Abstract
Mutations can be beneficial under conditions where genetic diversity is advantageous, such as somatic hypermutation and antibody generation, but they can also be lethal when they disrupt basic cellular processes or cause uncontrolled proliferation and cancer. Mutations arise from inaccurate processing of lesions generated by endogenous and exogenous DNA damaging agents, and the genome is particularly vulnerable to such damage during S phase. In this phase of the cell cycle, many lesions in the DNA template block replication. Such lesions must be bypassed in order to preserve fork stability and to ensure completion of DNA replication. Lesion bypass is carried out by a set of error-prone and error-free processes collectively referred to as DNA damage tolerance mechanisms. Here, we discuss how two types of DNA damage tolerance, translesion synthesis and template switching, are regulated at stalled replication forks by ubiquitination of PCNA, and the conditions under which they occur.
Introduction
Mutations provide genetic diversity that can be either deleterious or advantageous for survival. Evolutionarily speaking, mutagenesis is the mechanism through which natural selection acts, and it is the basis for adaptation and species diversification. It is also important for events such as somatic hypermutation and antibody generation1. At the cellular level, however, mutagenesis is a risky business. Although many mutations are silent or lead to apoptosis, some can cause aberrant cellular behavior and uncontrolled cellular proliferation, a hallmark of cancer.
Mutations can arise from improper processing of molecular lesions (Box 1), which can take the form of single- or double-strand DNA breaks, covalent adducts and missing or altered bases. Such lesions result from endogenous metabolic processes as well as exogenous DNA damaging agents. In eukaryotic cells, highly conserved pathways known as checkpoints coordinate many aspects of the DNA damage response by inducing cell cycle arrest, activating DNA repair pathways, stabilizing the damaged DNA, and in some cases initiating apoptotic pathways2,3.
Box 1: Key terms
ATR checkpoint pathway – a surveillance pathway that operates during S phase to sense DNA damage and coordinate multiple aspects of the DNA damage response.
DNA damage tolerance – mechanisms that allow the cell to bypass lesions which block DNA replication without their actual removal.
PCNA – a homotrimeric protein complex that encircles double-stranded DNA and functions as a processivity factor for replicative DNA polymerases.
Regressed or reversed fork – a replication fork structure formed by pairing of the newly synthesized DNA on the leading and lagging strand (see Fig. 1a). This structure may be formed to allow lesion bypass during template switching.
Replication fork – the region of unwound DNA where active replication takes place. A stalled fork is one in which DNA replication is blocked, for example by a lesion in the template that inhibits polymerase progression.
Replicative polymerases – high-fidelity DNA polymerases that replicate undamaged DNA during DNA replication. These polymerases (Polε, Polδ and Polα) stall at lesions in the DNA. Polα is responsible for priming replication by synthesizing a short RNA-DNA primer, while Polε and Polδ extend these primers primarily on the leading and lagging strand respectively.
Template switching – a form of error-free DNA damage tolerance which utilizes the newly synthesized, undamaged strand of the sister chromatid as the template for bypass replication.
Translesion synthesis (TLS) – a form of DNA damage tolerance which utilizes specialized, TLS polymerases to directly replicate over and past lesions in the damaged template.
TLS polymerases – low-fidelity DNA polymerases capable of replicating over a lesion in the DNA template. The Y family polymerases are a subset of the TLS polymerases that are regulated through the ubiquitination of PCNA. Members of this family include Polη, Polι, Polκ and Rev1, each of which can bypass different lesions in vitro.
In S phase, the genome is particularly vulnerable as many types of lesions block replication fork progression. These lesions must be repaired or bypassed in order to complete DNA replication. Moreover, prolonged stalling of replication forks can lead to fork collapse, double-strand DNA breaks and genetic instability. Thus, stabilization and rescue of stalled replication forks and ultimately completion of DNA replication is essential for cell survival and preservation of the genome2,4.
DNA damage tolerance mechanisms, or post-replication repair processes, allow the cell to replicate over polymerase-blocking lesions1,5. Although widely used, the term “post-replication repair” may be misleading since no repair occurs per se, and instead the DNA damage is left behind for repair at a later time. There are two main DNA damage tolerance (DDT) pathways that differ in their potential to cause mutations -- translesion synthesis (TLS) and template switching (Fig. 1a). During translesion synthesis, specialized DNA polymerases replicate directly past the lesion in either an error-prone or error-free fashion. In contrast, the lesion is avoided during template switching by using an alternate (undamaged) DNA template, and as such this process is error-free. Because DNA synthesized past lesions will ultimately be used as a template for subsequent rounds of replication, the fidelity of lesion bypass and the choice between these processes is of utmost importance.
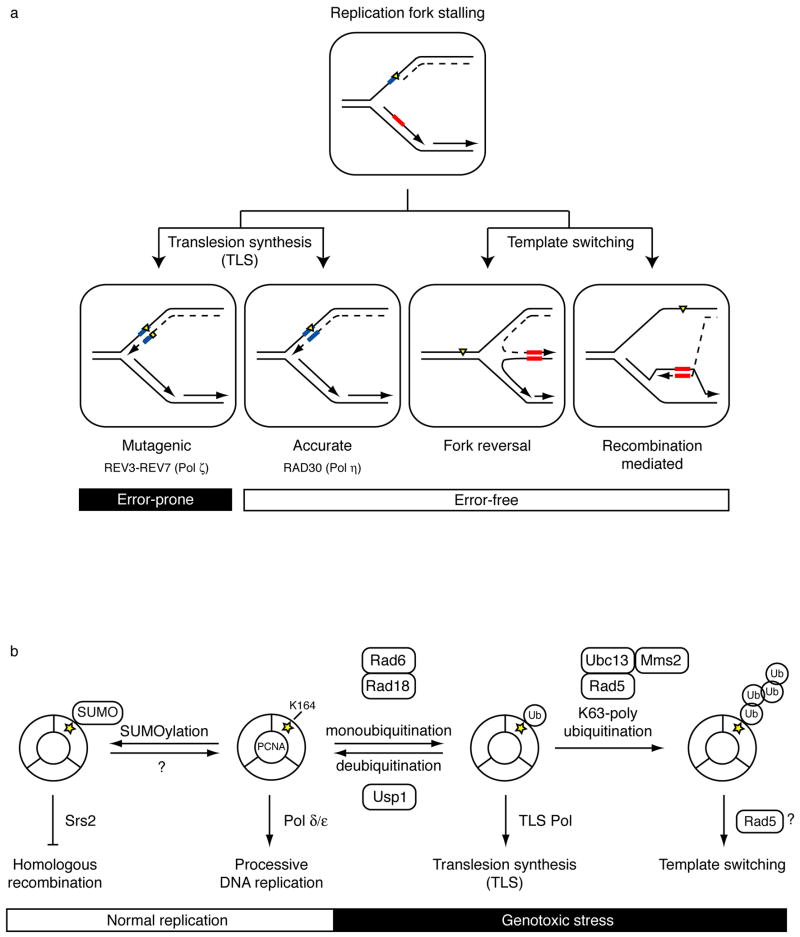
Figure 1. Overview of DNA damage tolerance pathways and PCNA ubiquitination.
