Abstract
Store-operated Ca2+ influx is mediated by store002Doperated Ca2+ channels (SOCs) and is a central component of receptor-evoked Ca2+ signals1. The Orai channels mediate SOCs2–4 and STIM1 is the ER-resident Ca2+ sensor that gates the channels5, 6. How STIM1 gates and regulates the Orai channels is unknown. Here, we report the molecular basis for gating of Orais by STIM1. All Orai channels are fully activated by the conserved STIM1(344–442), which we termed SOAR (the STIM1 Orai Activating Region). SOAR acts in combination with STIM1(450–485) to regulate the strength of interaction with Orai1. Orai1 activated by SOAR recapitulates all the entire kinetic properties of Orai1 activated by STIM1. Mutations of STIM1 within SOAR prevent activation of Orai1 without preventing co-clustering of STIM1 and Orai1 in response to Ca2+ store depletion, indicating that STIM1-Orai1 co-clustering is not sufficient for Orai1 activation. An intact C-terminus α-helicial region of Orai is required for activation by SOAR. Deleting most of Orai1 N terminus impaired Orai1 activation by STIM1, but (Δ1–73)Orai1 interacts with and is fully activated by SOAR. Accordingly, the characteristic inward rectification of Orai is mediated by an interaction between the polybasic STIM1(672–685) and a proline-rich region in the N terminus of Orai1. Hence, the essential properties of Orai1 function can be rationalized by interactions with discrete regions of STIM1.
Ca2+ influx is a critical step of receptor-evoked signaling, and is mediated by plasma membrane channels that are activated in response to release of Ca2+ from the endoplasmic reticulum (ER), and are thus named store-operated Ca2+ influx channels (SOCs)1. Recently, the molecular determinants of SOCs and their gating have been elucidated. STIM1 is a Ca2+ binding protein that functions as the ER Ca2+ sensor that activates SOCs5, 6. STIM1 is a multi-domain protein with N terminal EF hand and SAM domains that reside in the ER lumen and a cytoplasmic region composed of an ERM (Ezrin/radixin/moesin) domain, a S/P-rich domain, and a polybasic lysine- (K) rich domain5, 6. In response to Ca2+ release from the ER, Ca2+ dissociates from the EF hand, and STIM1 clusters next to the plasma membrane to activate the SOCs5–8.
Two channel families have been identified as STIM1-regulated SOCs; the TRPC (TRPCs)9 and Orai channels (Orais)2–4. The TRPCs function as non-selective, Ca2+ permeable channels10, whereas Orai1 mediates the Ca2+ selective Ca2+-release activated (CRAC) current2–4. The STIM1 ERM domain binds TRPCs7, allowing their heteromultimerization11, while STIM1(K684,685K) gats TRPCs by electrostatic interaction with two conserved negative charges within TRPCs TRP Box212.
How STIM1 gates and regulates the Orais is not known. STIM1 is obligatory for the Orais to function as channels4, 13, 14. Gating of the native CRAC current7 and of Orai115, 16 is mediated by the STIM1 C terminus, but the gating does not require the STIM1 S/P and K-domains12. A proline-rich region in the N terminus of Orai1 is suggested to be important for gating of Orai1 by STIM117. However, another study reported that deletion of the first 73 residues of Orai1, which includes the proline-rich region, does not prevent activation of Orai1 by STIM118. To better define gating and regulation of Orai1 by STIM1, we searched for the STIM1 domains that mediate gating and voltage-dependence of Orai1.
SOAR co-localize with and fully activates Orai1
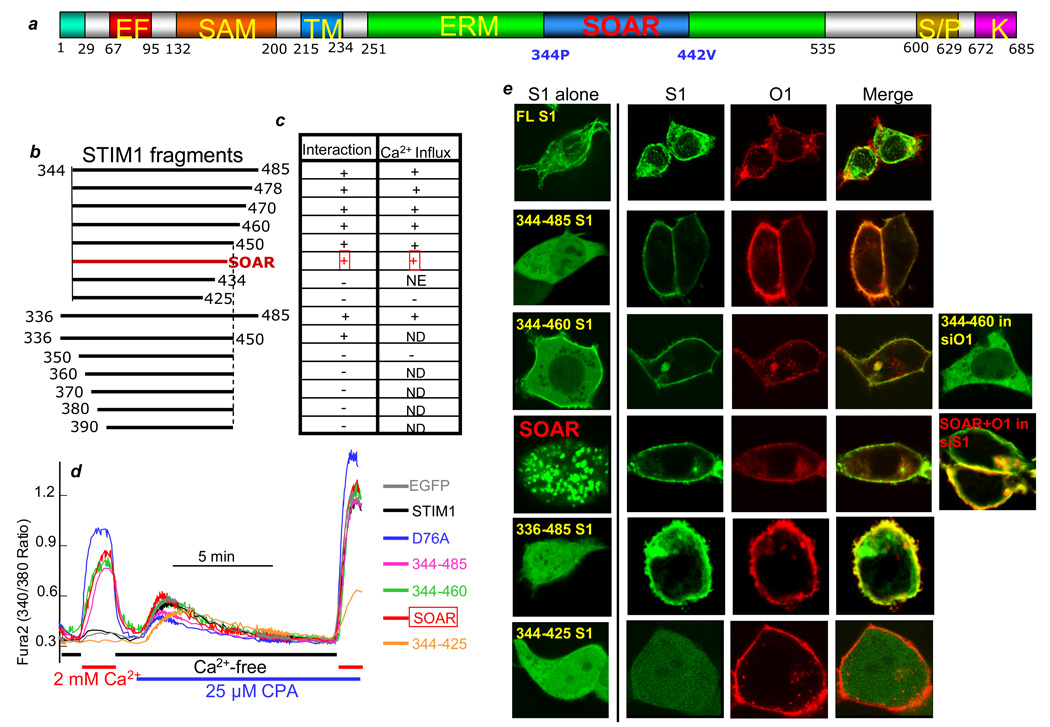
CRAC current7 and Orai115, 16 are activated by the STIM1 C terminus, but the S/P and polybasic K- domains are not required for activation of Orai112. We therefore searched for the minimal STIM1 domain that could fully activate Orai1. Drosophila STIM1 terminates at the equivalent human STIM1 residue 48519. Indeed, STIM1(235–485) activates native SOCs and Orai1. Analysis of systematic N and C terminal deletions identified STIM1(344–442) as the minimal sequence that fully activates Orai1. The effects of various STIM1 fragments evaluated in the work (Fig. 1c) on spontaneous Ca2+ influx are summarized in Fig. 1b, and example traces are shown in Fig. 1a. The STIM1 fragments had one of three effects: they activated spontaneous Ca2+ influx; inhibited SOCs; or had no effect. Store depletion by inhibition of SERCA with CPA showed that the STIM1 fragments did not deplete the stores. Since STIM1(344–442) is the minimal domain that activates Ca2+ influx (Fig. 1) and all Orai channels (see below), we termed it SOAR, for the STIM1 Orai Activating Region.
