Abstract
The NAD+-dependent histone deacetylase hSirT1 regulates cell survival and stress responses by inhibiting p53-, NF-κB-, and E2F1-dependent transcription. Here we show that the hSirT1/PCAF interaction controls the E2F1/p73 apoptotic pathway. hSirT1 represses E2F1-dependent P1p73 promoter activity in untreated cells and inhibits its activation in response to DNA damage. hSirT1, PCAF, and E2F1 are corecruited in vivo on theP1p73 promoter. hSirT1 deacetylates PCAF in vitro and modulates PCAF acetylation in vivo. In cells exposed to apoptotic DNA damage, nuclear NAD+ levels decrease and inactivate hSirT1 without altering the hSirT1 interaction with PCAF and hSirT1 binding to the P1p73 promoter. The reactivation of hSirT1 by pyruvate that increases the [NAD+]/[NADH] ratio completely abolished the DNA damage-induced activation of TAp73 expression, thus linking the modulation of chromatin-bound hSirT1 deacetylase activity by the intracellular redox state with P1p73 promoter activity. The release of PCAF from hSirT1 repression favors the assembly of transcriptionally active PCAF/E2F1 complexes onto the P1p73 promoter and p53-independent apoptosis. Our results identify hSirT1 and PCAF as potential targets to modulate tumor cell survival and chemoresistance irrespective of p53 status.
hSirT1, the mammalian homologue of Sir2 (silent information regulator 2), is a NAD-dependent class III deacetylase (15, 33) that regulates cell survival, stress responses, and metabolism by inhibiting p53 (3, 18, 19, 28)-, E2F1 (1, 30)-, NF-κB (31)-, and Forkhead (2)-dependent transcription. The role of hSirT1 in the regulation of mammalian cell survival in response to DNA damage is supported by several observations. hSirT1-deficient mice display increased levels of radiation-induced apoptosis and p53 hyperacetylation (4). hSirT1-dependent deacetylation attenuates the ability of p53 to trans-activate the cell cycle (p21) and apoptotic (bax) target genes (19, 28). The constitutive expression of the tumor suppressor hypermethylated in cancer 1 (Hic1) represses hSirT1 transcription, thereby allowing the accumulation of acetylated p53 species and the enhancement of p53-mediated growth arrest and apoptosis in response to DNA damage (3). hSirT1 deacetylase function also modulates p53-independent pathways involved in the DNA damage response. The targeted disruption of the SirT1 gene in p53-deficient cells strongly sensitizes cells to radiation-, cisplatin-, and etoposide-induced cell death (20). hSirT1 deacetylates the DNA damage repair protein Ku70, and deacetylated Ku70 prevents Bax translocation to mitochondria to initiate apoptosis (5). hSirT1 maintains the Nijmegen breakage syndrome protein (NBS1) hypoacetylated and susceptible to be phosphorylated by ATM in response to DNA damage (32). Finally, etoposide treatment results in the E2F1-dependent induction of hSirT1 expression, and the abrogation of hSirT1 expression sensitizes cells to E2F1-dependent apoptosis (30).
The E2F family of transcription factors has critical roles in the control of cell proliferation and apoptosis (7). E2F transcriptional activity is tightly regulated during the cell cycle through the association with pRb or the related pocket proteins p107 and p130, leading, in quiescent cells, to the recruitment of transcriptional corepressors, including histone deacetylases (HDACs), methyltransferases, and polycomb group proteins, onto the promoters of proliferation-associated E2F target genes (7). As cells progress into the cell cycle, cyclin-dependent kinases phosphorylate pRb, releasing free E2F and allowing it to interact with transcriptional coactivators, and directly transactivate genes required for S-phase entry (7). E2F1 also upregulates the transcription of several genes involved in the activation or execution of apoptosis, including the Apaf-1, caspase 7, and p73 genes (16, 21, 22, 24).
The E2F1/p73 pathway is thought to play a major role in DNA-damaging drug-induced apoptosis and tumor chemosensitivity (6, 8, 9, 13, 23). In response to DNA damage, E2F1 is phosphorylated by Chk2 (26) and acetylated by PCAF (22). These posttranslational modifications potentiate E2F1 apoptotic activity and direct its selective recruitment onto the P1p73 promoter (23). Preexisting and newly synthesized TAp73 is then phosphorylated by the nuclear tyrosine kinase cAbl (13), acetylated by p300 (6), and assembled with the WW domain protein YAP to activate the transcription of downstream apoptotic target genes (27). The importance of the E2F1/p73 pathway is reinforced by the strong reduction in p53-independent apoptosis when either PCAF, p73, or YAP expression is abrogated by specific small interfering RNAs (siRNAs) in cells exposed to DNA damage (23, 27).
Here we have investigated the mechanism of the hSirT1 regulation of different E2F1 apoptotic target genes, and we identified the functional interaction between PCAF and hSirT1 as an important determinant for E2F1-dependent p73-mediated apoptotic responses.
MATERIALS AND METHODS
Cell lines and DNA transfections.
U2Os and SaOs2 cells (osteosarcoma), Hep3B cells (HCC), HEK 293 cells, and mouse embryo C15 (E2F1−/−) and F6 fibroblasts (E2F1+/+) (a gift from L. Yamasaki) (23) were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Cell transfection with the indicated luciferase reporters, expression vectors, and 5 ng of the Renilla luciferase pRL null vector were performed using the Lipofectamine Plus reagent (Invitrogen). After 24 h, cells were either untreated or treated, as indicated, for an additional 24 h. Cell lysates were assayed for luciferase activity using the dual-luciferase assay system (Promega).
Antibodies, plasmids, siRNAs, and chemicals.
The following antibodies were used: anti-E2F1 (C20) (rabbit polyclonal immunoglobulin G [IgG]), anti-E2F1 (monoclonal antibody [MAb] KH95) (mouse monoclonal IgG2a), anti-SirT1 (C20) (goat polyclonal), antiactin (I19) (goat polyclonal IgG), and antihemagglutinin (anti-HA) (Y11) epitope (rabbit polyclonal IgG) from Santa Cruz Biotechnology, Inc.; anti-FLAG epitope (M2) (mouse monoclonal IgG1) from Sigma, Inc; anti-p73 MAb (mouse IgG) from Imgenex, Inc. (clone 1288); anti-active caspase-3 (rabbit polyclonal), anti-cleaved caspase 9 (rabbit polyclonal), and anti-cleaved poly(ADP-ribose) polymerase (PARP) (rabbit polyclonal) antibodies from Cell Signaling, Inc; anti-α-tubulin MAb (mouse monoclonal IgG1/k) from Neomarkers; anti-acetyl histone H4 (rabbit polyclonal), anti-HDAC1 (rabbit polyclonal), and anti-hSirT1 (mouse monoclonal IgG1) antibodies from Upstate Biotechnology, Inc.; anti-Myc epitope MAb (clone NE10) (mouse monoclonal IgG) from Invitrogen, Inc.; and anti-PCAF antibody (rabbit polyclonal), kindly provided by P. Nakatani (DFCI, Boston, MA).