(a) Lesions in the DNA template (yellow triangle) block processive DNA replication (dashed line). DNA damage tolerance mechanisms allow bypass of replication-blocking lesions by replicating over the damaged DNA (translesion synthesis, left) or using the undamaged sister chromatid (template switching, right). Template switching involves a structural rearrangement of the replication fork for which two models have been proposed. Fork reversal involves the formation of a four-way junction or “chicken-foot” intermediate (left) while recombination-mediated template switching involves D-loop formation and strand invasion (right). Templates used to bypass lesions and their complimentary sequences are boxed in blue for translesion synthesis and red for template switching. The mutagenic nature of the process is indicated.
(b) Overview of functions for post-translationally modified forms of PCNA and the enzymes that carry out the modifications. In the absence of DNA damage, S. cerevisiae PCNA is SUMOylated at a conserved site (K164, yellow star). This modification allows recruitment of the helicase Srs2, which inhibits homologous recombination during normal replication. Although SUMOylation is a reversible process, deSUMOylation of PCNA has not yet been characterized. Following genotoxic stress, PCNA is ubiquitinated at K164 to promote DNA damage tolerance pathways. Monoubiquitinated PCNA facilitates translesion synthesis (TLS) through recruitment of TLS polymerases, while K63-linked (nondegradable) polyubiquitinated PCNA is associated with template switching, possibly utilizing the helicase activity of Rad5. Each ubiquitination step is mediated by a distinct set of enzymes. Rad6 and Ubc13-Mms2 are E2 ubiquitin-conjugating enzymes (green). Rad18 and Rad5 are E3 ubiquitin ligases (red). Usp1 is a deubiquitinating enzyme.
Post-translational modification of PCNA (proliferating cell nuclear antigen) plays an important role in coordinating DNA replication and DNA damage tolerance processes5,6 (Fig. 1b). PCNA is a homotrimeric protein complex that forms a ring around double-stranded DNA. During normal replication, this sliding clamp functions as a processivity factor by tethering the replicative DNA polymerases to the DNA template6. Upon replication fork stalling, PCNA is ubiquitinated to promote DNA damage tolerance1,5. Here, we discuss recent advances in our understanding of the molecular mechanisms of PCNA ubiquitination, the role of PCNA modification in the regulation of translesion synthesis and template switching, and coordination between the DNA damage tolerance and checkpoint pathways in mediating the DNA damage response.
Error-prone and error-free modes of DNA damage tolerance
One type of DNA damage tolerance, translesion synthesis, is an evolutionarily conserved process that allows the replication machinery to bypass DNA lesions using a low-fidelity DNA polymerase1. Unlike the high-fidelity DNA polymerases, low-fidelity or TLS polymerases are non-processive, lack any proofreading capability, and contain larger active sites capable of accommodating distorted bases and base pair mismatches7–9 (Fig. 2a). The high-fidelity replicative polymerases such as Polα, Polδ and Polε belong to the classical B-family of DNA polymerases, while many TLS polymerases including Polη, Polκ, Polι and Rev1 belong to the Y-family. These two families of DNA polymerases share the same basic “fingers”, “thumb” and “palm” structure, but many Y-family TLS polymerases also contain a “little finger” domain that confers additional flexibility to the active site (Fig. 2a).
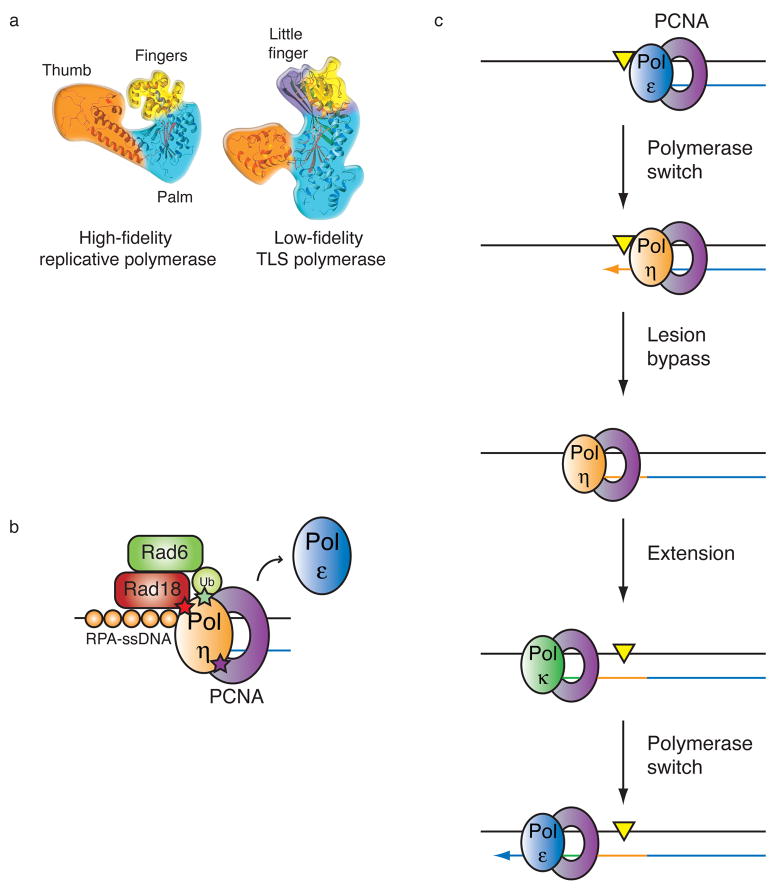
Figure 2. Translesion synthesis pathway.
