Abstract
Saccharomyces cerevisiae Pta1 is a component of the cleavage/polyadenylation factor (CPF) 3′-end processing complex and functions in pre-mRNA cleavage, poly(A) addition, and transcription termination. In this study, we investigated the role of the N-terminal region of Pta1 in transcription and processing. We report that a deletion of the first 75 amino acids (pta1-Δ75) causes thermosensitive growth, while the deletion of an additional 25 amino acids is lethal. The pta1-Δ75 mutant is defective for snoRNA termination, RNA polymerase II C-terminal domain Ser5-P dephosphorylation, and gene looping but is fully functional for mRNA 3′-end processing. Furthermore, different regions of Pta1 interact with the CPF subunits Ssu72, Pti1, and Ysh1, supporting the idea that Pta1 acts as a scaffold to organize CPF. The first 300 amino acids of Pta1 are sufficient for interactions with Ssu72, which is needed for pre-mRNA cleavage. By the degron-mediated depletion of Pta1, we show that the removal of this essential region leads to a loss of Ssu72, yet surprisingly, in vitro cleavage and polyadenylation remain efficient. In addition, a fragment containing amino acids 1 to 300 suppresses 3′-end processing in wild-type extracts. These findings suggest that the amino terminus of Pta1 has an inhibitory effect and that this effect can be neutralized through the interaction with Ssu72.
The synthesis of mature mRNA in eukaryotes and its utilization in the cytoplasm require cotranscriptional modifications of the pre-mRNA by capping at the 5′ end, removal of introns by splicing, and cleavage at the 3′ end followed by the addition of a poly(A) tail (4, 27, 32). mRNA 3′-end formation is an essential step in mRNA biogenesis and acts at many levels to influence gene expression. Its execution prevents readthrough transcription from interfering with DNA elements such as promoters, centromeres, and replication origins. Without a poly(A) tail, mRNA is targeted for degradation by nuclear surveillance mechanisms, is exported inefficiently from the nucleus, and is poorly translated in the cytoplasm. The maturation of mRNA 3′ ends also serves as an important point at which the cell can regulate the type and amount of mRNA derived from a particular gene. Furthermore, 3′-end processing has been linked to other essential processes such as chromosome segregation, DNA repair, and tissue-specific protein expression (23, 26, 46).
mRNA 3′-end formation in Saccharomyces cerevisiae requires the concerted action of two multisubunit factors, cleavage factor (CF) I and cleavage/polyadenylation factor (CPF), that recognize processing signals around the poly(A) site. These complexes are phylogenetically conserved and are comparable to mammalian CstF and CPSF, respectively (27). Cleavage requires CF I and CPF, while tail synthesis requires these factors plus the Pab1 or Nab2 poly(A) binding protein (17). CF I is composed of Rna14, Rna15, Pcf11, Clp1, and Hrp1/Nab4 (21, 22, 35). The holo-CPF complex can be separated into core CPF and the APT subcomplex (29). Core CPF includes Pta1, Cft1, Cft2, Mpe1, Pfs2, Fip1, Pap1, the poly(A) polymerase, and Ysh1/Brr5, the putative pre-mRNA endonuclease (10, 12, 15, 28, 29, 36, 45). The APT subcomplex of CPF includes Pta1, Pti1, Ref2, Swd2, Syc1, and the two phosphatases Ssu72 and Glc7 (29). Even though holo-CPF is important for optimal processing, traditional multistep chromatographic fractionation showed that a smaller complex, called CF II, was sufficient for cleavage in combination with CF I and contained only the Cft1, Cft2, Ysh1, and Pta1 subunits (48).
The essential Pta1 subunit was initially defined by a conditional growth mutation, pta1-1, that causes the accumulation of unspliced pre-tRNA in vivo (31). Pta1 is important for both cleavage and poly(A) addition (35, 48), and its phosphorylation inhibits the poly(A) addition step (16). Pta1 also has roles in redirecting the machinery to the 3′ end of nonadenylated snoRNA transcripts (29), in regulating the phosphorylated state of the RNA polymerase II (RNAP II) C-terminal domain (CTD) (25), and in the gene looping that juxtaposes the 5′ and 3′ ends of genes (1). Symplekin, the Pta1 homolog in higher eukaryotes, has been proposed to be a scaffold for assembling the mammalian 3′-end-processing complex (43). Symplekin has also been implicated in the formation of the cleaved, unadenylated ends of replication-dependent histone mRNAs (24), in the cytoplasmic polyadenylation of stored maternal mRNAs in preparation for their translation (2), and in the splicing of tRNA precursors (34).
Consistent with a scaffold function, the 90-kDa Pta1 interacts physically and/or genetically with the CPF subunits Ysh1 (the candidate nuclease), Pti1 (thought to suppress CPF's polyadenylation activity on snoRNA transcripts), Ssu72 (an RNAP II CTD serine-5 phosphatase and the only factor dedicated to cleavage), Glc7 (a phosphatase needed for the polyadenylation step), and Syc1 (a negative regulator of mRNA 3′-end formation) (9, 15, 16, 29, 49). However, it is unknown whether all of these contacts are made simultaneously; perhaps some occur sequentially during mRNA synthesis or only in certain types of complexes, while others may never happen inside the cell. The range of Pta1 interactions, especially with three of the enzymes of the complex (Ssu72, Ysh1, and Glc7) and functionally with the fourth, Pap1, suggests that Pta1 occupies a central position within the 3′-end processing complex and helps coordinate its various activities. However, little is known about how Pta1 performs this critical function.
In this study, we explore how Pta1 might act as a scaffold protein and show that it uses different regions to contact subunits that are essential to the function of CPF in mRNA 3′-end processing, snoRNA termination, CTD Ser5-P dephosphorylation, and gene looping. In addition, we identify new interactions between Pta1 and CF I, indicating that the organizational role of Pta1 includes making cross-factor connections. We also make the surprising observation that a Pta1 derivative that lacks the essential 300 amino acids at the N terminus and is incapable of interacting with and stabilizing Ssu72 is completely functional in cleavage and polyadenylation. These findings support a model in which the primary function of Ssu72 in mRNA 3′-end processing is to block an inhibitory activity of the Pta1 N-terminal domain.
MATERIALS AND METHODS
Yeast strains and culture.
The S. cerevisiae strains used in this study are as follows. Strain XH5 (MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 pta1::TRP1 [YCpURA3-PTA1]) was constructed by using a diploid strain with the chromosomal copy of PTA1 disrupted with TRP1 (pta1::TRP1) and transformed with a YCpURA3-PTA1 plasmid to then create the haploid strain by sporulation. The pta1-td, ssu72-td, and pJ69-4A strains were described previously (20, 25). The pta1-td strain was grown at 25°C in the presence of 0.1 mM CuSO4 to maintain the expression of the degron-tagged Pta1. To deplete cells of Pta1, cultures were shifted to medium lacking copper and incubated at 37°C. The ref2Δ strain (BY4741 background) was obtained from the American Type Culture Collection (ATCC). The PTA1 wild-type gene and the pta1 alleles encoding the N-terminal deletion derivatives were cloned into either the YCpLEU vector or the two-hybrid vector pGAD-C2 and introduced into strain XH5. The resident YCpURA3-PTA1 plasmid was then counterselected on 5-fluoroorotic acid (5-FOA) medium, as described previously (7). Growth properties were analyzed by growing the strains in liquid yeast extract-peptone-dextrose (YPD) medium at room temperature to an optical density at 600 nm of 1.0, spotting 5 μl of 10-fold serial dilutions onto YPD plates, and incubating the plates for 3 days at 16°C, 24°C, 30°C, 37°C, or 39°C.