HA-E2F1, HA-E2F3, HA-E2F4, FLAG-PCAF, and myc-SirT1 expression vectors and the Apaf-luc, DHFR-luc, and P1p73-luc reporter plasmids were previously described (2, 12, 23). Double-stranded Smart Pool siRNAs specific for either hSirT1 or PCAF and control siRNAs were purchased from Dharmacon Research Inc. and transfected using TransIT-TKO and TransIT-LT1 from Mirus, Inc.
Doxorubicin, nicotinamide (NAM), trichostatin (TSA), Valproate (VPA), resveratrol (RES), l-lactate, and pyruvate were all purchased from Sigma, Inc.
Immunoblotting and immunoprecipitations.
Cells were lysed in radioimmunoprecipitation assay buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium dodecyl sulfate [SDS], 0.1% deoxycholic acid, 140 mM NaCl, 1% Triton X-100, 1× protease inhibitor cocktail) for immunoblots and immunoprecipitations. NET buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Nonidet P-40, 1 mM EDTA [pH 8], 0.25% gelatin) was used for coimmunoprecipitation experiments. One milligram of cell extracts was immunoprecipitated overnight on a rocking platform at 4°C with the indicated antibodies (2 μg) and incubated with protein A or protein A/G Plus (Roche) (6) for 2 h at 4°C. The protein A/G-antigen-antibody complexes were washed three times with NET buffer, resuspended with LDL sample buffer (NuPAGE, Inc.) plus reducing agent (NuPAGE, Inc.), and heated at 70°C for 10 min. Samples were analyzed by electrophoresis with Tris-acetate or Bis-Tris minigels (NuPAGE, Inc.).
RT-PCR and qRT-PCR analysis.
Total cellular RNAs were extracted with TRIzol reagent (Gibco BRL), and 1 μg was reverse transcribed with the ThermoScript reverse transcription (RT)-PCR system (Invitrogen). cDNAs were PCR amplified using TAp73-, caspase 7-, and Bim-specific primers. PCR amplicons were collected at 25, 30, and 35 cycles and separated on 2% agarose gels. Real-time quantitative RT-PCR analysis (qRT-PCR) was performed using TaqMan DNA Master mix (Applied Biosystems). The following RT-PCR primers were used: TAp73 sense (S) primer 5′-TTGCTAGCATGGACGTCTTCCACCTGG-3′ and antisense (AS) primer 5′-GGCAAGCGTGCCTTCTAAGCGGCCGCAA-3′, Bim S primer 5′-ATGGCAAAGCAACCTTCTGA-3′ and AS primer 5′-TGTGGCTCTGTCTGTAGGGA-3′, and caspase 7 S primer 5′-AAGAGGACCATACAAATGCCG-3′ and AS primer 5′-TAGCCTGGAACCGTGGAATAGG-3′. Real-time qRT-PCR analysis for TAp73 expression was performed with the following primers: S primer 5′-TCTGGAGCTCTCTGGAACCA-3′, AS primer 5′-TCCATGGTGCTGCTCAGC-3′, and probe 5′-TCATGGCCCAGTTCAAT-3′.
ChIP assay.
Chromatin immunoprecipitation (ChIP) experiments were carried out as previously described (23). In sequential ChIP experiments, endogenous PCAF- or hSirT1-chromatin complexes from U2Os cells were immunoprecipitated using the corresponding antibodies, eluted by incubation for 30 min at 37°C in 10 mM dithiothreitol, diluted in re-ChIP buffer (1% Triton X, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.0]), and subjected to the second ChIP procedure.
The following ChIP oligonucleotides were used: P1p73 promoter S primer 5′-TGAGCCATGAAGATGTGCGAG-3′ and AS primer 5′-GCTGCTTATGGTCTGATGCTTATG-3′, TP73 control S primer 5′-AAGCAGCCCATCAAGGAGGAGTTC-3′ and AS primer 5′-GCAGTTTTGGACACACAGGAAGG-3′, Bim promoter S primer 5′-GCTGCTAAGGCTTGTGTCCGGA-3′ and AS primer 5′-TGCCCGCGTTCCCAATTGGT-3′, Bim control S primer 5′-TGACGCACTTACTACGACTGACGG-3′ and AS primer 5′-TTGCCCAGGACAGACTTCTTCG-3′, caspase 7 promoter (human) S primer 5′-TTTGGGCACTTGGAGCGCG-3′ and AS primer 5′-AAGAGCCCAAAGCGACCCGT-3′, and caspase 7 control S primer5′-TCCGTTTGTAGCAAGCAAGAGAC-3′ and AS primer 5′-CGGCGTCAGTGTCGGGAGTAAATA-3′.
Acetylation and deacetylation reactions.
FLAG-PCAF was purified from baculovirus-infected Sf9 cells as previously described (12). Purified active bacterial glutathione S-transferase (GST)-tagged human SirT1 (amino acids 193 to 741) and purified histones were purchased from Upstate Inc. Purified bovine serum albumin (BSA) was purchased from Sigma, Inc. PCAF autoacetylation and PCAF-mediated histone acetylation were performed, as described previously (12), prior to inactivating PCAF histone acetyltransferase activity by incubating the acetylation reaction mixture for 30 min at 55°C. Acetylated PCAF, acetylated BSA, and acetylated histones were used as substrates for GST-hSirT1 in the deacetylation reaction (1 h at 30°C in 50 mM Tris-HCl [pH 8.0], 10% glycerol, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 10 mM Na-butyrate with or without 0.5 mM NAD+).