Fig. 1. Interaction of SOAR and STIM1 fragments with Orai1 and activation of SOCs.
(a) A model of STIM1 domains showing the position of SOAR. (b) The STIM1 fragments analyzed in the present work. (c) Summary of the STIM1 fragments interaction with Orai1 (first column) and activation of spontaneous Ca2+ influx (second column). “+” indicates activation of spontaneous Ca2+ influx; “−“ indicates lack of activation of spontaneous Ca2+ influx and inhibition of the native SOC; NE indicates no effect on spontaneous Ca2+ influx or SOCs; and ND indicates not determined. (d) Example traces of HEK cells transfected with EGFP or the indicated EGFP-tagged STIM1 fragments that were incubated in Ca2+-free and then Ca2+-containing media to evaluate the spontaneous Ca2+ influx. The cells were then treated with 25 µM CPA in Ca2+-free media to estimate the ER Ca2+ content and finally the cells were incubated with 25 µM CPA and 2 mM Ca2+ to measure the fully activated SOCs. The traces are representative of 4 experiments, which were used to generate the activity in the table in (c). (e) Localization of the STIM1 fragments when transfected alone (first image in each series) and when co-transfected with Orai1. Also shown is expression of STIM1(344–460) in cells treated for 72 hrs with siOrai1 and co-expression of SOAR and Orai1 in cells treated for 72 hrs with siSTIM1. All STIM1 fragments and Orai1 are N terminally tagged with EGFP and FLAG-mCherry, respectively. Scale bars are 15 microns and here and in Fig. S2, the same expression pattern was observed in 5 separate transfections and in all cells examined within each cover slip (n>20).
To assess cell biological correlates of STIM1 activation of Orai1, we monitored the distribution of EGFP-STIM1 fragments and mCherry-Orai1 in HEK293 cells (Fig. 1e and Fig. S1). In all cases, STIM1 constructs that activate Orai1 co-localized with Orai1 at the plasma membrane. Several constructs, such as SOAR, redistributed from the cytosol to the plasma membrane when co-expressed with Orai1, independent of the native STIM1. Other constructs associated with the plasma membrane when expressed alone, but knock-down of native Orai1 prevented their plasma membrane enrichment (STIM1(344–460) in Fig. 1e). N and C terminal truncations of SOAR that failed to activate Orai also failed to co-localize with Orai1 (Figs. 1e and S1b). Depletion of internal Ca2+ stores had no further effect on their co-localization with Orai1 (not shown). This is consistent with a recent report showing that the STIM1 C terminus does not cluster with Orai120. These data indicate a precise correlation between the ability of STIM fragments to localize to the plasma membrane with Orai, and to activate the channel.
SOAR is a conserved α-helical domain
Fig. S2a shows that SOAR is conserved among vertebrate species ranging from human to Zebrafish and in invertebrates, but with lower sequence identity for C. elegans. Attempts to purify SOAR expressed in bacteria were not successful. However, we expressed and purified STIM1(336–485). The CD spectra of STIM1(336–485) (Fig. S2b) indicates a 45% α-helical structure, which compares with the predicted 62%. Both, gel filtration (red trace) and light scattering (blue trace) analyses indicate that STIM1(336–485) exists as a dimer in solution (Fig. S2c). Interestingly, recent work suggests that Orai1 functions as a tetramer21, 22 and assembles with two STIM1 molecules to form the functional channel21. The dimer formation of STIM1(336–485) is consistent with these findings.