(a) Structures of a high fidelity replicative DNA polymerase and a low fidelity TLS polymerase (reprinted with permission from Nature Reviews Molecular & Cell Biology)1.
The more restrictive nature of the active site in the replicative polymerase is apparent.
(b) Model for polymerase switching during TLS. Replicative DNA polymerases (blue) stall at lesions (yellow triangle) in the DNA template. In the first polymerase switch, a specialized TLS polymerase (tan) is recruited to the sliding clamp PCNA at the stalled fork and replicates over the lesion. This TLS “patch” is then extended by the same or another TLS polymerase (green). The final switch restores the replicative DNA polymerase to the template and processive DNA synthesis continues.
(c) Molecular interactions important for the switch from replicative to TLS polymerases. Following replication arrest, Rad6-Rad18 (E2–E3) ubiquitin enzymes are recruited to RPA bound single-stranded DNA. Following ubiquitination of PCNA by this complex, the TLS polymerase is recruited to the stalled fork. Polη interacts with three proteins at these sites via three different motifs: a ubiquitin binding domain (UBZ, green) which binds to the ubiquitin moiety on PCNA45, a PIP box (purple) which binds to the hydrophobic pocket between the subunits of PCNA6, and a carboxy-terminal domain (red) that mediates an interaction with Rad1838. Whether the replicative polymerase (Polε) is actually displaced from PCNA as shown or moves aside and remains bound to PCNA is not clear.
The process of TLS requires the exchange of one polymerase for another, an event that is thought to occur in a step-wise fashion involving at least two polymerase-switching events10 (Fig. 2b). In the first switch, the stalled replicative DNA polymerase is replaced by a TLS polymerase capable of replicating over the DNA lesion. The TLS “patch” is then extended by either the same or another TLS polymerase9,10. This extension step is necessary to allow the lesion to escape detection by the 3′ → 5′ exonuclease proofreading activity of the replicative DNA polymerase in vitro11,12, and extension can range from 5–60 nucleotides, depending on the lesion and polymerase involved11,12. Ultimately, a final switch restores a replicative DNA polymerase to the DNA template and processive DNA replication resumes.
TLS polymerases are often considered error-prone, as they display a higher frequency of misincorporation on undamaged templates than their replicative counterparts in vitro7,9. Furthermore, replication past certain lesions is often mutagenic, and necessarily so in certain cases, for example at abasic sites7,9. Consistent with this, genetic studies in S. cerevisiae have shown that loss of Rev1 or either of two subunits of Polζ, Rev3 or Rev7, results in decreased mutagenesis induced by DNA damage in vivo13,14. However, the “error-prone” label is somewhat misleading since several TLS polymerases have been shown to display proper base pairing opposite specific lesions7,9. For example, Polη preferentially inserts two “A”’s opposite a thymine dimer, a common UV photoproduct15,16, and Polκ has been shown to accurately bypass benzopyrene-induced guanine adducts17,18. Thus, translesion synthesis can be either mutagenic or accurate, depending on the lesion and which TLS polymerase is used (Fig. 1a). Underscoring the potential significance of TLS processes, mutation of the XPV gene, which encodes Polη in humans, results in a variant form of Xeroderma pigmentosum (XPV)15,19. Patients with XPV are hypersensitive to UV damage and are predisposed to cancer. The cell’s inability to appropriately substitute another TLS polymerase for Polη supports the idea that at least some of these specialized DNA polymerases are not functionally redundant but are specific for a particular type of DNA damage or lesion.
Much less is known about template switching, an error-free form of DNA damage tolerance that is genetically distinct from translesion synthesis. Evidence for this pathway is primarily based on epistasis studies in yeast showing that its loss results in increased mutagenesis, presumably due to an increased reliance on the more error-prone TLS for lesion bypass1,20–23. As the name implies, template switching is hypothesized to mediate lesion bypass by temporarily replacing the lesion-containing DNA template with an undamaged template, namely the newly synthesized daughter strand of the sister duplex. Two models for template switching have been proposed, one involving fork reversal using the nascent sister strand, and the other involving invasion of the sister duplex by a single-stranded gap in a manner reminiscent of homologous recombination1,24–27 (Fig. 1a). Although reversed forks have been observed by electron microscopy, there is some debate as to whether these structures are atypical or common intermediates in template switching28,29.
Linking DNA damage tolerance to PCNA ubiquitination
So how is DNA damage tolerance regulated at the fork? The sliding clamp PCNA is conveniently situated at the replication fork to coordinate DNA replication, DNA repair, and DNA damage tolerance pathways6, and over the past few years, post-translational modification of PCNA has emerged as a key regulatory mechanism controlling DNA damage tolerance (Fig. 1b). During normal replication in yeast, PCNA can be modified with SUMO (small ubiquitin-like modifier protein) at Lys164 to inhibit homologous recombination30,31. In response to DNA damage in yeast32–34, frog35–37, and human cells38,39, PCNA is ubiquitinated at the same conserved residue to facilitate the switch between DNA replication and DNA damage tolerance processes (Box 2). Of particular importance is the sequential manner in which PCNA ubiquitination occurs. PCNA is first monoubiquitinated to promote translesion synthesis33,34,38,39, and then this intermediate undergoes polyubiquitination to promote template switching22,34. This sequence of events places TLS first in line to perform lesion bypass and could provide an additional layer of regulation preceding template switching40–43 (Fig. 1b). Importantly, PCNA is polyubiquitinated through K63-linked chains22,34,43, and neither mono- nor polyubiquitination of PCNA appears to promote its degradation. Instead, these ubiquitination events function to initiate specific DNA damage tolerance pathways.