Biochemical evaluation of redox state.
hSirT1 enzymatic activity depends on NAD+ availability, and it is readily influenced by the [NAD+]/[NADH] ratio (12, 15). An accurate and direct measure of the free cytoplasmic/nuclear levels of NAD+ is technically unattainable (12). Since the free cytoplasmic [NAD+]/[NADH] ratio approximates the free nuclear ratio (33), we determined the values for cellular lactate and pyruvate as an indirect measurement of the free cytoplasmic/nuclear [NAD+]/[NADH] ratio. The [NAD+]/[NADH] ratio was calculated from the equilibrium constant of the lactate dehydrogenase (LDH) (KLDH) reaction as follows:
 |
where KLDH = 1.11 × 10 to 1.11 × 11 M and the pH was taken to be 7.2 in neutralized perchloric acid extracts of cell homogenates from untreated and doxorubicin-treated cells (12, 29). The rapid cessation of metabolic activity of cell cultures was accomplished by the addition of concentrated perchloric acid to a final concentration of about 1 M. Cells were scraped, transferred into a centrifuge tube, and frozen at −80°C until assay. After thawing, cell extracts were centrifuged, and supernatant aliquots were partially neutralized to pH 5 to 6 by the addition of potassium bicarbonate solution. Lactate and pyruvate concentrations were determined enzymatically by monitoring the appearance and disappearance, respectively, of NADH spectrophotometrically at 340 nm. For lactate determinations, the following concentrations were used in a reaction mixture volume of 100 μl before enzyme addition: 100 mM [AMP] (pH 9.9), 45 mM [glutamate] (pH 9.9), and 2.5 mM [NAD]. GPT and LDH were combined and spun, the ammonium sulfate was removed, and the pellet was dissolved in water (5 μl added 1.6 U and 0.625 U, respectively). For pyruvate determinations, the concentrations were as follows: 100 mM [imidazole] (pH 7.0) and 0.15 mM [NADH]. LDH was added as described above, but GPT was omitted. After reagents, extracts, and water were added, a 5-min kinetic read was taken to establish a baseline reading. After enzyme addition, a second kinetic read was taken to monitor the assay until completion. The lactate assay result was read for 20 min and the pyruvate assay result was read for 10 min at 20- and 10-s intervals, respectively. In each kinetic read, the final seven data points were averaged, and the difference in corrected absorbance (ΔA) was used to calculate the [lactate] and [pyruvate] in the neutralized extracts. The use of these concentrations along with the equilibrium constant for LDH allowed for the estimation of the cytosolic [NAD]/[NADH] ratio.
RESULTS AND DISCUSSION
hSirT1 regulates the E2F1 apoptotic target gene TP73.
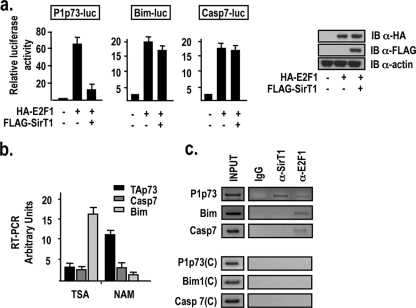
Expression profiling microarrays and ChIP-chip experiments have led to the identification of a large repertoire of genes activated at the transcriptional level by E2F1, which includes genes involved in DNA replication, cell proliferation, DNA repair, differentiation, development, and apoptosis (1, 24). The repertoire of E2F1 apoptotic target genes comprises the caspase 7, Bim, and p73 genes (16, 21, 22, 23, 24). Since hSirT1 has been shown to inhibit E2F1-dependent transcription and E2F1-dependent apoptosis (30), we aimed to compare the abilities of hSirT1 to repress the E2F1-dependent activation of different apoptotic E2F1 target genes. We found that exogenously expressed hSirT1 represses the E2F1-dependent activation of the P1p73 promoter in U2Os cells but that it has no effect on the ability of E2F1 to activate the Bim and the caspase 7 promoters (Fig. 1a). Similar results have been obtained using the osteosarcoma cell line SaOs2 and the hepatocytic cell line Hep3B (data not shown). Exogenously expressed hSirt1 does not affect E2F1 protein levels in the cotransfection experiments (Fig. 1a, right). The ability of hSirT1 to differentially affect E2F1 target genes was further supported by the ChIP analysis of E2F1 and hSirT1 recruitment onto the target promoters and the RT-PCR analysis of TAp73, caspase 7, and Bim transcripts in SaOs2 cells treated with the deacetylase inhibitors NAM and TSA. TAp73 expression was induced by NAM, a noncompetitive Nad+ antagonist that inhibits class III deacetylases, whereas Bim is strongly activated in cells treated by TSA, a potent inhibitor of class I and class II HDACs, and it is not significantly induced by NAM (Fig. 1b). Caspase 7 expression was only marginally induced by NAM and TSA. Furthermore, E2F1 is recruited onto the P1p73, Bim, and caspase 7 promoters in asynchronously growing U2Os cells, whereas hSirT1 is present on the P1p73 promoter but not on the promoters of the Bim and caspase 7 genes (Fig. 1c, top). The specificity of hSirT1 binding to the P1p73 promoter was confirmed by control reactions performed using distant primers that amplified total chromatin but did not amplify ChIP products with significant efficiency (Fig. 1c, bottom). These results identify p73 as a specific downstream target of the hSirT1-E2F1 pathway.
FIG. 1.
Selective modulation of E2F1 apoptotic target genes by hSirT1. (a) U2Os cells were transfected with the indicated reporter constructs and either the E2F1 or the hSirT1 expression vector. Luciferase activity, expressed as induction over the control, was normalized for transfection efficiency using the dual-luciferase assay system. Histograms show the means of data for three experiments, each performed in quadruplicate; bars indicate standard deviations. (Right) Exogenously expressed E2F1 and hSirt1 are detected by anti-HA (α-HA) and anti-FLAG immunoblotting (IB), respectively. (b) RNAs from SaOs2 cells exposed for 24 h to TSA (100 nm) or NAM (25 mM) were analyzed with primers specific for TAp73, caspase 7, and Bim transcripts by PCR. Results are expressed as arbitrary units compared to the normalized basal level of expression of each gene in untreated cells. (c, top) Chromatin from asynchronously growing U2Os cells was immunoprecipitated with either the relevant control IgG or the indicated antibodies and analyzed with primers amplifying the region containing E2F sites in P1p73, Bim, and caspase 7 by PCR. (Bottom) Control reactions using distant primers (C) did not amplify anti-E2F1 and hSirT1 ChIP products.
