. Talianidis, J. A. Gustafsson, and E. Treuter. 2007. Involvement of corepressor complex subunit GPS2 in transcriptional pathways governing human bile acid biosynthesis. Proc. Natl. Acad. Sci. USA 10415665-15670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz, R., J. Suominen, T. Varre, H. Hakovirta, M. Parvinen, J. Toppari, and M. Pelto-Huikko. 2003. Expression of aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator messenger ribonucleic acids and proteins in rat and human testis. Endocrinology 144767-776. [DOI] [PubMed] [Google Scholar]
- 43.Sugawara, T., N. Sakuragi, and H. Minakami. 2006. CREM confers cAMP responsiveness in human steroidogenic acute regulatory protein expression in NCI-H295R cells rather than SF-1/Ad4BP. J. Endocrinol. 191327-337. [DOI] [PubMed] [Google Scholar]
- 44.Tatematsu, K., N. Yoshimoto, T. Koyanagi, C. Tokunaga, T. Tachibana, Y. Yoneda, M. Yoshida, T. Okajima, K. Tanizawa, and S. Kuroda. 2005. Nuclear-cytoplasmic shuttling of a RING-IBR protein RBCK1 and its functional interaction with nuclear body proteins. J. Biol. Chem. 28022937-22944. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, H. G. R., J. W. Harris, L. Lin, and J. P. Brody. 2004. Identification of the protein Zibra, its genomic organization, regulation, and expression in breast cancer cells. Exp. Cell Res. 295448-459. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Z. J., B. Jeffs, M. Ito, J. C. Achermann, R. N. Yu, D. B. Hales, and J. L. Jameson. 2001. Aromatase (Cyp19) expression is up-regulated by targeted disruption of Dax1. Proc. Natl. Acad. Sci. USA 987988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe, M., and S. Nakajin. 2004. Forskolin up-regulates aromatase (CYP19) activity and gene transcripts in the human adrenocortical carcinoma cell line H295R. J. Endocrinol. 180125-133. [DOI] [PubMed] [Google Scholar]
- 48.Winnay, J. N., and G. D. Hammer. 2006. Adrenocorticotropic hormone-mediated signaling cascades coordinate a cyclic pattern of steroidogenic factor 1-dependent transcriptional activation. Mol. Endocrinol. 20147-166. [DOI] [PubMed] [Google Scholar]
- 49.Wu, R.-C., Q. Feng, D. M. Lonard, and B. W. O'Malley. 2007. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell 1291125-1140. [DOI] [PubMed] [Google Scholar]
- 50.Xirodimas, D., M. K. Saville, C. Edling, D. P. Lane, and S. Lain. 2001. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene 204972-4983. [DOI] [PubMed] [Google Scholar]
- 51.Zanaria, E., F. Muscatelli, B. Bardoni, T. M. Strom, S. Guioli, W. Guo, E. Lalli, C. Moser, A. P. Walker, E. R. McCabe, and et al. 1994. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature 372635-641. [DOI] [PubMed] [Google Scholar]
- 52.Zazopoulos, E., E. Lalli, D. M. Stocco, and P. Sassone-Corsi. 1997. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature 390311-315. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, H., J. S. Thomsen, L. Johansson, J. A. Gustafsson, and E. Treuter. 2000. DAX-1 functions as an LXXLL-containing corepressor for activated estrogen receptors. J. Biol. Chem. 27539855-39859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.