Box 2: Ubuqitin conjugation pathway
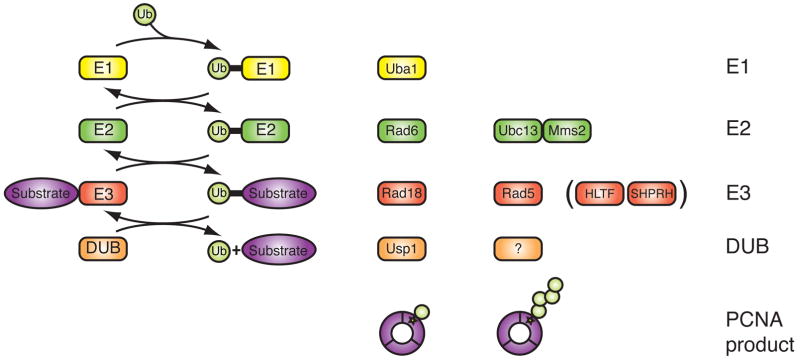
Ubiquitination is a highly conserved process comprised of three concerted reactions55 (Fig. 3). The 76-amino acid ubiquitin is first primed by a ubiquitin-activating enzyme (E1) via a thioester bond in an ATP-dependent manner. The ubiquitin is then transferred to an active site cysteine residue on a ubiquitin-conjugating enzyme (E2). The E2 interacts with its cognate ubiquitin ligase (E3) partner, which provides the target substrate. The E3 completes the transfer of ubiquitin to its designated substrate; conjugation occurs by formation of an isopeptide bond between the C-terminal glycine residue of ubiquitin and the ε-amino group of a lysine residue on the target protein. Monoubiquitinated proteins can undergo further ubiquitination cycles to form polyubiquitin chains, or they can be deubiquitinated by specialized deubiquitinating enzymes (DUBs) that cleave off conjugated ubiquitins, making ubiquitination a reversible process. In the former case, polyubiquitination can occur at one of seven lysine residues on ubiquitin. Until recently, polyubiquitination often referred to K48-linked polyubiquitin chains, which are associated with proteosomal degradation. Increasing evidence has shown that ubiquitin chains formed with alternative linkages, particularly K63-linked polyubiquitin chains, do not promote degradation but rather can direct signaling pathways ranging from DNA repair to chromatin remodeling, much like phosphorylation directs signaling55,86. In addition, polyubiquitin chains can be heterogeneous, containing mixed linkages and of varying lengths, adding to the complexity and diversity of ubiquitin-mediated signaling87. Conjugation of other protein modifiers such as SUMO occurs in a manner analogous to that of ubiquitin86.
Monoubiquitination of PCNA appears to promote DNA damage tolerance by recruiting Y-family TLS polymerases to stalled replication forks10,38,39,44–47 (Fig. 2c). This recruitment is mediated by ubiquitin-binding motifs characteristic of all Y-family TLS polymerases45, and direct interaction between several Y-family TLS polymerases and ubiquitinated PCNA or ubiquitin chains has been observed38,39,45–47. Importantly, these motifs are essential for the accumulation of TLS polymerases at sites of DNA replication, observed as replication foci44–47.
Underscoring the potential link between polymerase recruitment and function, monoubiquitination of PCNA was found to stimulate Polη - and Rev1-dependent bypass of abasic sites in vitro, but not the activity of Polζ, a B-family TLS polymerase that does not contain any known ubiquitin-binding motifs48. It should be noted, however, that a similar study did not see a significant effect of ubiquitination on TLS polymerase activity in vitro49, and the reasons for these differences are unclear. Interestingly, there is still some affinity of several TLS polymerases for unmodified PCNA, albeit weaker than the affinity for ubiquitinated PCNA47. Indeed, photobleaching studies suggest that TLS polymerases are constantly sampling chromatin, both in the absence and presence of modified PCNA, and that PCNA ubiquitination functions to prolong the interaction50. These dynamic properties of the TLS polymerases may be important for allowing replicative polymerases to regain access to the fork once bypass has occurred. Taken together, these observations provide the framework for a plausible model for how PCNA ubiquitination mediates the switch from replicative to TLS polymerases and back10.
One open question is when exactly lesion bypass occurs. One possibility is that bypass is coupled with ongoing replication and occurs immediately after encounter of the lesion by the replication fork. Alternatively, lesion bypass mechanisms may function to fill in gaps during late S phase or early G2, when the majority of replication is complete. Consistent with this idea, ubiquitinated PCNA remains stably bound to chromatin even after the lesions have been removed51, and Rev1, a protein involved in translesion synthesis, is highly expressed during G252.
Given that the ubiquitin-binding motifs found in the Y-family TLS polymerases can interact with monoubiquitinated PCNA as well as polyubiquitin chains, how is binding specificity achieved with countless other ubiquitinated proteins residing in the cell? Part of the answer may lie in other protein-protein interactions (Fig. 2c). Several TLS polymerases contain a PIP (PCNA-interacting protein) box sequence, a motif that mediates the interaction of many proteins with PCNA6. In the case of Polη, access to the DNA template is also facilitated by an interaction with Rad18, the E3 ubiquitin ligase required for PCNA monoubiquitination38. Polκ has also been shown to interact with Rad1844, indicating that this may also be a more general mechanism. Thus, there may be at least three binding interactions that regulate TLS recruitment to stalled forks (Fig. 2c). Another potentially important set of interactions involves Rev1, a TLS polymerase recently shown to bind ssDNA and primer termini53. Interestingly, Rev1 is capable of specifically interacting with several TLS polymerases, suggesting it may aid in the selection and localization of TLS polymerases at stalled replication forks1,9,54.
Despite strong genetic evidence linking polyubiquitination of PCNA to template switching modes of lesion bypass, how PCNA polyubiquitination promotes template switching at the molecular level is poorly defined. One possibility is that the polyubiquitin chains prevent TLS. Specifically, these chains may block access of the TLS polymerases to the DNA template or even lure the TLS polymerases away through direct interaction with ubiquitin-binding motifs in the polymerases45,47. This could allow access of other proteins to the DNA, thereby allowing template switching to occur. Another possibility is that polyubiquitinated PCNA functions as a scaffold by recruiting key effectors or enzymes that carry out template switching processes. Clearly, further investigation is needed to identify the key players and events involved in this error-free mode of DNA damage tolerance.
Enzymes mediating PCNA ubiquitination
The ubiquitination of PCNA requires the proper coordination and assembly of a number of enzymes. All ubiquitination events involve three different classes of enzymes, an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme and an E3 ubiquitin ligase. These enzymes act in a sequential manner to activate and transfer ubiquitin to its substrate55 (Box 2, Fig. 3). Mono- and polyubiquitination of PCNA are mediated by two distinct sets of E2–E3 enzymes that operate in a linear fashion5 (Fig. 1b). PCNA is monoubiquitinated by Rad6, an E2 protein, and the E3 ligase Rad18, which contains a RING-domain necessary for ubiquitination33,34,38,39,48. After monoubiquitination of PCNA by Rad6–Rad18, the lone ubiquitin can be further extended though K63-linkages. This is carried out by an E2 heterodimer, Ubc13-Mms2, in complex with another RING-domain containing E3 ligase known as Rad5 in S. cerevisiae34. In humans, K63-linked polyubiquitination of PCNA appears to involve two E3 ubiquitin ligases, HLTF (helicase-like transcription factor)40,43 and SHPRH (SNF2 histone linker PHD RING helicase)41,42. HLTF and SHPRH share a similar domain architecture with S. cerevisiae Rad5 (Fig. 4a). Consistent with biochemical studies involving S. cerevisiae Rad5, HLTF and SHPRH both interact with the Ubc13 and Mms2 (E2) heterodimer and with Rad18 (E3)40–43,56.