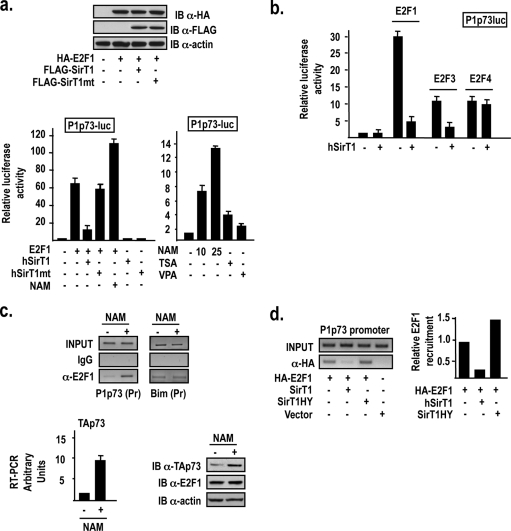
hSirT1 inhibition of the E2F1-driven P1p73 promoter activity requires hSirT1 enzymatic activity. Indeed, the deacetylase-deficient hSirT1 H355Y point mutant is significantly impaired in its ability to repress E2F1-dependent P1p73 activation (Fig. 2a, left). Conversely, the inhibition of hSirT1 activity by NAM treatment results in a dose-dependent activation of the P1p73 promoter (Fig. 2a, right). Interestingly, hSirT1 also inhibits the ability of E2F3 to activate the P1p73 promoter, but it has no effect on E2F4-dependent P1p73 activation (Fig. 2b).
FIG. 2.
hSirT1 modulates E2F1 recruitment and transcriptional activity on the P1p73 promoter. (a) U2Os cells were transfected with the P1p73 reporter and the indicated expression vectors and exposed for 24 h to NAM (10 to 25 mM), TSA (100 nm), and VPA (10 mM). (Top) Representative anti-HA (α-HA) and anti-FLAG immunoblot (IB) of exogenously expressed E2F1 and hSirt1/mthSirt1. (b) U2Os cells were transfected with the P1p73 reporter and the indicated expression vectors or exposed for 24 h to NAM (10 mM). (c, top left) Chromatin from untreated and NAM (10 mM)-treated U2Os cells was immunoprecipitated with either the relevant control IgG or anti-E2F1 antibodies and analyzed with P1p73 primers by PCR. Control reactions using distant primers (C) did not amplify anti-E2F1 ChIP products from either untreated or NAM-treated cells (data not shown). (Top right) Chromatin from untreated and NAM (10 mM)-treated U2Os cells was immunoprecipitated with either the relevant control IgG or anti-E2F1 antibodies and analyzed with the Bim promoter primers by PCR. Control reactions using distant primers (C) did not amplify anti-E2F1 ChIP products from either untreated or NAM-treated cells (data not shown). (Bottom left) TAp73 transcripts from untreated and NAM-treated U2Os cells were quantitated by qRT-PCR as described in the legend to Fig. 1B. (Bottom right) TAp73 and E2F1 protein levels detected by immunoblotting using specific antibodies. (d, left) Modulation of E2F1 binding to the P1p73 promoter by the hSirT1 deacetylase activity. HA-E2F1, wild-type hSirT1, and deacetylase-defective hSirT1-H355Y were exogenously expressed in U2Os cells and immunoprecipitated in a ChIP assay using a polyclonal anti-HA antibody. (Right) Densitometric quantification with Image J software.
Next, we investigated the mechanism by which hSirT1 might inhibit the E2F1-dependent expression of p73. hSirT1 deacetylates E2F1 both in vitro and in vivo (30), and acetylation was previously shown by us and by others to increase the ability of E2F1 to bind in vitro and to be recruited in vivo onto the apoptotic target promoters P1p73 and Bim (23, 35). We found that the inhibition of hSirT1 in U2Os cells treated with NAM results in a strong increase in levels of E2F1 recruitment onto the P1p73 promoter (Fig. 2c, top), in an increase in levels of TAp73 transcripts (Fig. 2c, left), and in an accumulation of the TAp73 protein (Fig. 2c, right) without affecting E2F1 protein levels (Fig. 2c, right). Interestingly, NAM does not modulate E2F1 recruitment onto the Bim promoter (Fig. 2c, top), whereas in TSA-treated cells, E2F1 is readily recruited onto the Bim promoter (35; data not shown). Accordingly, the induction of TAp73 transcription in response to NAM is blunted in the E2F1−/− C15 fibroblasts compared to their F6 wild-type counterparts (Fig. 3c, left). The role of hSirT1 deacetylase activity in the regulation of E2F1 binding to the P1p73 promoter was further confirmed by the observation that exogenously expressed wild-type hSirT1 reduced the binding of coexpressed E2F1 to the P1p73 promoter, whereas the coexpression of hSirT1HY, a deacetylase mutant that also acts as a dominant negative for endogenous hSirT1, resulted in an increased level of E2F1 binding (Fig. 2d). Altogether, these results indicate that hSirT1 differentially modulates E2F1 recruitment and transcriptional activity on its target genes and strongly suggest that different deacetylases are at work to modulate E2F1 binding to different apoptotic target promoters.
FIG. 3.
hSirT1 regulates P1p73 activity in cells exposed to DNA damage. (a) U2Os cells transfected with the P1p73 reporter and treated for 24 h with doxorubicin (DOXO) (2 μM) were either cotransfected with control and hSirT1-specific siRNAs (left) or the hSirT1 expression vector (right) or treated with RES (30 μM), NAM (10 mM), TSA (100 nm), and VPA (10 mM) (right). (b, left) RNAs from U2Os cells exposed for 24 h to doxorubicin (2 μM), NAM (25 mM), or the combination of doxorubicin and NAM were analyzed by RT-PCR as described in the legend of Fig. 1b. (Top right) Representative immunoblotting analysis of TAp73 and E2F1 protein levels in untreated and doxorubicin-, nicotinamide-, and doxorubicin-plus-nicotinamide (D/N)-treated cells. (Bottom right) Immunoblotting (IB) densitometric quantification using Image J software with data from three independent experiments. (c, left) RNAs from F6 (E2F1+/+) and C15 (E2F1−/−) cells exposed for 24 h to doxorubicin (2 mM), NAM (25 mM), or the combination of doxorubicin and NAM were analyzed by RT-PCR as described in the legend to Fig. 1b. (Right) TAp73 protein levels in untreated and doxorubicin-, nicotinamide-, and doxorubicin-plus-nicotinamide-treated F6 (E2F1+/+) and C15 (E2F1−/−) cells.
The E2F1-TAp73 apoptotic pathway is regulated by hSirT1 after DNA damage.