Western blot analyses.
Steady-state levels of specific proteins were determined by Western blotting using mid-log-phase, whole-cell extracts as described previously using either 30 μg (see Fig. 1A, 2B, 4B, and 5B) (16) or 10 μg (see Fig. 3C) of total protein (33). Extracts were resolved on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel, and proteins were transferred onto either Immobilon-P polyvinylidene difluoride (Millipore) or nitrocellulose (Bio-Rad) membranes. Commercially available monoclonal antibodies were used to detect RNAP IIA (8WG16; Covance), CTD Ser5-P (H14; Covance), and the Rpb3 subunit of RNAP II (Neoclone). For the detection of differences in CTD Ser5-P levels, we have found that it is critical to use the H14 and 8WG16 antibodies at a 1/5,000 dilution. Monoclonal antibody against Pta1 and polyclonal antibodies against Ssu72 and other CPF and CF I subunits were described previously (25, 38, 47). Glutathione S-transferase (GST) antibody was obtained from BD Biosciences-Pharmingen, and GAL4-DBD (RK5C1) antibody was obtained from Santa Cruz Biotechnology. Rpa1 antibody was a gift from Steve Brill (Rutgers University).
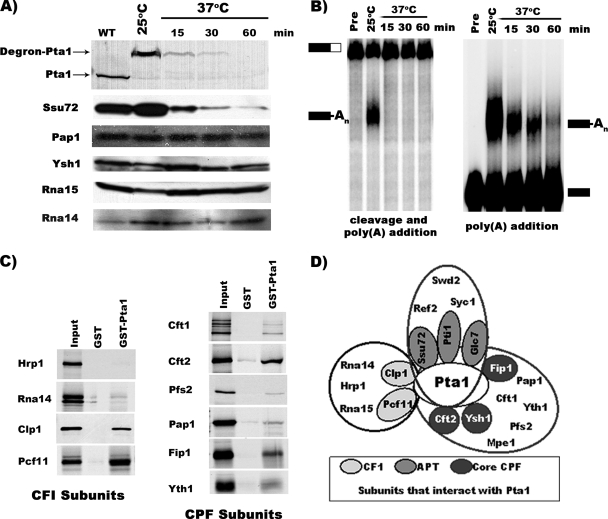
FIG. 1.
In vivo depletion of Pta1 affects pre-mRNA 3′-end processing in vitro. (A) Western blot analysis. Whole-cell extracts were prepared from pta1-td cells grown at 25°C or shifted to 37°C for the indicated times or from cells expressing normal Pta1 (wild type [WT]). The blot was probed with antibodies against the indicated proteins. (B) In vitro cleavage and poly(A) addition assays. For coupled cleavage-polyadenylation assays (left), the extracts used in A were incubated with ATP and 32P-labeled full-length GAL7-1 RNA (Pre, unreacted precursor) for 20 min at 30°C. The same conditions were used for poly(A) addition assays (right) except that the precleaved RNA GAL7-9 was used as a precursor. Products were resolved on a denaturing 5% polyacrylamide gel and visualized with a PhosphorImager apparatus. Positions of substrate and product are indicated on the left and right. (C) Pta1 interacts with different CF I and CPF subunits. 35S-labeled in vitro-translated subunits of 3′-end processing factors were incubated with GST or GST-Pta1 bound to glutathione-Sepharose beads. The proteins bound to GST or GST-Pta1, and 10% of the input was separated on a 10% polyacrylamide-SDS gel and detected by autoradiography. (D) Model summarizing interactions between Pta1 and different CF I and CPF subunits.
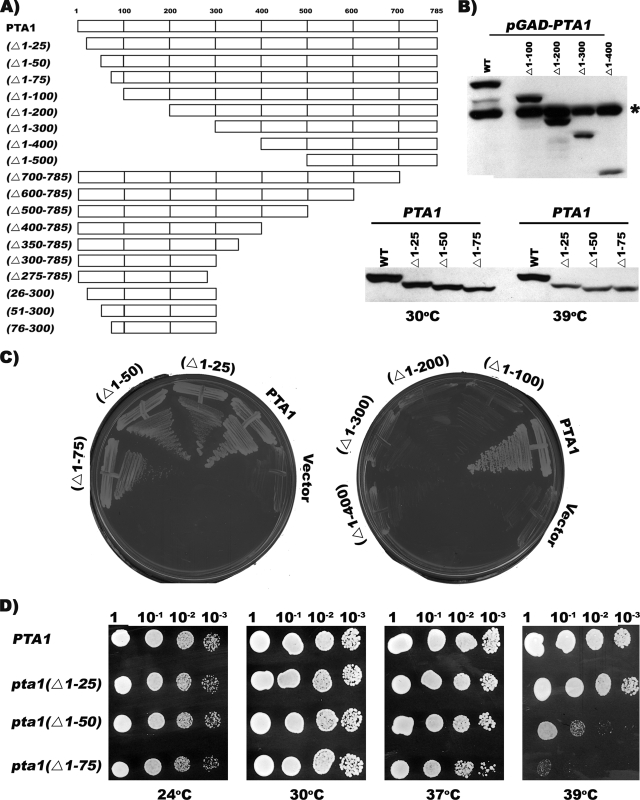
FIG. 2.
Growth properties of pta1 N-terminus mutants. (A) Pta1 deletion mutants. Amino acid sequences encoded by the pta1 alleles are indicated by bars. (B) In vivo expression of truncated forms of Pta1. Whole-cell extracts were prepared from strains expressing either full-length Pta1 or the indicated deletion derivatives and analyzed by Western blotting using a Pta1 monoclonal antibody. The nonviable truncations (top) were expressed as fusions to the GAL4 activation domain two-hybrid tag. (* indicates wild-type [WT] Pta1 of strain XH5.) (C) Plasmid shuffle complementation assay. The XH5 [YCpURA3-PTA1] plasmid shuffle strain expressing epitope-tagged versions of wild-type Pta1 or the indicated deletion derivatives were streaked onto 5-FOA medium to counterselect the YCpURA3-PTA1 plasmid. Plates were photographed following incubation for 4 days at 24°C. (D) Relative growth rates at different temperatures. Strains were grown in liquid YPD medium, and 10-fold serial dilutions were spotted onto YPD plates and incubated for 2 to 3 days at the indicated temperatures.
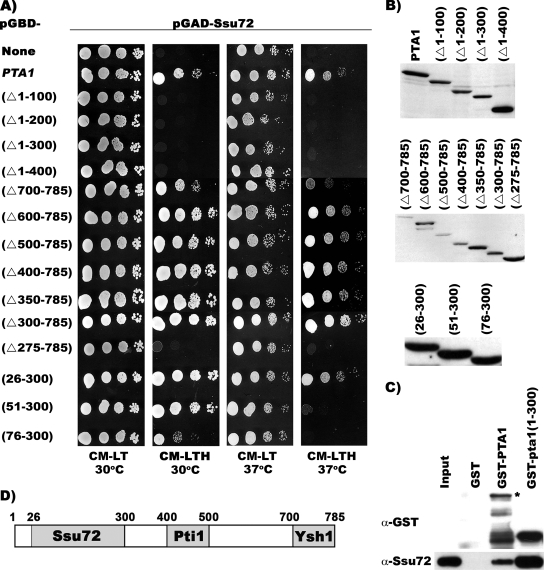
FIG. 4.