Figure 3. Ubiquitin conjugation pathway and enzymes involved in PCNA ubiquitination.
Cartoon depiction of the ubiquitin conjugation pathway. Ubiquitination occurs through at least three concerted reactions. Enzymes responsible for mono- and polyubiquitination of PCNA are as indicated. Human homologs of yeast Rad5 are in parenthesis. E1 = ubiquitin activating enzyme (yellow). E2 = ubiquitin conjugating enzyme (green). E3 = ubiquitin ligase (red). DUB = deubiquitinating enzyme (orange). Ub = ubiquitin. Substrate is shown in purple.
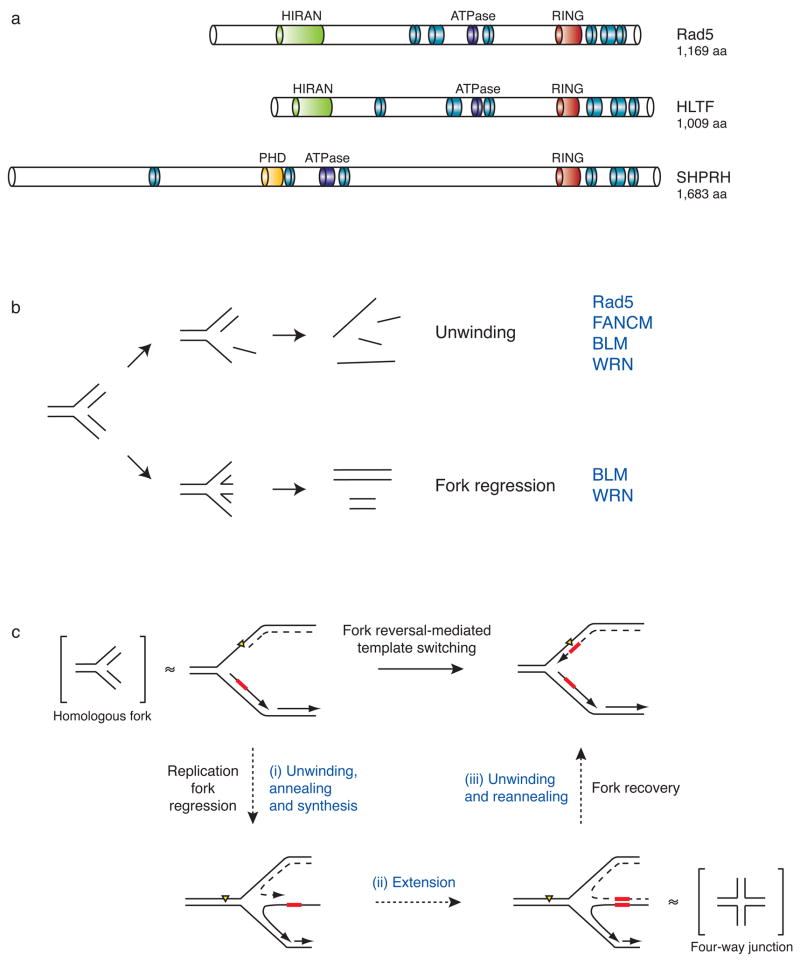
Figure 4. Domain architecture and functions of the E3 ubiquitin ligase Rad5 and its putative orthologs SHPRH and HLTF.
(a) Structural comparisons between S. cerevisiae Rad5 and its putative human orthologs, HLTF and SHPRH. Blue and purple modules represent the seven helicase motifs characteristic of the SWI/SNF2 family of ATP-driven motor proteins. Although these motifs are spread over the length of the protein, they are collectively referred to as a helicase domain. Other domain names and descriptions are as listed through the NCBI conserved domain database and are described in the text.
(b) The possible effects of helicase activity on a model homologous fork substrate. Unwinding and annealing of the nascent and parental DNA strands (bottom) results in fork regression and double-stranded DNA products, while unwinding alone (top) leads to single-stranded DNA products. Helicases known to exhibit these activities are listed in blue.
(c) A possible mechanism for template switching mediated by fork reversal. Fork regression requires (i) concerted unwinding and annealing of the newly synthesized DNA strands, (ii) extension of the strand formed by the stalled polymerase past the sequence where the lesion is found on the parental template, and (iii) unwinding of the newly formed duplex so that the nascent strands can reanneal to their original templates and restore the fork to its proper conformation. Model four-way junction and homologous fork structures, which are known substrates of Rad5 in vitro62, are placed in brackets adjacent to the fork structures they are thought to mimic. Yellow triangle = replication-blocking lesion. Dashed line = leading strand. Red box = template.
Monoubiquitinated PCNA can also be deubiquitinated by the cysteine protease Usp1 (ubiquitin specific protease 1)57 (Fig. 1b). In response to UV, Usp1 undergoes autocleavage and subsequent degradation, thereby allowing monoubiquitinated PCNA to persist57. Moreover, knockdown of Usp1 leads to increased UV-induced mutagenesis57, presumably due to increased TLS polymerase activity. Thus, TLS may be regulated both at the level of ubiquitination and deubiquitination. Interestingly, Usp1 degradation is observed following UV treatment but not treatment with methyl methanesulfonate (MMS) or mitomycin C (MMC), suggesting that deubiquitination may be a UV-specific mode of regulating TLS51.