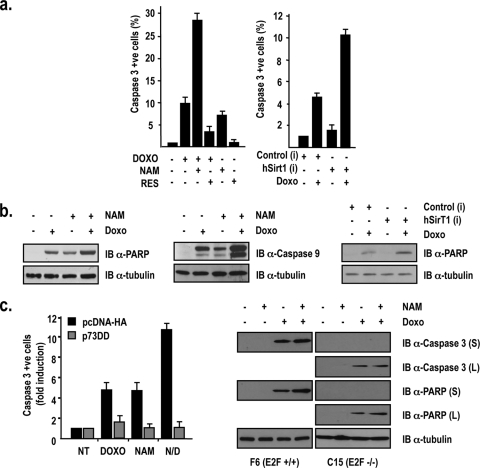
The TP73 gene is both a cell cycle-regulated gene activated by E2F1 during the S phase of the cell cycle (11, 16) and a prominent E2F1 apoptotic target gene in the DNA damage response (17, 23). As shown in Fig. 3a, both the abrogation of hSirT1 expression by specific siRNAs (left) and the inhibition of hSirT1 catalytic activity by NAM (right) potentiate doxorubicin-induced P1p73-driven transcription. Neither TSA nor VPA treatment significantly affected doxorubicin-dependent activation (Fig. 3a, right). Conversely, both hSirT1 overexpression and treatment with the specific hSirT1 activator RES almost completely abolished doxorubicin-induced P1p73 activity (Fig. 3a, right). RT-PCR analysis of TAp73, caspase 7, and Bim transcripts in SaOs2 cells treated with either doxorubicin or NAM (Fig. 3b) showed that TAp73 expression was induced by both doxorubicin (23) and NAM and further potentiated by the combined treatment of doxorubicin plus NAM. Importantly, both TAp73 transcription (Fig. 3c, left) and TAp73 protein accumulation (Fig. 3c, right) were almost abolished in C15 mouse fibroblasts lacking E2F1 exposed to NAM, doxorubicin, or their combination. Altogether, these results indicate that hSirT1 and E2F1 regulate TAp73 expression both in untreated cells and in response to DNA damage.
Next, we evaluated the impact of hSirT1 modulation on DNA damage-induced p53-independent apoptosis. In SaOs2 cells exposed to doxorubicin, apoptosis is mediated by the activation of the E2F1/p73 pathway (6, 23), and hSirT1 inhibition by NAM results in significant increases in the numbers of cleaved-caspase 3-positive cells (Fig. 4a, left), in levels of PARP cleavage (Fig. 4b, left), and in levels of caspase 9 accumulation (Fig. 4b, middle). p53 null Hep3B hepatocellular carcinoma cells are more resistant to DNA-damaging drugs (Fig. 4a, right) and display high levels of hSirT1 transcripts and protein (data not shown). The abrogation of SirT1 expression by specific siRNAs potentiated doxorubicin-induced apoptosis, assessed as both the number of cleaved-caspase 3-positive cells (Fig. 4a, right) and levels of PARP cleavage (Fig. 4b, right). The inhibition of p73 function by exogenously expressed p73DD, a p73-specific dominant negative (17, 23), abolished Hep3B apoptosis in response to doxorubicin, NAM, and their combination, indicating that the effect of hSirT1 on Hep3B cell death is mediated by p73 (Fig. 4c, left). As shown in Fig. 4c (right), E2F1+/+ mouse embryonic fibroblasts display a significantly stronger apoptotic response to doxorubicin treatment than do E2F1−/− mouse embryonic fibroblasts. Although the contribution of additional proapoptotic mechanisms cannot be excluded, these results reinforce the role of hSirt1, E2F1, and TAp73 acting as a pathway in mediating doxorubicin-induced apoptosis in p53 null cell environments.
FIG. 4.
hSirT1 regulates p53-independent apoptosis in response to DNA damage. (a) Inhibition of hSirT1 activity potentiates doxorubicin-induced apoptosis in p53 null cells. (Left) SaOs2 cells were exposed for 24 h to doxorubicin (DOXO) (2 μM), NAM (10 mM), and RES (30 μM). (Right) Hep3B cells transfected with control and hSirT1-specific siRNAs were exposed for 24 h to doxorubicin (2 μM). Active caspase 3-positive (apoptotic) cells were identified by indirect immunofluorescence and counted. Results are expressed as induction compared to basal caspase 3 activity measured in untreated cells. (b, left and middle) Whole-cell extracts from untreated and doxorubicin (2 μM)-, NAM (10 mM)-, and doxorubicin-plus-NAM-treated SaOs2 cells were immunoblotted (IB) with anti-cleaved PARP (left) and anti-cleaved caspase 9 (middle). (Right) Hep3B cells were treated as described above (a), and whole-cell extracts were immunoblotted with anti-cleaved PARP. (c) p73 mediates hSirT1 inhibition of p53-independent apoptosis in Hep3B cells exposed to DNA damage. p53 null Hep3B cells transfected with either control PCDNA3-HA vector or the pCDNA3-p73DD-HA expression vector were treated for 24 h with doxorubicin (2 mM), NAM (10 mM), and the combination of NAM plus Doxo (N/D). Active caspase 3-positive (apoptotic) cells were identified by indirect immunofluorescence and counted. Results are expressed as induction compared to that of untreated control and p73DD-transfected cells. The efficiencies of transfection ranged between 75 and 80% among experiments. The numbers of apoptotic cells were consistently between 30 and 40% apoptotic cells in doxorubicin- or NAM-treated cultures and >80% in cells treated with the combination of NAM and doxorubicin. NT, not treated. (Right) Whole extracts from F6 (E2F1+/+) and C15 (E2F1−/−) cells exposed for 24 h to doxorubicin (2 μM), NAM (25 mM), or the combination of doxorubicin and NAM were immunoblotted with anti-cleaved caspase 3 and anti-cleaved PARP. In the case of C15 (E2F1−/−) cells, data for both a short ECL exposure (2 min) and a longer exposure (2 h) are provided.
hSirT1, PCAF, and E2F1 are corecruited onto the P1p73 promoter.