Different regions are required for Pta1 interactions with Ssu72, Ysh1, and Pti1. (A) Yeast two-hybrid analysis. Plasmids carrying SSU72 fused to the GAL4 activation domain (pGAD-Ssu72) and plasmids carrying pta1 variants fused to the GAL4 DNA binding domain were cotransformed into strain PJ69-4A. Tenfold serial dilutions of the resulting strains were spotted onto CM−LT, as indicated. Activation of HIS3 expression was tested by spotting the same strains onto CM-LTH and incubation for 2 to 3 days at the indicated temperatures. As a control, vector pGAD-Ssu72 was tested in the presence of empty pGBD vector (none). (B) Western blot analysis. Whole-cell extracts were prepared from the different two-hybrid strains grown at 30°C and analyzed by Western blotting using a monoclonal antibody recognizing the GAL4 DNA binding domain. (C) GST-pull down assays. Full-length recombinant Pta1 (GST-Pta1), truncated pta1 [GST-pta1(1-300)], or GST alone was bound to glutathione-Sepharose beads and then incubated with recombinant Ssu72. Proteins bound to the beads and 10% of the recombinant Ssu72 input were separated on a 10% polyacrylamide-SDS gel and detected by Western blot analysis using antibodies directed against GST and Ssu72. “*” indicates full-length GST-Pta1. (D) Pta1 regions. Schematic diagram of Pta1 summarizing the regions needed for wild-type interactions with Ssu72, Ysh1, and Pti1.
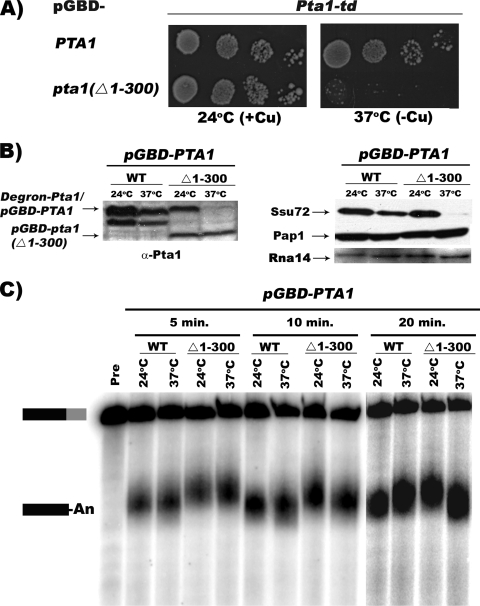
FIG. 5.
Pta1 interaction with Ssu72 is not required for cleavage and polyadenylation when the first 300 amino acids of Pta1 are missing. (A) Rescue of degron-mediated Pta1 depletion by pGBD-Pta1. Tenfold serial dilutions of the pta1-td strain transformed with either pGBD-PTA1 or pGBD-pta1 Δ1-300 were spotted onto complete medium lacking uracil and tryptophan in the presence or absence of 0.1 mM CuSO4 and incubated for 2 or 3 days at 24°C or 37°C. (B) Western blot analysis. Whole-cell extracts were prepared from pta1-td strains transformed with either the wild type (WT) or the pta1 Δ1-300 plasmid grown at 24°C or shifted to 37°C for 40 min and analyzed by Western blotting using antibodies against the indicated proteins. (C) In vitro cleavage and poly(A) addition assays. The extracts used in B were incubated with ATP and 32P-labeled full-length GAL7-1 RNA (Pre, unreacted precursor) for 5, 10, and 20 min at 30°C. Products were resolved on a denaturing 5% polyacrylamide gel and visualized with a PhosphorImager apparatus. Positions of substrate and product are indicated on the left.
FIG. 3.
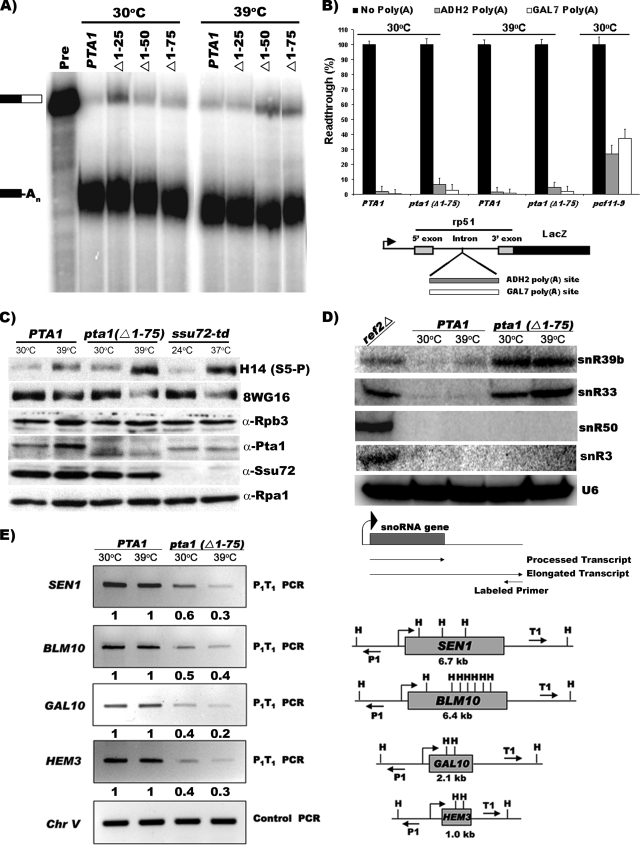
The Pta1 N-terminal 75 amino acids are required for snoRNA termination, CTD-serine-5 phosphorylation, and gene looping but not for mRNA 3′-end formation. (A) In vitro 3′-end processing assay. Extracts prepared from the strains containing the wild type (WT) and the indicated pta1 mutants, grown at 30°C or shifted to 39°C for 60 min, were incubated with ATP and 32P-labeled full-length GAL7-1 RNA (Pre, unreacted precursor) for 20 min at 30°C. Products were resolved on a denaturing 5% polyacrylamide gel and visualized with a PhosphorImager apparatus. Positions of substrate and product are indicated on the left. (B) In vivo 3′-end processing and termination assay. Quantitative measurements of β-galactosidase activity were performed using wild-type or mutant strains transformed with reporter vectors lacking or containing an efficient polyadenylation signal upstream of the lacZ gene, as illustrated at the bottom. The amount of readthrough is expressed as a percentage of that obtained from vector lacking the poly(A) site. The data are the averages of data for 48 independent assays of each strain. (C) Analysis of RNAP II CTD serine-5-P levels. Western blots of whole-cell extracts from wild-type (PTA1) or pta1 Δ1-75 strains grown at 30°C or shifted to 39°C for 60 min were probed using antibodies directed against the unphosphorylated form of RNAP II (8WG16), RNAP II CTD serine-5-P (H14), the RNAP II Rpb3 subunit, Pta1, Ssu72, or Rpa1 (loading control). The ssu72-td degron strain grown at 24°C in the presence of 0.1 mM CuSO4, or shifted to 37°C in the absence of CuSO4 for 60 min, was included as a control for the accumulation of CTD serine-5-P (25). No Ssu72 protein was detected in the ssu72-td strain at 24°C because the degron-encoded hemagglutinin (HA) epitope tag causes Ssu72-HA to comigrate with Rpb3; however, previous experiments established that Ssu72-HA accumulates in the ssu72-td strain at 24°C but is depleted at 37°C (25). (D) Detection of extended transcripts from snoRNA loci. Total RNA was prepared from strains containing the wild type (PTA1) or the pta1Δ1-75 mutant grown at 30°C or shifted to 39°C for 60 min or the ref2Δ strain grown at 30°C. Extended transcripts were detected by reverse transcription with 5′-end-labeled primers complementary to regions downstream of each snoRNA 3′ end, as depicted at the bottom. Primer extension of U6 RNA was used as a normalization control. (E) Effects of pta1-Δ75 on gene looping at the SEN1, BLM10, GAL10, and HEM3 genes. (Right) Schematic representation of the genes used in the 3C analysis indicating positions of the HindIII sites, drawn approximately to scale, and the divergent P1 and T1 primer pairs. Open reading frame lengths are indicated under each gene. (Left) Gene looping was assayed in the wild-type or pta1 Δ1-75 strains grown at 30°C or shifted to 39°C for 60 min. PCR products derived from the P1-T1 primer pair by 3C analysis were resolved by gel electrophoresis. Control PCR represents an intergenic region of chromosome V generated using convergent primers. P1-T1 PCR products were quantified by dividing P1-T1 PCR signals by control PCR signals for each sample; these ratios were then divided by the ratio of the wild-type 24°C sample for each gene to yield the number depicted beneath each lane.