SOAR recapitulates activation of Orai1 by STIM1
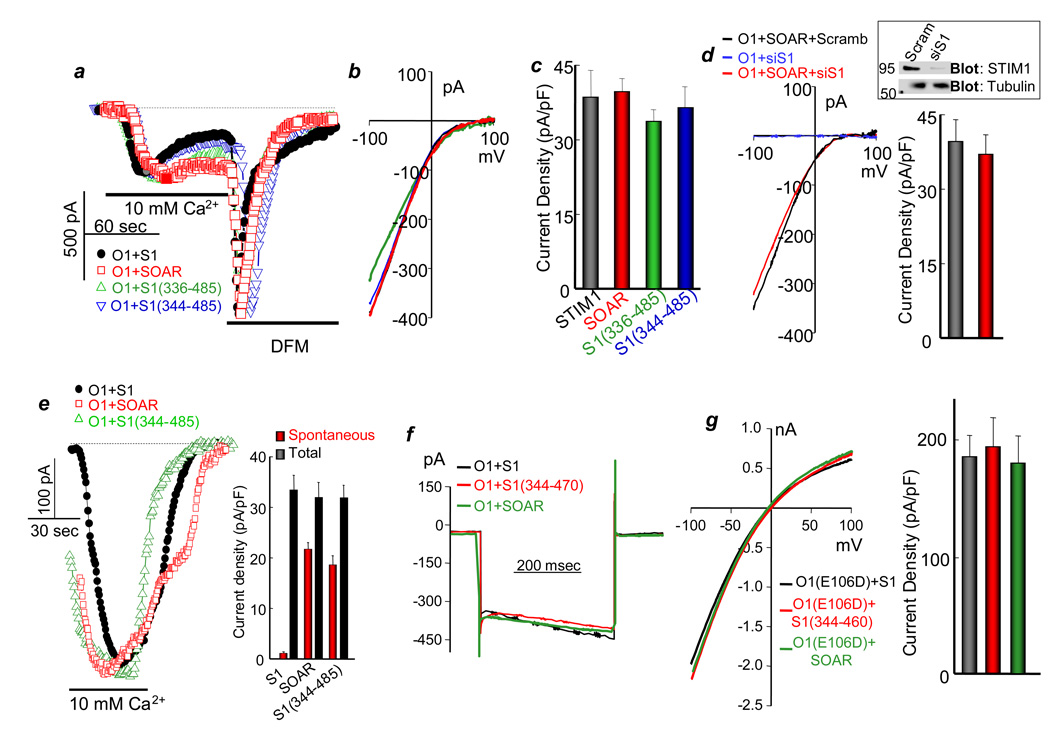
The effect of SOAR on Orai1 current was measured at a 1SOAR:1Orai1 expression ratio, which is optimal for maximal activation of Orai1 (Fig. S3). Fig. 2 shows that SOAR and the cytosolic STIM1 fragments that co-localize with Orai1 fully activate Orai1, and SOAR activates Orai1 independent the native STIM1 (Fig. 2d). Current measurement starting within 10 sec of break-in shows that about 50% of the Orai1 current induced by the STIM1 fragments is spontaneously active (Fig. 2e), with further activation likely due to chelation of cytoplasmic Ca2+ by the slowly diffusing BAPTA. Importantly, all kinetic parameters of the Orai1 current activated by SOAR are identical to the parameters of Orai1 current activated by STIM1 (Figs. 2a, 2b, 2f), and STIM1 and SOAR similarly activated the Na+ current of the pore mutant Orai1(E106D) (Figs. 2g).
Fig. 2. The properties of activation of Orai1 by SOAR and the STIM1 fragments.
(a) The CRAC current was measured in HEK cells expressing Orai1 and the indicated STIM1 fragments. In these and all other current measurements, Orai1 is tagged with FLAG-mCherry and the STIM1 fragments with EGFP. Preliminary experiments showed that the tags had no effect on the activity of the proteins. The cells were dialyzed with a pipette solution containing 10 mM BAPTA and external solution containing 10 mM Ca2+. Then the cells were perfused with divalent-free media (DFM). Here and in all Figs., dashed lines show the zero current. (b) shows the I/V for each condition at the peak current, as marked by the filled symbols in (a). (c) Shows the mean±s.e.m of 4 experiments in each condition. (d) The native STIM1 is not required for activation of Orai1 by SOAR. HEK cells treated with scrambled or siSTIM1 (the blot shows efficiency of the knock-down, full blot is shown in Fig. S7) were transfected with Orai1 alone (control) or Orai1 and SOAR and used to measure the CRAC current. The columns show the mean±s.e.m of 4 experiments. (e) Current measurement started within 10 sec of break-in using the recording conditions in (a) to evaluate the spontaneous current before substantial diffusion of BAPTA into the cells. The columns show the mean±s.e.m of 4 experiments. (f) Fast Ca2+-mediated inactivation of CRAC current was measured in cells expressing Orai1 and STIM1 or Orai1 and SOAR or STIM1(344–470). After maximal activation of the current, the membrane potential was stepped from a holding potential at 0 to −120 mV for 400 msec. (g) Activation of Na+ current by Orai1(E106D) was measured by dialyzing the cells with pipette solution containing 140 mM Cs+ and 10 mM BAPTA and bath solution containing 140 mM Na+. The cells were transfected with Orai1 and STIM1 or SOAR. The columns show the mean±s.e.m of 4 experiments.
SOAR activates all Orai isoforms
The three Orais are activated by STIM1 but show different conductance with Orai1≫Orai2>Orai314, 23 and Figs. S4a–S4c show that SOAR activates Orai2 and Orai3 with the same current density as STIM1.
The role of Orai1 C and N termini in the interaction and activation by SOAR
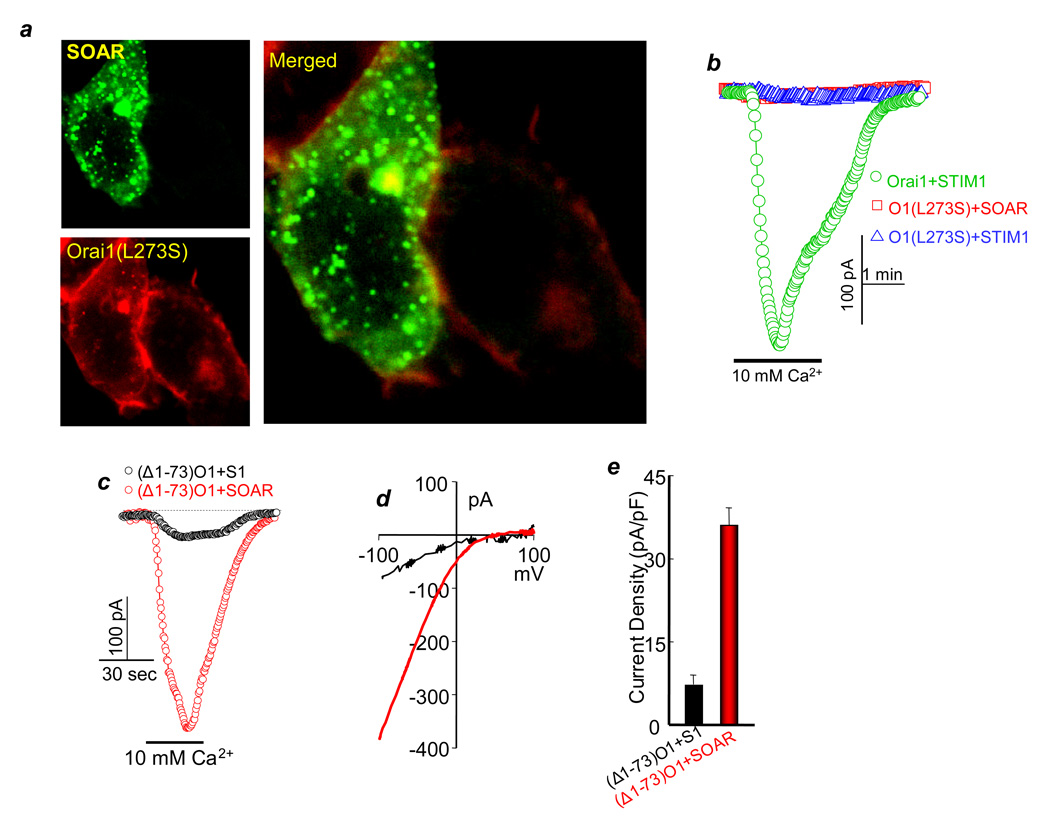
Disruption of the Orai1 C-terminus α-helix by the Orai1(L273S) mutation inhibits interaction with and activation of Orai1 by STIM115. Similarly, SOAR does not co-localize with Orai1(L273S) at the plasma membrane (Fig. 3a), and STIM1 and SOAR do not activate Orai1(L273S) (Fig. 3b). Hence, activation of Orai1 requires interaction of SOAR with the Orai1 C terminus.