Rad5: Not just your ordinary E3 ubiquitin ligase
Perhaps one of the most interesting questions surrounding the ubiquitination machinery is why there appear to be two orthologs of Rad5 in humans (Fig. 4a). Both HLTF and SHPRH are capable of polyubiquitinating PCNA in vitro40,41. Moreover, overexpression of either protein in human cells leads to increased PCNA polyubiquitination in vivo40,42,43. These observations suggest that HLTF and SHPRH are capable of eliciting the same response polyubiquitination of PCNA in the absence of the other, raising the possibility that they are redundant for this function. However, loss of either HLTF or SHPRH alone was shown to result in a reduction in PCNA ubiquitination, increased sensitivity to MMS, and increased numbers of chromosome breaks following MMS40,42,43, indicating that HLTF and SHPRH have non-overlapping functions.
One possible reason for the divergence of Rad5 into two separate proteins is that both proteins have substrates other than PCNA, and/or roles in DNA repair or chromosome stability that are independent of PCNA ubiquitination. Several observations are consistent with this idea58–60. For example, Rad5 was shown to have a role in double-strand DNA break repair that is dependent on its ATPase activity but independent of its ubiquitin function60. Why HLTF and SHPRH are both required for PCNA ubiquitination is less clear. It is possible that both proteins work together to promote more efficient ubiquitination. Indeed, overexpression of HLTF or SHPRH together with the E2, Ubc13-Mms2, leads to more efficient polyubiquitination43. In addition, both proteins interact with each other43. In this context, it should also be pointed out that current methods for assessing polyubiquitination are crude and nonspecific. Thus, it remains possible that the polyubiquitin chains produced by SHPRH and HLTF in vivo are dissimilar and that these differences affect their function and the type of template switching.
The idea that SHPRH and HLTF might have other functions is consistent with the structure of these proteins, which is more complex than a simple RING-finger-containing protein (Fig. 4a). One intriguing domain found in HLTF and SHPRH, as well as Rad5, is the conserved helicase-like domain shared by members of the SWI2/SNF2 family of chromatin remodeling proteins61. This helicase domain spans most of the length of each protein and embedded within it are both an ATPase domain and a RING domain.
Interestingly, Rad5 was recently shown to exhibit helicase activity on four way-junctions and fork structures with homologous arms, simultaneously unwinding and reannealing these structures to generate double-stranded products62 (Fig. 4b). In eukaryotes several other DNA helicases, including the Bloom (BLM) and Werner (WRN) helicases of the RecQ family63–66, as well as the Fanconi anemia protein FANCM67–69, are also capable of regressing fork structures with homologous arms in vitro. However, both BLM and WRN also generate single-stranded products65,70 (Fig. 4b). The ability of Rad5 to unwind and reanneal homologous fork structures in a concerted fashion has led to the model that this activity may be needed at replication forks during fork regression and template switching62 (Fig. 4c). Whether any or all of these helicase activities are important for DNA damage tolerance and whether HLTF and SHPRH also have helicase activity is not yet clear.
HLTF and SHPRH also contain other domains that may be involved in chromatin remodeling and DNA binding (Fig. 4a). A PHD (plant homeodomain) finger motif found in SHPRH has been shown to recognize specific methyl-lysine residues in histone tails as well phosphoinositides71. Further, the PHD finger is known to function as a ligase for both ubiquitin and SUMO72. The HIRAN domain found in Rad5 and HLTF is a hypothetical domain found in a group of known chromatin/DNA binding proteins, suggesting it could be important for recognition of damaged DNA or stalled replication forks73. Together, the diversity of functions represented by these many domains raises questions about possible roles for HLTF and SHPRH in DNA damage tolerance, DNA repair and chromatin remodeling pathways, and how these functions are regulated and coordinated.
Molecular requirements for PCNA ubiquitination
Much of what is known about the regulation of DNA damage tolerance pathways is based on studies characterizing the requirements for PCNA ubiquitination. PCNA is monoubiquitinated in response to several different types of genotoxic agents34,74, and what the strong effectors of PCNA ubiquitination have in common is the ability to block replication fork progression. Replication fork stalling uncouples the activities of the replicative helicase and polymerase, allowing the helicase to continue to unwind DNA while the polymerase remains stalled75,76. The result is the accumulation of primed single-stranded DNA (ssDNA). These observations suggest that ssDNA may be the structure recognized by the PCNA ubiquitination machinery during DNA replication. Indeed, primed-ssDNA is both necessary and sufficient to induce PCNA ubiquitination in Xenopus egg extracts36 and in vitro with purified yeast proteins48. Moreover, when ssDNA formation at the fork is blocked by preventing helicase-polymerase uncoupling, PCNA ubiquitination does not occur36.
So how is ssDNA recognized? A complex of Rad18 and Rad6 is known to bind ssDNA and stalled fork structures in vitro77,78. More relevant perhaps is the finding that Rad18 is recruited to ssDNA by replication protein A (RPA)74, a ssDNA binding protein that coats DNA during replication and accumulates at stalled forks79. Thus, Rad18 functions as a DNA damage sensor protein that is recruited along with Rad6 to RPA-ssDNA at stalled forks, where it monoubiquitinates PCNA (Fig. 5).
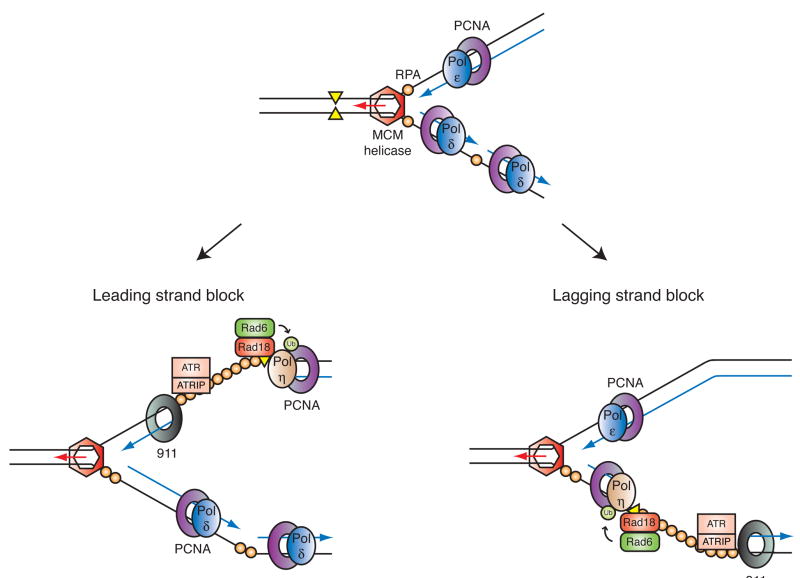
Figure 5. Coordinated activation of PCNA ubiquitination and the ATR-dependent checkpoint response at a stalled replication fork.