Yeast two-hybrid analysis.
Derivatives of vector pGAD-C2 expressing the Gal4 activation domain fused to full-length Ssu72, Pti1, or Ysh1 were introduced into host strain PJ69-4A. The resulting strains were transformed with vector pGBD-C2 expressing the Gal4 DNA binding domain fused to full-length Pta1 or its deletion derivatives. Transformants were selected on complete medium lacking leucine and tryptophan (CM−LT) to ensure that both the GAL4 activation domain and GAL4 binding domain plasmids were present. Protein-protein interactions were scored by the ability of cells to grow on complete medium lacking leucine, tryptophan, and histidine (CM−LTH). Tenfold serial dilutions of the cultures were made, and 5 μl of each dilution was plated onto CM−LT as a growth control or CM−LTH for scoring two-hybrid interactions. Plates were incubated at 30°C or 37°C for 2 to 3 days.
Recombinant proteins and in vitro protein-protein interaction assays.
Plasmids expressing GST-Pta1 or Ssu72-His6 were described previously (15). Plasmid GST-Pta1(1-300) was generated by inserting the first 300 amino acids of Pta1 into the SpeI/NotI sites of pGEX-6P. Expression and purification of recombinant proteins from Escherichia coli Rosetta (DE3) cells as well as in vitro binding experiments were carried out as described previously (14, 18). Radiolabeled proteins used in this study were generated in vitro with the TNT rabbit reticulocyte lysate system (Promega) in the presence of [35S]methionine in a total volume of 50 μl. GST-Pta1 or GST was incubated with 20 μl of bed volume of glutathione-Sepharose beads in 200 μl of IP-150 buffer for 1 h at 4°C. After extensive washing, proteins bound to the beads were incubated with radiolabeled proteins (1 to 5 μl of the in vitro translation reaction mixture) for 2 h at 4°C. Beads were pelleted and washed four times with 400 μl of IP-150 buffer with 0.05% NP-40. Proteins were eluted in 15 μl of SDS sample buffer and resolved by SDS-10% polyacrylamide gel electrophoresis.
In vitro and in vivo RNA analyses.
Preparation of yeast cell extracts, transcription of [α-32P]UTP-labeled full-length GAL7-1 RNA or precleaved GAL7-9 RNA, and processing assays were described previously by Zhao et al. (47). Total yeast RNA was prepared by using the acid-phenol extraction method (40). Primer extension analysis was carried out as described previously by Nedea et al. (29) using primers specific for sequences downstream of the mature snoRNAs and for U6 as described previously (8, 42). The amount of reverse-transcribed U6 loaded was adjusted to match the signal given by snoRNA cDNAs.
β-Galactosidase assay.
Wild-type and mutant strains were transformed with various reporter vectors [pHZ18Δ2, no poly(A); pL101, ADH2 poly(A); pL501, GAL7 poly(A)] (19). Cells were grown in synthetic medium lacking uracil and containing 2% galactose to an optical density at 600 nm of 0.2 and assayed by using the Gal-Screen system (Applied Biosystems).
Capturing chromosome conformation.
Gene looping was analyzed by the capturing chromosome conformation (3C) method as described previously (1, 39).
RESULTS
Pta1 is required for 3′-end processing in vivo and in vitro.
We previously reported that Pta1 mutants are defective for both cleavage and poly(A) addition in vitro (15, 47), but Preker et al. previously observed a defect in the poly(A) addition step only (35). To clarify the role of Pta1, we examined the effects of depletion of Pta1 on mRNA 3′-end processing in vitro. Pta1 was depleted using the degron strategy (11), which places PTA1 expression under the control of the copper-inducible CUP1 promoter and integrates a heat-inducible degradation signal in frame with the Pta1 amino terminus at the PTA1 chromosomal locus. As shown previously (25), Pta1 was barely detectable after shifting this pta1-td strain to 37°C for 30 min and was totally degraded after 60 min (Fig. 1A). The depletion of Pta1 did not change the protein level of the tested CF I and CPF subunits except for that of Ssu72, which was significantly decreased upon the loss of Pta1 (Fig. 1A) (25). These results are reminiscent of the loss of Pta1 upon Ssu72 depletion, which we previously reported (15), and are in agreement with the physical and the genetic interactions that we had found between the two proteins. Thus, the levels of Pta1 and Ssu72 are tightly balanced in vivo.
Extracts were prepared from the pta1-td strain at different times after the shift to 37°C and assayed for in vitro processing activity using precursor RNA containing the GAL7 poly(A) site. Extracts from cells grown at 25°C efficiently cleaved and polyadenylated the RNA, but extracts from cells after the shift to 37°C were not functional in this coupled reaction (Fig. 1B, left). The poly(A) addition step can be studied separately from cleavage using RNA that ends at the GAL7 poly(A) site. Compared to cells grown at 25°C, levels of polyadenylation markedly decreased in extracts prepared from cells 15 min after the shift to 37°C, and very little adenylated product could be detected after 60 min (Fig. 1B, right). We conclude that Pta1 is important for both cleavage and poly(A) addition.
To further understand how Pta1 works in 3′-end formation of RNAP II transcripts, we next asked if Pta1 interacts directly with other CPF subunits or with components of CF I. By using a GST-Pta1 fusion protein in pull-down assays, we detected clear interactions with CF I components Clp1 and Pcf11 and CPF components Cft2, Fip1, and Yth1; weaker interactions with Cft1, Pfs2, and Pap1; and no interactions above background with Hrp1 and Rna14 (Fig. 1C) or Rna15 (49). This analysis, in addition to the previously published data on Pta1 interactions (9, 15, 16, 29), suggests that Pta1 serves as a scaffold to organize CPF as well as make cross-factor connections to CF I (Fig. 1D).
A Pta1 N-terminal deletion causes defects in snoRNA termination, CTD Ser5-P dephosphorylation, and gene looping.