Fig. 3. Effect of SOAR on Orai1(L273S) and (Δ1–73)Orai1.
(a) The cells were co-transfected with Orai1(L273S) and SOAR. The images represent 50 analyzed cells from 3 transfections. Note that Orai1(L273S) failed to recruit SOAR to the plasma membrane. (e) Orai1(L273S) is not activated by STIM1 or SOAR. The traces are representative of 4 experiments. (c–e) CRAC current was measured in cells transfected with (Δ1–73)Orai1 and either STIM1 (black traces) or SOAR (red traces), (b) is the I/Vs and (c) is the mean±s.e.m. of 4 experiments with STIM1 and 5 experiments with SOAR.
The Orai1 N terminus encompasses aa 1–86. Deletion of aa 1–73 reduces activation of Orai1 by STIM118. Fig. 3c–e shows that STIM1 poorly activates (Δ1–73)Orai1. Most strikingly, SOAR fully activates (Δ1–73)Orai1, and (Δ1–73)Orai1 activated by SOAR has the same properties as Orai1 activated by SOAR or STIM1. Fig. S4d shows that (Δ1–73)Orai1 recruits SOAR to the plasma membrane and, in response to store depletion, co-clusters with STIM1 at the plasma membrane. SOAR and STIM1(344–460) co-IP equally well with Orai1 and with (Δ1–73)Orai1, but not with Orai1(L273S) (Fig. S4e). The specificity of the co-IP is confirmed by the lack of co-IP of STIM1(344–425) and Orai1 or (Δ1–73)Orai1. SOAR and STIM1(344–460) did not co-IP with an Orai1 mutant lacking the C-terminus, however, Orai1(ΔC) was mostly retained in the ER which makes this construct less informative (not shown). Hence, SOAR binding and activation of Orai1 requires the C-terminus but not the N-terminus of Orai1.
Functional SOAR is required for activation of Orai1 by STIM1, but not for co-clustering of Orai1 and STIM1
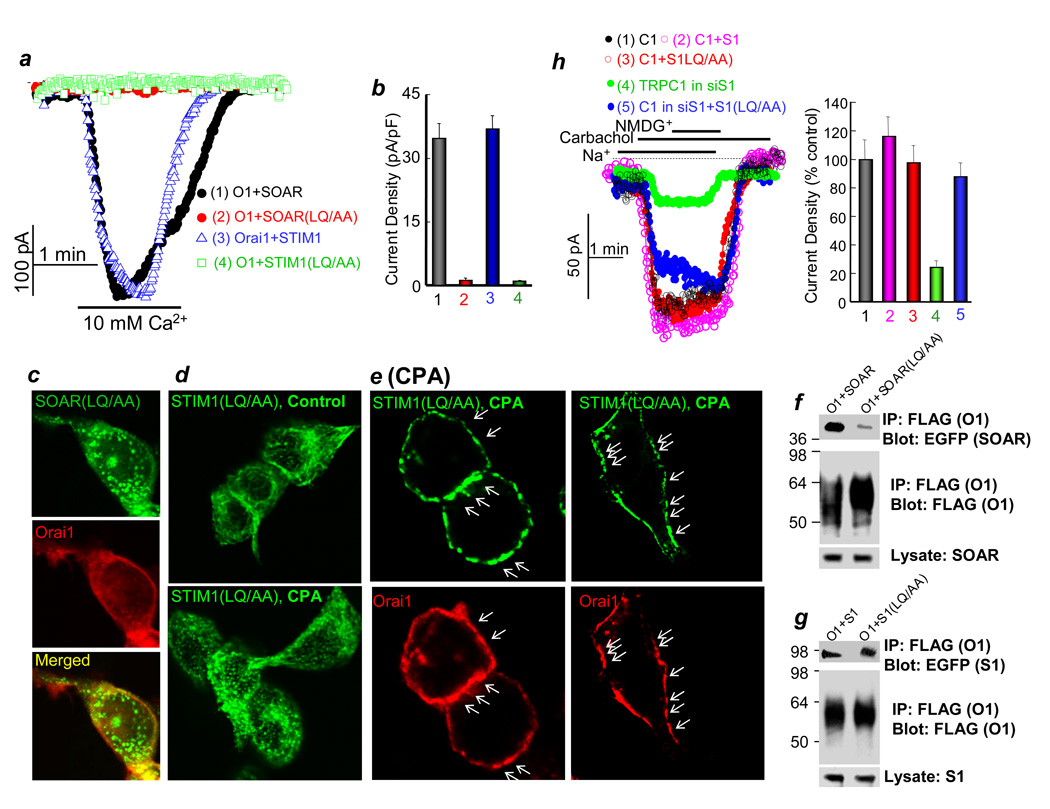
Precedence from studies of STIM1 activation of TRPC1 indicates that STIM1 binding and gating is mediated by distinct regions of the molecule12. We searched for mutants of STIM1 that might distinguish these properties for Orai. Residues 344–350 of SOAR are essential for activation of Orai1 (Fig. 1b), and Figure 4 shows that STIM1(LQ347/348AA) and SOAR(LQ347/348AA) do not activate Orai1. SOAR(LQ347/348AA) does not co-localize or co-IP with Orai1 (Fig. 4c and 4f). By contract, when expressed alone, STIM1(LQ347/348AA) is found in tubular-like structures in untreated cells and it clusters into puncta in response to store depletion (Fig. 4d). Moreover, when co-expressed with Orai1, STIM1(LQ347/348AA) co-clusters with Orai1 in cells with depleted stores (arrows in Fig. 4e), and co-IPs with Orai1 (Fig. 4f). Importantly, STIM1(LQ347/348AA) is functional towards TRPC1 (Fig. 4h), suggesting that the overall structure of STIM1(LQ347/348AA) is not disrupted.