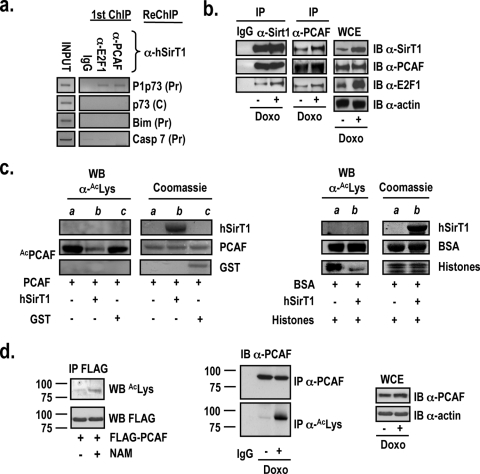
In cells exposed to DNA-damaging agents, E2F1 is phosphorylated by Chk2 (26) and acetylated by PCAF (23). As a result, E2F1 accumulates, and it is selectively recruited, together with PCAF, onto the P1p73 promoter to elevate TAp73 protein levels in the cell and to induce apoptosis (23). Since hSirT1 deacetylates E2F1 (30), binds to the P1p73 promoter (Fig. 1c), and modulates E2F1 recruitment onto the P1p73 promoter (Fig. 2c), we tested whether hSirT1 colocalizes onto the P1p73 promoter in vivo with PCAF and E2F1. Sequential ChIP assays (E2F1 and PCAF ChIPs followed by hSirT1 ChIP) confirmed that hSirT1, PCAF, and E2F1 are corecruited onto the P1p73 promoter in vivo (Fig. 5a) and that endogenous hSirT1, PCAF, and E2F1 are coprecipitated with both anti-hSirT1 and anti-PCAF antibodies (Fig. 5b). The treatment of cell extracts with ethidium bromide in order to disrupt potential DNA-protein interactions did not affect hSirT1, PCAF, and E2F1 coimmunoprecipitations, thus suggesting that their interaction, which is likely to occur at the chromatin level (Fig. 5a), does not necessarily require DNA (data not shown). Altogether, these results indicate that E2F1, PCAF, and SirT1 do interact in vivo and strongly support the notion that E2F1-PCAF-SirT1 complexes can be corecruited on E2F1-responsive promoters in vivo, thus providing the basis for their functional interactions in the context of chromatin.
FIG. 5.
hSirT1, PCAF, and E2F1 interact and are corecruited onto the P1p73 promoter. (a) Chromatin from asynchronously growing U2Os cells was analyzed by sequential ChIP using either anti-E2F1 or anti-PCAF (first immunoprecipitation) and anti-hSirT1 (second immunoprecipitation). IgG, control immunoprecipitation with the relevant control IgGs. ChIP DNA was amplified with primers specific for the P1p73, Bim, and caspase 7 promoter regions containing E2F1 sites (Pr) and with the corresponding control distant oligonucleotides (C) (data not shown). (b) Extracts from asynchronously growing or doxorubicin (Doxo) (2 μM)-treated U2Os cells were immunoprecipitated (IP) with control IgG, anti-hSirT1, and anti-PCAF antibodies and immunoblotted (IB) with anti-hSirT1 (α-SirT1), anti-PCAF, and anti-E2F1 antibodies. (c) hSirT1 deacetylates PCAF in vitro. (Left) Baculovirus affinity-purified full-length FLAG-PCAF (kindly provided by V. Sartorelli, NIAMS, NIH) and purified active bacterial GST and GST-tagged human SirT1 (amino acids 193 to 741) were employed in the acetylation (lane a) and deacetylation (lanes b and c) reactions in vitro. Reaction products were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with the anti-acetyl-Lys antibody. Lane a shows in vitro PCAF autoacetylation. (Right) Acetylated BSA and acetylated histones (lane a) were incubated with GST-tagged human SirT1 (amino acids 193 to 741) (Upstate Inc.) in the deacetylation reaction (see b). Coomassie-stained gels show the input proteins used in the reactions. WB, Western blot. (d) hSirT1 regulates PCAF acetylation in vivo. (Left) 293 cells were transfected with the FLAG-PCAF vector and exposed to NAM (10 mM). Extracts were immunoprecipitated with the M2 FLAG antibody and immunoblotted with anti-acetyl-Lys antibody. (Middle) PCAF acetylation increases in response to DNA damage. Extracts from untreated and doxorubicin (2 μM)-treated U2Os cells were immunoprecipitated with either anti-PCAF or anti-acetyl-Lys antibody and immunoblotted with the anti-PCAF antibody. (Right) Total PCAF protein levels were analyzed by immunoblotting in whole-cell extracts (WCE) from untreated and doxorubicin-treated cells. Molecular weight positions (in thousands) in the SDS-polyacrylamide gels are shown.
hSirT1 modulates PCAF acetylation in vivo.
The observation that both PCAF (Fig. 5a) and hSirT1 (Fig. 1c) are recruited onto the P1p73 promoter in vivo in asynchronously proliferating cells suggests that PCAF might also be a target of hSirT1 regulation on the P1p73 promoter. Indeed, in undifferentiated myoblasts, PCAF forms complexes with the myogenic transcription factor MyoD that are inactive due to the interaction with Sir2, and Sir2 inhibitors promote the acetylation of promoter-bound MyoD, myogenic transcription, and differentiation (12). We found that hSirT1, in addition to significantly decreasing baculovirus PCAF autoacetylation and PCAF-dependent H3/H4 histone acetylation in vitro (Fig. 5c), influences PCAF activity in vivo by modulating its acetylation. PCAF is both autoacetylated and a target for p300, but not for CBP, acetylation in vivo (25), and autoacetylation is required to stabilize PCAF histone acetyltransferase activity in vitro (14). We showed that both NAM treatment (Fig. 5d, left) and the exposure to apoptotic (2 μM) dosages of doxorubicin (Fig. 5d, middle and right) increase levels of PCAF acetylation in vivo. These results identify PCAF as a target of hSirT1 in the regulation of TAp73 expression and further implicate hSirT1 as an important determinant for E2F1-dependent p73-mediated apoptotic responses.
DNA damage modulates chromatin-bound hSirT1 deacetylase activity on the P1p73 promoter.