The C terminus of Pta1, which is conserved across eukaryotes, is involved in linking Pta1 to core CPF and is essential for mRNA cleavage and polyadenylation (29, 47). However, the role of its N terminus has not been defined. To address this question, we tested a series of N-terminal truncations (Fig. 2A) for their ability to support cell growth using the plasmid shuffle assay. Mutants lacking the first 25 (pta1 Δ1-25), 50 (pta1 Δ1-50), or 75 (pta1 Δ1-75) amino acids were viable when the wild-type PTA1 plasmid was counterselected on 5-FOA medium (Fig. 2C, left), but those mutants lacking the first 100 (pta1 Δ1-100), 200 (pta1 Δ1-200), 300 (pta1 Δ1-300), or 400 (pta1 Δ1-400) amino acids were not (Fig. 2C, right). When tested at different temperatures, the growth rates of the viable mutants were similar to those of the wild type at 16°C, 24°C, 30°C, and 37°C, but the pta1 Δ1-50 and pta1 Δ1-75 mutants exhibited reduced growth at 39°C (Fig. 2D and data not shown). Western blot analysis confirmed that the growth phenotypes were not due to an instability of the mutant Pta1 proteins (Fig. 2B and 3C) or to a loss of Ssu72 (Fig. 3C). These results suggest an important role for the N terminus in Pta1 function.
To more directly examine the cause of the thermosensitive growth phenotype, we tested the activity of these pta1 alleles in cleavage, polyadenylation, snoRNA termination, CTD Ser-5 phosphorylation, and gene looping. Extracts prepared from cells containing wild-type Pta1, pta1 Δ1-25, pta1 Δ1-50, and pta1 Δ1-75 grown at 30°C or shifted to 39°C for 60 min were examined for their activities in mRNA 3′-end processing. Surprisingly, the truncations had no effect on coupled in vitro cleavage/polyadenylation assays, even when cells were incubated at the nonpermissive temperature (Fig. 3A), and no difference was observed when extracts prepared at 30°C were incubated at 37°C for 30 min prior to performing the processing reactions (data not shown). We also saw no change when reactions were performed for shorter times (data not shown), indicating that the rate of processing had not slowed. These results show that the first 75 amino acids of Pta1 do not have a direct role in cleavage and polyadenylation in vitro.
To test whether the N-terminal 75 amino acids are involved in 3′-end formation in vivo, we used a previously described system in which a poly(A) site is inserted into an intron upstream of the lacZ gene (19). Cells containing wild-type Pta1 or pta1 Δ1-75 were transformed with reporter plasmids that had either the 3′ end of the ADH2 or GAL7 gene inserted into the intron or, as a control, one with no insertion, and levels of β-galactosidase activity were determined. As expected, in wild-type cells, the presence of either poly(A) site blocked readthrough transcription into the lacZ gene (Fig. 3B). The pta1 Δ1-75 strain showed a slight increase in the level of readthrough compared to that of the wild type for the ADH2 and GAL7 polyadenylation signals, respectively, when cells were grown at 30°C or shifted to 39°C for 1 h prior to performance of the assays (Fig. 3B). As a control, we used the pcf11-9 allele, which was previously shown to have cleavage and transcription termination defects (6, 37). This mutation caused a much greater readthrough than did the Pta1 truncation (Fig. 3B). These results indicate that the removal of the first 75 amino acids of Pta1 has only a mild effect on 3′-end processing and termination of mRNA transcripts in vivo.
The Pti1, Pta1, Ssu72, Ref2, Swd2, and Glc7 components of the APT complex function in snoRNA 3′-end formation (8, 9, 29, 30, 41, 42). For example, the deletion of the last 200 amino acids of Pta1 causes significant readthrough transcription at the snR33 locus compared to that of the wild type (29). Consistent with data from previous studies (9, 29), we detected little or no transcription downstream of the SNR39b, SNR33, SNR50, and SNR3 termination sites in wild-type cells by primer extension but significant readthrough in the ref2Δ strain (Fig. 3D). By the same method, we found that the pta1 Δ1-75 allele showed noticeable readthrough compared to that of the wild type at the SNR33, SNR71, and SNR39b loci (Fig. 3D and data not shown). However, for the SNR3 and SNR50 snoRNA genes, which are affected by a mutation of the NRD1, PTI1, SEN1, SSU72, or SWD2 gene (8, 9, 29, 42), no extended transcripts were detected (Fig. 3D). These results support the previous finding that Pta1 is necessary for efficient termination in some snoRNA genes, and we conclude that regions at both ends of the protein are important for this function.
We have previously shown that physical and genetic interactions between Pta1 and the CTD phosphatase Ssu72 exist (15) and that Ssu72 catalyzes CTD Ser-5-P dephosphorylation in association with Pta1 (25). To test if the first 75 amino acids of Pta1 are required for this enzymatic function of Ssu72, the levels of CTD Ser-5-P were analyzed using whole-cell extracts prepared from cells containing wild-type Pta1 and the pta1 Δ1-75 mutant grown at 30°C or shifted to 39°C for 60 min. Western blot analysis revealed that the Ser5-P level (H-14) increased in the pta1-Δ75 mutant at 39°C, with a commensurate decrease in hypophosphorylated RNAP II levels (8WG16) (Fig. 3C). Essentially identical results were observed using the ssu72-td strain at 24°C and 37°C (Fig. 3C), consistent with our previously reported results (25). Moreover, there were no changes in the levels of the Rpb3 subunit of RNAP II, Pta1, or Ssu72 in the wild-type or mutant strains at either permissive (30°C) or restrictive (37°C or 39°C) temperatures (Fig. 3C). Thus, the N-terminal 75 amino acids of Pta1 are critical for the CTD phosphatase activity of Ssu72.
Pta1 also plays a role in gene looping (1). This model for RNAP II transcription proposes that promoter and terminator regions are juxtaposed in a way that might facilitate transcription reinitiation. To determine whether the N terminus of Pta1 is required for looping, we assayed loop formation at the SEN1, BLM10, GAL10, and HEM3 genes by the 3C assay (1, 39) using P1 and T1 PCR primer pairs specific for each gene (Fig. 3E, right). Consistent with previous results (39), P1-T1 PCR signals were detected for all four genes, and the intensities of these signals were identical for wild-type cells grown at either 30°C or 39°C. However, levels of the P1-T1 PCR products were decreased for all of the tested genes in the pta1 Δ1-75 mutant grown at 30°C and further diminished upon incubation at the restrictive temperature (Fig. 3E). These results indicate that the N-terminal 75 amino acids of Pta1 are required for efficient gene looping.
Different regions on Pta1 are required for the interaction with Ssu72, Pti1, and Ysh1.
To determine the molecular basis for the effects of Pta1 amino-terminal truncations on cell viability, snoRNA termination, and gene looping, we used the GAL4-based two-hybrid system and the amino- and carboxy-terminal truncations of Pta1 described in the legend of Fig. 2A to identify the regions of Pta1 that interact with specific CPF subunits. For this analysis, we tested Ysh1/Brr5, the nuclease thought to cleave the precursor at the poly(A) site; Pti1, which is needed for snoRNA termination; and Ssu72, the CTD serine-5 phosphatase that also affects cleavage, snoRNA termination, and gene looping.
Plasmids carrying the GAL4 DNA binding domain fused to PTA1 (pGBD-Pta1) or to the different pta1 mutants and plasmids carrying the GAL4 activation domain fused to test genes (pGAD-Ssu72, pGAD-Pti1, and pGAD-Ysh1) were cotransformed into the two-hybrid tester strain PJ69-4A. Growth at 30°C and 37°C was examined. All transformants grew well on media that selected for both plasmids, indicating that none of the fusion proteins was dominant negative (Fig. 4A). In addition, all of the GBD-Pta1 fusions could be detected by Western blot analysis using antibodies specific to the GAL4 binding domain (Fig. 4B). Positive two-hybrid interactions activate the transcription of a HIS3 reporter gene and allow growth on CM−LTH. pGAD-Ssu72, pGAD-Pti1, and pGAD-Ysh1 did not promote growth in the presence of pGBD vector alone (Fig. 4A and Table 1), indicating that they would be usable as prey in the two-hybrid analysis.