Fig. 4. SOAR gates Orai1 but does not mediate clustering of STIM1 or co-clustering of STIM1-Orai1.
(a–b) CRAC current was measured in cells transfected with Orai1 and either SOAR (black), SOAR(LQ/AA) (red), STIM1 (blue) or STIM1(LQ/AA) (green). (b) is the mean±s.e.m. of 4 experiments. In (c), cells were co-transfected with mCherry-Orai1 and EGFP-SOAR(LQ/AA). In (d), cells were transfected with STIM1(LQ/AA) alone and were untreated (upper image) or treated with 25 µM CPA in Ca2+- free media for 10 min. (e) shows two examples of cells co-transfected with mCherry-Orai1 and EGFP-STIM1(LQ/AA) and treated with CPA for 10 min. Arrows in (e) point to co-localized Orai1-STIM1 in puncta. Each set of images represent at least 50 analyzed cells from 2 transfections. In (f, g), cells were transfected with Orai1 and SOAR or SOAR(LQ/AA) (f) or Orai1 and STIM1 or STIM1(LQ/AA) (g) and used to IP Orai1 with anti FLAG and probe for co-IP of SOAR or STIM1 with anti-GFP. Fig. S7b shows full blot of Orai1 and Fig. S7c shows full blot of EGFP-STIM1 and EGFP-SOAR. In addition, Fig. S7c is the control for the co-IP in panels (f, g). In (h), TRPC1 was expressed in control cells alone (condition 1, black) or together with STIM1 (condition 2, magenta), STIM1(LQ/AA) (condition 3, red), or in siSTIM1-treated cells alone (condition 4, green) or together with STIM1(LQ/AA) (condition 5, blue), and the carbachol-activated TRPC1-mediated Na+ current was measured. The columns show the mean±s.e.m. of the % current in 4 experiments.
Together, the results in Fig. 4 indicate that SOAR is essential for activation of Orai1 by STIM1. Furthermore, the interaction and co-clustering of STIM1 with Orai1 is not sufficient for activation of Orai1 by STIM1. This implies that SOAR within STIM1 actively gates Orai1 to open the channel.
The proline-rich domain in Orai1 N terminus regulates activation by STIM1
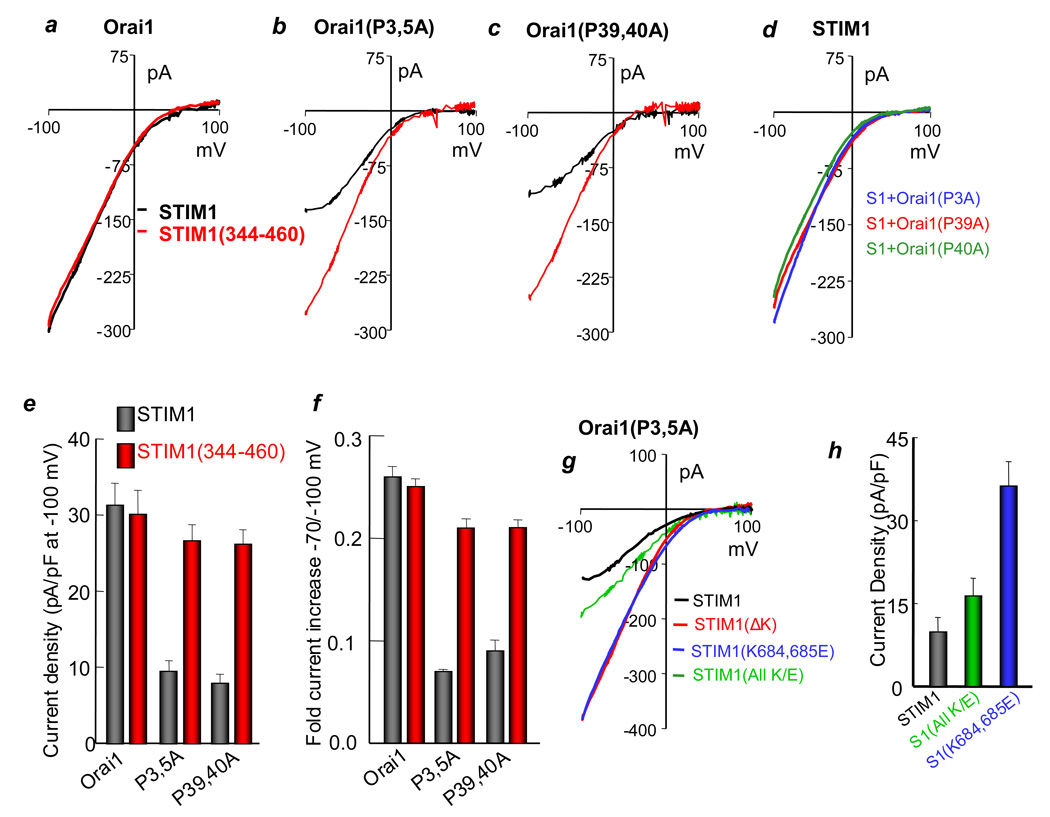
The differential action of SOAR versus STIM1 in activating (Δ1–73)Orai1 (Fig 3) suggests that another domain in STIM1 normally interacts with the N terminus of Orai1 and this is required for the action of SOAR when it is part of STIM1. To examine this hypothesis, we noted that the N-terminus of Orai contains prolines grouped at positions 3, 5; 7–9; 14; 17, 18; 39, 40 and 43–47. Mutation of prolines 14; 17, 18; 43, 44; and 45–47 to alanines had no effect on activation of Orai1 by STIM1 or SOAR. Mutation of prolines 7–9 resulted in a mutant with low channel activity with STIM1 and SOAR (not shown), independent of surface expression (Fig. S6a). However, Orai1(P3,5A) and Orai1(P39,40A) behave similar to (Δ1–73)Orai1 with respect to activation by STIM1 and SOAR (Fig. 5a– 5c). The I/V plots for activation of Orai1(P3,5A) and Orai1(P39,40A) by STIM1 are flattened at negative membrane potentials, with the channel becoming both, inwardly and outwardly rectifying. This is illustrated by calculating the % increase in current on changing the membrane potential from −70 to −100 mV (Fig. 5f), which shows about 25% increase for Orai1 and only 5–7% increase for Orai1(P3,5A) and Orai1(P39,40A). The pairs of prolines (3,5) and (39,40) are required to observe the change in the Orai1 I/V, since the single mutants Orai1(P3A), Orai1(P39A) and Orai1(P40A) behave like wild-type Orai1 when activated by STIM1 (Fig. 5d) or by SOAR (not shown).
Fig. 5. The role of Orai1 N terminal proline-rich domain and STIM1 K-domains in Orai1 function.