Primed ssDNA accumulates at stalled replication forks when a polymerase stalls. In apparently independent processes, the resulting structure supports assembly of the proteins required for PCNA ubiquitination (Rad6 and Rad18) as well as the proteins required for activation of the ATR-dependent checkpoint pathway. For simplicity, only a few checkpoint proteins are depicted here: the 911 checkpoint clamp and the ATR-ATRIP heterodimer. Proper assembly of these proteins leads to phosphorylation of the downstream effector kinase Chk1 and cell cycle arrest. Shown are the presumed effects of stalling the leading (left) and lagging (right) strand polymerases. On the leading strand, ssDNA accumulates upon functional uncoupling of MCM helicase and replicative DNA polymerase activities, and in this case, the 5′-primer end thought to be required for checkpoint activation could be provided by the adjacent origin, or possibly replication restart. Note how a single stalled fork (i.e. gap) can be detected simultaneously by the two PCNA and 911 clamps.
What then triggers the switch from mono- to polyubiquitination of PCNA and, by extension, translesion synthesis to template switching? Polyubiquitination of PCNA is observed with increasing amounts of DNA damage36,43, and in Xenopus egg extracts, it is correlated with the accumulation of RPA-coated ssDNA36. In addition, Rad18 and Rad5 interact in yeast through the same domains required for their homodimerization (or multimerization)56. Based on this and other observations, the interaction between Rad5 and Rad18 has been hypothesized to coordinate the switch from mono- to polyubiquitination of PCNA through a physical coupling of the two sets of ubiquitin enzymes56. One intriguing model that could then link template switching to the accumulation of RPA is that the amount of Rad18 bound to RPA-ssDNA could affect the binding of Rad5 to Rad18. Accumulation of RPA-ssDNA may result in more Rad18-RPA interactions and less Rad18 homodimerization, allowing Rad5 to better compete for binding to Rad18 and to promote polyubiquitination of PCNA. Although SHPRH and HLTF each interact with Rad1840–43 and at least SHRPH can interact with itself42, further studies are needed to address whether these interactions occur in a similar fashion to yeast Rad5 and Rad18.
Coordination of DNA damage responses at the replication fork
The ability of RPA-coated ssDNA to induce ubiquitination of PCNA raises another interesting problem, namely how checkpoint and DNA damage tolerance processes are coordinated at the fork. A number of studies suggest RPA-ssDNA is a central component of the checkpoint activating signal79 and primed RPA-coated ssDNA is sufficient for activation of the ATR checkpoint pathway in Xenopus egg extracts80. The obvious question becomes whether and to what degree checkpoint activation and DNA damage tolerance are linked. Despite similarities in the structure of the activating signal, ATR function is not required for PCNA ubiquitination in yeast32,74 or Xenopus laevis35,36 and most studies in human cells are consistent with this idea44,51,81. However, other checkpoint proteins including Claspin and the effector kinase Chk1 are required for maximal PCNA ubiquitination81. Whether or not these checkpoint proteins directly or indirectly affect the recruitment of Rad18 to RPA-ssDNA is not clear. Conversely, loss of PCNA ubiquitination does not affect activation of the ATR pathway32,74. Thus, it seems that primed ssDNA initiates two DNA damage response pathways in parallel: the ATR-dependent checkpoint and PCNA-mediated DNA damage tolerance.
How then do the factors that mediate checkpoint activation and DNA damage tolerance compete for the same activating structure -- primed RPA-ssDNA? Each pathway requires proper assembly of its own ssDNA-binding protein and clamp: Rad18 and PCNA are required for PCNA ubiquitination5 while the RPA-binding protein ATRIP (ATR-interacting protein) and the Rad9-Hus1-Rad1 (911) checkpoint clamp are necessary for ATR activation2. At least part of the solution may lie in clamp specificity, as it appears that the 911 complex loads preferentially onto the 5′ end of the primer when RPA is present and the checkpoint is activated79, while PCNA occupies the 3′ end during processive DNA replication6 (Fig. 5). Thus, a single stalled fork may be able to accommodate both complexes simultaneously.
Although ATR activity is dispensable for PCNA ubiquitination, this does not preclude a role for the checkpoint pathway in directing other steps in the DNA damage tolerance process. Indeed, genetic studies in yeast suggest a role for the ATR-mediated checkpoint pathway in regulating TLS polymerase activity independently of PCNA monoubiquitination50,82–84. Of particular interest is the observation that the 911 checkpoint clamp physically interacts with two TLS polymerases82,84. It is also worth noting that Rad1885 and possibly also HLTF and SHPRH may be substrates of the checkpoint kinases. Clearly, further work is needed to delineate the connections between the checkpoint and DNA damage tolerance pathways, in particular how the checkpoint may regulate TLS polymerase recruitment and activity and the switch between error-free and error-prone pathways.
Concluding remarks
Proper regulation of DNA damage tolerance and mutagenic DNA synthesis is essential for maintaining genome stability. Increased use of a mutagenic pathway over an error-free pathway would increase the frequency of mutations, while a failure to bypass lesions could lead to replication fork collapse and chromosomal translocations. Recent insights regarding the role of PCNA ubiquitination in the control of DNA damage tolerance have brought a more molecular perspective to what was only recently a genetic phenomenon, but they also raise many more questions.
For instance, the sequential nature by which translesion synthesis precedes template switching raises the issue of why the cell would choose a more error-prone form of DNA damage tolerance as its first line of defense at stalled replication forks. The initial use of pro-mutagenic TLS polymerases over error-free template switching is somewhat counterintuitive. It is also not clear how the different TLS polymerases are paired with a lesion if TLS polymerases are recruited by the monoubiquitinated form of PCNA, how is the most appropriate polymerase chosen? Related to this issue is how the structure of PCNA affects DNA damage tolerance. The trimeric nature of this protein raises the possibility that each of the three subunits could undergo a different modification to coordinate processive DNA replication, translesion synthesis, and template switching. Another question is how the cell decides when to move from TLS to template switching, and whether the decision is based on problems at the level of a single fork versus multiple forks. Finally, the biggest mystery may be how polyubiquitination of PCNA directs template switching. We have virtually no understanding of the key players and events involved in this error-free mode of lesion bypass, and this “black box” of DNA damage tolerance is sure to keep many researchers busy in the years to come.