TABLE 1.
Summary of the yeast two-hybrid data analyzing the interaction between Pta1 and Ssu72, Ysh1, and Pti1a
| Pta1 derivative | Interaction with:
|
||
|---|---|---|---|
| Ssu72 | Ysh1 | Pti1 | |
| None | − | − | − |
| WT | +++ | +++ | +++ |
| Pta1 Δ1-300 | − | +++ | +++ |
| Pta1 Δ1-400 | − | +++ | +++ |
| Pta1 Δ1-500 | − | +++ | − |
| Pta1 Δ700-785 | +++ | − | +++ |
| Pta1 Δ600-785 | +++ | − | +++ |
| Pta1 Δ500-785 | +++ | − | +++ |
| Pta1 Δ400-785 | +++ | − | − |
| Pta1 Δ350-785 | +++ | − | − |
| Pta1 Δ300-785 | +++ | − | − |
| Pta1 Δ275-785 | − | − | − |
| Pta1 26-300 | +++ | − | − |
| Pta1 51-300 | +++ | − | − |
| Pta1 76-300 | + | − | − |
Interactions are based on growth on CM−LTH at 30°C. “+,” weak growth; “+++,” strong growth on CM−LTH equivalent to that seen with full-length Pta1; “−,” no growth.
When pGAD-Ssu72 was tested for interactions with the different pGBD-Pta1 constructs, we found that in contrast to wild-type Pta1, the N-terminal deletions pta1 Δ1-100, pta1 Δ1-200, pta1 Δ1-300, pta1 Δ1-400, and pta1 Δ1-500 did not grow in the absence of histidine (Fig. 4 and Table 1). Deletions from the C terminus (pta1 Δ700-785, pta1 Δ600-785, pta1 Δ500-785, pta1 Δ400-785, pta1 Δ350-785, and pta1 Δ300-785) all showed a His+ phenotype at both 30°C and 37°C. However, the deletion of an additional 25 amino acids (pta1 Δ275-785) gives a His− phenotype. This analysis defined Pta1 amino acids 1 to 300 as being important for Ssu72 interactions. A GST pull-down experiment confirmed that this region of Pta1 interacts directly with Ssu72 (Fig. 4C) and is both necessary and sufficient for Ssu72 interactions.
To delimit the N-terminal boundary of the Ssu72 interaction, smaller deletions of 25 amino acids were used. While the region containing amino acids 26 to 300 shows a His+ phenotype at 30°C and 37°C, regions containing amino acids 51 to 300 or 76 to 300 give a His+ phenotype at 30°C but a His− phenotype at 37°C (Fig. 4A). The interaction of Ssu72 with pta1(76−300) is also weaker at 30°C than the interaction with full-length Pta1 or pta1 Δ300-785. These results indicate that amino acids 26 to 300 of Pta1 are required for the optimal Ssu72 interaction but that some interaction is still maintained with a Pta1 fragment containing amino acids 76 to 300 (Fig. 4A and D and Table 1).
The same Pta1 constructs were used to map Pta1 interactions with Ysh1 and Pti1 (Table 1). Deletions from both ends of Pta1 showed that the amino acid region between residues 400 and 500 is required for the Pti1 interaction. When we applied a similar analysis to Ysh1, we found that the interaction was preserved with a Pta1 truncation mutant lacking the first 500 amino acids but was lost when only 85 amino acids were removed from the C terminus of Pta1 (Table 1 and Fig. 4D).
Removal of the first 300 amino acids of Pta1 causes a loss of Ssu72 but does not affect 3′-end processing in vitro.
The results of Fig. 3 and 4 show that the first 75 amino acids of Pta1 are required for efficient Ssu72 interactions and other Pta1 functions but are not required for cleavage and polyadenylation. The Pta1 depletion shown in Fig. 1 left open the possibility that Pta1 plays no direct role in 3′-end formation other than to maintain the level of Ssu72. To determine the consequence of the complete loss of the Ssu72 interaction region on cleavage and poly(A) addition, we tested the effect of deleting the first 300 amino acids of Pta1 on 3′-end processing in vitro. For this purpose, we used the degron strategy to replace full-length Pta1 with the nonviable pta1 Δ1-300 derivative.
The pta1-td strain was transformed with plasmid DNA expressing either pGBD-Pta1 or pta1 Δ1-300. When the degron-tagged Pta1 strain was grown at 37°C, only the strain with pGBD-Pta1 was able to support cell growth (Fig. 5A), consistent with the inviability of the pta1 Δ1-300 mutant (Fig. 2C). The levels of Pta1, Ssu72, Pap1, and Rna14 were tested in extracts prepared from the different strains grown at 24°C or after shifting to 37°C for 40 min to deplete Pta1. Degron-Pta1 was efficiently removed after 40 min at 37°C, and both GBD-Pta1 (which comigrates with degron-Pta1) and GBD-pta1 Δ1-300 expressed well at 24°C and 37°C (Fig. 5B). The levels of the core CPF subunit Pap1 and the CF IA subunit Rna14 (CF IA) were unchanged. In contrast, Ssu72 was almost undetectable at 37°C in the strain expressing pGBD-Pta1 Δ1-300, mirroring what we had seen upon the total depletion of full-length Pta1 (Fig. 1A). Thus, the interaction with Pta1 is essential for Ssu72 stability.
The same extracts were examined for their activity in mRNA 3′-end processing. Surprisingly, the deletion of the first 300 amino acids of Pta1, and the accompanying loss of Ssu72, had no effect on the efficiency of cleavage and polyadenylation in vitro in extracts containing only this form of Pta1 (Fig. 5C). No change was observed at a reaction time of 20 min, when processing is almost complete, or at shorter times, when less precursor has been consumed. These results indicate that the interaction of Pta1 with Ssu72, and Ssu72 itself, is not required for 3′-end processing if the first 300 amino acids of Pta1 are absent. The lethality of this Pta1 truncation is more likely due to the loss of Ssu72, which is no longer available for its roles in transcription.
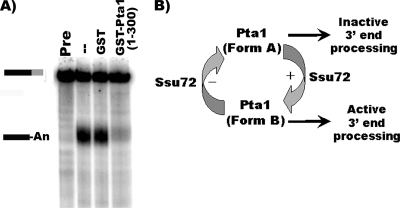
To further address the role of the first 300 amino acids of Pta1 in 3′-end processing, GST-pta1(1-300) or GST alone was added to processing reaction mixtures with wild-type extract. Cleavage and polyadenylation were equally efficient for wild-type extract or wild-type extract supplemented with GST only but significantly reduced by the addition of the first 300 amino acids of Pta1 (Fig. 6A). Together, these data suggest that the N terminus of Pta1 inhibits mRNA 3′-end processing and that this inhibition can be masked by the binding of Ssu72.
FIG. 6.
The first 300 amino acids of Pta1 inhibit pre-mRNA 3′-end processing in vitro. (A) In vitro cleavage and poly(A) addition assay. Extract prepared from wild-type cells was supplemented with equal amounts of GST alone or GST-Pta1(1-300) and incubated with ATP and 32P-labeled full-length GAL7-1 RNA (Pre, unreacted precursor) for 20 min at 30°C. Products were resolved on a denaturing 5% polyacrylamide gel and visualized with a PhosphorImager apparatus. Positions of substrate and product are indicated on the left. (B) Model for the role of the first 300 amino acids of Pta1 and Ssu72 in modulating Pta1 activity during mRNA 3′-end processing.