(a–c) CRAC current was measured in HEK cells transfected with STIM1 (black traces) or STIM1(344–460) (red traces) and Orai1 (a), Orai1(P3,5A) (b) or Orai1(P7–9A) (c). (d) Current was measured in cells transfected with wild-type STIM1 and the single proline mutants Orai1(P3A) (blue trace), Orai1(P39A) (red trace) or Orai1(P40A) (green trace). The traces represent 4 experiments with similar results. (e, f) are the mean±s.e.m. (n=4) of the current density (e) and the fold increase in current between −70 and −100 mV (f) of wild-type Orai1 and the Orai1(P3,5A) and Orai1(P39,40A) mutants. (g) Current was measured in cells transfected with Orai1(P3,5A) and either STIM1 (black trace, n=3), STIM1(ΔK) (red trace), STIM1(K684,684E) (blue trace, n=5), or STIM1(all K/E) (green trace, n=5). The mean±s.e.m. is shown in (h).
The reduced activation of Orai1(P3,5A) and Orai1(P39,40A) by STIM1 is not due to reduced surface expression (Fig. S6a). To the extent tested, the properties of the Orai1(P3,5A) and Orai1(P39,40A) activated by STIM1(344–460) are similar to those of Orai1 activated by STIM1 (Figs. 5b and 5d and Fig. S5d). The pore mutation E106D in the context of Orai1(P3,5A) and Orai1(P39,40A) activated by STIM1 or STIM1(344–460) resulted in a large monovalent current, but with a small current with STIM1 (Figs. S5a–S5c).
The polybasic K-domain mediates inhibition of SOAR within STIM1
The preceding analysis supports a model in which the Orai1 N-terminus interacts with a region of STIM1 that normally inhibits the action of SOAR. We examined a series of mutants and pinpointed the polybasic K-domain. Fig. 5g and 5h and Fig. S6b show that STIM1(ΔK) and point mutations of lysines in the K-domain rescue the reduced activation of Orai1(P3,5A) by STIM1. The same results were obtained with all the STIM1 K-domain mutants with Orai1(P39,40A) (not shown). However, the mutant in which all seven K-domain lysines were substituted by glutamates did not activate Orai1(P3,5) (Fig. 5h). Hence, intact K-domain is required for its interaction with the Orai1 mutants. These observations indicate that the positive charges within the K-domain regulate the ability of SOAR in the context of wild-type STIM1 to activate Orai1. Consistent with this model, deletion of the K domain strongly enhances STIM1 binding to Orai1 (Fig. S5e).
The present study reveals the molecular mechanism by which STIM1 gates the Orai channels. SOAR within STIM1 is required and sufficient for full activation of all Orai channels by interacting with the C terminus of Orai1. The STIM1 C- terminus multimerizes two Orai1 dimers to form the active channel20. STIM1(336–485) exists as a dimer in solution (Fig. S2c). A potential mechanism for gating of the Orais by STIM1 is that the Orai C termini prevent multimerization and assembly of functional channels. Binding of STIM1 molecules through their SOAR domains to the C termini of Orais can serve to deflect the C termini, thereby allowing assembly and opening of the channels. Hence, SOAR within STIM1 actively gates the Orai channels. Thus, clustering of Orai1 is neither sufficient nor necessary for activation of the channel. This is also supported by the function of STIM1(LQ/AA).
Another STIM1 domain, the polybasic K-rich domain, regulates Orai1 and is separate from opening of Orai1 by SOAR. The K-domain regulates the activity of Orai1 at negative membrane potential by communicating with the Orai1 N terminal proline-rich domain. It appears that the K-domain retards interaction of STIM1 with Orai1. In one model, the K-domain interacts with a region of Orai1 or STIM1 to prevent SOAR binding and this is reversed when the K-domain interacts with Orai1 N terminus.