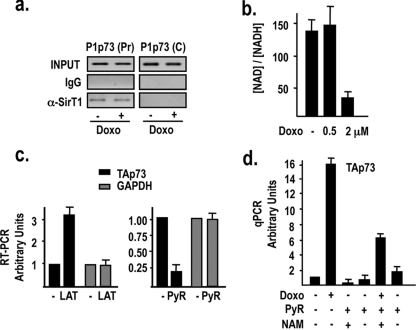
Since hSirT1 binds to the P1p73 promoter in proliferating cells (Fig. 1c) and regulates E2F1 binding to the promoter (Fig. 2c and d), we postulated that under apoptotic conditions, hSirT1 would be released to favor the recruitment of transcriptionally active complexes containing E2F1 and PCAF. Unexpectedly, we found that hSirT1 is not released from the P1p73 promoter in vivo in doxorubicin-treated cells (Fig. 6a). We then investigated how the repression of the P1p73 promoter exerted by chromatin-bound hSirT1 might be relieved in response to DNA damage. hSirT1 enzymatic activity depends on NAD+ availability, and it is readily influenced by the [NAD+]/[NADH] ratio. As an indirect measurement of the free [NAD+]/[NADH] ratio, we determined lactate and pyruvate levels in untreated and doxorubicin-treated cells. As shown in Fig. 6b, the NAD+/NADH+ ratio sharply decreases in cells exposed to apoptotic doses of doxorubicin, but it is not affected when lower nonapoptotic dosages of doxorubicin, which are unable to activate P1p73 transcription (23), are used. We next modulated the [NAD+]/[NADH] ratio by culturing cells in the presence of either l-lactate (to decrease the [NAD+]/[NADH] ratio) or pyruvate (to increase the [NAD+]/[NADH] ratio) and evaluated TAp73 expression. As shown in Fig. 6c, TAp73 mRNA levels were increased by increasing the concentration of l-lactate (left), and the opposite was observed when cells were treated with pyruvate (right). Furthermore, the reactivation of hSirT1 deacetylase activity by pyruvate completely abolishes the DNA damage-induced activation of the endogenous P1p73 promoter and TAp73 expression (Fig. 6d), and NAM counteracts, at least in part, the inhibitory effect of pyruvate on the doxorubicin-induced activation of TA-p73 transcription (Fig. 6d). Altogether, these results link the modulation of chromatin-bound hSirT1 deacetylase activity by the intracellular redox state with the transcriptional activity of the P1p73 promoter and support a model in which the release of hSirT1 repression on the P1p73 promoter is mediated, in the context of the DNA damage response, by a modulation of promoter-bound hSirT1 deacetylase activity rather than by a “protein complex shifting.”
FIG. 6.
DNA damage modulates chromatin-bound hSirT1 deacetylase activity on the P1p73 promoter. (a) hSirT1 occupancy of the P1p73 promoter is unaffected by DNA damage. Chromatin from untreated and doxorubicin (2 μM)-treated U2Os cells was immunoprecipitated with either relevant control IgGs or anti-hSirT1 (α-SirT1) antibody and analyzed by P1p73 (Pr) and C PCR primers. (b) DNA damage modulates hSirT1 enzymatic activity by reducing [NAD+] levels. Free [NAD+] was calculated by measurements of [lactate] and [pyruvate] (PyR) (data not shown) in extracts from asynchronously growing U2Os cells and from U2Os cells exposed to a low nonapoptotic (0.5 μM) or apoptotic (2 μM) dosage of doxorubicin (Doxo) for 18 h. (c) Modulation of the [NAD+]/[NADH] ratio affects TAp73 expression. Cells were cultured either in the absence or in the presence of l-lactate (10 mM) or pyruvate (30 mM) for 24 h, and both TAp73 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA levels were analyzed by RT-PCR as described in the legend of Fig. 1b. (d) Reactivation of hSirT1 deacetylase activity by pyruvate treatment abolishes the DNA damage-induced activation of the endogenous P1p73 promoter. Untreated and doxorubicin (2 μM)-treated cells were cultured either in the absence or in the presence of pyruvate (30 mM) and the combination of pyruvate and NAM (10 mM) for 24 h, and TAp73 mRNA levels were analyzed by real-time quantitative PCR (qPCR).
Our results identify the functional interaction between PCAF and hSirT1 as being an important determinant for E2F1-dependent p73-mediated apoptotic responses. We show that hSirT1, PCAF, and E2F1 coimmunoprecipitate and are corecruited in vivo onto the P1p73 promoter. Moreover, hSirT1 deacetylates PCAF in vitro and modulates PCAF autoacetylation in vivo. Both autoacetylation and p300-mediated acetylation of PCAF were previously shown to increase its acetyltransferase activity (25). The ability of Sir2 to modulate PCAF acetyltransferase activity was also previously established (12). Alternative mechanisms may mediate the repressive activity of hSirt1, including E2F1 deacetylation or changes in the global level of acetylation of core histone tails, and we cannot exclude their involvement in P1p73 regulation in response to DNA damage. Indeed, E2F1 transcriptional activity is regulated by its acetylation (23), and it is known that Sirt1 controls E2F1 acetylation in response to DNA damage (30). However, we clearly show that the modulation of hSirt1 activity by NAM results in an increased level of PCAF acetylation and that the level of PCAF acetylation also increases in response to doxorubicin treatment. Thus, both our results and previously published evidence suggest that one mechanism by which hSirT1 controls the activity of the P1p73 target promoter involves PCAF. We also found that under apoptotic conditions in response to DNA damage, the amount of hSirT1 bound to the P1p73 promoter does not change, but intracellular NAD+ levels decrease, and the enzymatic activity of hSirT1 is inhibited, thus allowing PCAF activation and E2F1 acetylation, leading to a further recruitment of E2F1/PCAF transcriptionally active complexes. It is tempting to speculate that the persisting binding of hSirT1 to the P1p73 promoter may serve as a failsafe mechanism to limit and/or delay p73 transcription and apoptosis before the final decision on cell fate is made.
Although it was widely reported that hSirT1 downregulates the stress-induced activation of p53 (18, 19, 28) and the p53-dependent DNA damage responses (3, 34), the knockdown of hSirT1 increases sensitivity to DNA-damaging drugs in both wild-type and null p53 cells (30; this paper). Moreover, it was recently shown that apoptosis in response to hSirT1 silencing in epithelial transformed cells has a delayed kinetics and that it is independent of p53 (10). These results and the demonstration that hSirT1 controls the E2F1/p73 apoptotic pathway identify hSirT1 and PCAF as being potential targets to modulate tumor cell survival and chemoresistance irrespective of p53 status.
Acknowledgments
This work was supported by grants from the AIRC and MIUR-FIRB to M.L. and M.F., the European Community (LSHC-CT-2004-503576) to M.L., MIUR-Cofin to M.L., and Fondazione Cenci Bolognetti, Istituto Pasteur, and Schering-Plough to M.L. N.P. has been supported by a fellowship from the Fondazione Adziano Buzzati Traverso. L.B., C.S., N.P., and S.V. are supported by fellowships from the Fondazione A. Cesalpino.
Footnotes
Published ahead of print on 2 February 2009.