More defined approaches at the single-molecule and single-fork levels would provide much needed insight into how PCNA coordinates multiple roles in the DNA damage response. Current methods are limited to the analysis of heterogeneous populations of PCNA molecules and stalled forks. Functional read-outs are also crude and non-specific, relying solely on changes in mutation frequency without any way to distinguish between accurate translesion synthesis events and template switching. The development of an in vitro system that allows one to study and manipulate the regulation of these processes and the nature of PCNA modifications would thus be useful, as would more high-throughput and more information-rich ways of assessing DNA damage tolerance. Finally, it seems likely that a better knowledge of the key players and events involved could be used to design molecules which control which mode of DNA damage tolerance the cell uses. For instance, specific inhibitors of different ubiquitin ligases or helicases could turn off template switching modes of lesion bypass or DNA damage tolerance altogether. An increased reliance on pro-mutagenic TLS polymerases or the absence of tolerance pathways might be useful in the context of cancer as it may cause a cancer cell to make more mistakes than it can tolerate, effectively pushing it towards apoptosis.
This past decade has reshaped our perspective of mutagenesis. The discovery of pro-mutagenic enzymes and PCNA’s role in directing DNA damage tolerance has opened up new and exciting avenues of research. The next decade is sure to offer many more surprises in unraveling the mystery of template switching, as well as some insight into how and why the cell chooses to make mistakes.
Acknowledgments
The authors would like to thank Dr. Thomas Wandless, Angela Hahn, Jia-Ren Lin and Michelle Zeman for helpful discussions and critical reading of the manuscript.
Footnotes
Competing Financial Interests Statement
The authors declare they have no competing financial interests.
References
- 1.Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 2.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 4.Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- 6.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 7.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 11.Fujii S, Fuchs RP. Interplay among replicative and specialized DNA polymerases determines failure or success of translesion synthesis pathways. J Mol Biol. 2007;372:883–893. doi: 10.1016/j.jmb.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 12.Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence CW, Christensen R. UV mutagenesis in radiation-sensitive strains of yeast. Genetics. 1976;82:207–232. doi: 10.1093/genetics/82.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemontt JF. Mutants of Yeast Defective in Mutation Induced by Ultraviolet Light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 16.McCulloch SD, et al. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci USA. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 20.Broomfield S, Chow BL, Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc Natl Acad Sci USA. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broomfield S, Hryciw T, Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res. 2001;486:167–184. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 22.Chiu RK, et al. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2006;2:e116. doi: 10.1371/journal.pgen.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao W, Chow BL, Broomfield S, Hanna M. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics. 2000;155:1633–1641. doi: 10.1093/genetics/155.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J Mol Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann AR. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J Mol Biol. 1972;66:319–337. doi: 10.1016/0022-2836(72)90418-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara Y, Tatsumi M. Replicative bypass repair of ultraviolet damage to DNA of mammalian cells: caffeine sensitive and caffeine resistant mechanisms. Mutat Res. 1976;37:91–110. doi: 10.1016/0027-5107(76)90058-0. [DOI] [PubMed] [Google Scholar]
- 28.Cotta-Ramusino C, et al. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 29.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 32.Frampton J, et al. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:2976–2985. doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 34.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 35.Gohler T, Munoz IM, Rouse J, Blow JJ. PTIP/Swift is required for efficient PCNA ubiquitination in response to DNA damage. DNA Repair (Amst) 2008;7:775–787. doi: 10.1016/j.dnarep.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J Biol Chem. 2006;281:32081–32088. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- 37.Leach CA, Michael WM. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J Cell Biol. 2005;171:947–954. doi: 10.1083/jcb.200508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe K, et al. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 40.Unk I, et al. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci USA. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unk I, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motegi A, et al. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol. 2006;175:703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motegi A, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci USA. 2008;105:12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bi X, et al. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 46.Guo C, Tang TS, Bienko M, Dikic I, Friedberg EC. Requirements for the interaction of mouse Polkappa with ubiquitin and its biological significance. J Biol Chem. 2008;283:4658–4664. doi: 10.1074/jbc.M709275200. [DOI] [PubMed] [Google Scholar]
- 47.Plosky BS, et al. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc Natl Acad Sci USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabbioneda S, et al. Effect of proliferating cell nuclear antigen ubiquitination and chromatin structure on the dynamic properties of the Y-family DNA polymerases. Mol Biol Cell. 2008;19:5193–5202. doi: 10.1091/mbc.E08-07-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niimi A, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc Natl Acad Sci USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masuda Y, Kamiya K. Role of single-stranded DNA in targeting REV1 to primer termini. J Biol Chem. 2006;281:24314–24321. doi: 10.1074/jbc.M602967200. [DOI] [PubMed] [Google Scholar]
- 54.Tissier A, et al. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair (Amst) 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 55.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 56.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 58.Gangavarapu V, et al. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:7783–7790. doi: 10.1128/MCB.01260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pages V, et al. Requirement of Rad5 for DNA polymerase zeta-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2008;180:73–82. doi: 10.1534/genetics.108.091066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S, Davies AA, Sagan D, Ulrich HD. The RING finger ATPase Rad5p of Saccharomyces cerevisiae contributes to DNA double-strand break repair in a ubiquitin-independent manner. Nucleic Acids Res. 2005;33:5878–5886. doi: 10.1093/nar/gki902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blastyak A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Constantinou A, et al. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 66.Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 67.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 68.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci USA. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun W, et al. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 72.Peng J, Wysocka J. It takes a PHD to SUMO. Trends Biochem Sci. 2008;33:191–194. doi: 10.1016/j.tibs.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 73.Iyer LM, Babu MM, Aravind L. The HIRAN domain and recruitment of chromatin remodeling and repair activities to damaged DNA. Cell Cycle. 2006;5:775–782. doi: 10.4161/cc.5.7.2629. [DOI] [PubMed] [Google Scholar]
- 74.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 78.Tsuji Y, et al. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 2008;13:343–354. doi: 10.1111/j.1365-2443.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- 79.Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–885. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- 80.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–1152. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sabbioneda S, et al. The 9-1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous polzeta-dependent mutagenesis in Saccharomyces cerevisiae. J Biol Chem. 2005;280:38657–38665. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- 83.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16:586–590. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kai M, Wang TS. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 86.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 87.Kim HT, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]