SOAR and the K-domain appear to have different roles in gating the TRPC and Orai channels. SOAR within the STIM1 ERM domain participates in binding of STIM1 to TRPCs, but is not sufficient to activate TRPC1 (Fig. S6c). The STIM1 K-domain is essential for opening of TRPCs12, but inhibits Orai1. Under physiological conditions, the balance of association of the K-domain and SOAR with the two channel types will determine the extent of regulation of Orai1 and TRPC channels by STIM1 and Ca2+ influx.
Methods
Solutions, reagents, and clones
The TRPC1, STIM1 (wild-type, ΔK, All K/E, 681X), and Orai1 clones were described previously 7, 11. The human Orai2 and Orai3 clones were obtained from Open Biosystems (Orai2 clone #: BC069270 and Orai3 clone #: BC015555). Orai2 and Orai3 were cloned into mCherry-Red-p3XFLAG vector12 using NotI(5’) and SalI(3’). The mCherry-Red is downstream and next to the 3X FLAG and was cloned into the HindIII site of the multi-cloning region of the p3XFLAG vector. SOAR and the various STIM1 fragments were generated by PCR and cloned into the pEGFP-C1 (Clontech) vector using EcoRI (5’ and 3’). Hence, all STIM1 fragments are tagged with EGFP at their N terminus. All point mutations on STIM1 and Orai1 were generated using the site-directed mutagenesis kit (Stratagene). The antibodies used were monoclonal anti-STIM1 (BD Biosciences), polyclonal anti-tubulin (Cell Signaling Technology), monoclonal HRP-conjugated anti-GFP (Santa Cruz Biotech) and monoclonal anti-FLAG and HRP-conjugated anti-FLAG (Sigma-Aldrich). Anti-FLAG antibodies were used for co-IP, while HRP-conjugated anti-GFP and anti-FLAG antibodies were used for Western blotting. The siRNA sequence used to knockdown human Orai1 is: 5’-GCCAUAAGACUGACCGACAGUUCCA-3’. A 6-hour siRNA transfection of HEK293 cells was done using Lipofectamine 2000 (day 1). The amount of siRNA used was 0.8 µg per 12-well with HEK cells at 80–90% confluency. After transferring the cells containing siRNA to 35mm dish on day 2, they were transfected with plasmid for 6 hours on day 3. The total amount of cDNA used per 35 mm dish was 0.5 µg/ml. Localization assays using confocal microscopy was done, current was measured, or cells were harvested and extracted for co-IP analysis the following day (day 4). Thus, the total time for cells exposed to siRNA was 72 hours.
Western blot and co-IP
Transfected cells were harvested and lysed using 500 µL of binding buffer: 1× PBS buffer containing 1 mM NaVO3, 10 mM NaPyrophosphate, 50 mM NaF [pH 7.4], and 1% Triton X-100. The cell extracts were sonicated, and insoluble material was spun down at 30,000 × g for 20 min. For the co-IP experiments, 1 µg of myc antibody was added to 100 µL of cell extract and incubated for 1 hr at 4 C. Then, 50 µL of 1:1 slurry of protein G sepharose 4B beads were added to the antibody-extract mix and incubated for an additional hr at 4 C. Beads were washed 3 × 10 min with binding buffer, proteins were released from the beads with 50 µL of SDS-loading buffer. 25 µL was loaded onto 8% tris-glycine SDS-PAGE gels. Gels were transferred onto PVDF membrane, and Western blot analysis was done.
Biotinylation
Transfected cells were washed once with 1X PBS on ice. 0.5 mg/mL of EZ-Link Sulfo-NHS-SS-Biotin (Pierce) was added to the cells for 30 min on ice. Afterwards, the biotin was quenched with 50 mM glycine on ice for 10–15 min. The cells were then processed as described above to make cell extract. 50 µL of 1:1 slurry of immobilized avidin beads(Pierce) were added to 100 µL of cell extract and incubated for 2 hrs at 4 C. Beads were washed 3 × 10 min with binding buffer, proteins were released from the beads with 50 µL of SDS-loading buffer. 25 µL was loaded onto 8% tris-glycine SDS-PAGE gels. Gels were transferred onto PVDF membrane, and Western blot analysis was done.
Measurement of [Ca2+]i
[Ca2+]i was measured about 24 hrs post transfection using a PTI image acquisition system. [Ca2+]i was measured by loading the cells with Fura2 and recording Fura2 fluorescence at excitation wavelengths of 340 and 380 nm and collecting the light emitted at wavelength above 500 nm. [Ca2+]i is expressed as the 340/380 ratio.
Confocal imaging
After HEK cells were transfected with the chosen EGFP-STIM1 fragment +/− mCherry Red-Orai1, the cells were fixed with 4% paraformaldehyde and mounted onto microscope slides. Images were collected under 400X magnification using the LaserSharp 2000 (Bio-Rad) software and a Bio-Rad confocal microscope.
Purification of STIM1(336–485)
The STIM1(336–485) sequence was cloned into the pET 28 vector and expressed as an N-terminal Hisx6-fusion protein in BL21 Codon Plus cells (Stratagene) following the manufacture’s guidelines. The fusion protein was purified using TALON metal affinity resin (Clontech) following the manufacturer’s protocol, and the Hisx6 tag was removed using Thrombin (Roche). The STIM1(336–485) fragment was concentrated using Amicon Ultra 10K concentrators (Millipore) and run over a Superdex 200 gel filtration column (GE Healthcare). Light scattering measurements of the STIM1(336–485) fragment were collected with a miniDAWN TREOS (Wyatt Technology) while eluting off of the Superdex 200 gel filtration column in 20 mM Tris pH 8.0, 100 mM NaCl. ASTRA V software (Wyatt Technlogy) was used to analyze the data. The STIM1(336–485) CD spectrum was collected on a Jasco J-810 spectropolarimeter in 10 mM HEPES pH 7.4, 140 mM KCl. The helical content of STIM1(336–485) was calculated using the Greenfield and Fasman method fh = (([Θ]222–[Θ0]222)/[Θ100]222) where [Θ]222 is the observed molar elipticity at 222 nm, [Θ0]222 is the mean molar elipticity of a model peptide with no alpha helix and/[Θ100]222 which is completely helical 24. The 0% and 100% helix estimates used were 2,000 and 30,000 deg·cm2/dmol 25, 26.
