Abstract
Methods to simultaneously localize the positions of multiple single fluorophores by precisely determining their individual positions are now yielding impressive gains in fluorescence microscopy resolution.
Optical microscopy has been a mainstay of cellular biology ever since van Leeuwenhoek used Hooke’s microscope to observe the strange biological objects he called “animal-cules.” Using light for biological microscopy takes advantage of its relatively noninvasive character, and with high magnification one can work at a distance, observing small objects. Fluorescence microscopy depends upon a variety of labeling techniques to light up different structures in cells, but the price often paid for using visible light is the relatively poor resolution compared to X-ray or electron microscopy. The basic problem is that fundamental diffraction effects limit the resolution of standard optical microscopy to a dimension of roughly the optical wave-length divided by two, λ/2, that is, ∼250 nm for visible light of 500 nm wavelength. In recent work, the light from single fluorescent molecules about 1–2 nm in size has provided a way around this problem.
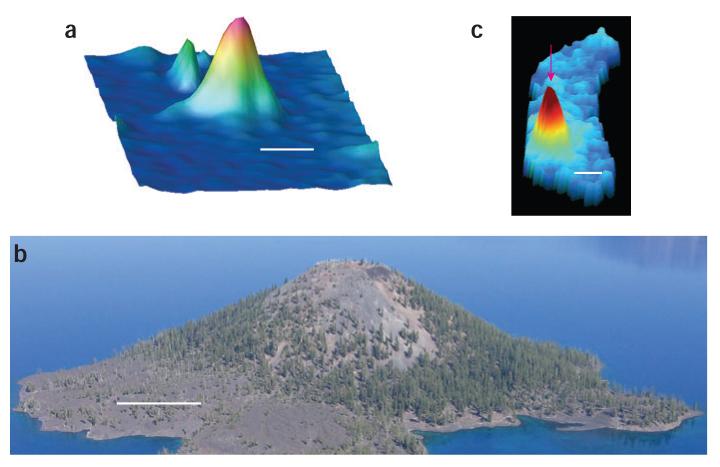
How can single molecules help? Since 1989, researchers worldwide have been using optics to detect and measure individual molecules in solids, liquids and biological systems. For example, Figure 1a is a 1991 image of single molecules of pentacene in a crystal1. Each single molecule acts as a nanoscale light source, where the light is generated by pumping the pentacene molecules one at a time, each of which is about 2 nm in size. But owing to diffraction effects (and the large laser spot in a cryostat), scanning a laser spot over a single molecule yielded the much wider mountain-like peaks shown and actually provided a way to measure the laser spot size (here about 5 μm).
Figure 1.
Measuring mountain peaks for superresolution imaging. (a) Position-frequency image of single molecules of pentacene in a crystal1, as a pumping laser beam is moved across the sample and the fluorescence is recorded. (b) It is easily possible to walk to the top of the cinder cone in Crater Lake to find the position of the peak accurately. (c) Similarly, in a living bacterial cell (blue)2, the image of a can determine its position (pink arrow) to much less than the width of the “mountain.” By turning single-molecule light sources like this on and off in a cell, researchers have recently obtained superresolution single fluorescent protein molecule appears as a wide spot due to fundamental diffraction effects, but one images. Scale bars: a, 5 × 103 nm; b, ∼120 × 109 nm; c, 250 nm.
Recently, several researchers have begun to take advantage of the nanoscale size of single-molecule emitters more directly. The basic idea is illustrated in Figure 1b. Everyone knows that a hiker can walk to the top of a mountain with a global positioning system device to nail down the position of the peak quite accurately. In optical microscopy, the observed ‘peak’ from a single point source of light is called the point-spread function (PSF) of the microscope. Figure 1c illustrates this PSF for emission from a single molecule of the bacterial actin protein MreB (the mountain, labeled by fusion to yellow fluorescent protein) in a bacterial cell2. Simply by measuring the shape of the PSF, the position of its center can be determined much more accurately than its width. This idea, digitizing the PSF, a form of simple deconvolution, has been known for many years, but deconvolution alone can generate spurious features in the presence of noise. The knowledge that a single small object is producing the PSF helps tremendously in determining the centroid position with nanometer-scale accuracy, and this idea was first applied with single beads3 and then to single-molecule images4–6.