REFERENCES
- 1.Bieda, M., X. Xu, M. A. Singer, R. Green, and P. J. Farnham. 2006. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 3032011-2015. [DOI] [PubMed] [Google Scholar]
- 3.Chen, W. Y., D. H. Wang, R. C. Yen, J. Luo, W. Gu, and S. B. Baylin. 2005. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123437-448. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, H. L., R. Mostoslavsky, S. Saito, J. P. Manis, Y. Gu, P. Patel, R. Bronson, E. Appella, F. W. Alt, and K. F. Chua. 2003. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 10010794-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, H. Y., C. Miller, K. J. Bitterman, N. R. Wall, B. Hekking, B. Kessler, K. T. Howitz, M. Gorospe, R. de Cabo, and D. A. Sinclair. 2004. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305390-392. [DOI] [PubMed] [Google Scholar]
- 6.Costanzo, A., P. Merlo, N. Pediconi, M. Fulco, V. Sartorelli, P. A. Cole, G. Fontemaggi, M. Fanciulli, L. Schiltz, G. Blandino, C. Balsano, and M. Levrero. 2002. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell 9175-186. [DOI] [PubMed] [Google Scholar]
- 7.DeGregori, J., and D. G. Johnson. 2006. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 6739-748. [DOI] [PubMed] [Google Scholar]
- 8.Flores, E. R., S. Sengupta, J. B. Miller, J. J. Newman, R. Bronson, D. Crowley, A. Yang, F. McKeon, and T. Jacks. 2005. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7363-373. [DOI] [PubMed] [Google Scholar]
- 9.Flores, E. R., K. Y. Tsai, D. Crowley, S. Sengupta, A. Yang, F. McKeon, and T. Jacks. 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416560-564 [DOI] [PubMed] [Google Scholar]
- 10.Ford, J., M. Jiang, and J. Milner. 2005. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 6510457-10463. [DOI] [PubMed] [Google Scholar]
- 11.Fulco, M., A. Costanzo, P. Merlo, R. Mangiacasale, S. Strano, G. Blandino, C. Balsano, P. Lavia, and M. Levrero. 2003. p73 is regulated by phosphorylation at the G2/M transition. J. Biol. Chem. 27849196-49202. [DOI] [PubMed] [Google Scholar]
- 12.Fulco, M., R. L. Schiltz, S. Iezzi, M. T. King, P. Zhao, Y. Kashiwaya, E. Hoffman, R. L. Veech, and V. Sartorelli. 2003. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 1251-62. [DOI] [PubMed] [Google Scholar]
- 13.Gong, J. G., A. Costanzo, H. Q. Yang, G. Melino, W. G. Kaelin, Jr., M. Levrero, and J. Y. Wang. 1999. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399806-809. [DOI] [PubMed] [Google Scholar]
- 14.Herrera, J. E., K. Sakaguchi, M. Bergel, L. Trieschmann, Y. Nakatani, and M. Bustin. 1999. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol. Cell. Biol. 193466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403795-800. [DOI] [PubMed] [Google Scholar]
- 16.Irwin, M., M. C. Marin, A. C. Phillips, R. S. Seelan, D. I. Smith, W. Liu, E. R. Flores, K. Y. Tsai, T. Jacks, K. H. Vousden, and W. G. Kaelin, Jr. 2000. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407645-648. [DOI] [PubMed] [Google Scholar]
- 17.Irwin, M. S., K. Kondo, M. C. Marin, L. S. Cheng, W. C. Hahn, and W. G. Kaelin, Jr. 2003. Chemosensitivity linked to p73 function. Cancer Cell 3403-410. [DOI] [PubMed] [Google Scholar]
- 18.Langley, E., M. Pearson, M. Faretta, U. M. Bauer, R. A. Frye, S. Minucci, P. G. Pelicci, and T. Kouzarides. 2002. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 212383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107137-148. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita, N., Y. Takami, M. Kimura, S. Tachiiri, M. Ishiai, T. Nakayama, and M. Takata. 2005. Role of NAD-dependent deacetylases SIRT1 and SIRT2 in radiation and cisplatin-induced cell death in vertebrate cells. Genes Cells 10321-332. [DOI] [PubMed] [Google Scholar]
- 21.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nahle, Z., J. Polakoff, R. V. Davuluri, M. E. McCurrach, M. D. Jacobson, M. Narita, M. Q. Zhang, Y. Lazebnik, D. Bar-Sagi, and S. W. Lowe. 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4859-864. [DOI] [PubMed] [Google Scholar]
- 23.Pediconi, N., A. Ianari, A. Costanzo, L. Belloni, R. Gallo, L. Cimino, A. Porcellini, I. Screpanti, C. Balsano, E. Alesse, A. Gulino, and M. Levrero. 2003. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 5552-558. [DOI] [PubMed] [Google Scholar]
- 24.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos-Rosa, H., E. Valls, T. Kouzarides, and M. Martinez-Balbas. 2003. Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res. 314285-4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens, C., L. Smith, and N. B. La Thangue. 2003. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5401-409. [DOI] [PubMed] [Google Scholar]
- 27.Strano, S., O. Monti, N. Pediconi, A. Baccarini, G. Fontemaggi, E. Lapi, F. Mantovani, A. Damalas, G. Citro, A. Sacchi, G. Del Sal, M. Levrero, and G. Blandino. 2005. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA damage. Mol. Cell 18447-459. [DOI] [PubMed] [Google Scholar]
- 28.Vaziri, H., S. K. Dessain, E. N. Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2 (SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107149-159. [DOI] [PubMed] [Google Scholar]
- 29.Veech, R. L., L. V. Eggleston, and H. A. Krebs. 1969. The redox state of free nicotinamide-adenine dinucleotide phosphate in the cytoplasm of rat liver. Biochem. J. 115609-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, C., L. Chen, X. Hou, Z. Li, N. Kabra, Y. Ma, S. Nemoto, T. Finkel, W. Gu, W. D. Cress, and J. Chen. 2006. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 81025-1031. [DOI] [PubMed] [Google Scholar]
- 31.Yeung, F., J. E. Hoberg, C. S. Ramsey, M. D. Keller, D. R. Jones, R. A. Frye, and M. W. Mayo. 2004. Modulation of NF-kappa-B-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 232369-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan, Z., X. Zhang, N. Sengupta, W. S. Lane, and E. Seto. 2007. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol. Cell 27149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Q., D. W. Piston, and R. H. Goodman. 2002. Regulation of repressor function by nuclear NADH. Science 2951895-1897. [DOI] [PubMed] [Google Scholar]
- 34.Zhao, W., J. P. Kruse, Y. Tang, S. Y. Jung, J. Qin, and W. Gu. 2008. Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451587-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao, Y., J. Tan, L. Zhuang, X. Jiang, E. T. Liu, and Q. Yu. 2005. Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc. Natl. Acad. Sci. USA 10216090-16095. [DOI] [PMC free article] [PubMed] [Google Scholar]