Current measurements
The whole cell configuration was used to measure the Orai1 CRAC current in HEK cells co-transfected with Orai1, the Orai1 mutants and with STIM1, SOAR or the STIM1 fragments as detailed before 7, 11, 12. The standard pipette solution contained (in mM): 140 Cs aspartate, 6 MgCl2, 10 BAPTA, and 10 Hepes (pH 7.2 with CsOH). The standard bath solution contained (in mM): 130 NaCl, 5 KCl, 10 CaCl2, 1 MgCl2, and 10 Hepes (pH 7.4 with NaOH). The divalent-free (DVF) solution contained (in mM): 150 NaCl, 10 EDTA, and 10 Hepes (pH 7.4 with NaOH). The current was recorded by 400 ms rapid alterations of membrane potential (RAMPs) from −100 to +100 mV from a holding potential of 0 mV. The current recorded at −100 mV was used to plot the time course of current development and to calculate current density in pA/pF. The averages of multiple experiments are given as mean ±s.e.m of the number of experiments performed.
TRPC1 current was measured in transiently transfected HEK cells by whole current recording, as described previously 11, 12, 27. The pipette solution contained (in mM) 140 CsCl, 2 MgCl2, 1 ATP, 5 EGTA, 1.5 CaCl2 (free Ca2+ 70 nM) and 10 HEPES at pH 7.2 with CsOH. The bath solution contained (in mM) 140 NaCl or 140 NMDG-Cl, 5 KCl, 0.5 EGTA and 10 HEPES at pH 7.4 with NaOH or NMDG-OH−). Cells were transfected with TRPC1 and empty vector or TRPC1 and SOAR. RAMPs of −100 to +100 mV were used to record the TRPC1current. The cells were also transfected with the M3 receptor so that the current can be consistently activated by stimulating the cells with 100 µM carbachol. The current recorded at −100 mV was used to calculate current density as pA/pF.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health Grants DE12309 and DK38938 and the Ruth S. Harrell Professorship in Medical Research to S. M. and by the National Institute on Drug Abuse (NIDA; DA00266, DA10309) and the National Institute of Mental Health (NIMH; MH068830) to P. F. W.
References
- 1.Parekh AB, Putney JW., Jr Store-operated calcium channels. Physiol Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- 2.Feske S, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 3.Vig M, et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang SL, et al. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc Natl Acad Sci U S A. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liou J, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roos J, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang GN, et al. STIM1 carboxyl-terminus activates native SOC, I(crac) and TRPC1 channels. Nat Cell Biol. 2006;8:1003–1010. doi: 10.1038/ncb1454. [DOI] [PubMed] [Google Scholar]
- 8.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worley PF, et al. TRPC channels as STIM1-regulated store-operated channels. Cell calcium. 2007;42:205–211. doi: 10.1016/j.ceca.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilius B, Owsianik G, Voets T, Peters JA. Transient receptor potential cation channels in disease. Physiol Rev. 2007;87:165–217. doi: 10.1152/physrev.00021.2006. [DOI] [PubMed] [Google Scholar]
- 11.Yuan JP, Zeng W, Huang GN, Worley PF, Muallem S. STIM1 heteromultimerizes TRPC channels to determine their function as store-operated channels. Nat Cell Biol. 2007;9:636–645. doi: 10.1038/ncb1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng W, et al. STIM1 gates TRPC channels, but not Orai1, by electrostatic interaction. Mol Cell. 2008;32:439–448. doi: 10.1016/j.molcel.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peinelt C, et al. Amplification of CRAC current by STIM1 and CRACM1 (Orai1) Nat Cell Biol. 2006;8:771–773. doi: 10.1038/ncb1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer JC, et al. Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. The Journal of biological chemistry. 2006;281:24979–24990. doi: 10.1074/jbc.M604589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muik M, et al. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. The Journal of biological chemistry. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang SL, et al. Store-dependent and -independent Modes Regulating Ca2+ Release-activated Ca2+ Channel Activity of Human Orai1 and Orai3. The Journal of biological chemistry. 2008;283:17662–17671. doi: 10.1074/jbc.M801536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi Y, et al. Essential role of the N-terminus of murine Orai1 in store-operated Ca(2+) entry. Biochem Biophys Res Commun. 2007;356:45–52. doi: 10.1016/j.bbrc.2007.02.107. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, et al. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. The Journal of biological chemistry. 2007;282:29448–29456. doi: 10.1074/jbc.M703573200. [DOI] [PubMed] [Google Scholar]
- 19.Dziadek MA, Johnstone LS. Biochemical properties and cellular localisation of STIM proteins. Cell calcium. 2007;42:123–132. doi: 10.1016/j.ceca.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Penna A, et al. The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature. 2008;456:116–120. doi: 10.1038/nature07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji W, et al. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13668–13673. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mignen O, Thompson JL, Shuttleworth TJ. Orai1 subunit stoichiometry of the mammalian CRAC channel pore. The Journal of physiology. 2008;586:419–425. doi: 10.1113/jphysiol.2007.147249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross SA, et al. Murine ORAI2 splice variants form functional Ca2+ release-activated Ca2+ (CRAC) channels. The Journal of biological chemistry. 2007;282:19375–19384. doi: 10.1074/jbc.M701962200. [DOI] [PubMed] [Google Scholar]
- 24.Greenfield N, Fasman GD. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 25.Wu CS, Ikeda K, Yang JT. Ordered conformation of polypeptides and proteins in acidic dodecyl sulfate solution. Biochemistry. 1981;20:566–570. doi: 10.1021/bi00506a019. [DOI] [PubMed] [Google Scholar]
- 26.Chen YH, Yang JT, Chau KH. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 27.Yuan JP, et al. Homer binds TRPC family channels and is required for gating of TRPC1 by IP3 receptors. Cell. 2003;114:777–789. doi: 10.1016/s0092-8674(03)00716-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.