In the last two years, a plethora of acronyms has been proposed for biological imaging beyond the diffraction limit based on digitizing the PSF. In Selvin’s FIONA7, the position of single labeled myosin molecules was determined to an accuracy as small as a few nanometers, limited now by the number of photons before photobleaching (which controls the signal-to-noise ratio) rather than diffraction effects. But there is still a problem for imaging of complex structures in cells: keeping the single molecules more than ∼500 nm apart so that their mountains do not overlap.
The solution to this (apparently arising from work both from Hell et al.8 and Qu et al.9 in the method termed NALMS) was to photoswitch the little single-molecule emitters on and off in various ways. In the PALM method of Betzig et al.10, light-induced photoactivation of GFP mutant fusions was used to randomly turn on only a few single molecules at a time in fixed cell sections or in fixed cells. In a tour-de-force effort, a few photoactivated molecular PSFs were imaged in detail to find their positions to ∼20 nm, then they were photobleached so that others could be turned on, and so on; after 2–12 h of imaging and many thousands of molecules, a high-resolution image was extracted that correlated well with a TEM image.
Lidke et al.11 showed that superresolution beyond the diffraction limit can also be achieved with the blinking of fluorescent semiconductor quantum dots, and these emitters have allowed observation of timedependent, nanometer-sized steps in cells by the Xie group12. In another approach, the STORM method of Zhuang13, a single photo-switchable molecule on DNA or an antibody is turned on and off, again and again, to measure its position accurately with ∼20 nm resolution. This method uses a Cy3-Cy5 pair in close proximity, which shows a novel property: restoration of Cy5’s bleached emission by brief pumping of the Cy3 molecule. The controllability of a reversible photoswitch based on these good single-molecule emitters is appealing, but since these molecules can-not be genetically encoded in cells, the way autofluorescent proteins are encoded, other methods of implanting properly formed Cy3-Cy5 pairs in cells need to be developed.
Finally, in an alternative approach, Hell’s group is using a powerful but sophisticated laser method called STED microscopy14 to make the PSF itself much smaller, a step that will improve, in principle, all the other super-resolution methods at the cost of somewhat higher complexity.
All of these tantalizing new approaches to superresolution in biological imaging have advantages and disadvantages, and which method will achieve widespread use and useful time resolution to observe nanoscale cellular dynamics is yet to be determined. Better single-molecule emitters for cellular labeling would certainly help, as all superresolution methods have accuracy limitations arising from the finite number of photons available from a single molecule before photobleaching. It is an exciting time for single molecules as nanoscale light sources, indeed!
References
- 1.Ambrose WP, Basché T, Moerner WE. J. Chem. Phys. 1991;95:7150–7163. [Google Scholar]
- 2.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Proc. Natl. Acad. Sci. USA. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelles J, Schnapp BJ, Sheetz MP. Nature. 1988;331:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 4.Güttler F, Irngartinger T, Plakhotnik T, Renn A, Wild UP. Chem. Phys. Lett. 1994;217:393–397. [Google Scholar]
- 5.van Oijen AM, Kohler J, Schmidt J, Muller M, Brakenhoff GJ. Chem. Phys. Lett. 1998;292:183–187. [Google Scholar]
- 6.Thompson RE, Larson DR, Webb WW. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yildiz A, et al. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 8.Hell SW, Jakobs S, Kastrup L. Appl. Phys. A. 2003;77:859–860. [Google Scholar]
- 9.Qu X, Wu D, Mets L, Scherer NF. Proc. Natl. Acad. Sci. USA. 2003;101:11298–11303. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betzig E, et al. Science Express. 2006 August 10; published online. [Google Scholar]
- 11.Lidke KA, Rieger B, Jovin TM, Heintzmann R. Opt. Express. 2005;13:7052–7062. doi: 10.1364/opex.13.007052. [DOI] [PubMed] [Google Scholar]
- 12.Na X, Sims PA, Chen P, Xie XS. J. Phys. Chem. B. 2005;109:24220–24224. doi: 10.1021/jp056360w. [DOI] [PubMed] [Google Scholar]
- 13.Rust MJ, Bates M, Zhuang X. Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willig KI, et al. Nat. Methods. 2006;3:721–723. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]