DISCUSSION
The formation of the polyadenylated 3′ ends of eukaryotic mRNA requires that several different factors come together to recognize the polyadenylation signals on the mRNA precursor and, in temporal sequence, cleave at the poly(A) site and then synthesize a poly(A) tail of the correct length. In addition, the activity of this large complex must be coordinated with that of other nuclear complexes involved in transcription, splicing, acquisition of export factors, and confirmation that the final mRNP has been correctly processed and assembled. CPFs are also needed for activities such as the cleavage of nonadenylated metazoan histone mRNAs, readenylation of cytoplasmic mRNAs, termination of RNAP II transcription, and facilitating transcription reinitiation by the interaction of the 5′ and 3′ ends of genes (1, 5, 6, 13, 39). The execution of these diverse functions requires that the CPFs interact differently with each other and with other factors depending on the job needed to be done. One of the proteins proposed to mediate these different interactions is the Pta1 subunit of the yeast CPFs and its metazoan homolog Symplekin.
A scaffold role for Pta1 and Symplekin.
Several studies suggested that Pta1 acts to organize the yeast CPF, which is depicted in Fig. 1D. Our previous work showed that Pta1 interacts directly with the C terminus of Ysh1, which lies outside of the RNase domain and is needed for polyadenylation as well as cleavage (49). The interaction with core CPF and function in 3′-end processing require the last 200 amino acids of Pta1 (15, 29). Our two-hybrid analysis showed that the removal of as little as 85 amino acids from the C terminus of Pta1 destroys the Ysh1 interaction (Fig. 4D and Table 1) and 3′-end processing in vitro (data not shown). Thus, a contact with Ysh1 may be sufficient to account for Pta1 acting at both steps of 3′-end processing. However, our pull-down assays suggest that direct contacts with other CPF subunits involved only in poly(A) addition (such as Fip1) or in both steps (such as Cft2 and Yth1) may also be involved.
Pta1 also connects the APT subcomplex to core CPF (Fig. 1D) (15, 29, 49). Unlike core CPF, the primary function of APT appears to be in snoRNA termination rather than mRNA processing. Pta1 is known to physically interact with the Ssu72, Pti1, Syc1, and Glc7 subunits of APT (15, 16, 29, 49), but the domains of Pta1 responsible for these interactions had not been localized. The work described here adds to our knowledge of the APT architecture by showing that distinct, nonoverlapping regions of Pta1 are involved in the Ssu72 (amino acids 26 to 300) and the Pti1 (amino acids 400 to 500) interactions. Furthermore, the Syc1 protein is almost identical to the C terminus of Ysh1 and can functionally substitute for this essential region (49). Our data suggest that Syc1 and Ysh1 both contact the C-terminal domain of Pta1, and as originally proposed by Nedea et al. (29), the simplest way to account for both proteins in holo-CPF would be the presence of two copies of Pta1 in the complex. In addition, we now show that Pta1 also interacts with CF I subunits Clp1 and Pcf11, suggesting that Pta1 acts as a scaffold to organize CPF as well as make cross-factor connections to CF I. In this way, Pta1 could bring CF I and holo-CPF together to work at the ends of both snoRNA and protein-encoding RNAs.
Symplekin, the metazoan homolog of Pta1, is also thought to act as a scaffold protein (43). Symplekin is significantly larger than its yeast counterpart, with little similarity except to the C terminus of Pta1 (45% similar). Thus, Pta1 has yeast-specific domains that interact with proteins such as Ssu72 or complexes such as APT that have not been described for metazoan cells. Unlike Pta1, symplekin is not tightly associated with any of the purified mammalian factors but clearly has a similar function in assembling diverse smaller complexes into larger ones. Symplekin is needed for the cleavage of the nonadenylated histone transcripts and performs this function as a component of heat-labile factor, a large complex that shares subunits with the CPFs CPSF and CstF (24). Symplekin in Xenopus oocytes is important for cytoplasmic polyadenylation directed by the xGLD-2 poly(A) polymerase (2). For these two activities, it is likely that critical contacts will be mediated by domains not found in Pta1.
Function of the Pta1 N terminus.
While the C terminus of Pta1 is clearly important for mRNA cleavage and polyadenylation (35, 47), the role of the N terminus had not been characterized. In this report, we found that the first 300 amino acids of Pta1 define an essential region needed for Ssu72 interactions and stability (Fig. 4A and 5B), although as shown by the deletion of amino acids 1 to 75 of Pta1, this interaction can be weakened without affecting Ssu72 levels (Fig. 3C and 4A). Ssu72 is a CTD Ser-5 phosphatase, but this activity is not needed for Ssu72's function in pre-mRNA cleavage (25). The deletion of the first 75 amino acids of Pta1 also impaired growth at 39°C (Fig. 2D) but had no effect on mRNA 3′-end processing (Fig. 3A). However, this deletion caused a gene-specific defect in snoRNA termination (Fig. 3D), accumulation of the CTD Ser5-P form of RNAP II (Fig. 3C), and a defect in gene looping (Fig. 3E). These results suggest that a strong Pta1-Ssu72 interaction is required for these transcription-related activities.
By examining the activity of the first 300 amino acids of Pta1 and Pta1 lacking this Ssu72 interaction domain, we were able to clarify the role of Pta1 and Ssu72 in mRNA 3′-end processing. Surprisingly, extracts from cells expressing only Pta1 lacking the Ssu72 interaction domain are fully active for cleavage and polyadenylation but show a loss of Ssu72 protein similar to that observed for the total depletion of Pta1 (Fig. 5). However, the addition of the first 300 amino acids of Pta1 reduces 3′-end processing in vitro (Fig. 6A). Furthermore, previous studies have shown that the ssu72-2 mutant is defective for Pta1 interactions by two-hybrid analysis and in vitro pull-downs (10) and is impaired for cleavage (15). Based on these findings, we propose a model to explain the function of the Pta1-Ssu72 interaction in 3′-end processing (Fig. 6B), with Ssu72 acting as a positive regulator. When Ssu72 interacts with the first 300 amino acids of Pta1, it masks an inhibitory effect of this region (perhaps by displacing other factors or by making the C terminus of Pta1 accessible for interactions with the nuclease Ysh1) and promotes a Pta1 conformation (Form B) that favors cleavage and polyadenylation. The absence of Ssu72 leads to a different conformation (form A) that is inactive for 3′-end processing.
Forming and breaking of the Pta1-Ssu72 contact may also work in concert with posttranslational modifications of Pta1. For example, Pta1 phosphorylation by CKII and dephosphorylation by Glc7 modulate its function in poly(A) addition (16). Similarly, it has been reported that the sumoylation of Symplekin and/or CPSF-73 affects the assembly and the activity of the cleavage and polyadenylation complex (44). Ubiquitination of the splicing complex controls the assembly and disassembly of the spliceosome (3), suggesting a common regulatory theme for complexes mediating multistep reactions.
In summary, we have shown that the N terminus of Pta1 is important for CTD Ser5-P dephosphorylation, gene looping, snoRNA termination, and Ssu72 stability, and through the interaction with Ssu72, it also modulates the function of Pta1 in mRNA processing. These findings support a role for Pta1 in organizing the cleavage/polyadenylation complex and directing its activity.
Acknowledgments
We are grateful to Krishnamurthy Shankarling, Jason Kuehner, and Xiangping Qu for valuable discussions.
This work was supported by NIH grants GM39484 (to M.H.), GM41752 (to C.M.), and GM68887 (to C.M. and M.H.).
Footnotes
Published ahead of print on 2 February 2009.
REFERENCES
- 1.Ansari, A., and M. Hampsey. 2005. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 192969-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard, D. C., K. Ryan, J. L. Manley, and J. D. Richter. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119641-651. [DOI] [PubMed] [Google Scholar]
- 3.Bellare, P., E. C. Small, X. Huang, J. A. Wohlschlegel, J. P. Staley, and E. J. Sontheimer. 2008. A role for ubiquitin in the spliceosome assembly pathway. Nat. Struct. Mol. Biol. 15444-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17251-256. [DOI] [PubMed] [Google Scholar]
- 5.Bilger, A., C. A. Fox, E. Wahle, and M. Wickens. 1994. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes Dev. 81106-1116. [DOI] [PubMed] [Google Scholar]
- 6.Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280298-301. [DOI] [PubMed] [Google Scholar]
- 7.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154164-175. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, H., X. He, and C. Moore. 2004. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol. Cell. Biol. 242932-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dheur, S., L. T. A. Vo, F. Voisinet-Hakil, M. Minet, J. M. Schmitter, F. Lacroute, F. Wyers, and L. Minvielle-Sebastia. 2003. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 222831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dichtl, B., D. Blank, M. Ohnacker, A. Friedlein, D. Roeder, H. Langen, and W. Keller. 2002. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 101139-1150. [DOI] [PubMed] [Google Scholar]
- 11.Dohmen, R. J., P. Wu, and A. Varshavsky. 1994. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 2631273-1276. [DOI] [PubMed] [Google Scholar]
- 12.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415141-147. [DOI] [PubMed] [Google Scholar]
- 13.Gilmartin, G. M. 2005. Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev. 192517-2521. [DOI] [PubMed] [Google Scholar]
- 14.Gross, S., and C. Moore. 2001. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. USA 986080-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, X., A. U. Khan, H. Cheng, D. L. Pappas, Jr., M. Hampsey, and C. L. Moore. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 171030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, X., and C. Moore. 2005. Regulation of yeast mRNA 3′ end processing by phosphorylation. Mol. Cell 19619-629. [DOI] [PubMed] [Google Scholar]
- 17.Hector, R. E., K. R. Nykamp, S. Dheur, J. T. Anderson, P. J. Non, C. R. Urbinati, S. M. Wilson, L. Minvielle-Sebastia, and M. S. Swanson. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 211800-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmling, S., A. Zhelkovsky, and C. L. Moore. 2001. Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Mol. Cell. Biol. 212026-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyman, L. E., and C. L. Moore. 1993. Termination and pausing of RNA polymerase II downstream of yeast polyadenylation sites. Mol. Cell. Biol. 135159-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler, M. M., M. F. Henry, E. Shen, J. Zhao, S. Gross, P. A. Silver, and C. L. Moore. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 112545-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kessler, M. M., J. Zhao, and C. L. Moore. 1996. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3′ ends. J. Biol. Chem. 27127167-27175. [DOI] [PubMed] [Google Scholar]
- 23.Kleiman, F. E., F. Wu-Baer, D. Fonseca, S. Kaneko, R. Baer, and J. L. Manley. 2005. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev. 191227-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolev, N. G., and J. A. Steitz. 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 192583-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnamurthy, S., X. He, M. Reyes-Reyes, C. Moore, and M. Hampsey. 2004. Ssu72 is an RNA polymerase II CTD phosphatase. Mol. Cell 14387-394. [DOI] [PubMed] [Google Scholar]
- 26.Lutz, C. S. 2008. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem. Biol. 3609-617. [DOI] [PubMed] [Google Scholar]
- 27.Mandel, C. R., Y. Bai, and L. Tong. 2008. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 651099-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandel, C. R., S. Kaneko, H. Zhang, D. Gebauer, V. Vethantham, J. L. Manley, and L. Tong. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nedea, E., X. He, M. Kim, J. Pootoolal, G. Zhong, V. Canadien, T. Hughes, S. Buratowski, C. L. Moore, and J. Greenblatt. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 27833000-33010. [DOI] [PubMed] [Google Scholar]
- 30.Nedea, E., D. Nalbant, D. Xia, N. T. Theoharis, B. Suter, C. J. Richardson, K. Tatchell, T. Kislinger, J. F. Greenblatt, and P. L. Nagy. 2008. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell 29577-587. [DOI] [PubMed] [Google Scholar]
- 31.O'Connor, J. P., and C. L. Peebles. 1992. PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol. Cell. Biol. 123843-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108439-451. [DOI] [PubMed] [Google Scholar]
- 33.Patturajan, M., R. J. Schulte, B. M. Sefton, R. Berezney, M. Vincent, O. Bensaude, S. L. Warren, and J. L. Corden. 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 2734689-4694. [DOI] [PubMed] [Google Scholar]
- 34.Paushkin, S. V., M. Patel, B. S. Furia, S. W. Peltz, and C. R. Trotta. 2004. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117311-321. [DOI] [PubMed] [Google Scholar]
- 35.Preker, P. J., M. Ohnacker, L. Minvielle-Sebastia, and W. Keller. 1997. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 164727-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roguev, A., A. Shevchenko, D. Schaft, H. Thomas, A. F. Stewart, and A. Shevchenko. 2004. A comparative analysis of an orthologous proteomic environment in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol. Cell. Proteomics 3125-132. [DOI] [PubMed] [Google Scholar]
- 37.Sadowski, M., B. Dichtl, W. Hubner, and W. Keller. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 222167-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saguez, C., M. Schmid, J. R. Olesen, M. A. Ghazy, X. Qu, M. B. Poulsen, T. Nasser, C. Moore, and T. H. Jensen. 2008. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol. Cell 3191-103. [DOI] [PubMed] [Google Scholar]
- 39.Singh, B. N., and M. Hampsey. 2007. A transcription-independent role for TFIIB in gene looping. Mol. Cell 27806-816. [DOI] [PubMed] [Google Scholar]
- 40.Sparks, K. A., and C. L. Dieckmann. 1998. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 264676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinmetz, E. J., and D. A. Brow. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 236339-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinmetz, E. J., N. K. Conrad, D. A. Brow, and J. L. Corden. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413327-331. [DOI] [PubMed] [Google Scholar]
- 43.Takagaki, Y., and J. L. Manley. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 201515-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vethantham, V., N. Rao, and J. L. Manley. 2007. Sumoylation modulates the assembly and activity of the pre-mRNA 3′ processing complex. Mol. Cell. Biol. 278848-8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh, E. P., D. J. Lamont, K. A. Beattie, and M. J. Stark. 2002. Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectrometry. Biochemistry 412409-2420. [DOI] [PubMed] [Google Scholar]
- 46.Wang, S. W., K. Asakawa, T. Z. Win, T. Toda, and C. J. Norbury. 2005. Inactivation of the pre-mRNA cleavage and polyadenylation factor Pfs2 in fission yeast causes lethal cell cycle defects. Mol. Cell. Biol. 252288-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao, J., M. Kessler, S. Helmling, J. P. O'Connor, and C. Moore. 1999. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol. 197733-7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao, J., M. M. Kessler, and C. L. Moore. 1997. Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J. Biol. Chem. 27210831-10838. [DOI] [PubMed] [Google Scholar]
- 49.Zhelkovsky, A., Y. Tacahashi, T. Nasser, X. He, U. Sterzer, T. H. Jensen, H. Domdey, and C. Moore. 2006. The role of the Brr5/Ysh1 C-terminal domain and its homolog Syc1 in mRNA 3′-end processing in Saccharomyces cerevisiae. RNA 12435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]