Abstract
Targeting genomic loci by massively parallel sequencing requires new methods to enrich templates to be sequenced. We developed a capture method that uses biotinylated RNA “baits” to “fish” targets out of a “pond” of DNA fragments. The RNA is transcribed from PCR-amplified oligodeoxynucleotides originally synthesized on a microarray, generating sufficient bait for multiple captures at concentrations high enough to drive the hybridization. We tested this method with 170-mer baits that target >15,000 coding exons (2.5 Mb) and four regions (1.7 Mb total) using Illumina sequencing as read-out. About 90% of uniquely aligning bases fell on or near bait sequence; up to 50% lay on exons proper. The uniformity was such that ~60% of target bases in the exonic “catch”, and ~80% in the regional catch, had at least half the mean coverage. One lane of Illumina sequence was sufficient to call high-confidence genotypes for 89% of the targeted exon space.
The development and commercialization of a new generation of increasingly powerful sequencing methodologies and instruments1–4 has lowered the cost per nucleotide of sequencing data by several orders of magnitude. Within a short time, several individual human genomes have been sequenced on “next-generation” instruments3,5–7, with plans and funding in place to sequence more (www.1000genomes.org).
Sequencing entire human genomes will be an important application of next-generation sequencing. However, many research and diagnostic goals may be achieved by sequencing a specific subset of the genome in large numbers of individual samples. For example, there may be substantial economy in targeting the protein-coding fraction, the “exome”, which represents only ~1% of the human genome. The economy is even greater for many key resequencing targets, such as genomic regions implicated by whole-genome association scans and the exons of sets of protein-coding genes implicated in specific diseases. Efficient and cost-effective targeting of a specific fraction of the genome could substantially lower the sequencing costs of a project, independent of the sequencing technology used.
Sequencing targeted regions on massively parallel sequencing instruments requires developing methods for massively parallel enrichment of the templates to be sequenced. Recognizing the inadequacy of traditional single- or multiplex PCR for this purpose, several groups have developed “genome-partitioning” methods for preparing complex mixtures of sequencing templates that are highly enriched for targets of interest8–15. Only two of these methods have been tested on target sets complex enough to match the scale of current next-generation sequencing instruments.
The first method, microarray capture9,12,13, uses hybridization to arrays containing synthetic oligonucleotides matching the target sequence to capture templates from randomly sheared, adapter-ligated genomic DNA; it has been applied to more than 200,000 coding exons12. Array capture works best for genomic DNA fragments that are ~500 bases long12, thereby limiting the enrichment and sequencing efficiency for very short dispersed targets such as human protein-coding exons that have a median size of 120 bp16.
The second method, multiplex amplification14, uses oligonucleotides that are synthesized on a microarray, subsequently cleaved off and PCR-amplified, to perform a padlock and molecular-inversion reaction17,18 in solution where the probes are extended and circularized to copy rather than directly capture the targets. Uncoupling the synthesis and reaction formats in this manner is an advantage in that it allows re-using and quality testing of a single lot of oligonucleotide probes. However, the padlock reaction is far less understood than a simple hybridization and has not been properly optimized for this purpose. At the time of publication14, multiplex amplification missed more than 80% of the targeted exons in any single reaction and showed highly uneven representation of sequencing targets, poor reproducibility between technical replicates, and uneven recovery of alleles. A more recent non-sequencing-based study using a similar approach suggests that the uniformity, reproducibility and efficiency of multiplex amplification can be improved15.
Here we describe a new method, developed independently, that overcomes some of the weaknesses of previous methods. It combines the simplicity and robust performance of oligonucleotide hybridization with the advantages of amplifying array-synthesized oligonucleotides and performing the selection reaction in solution.
RESULTS
Hybrid selection method
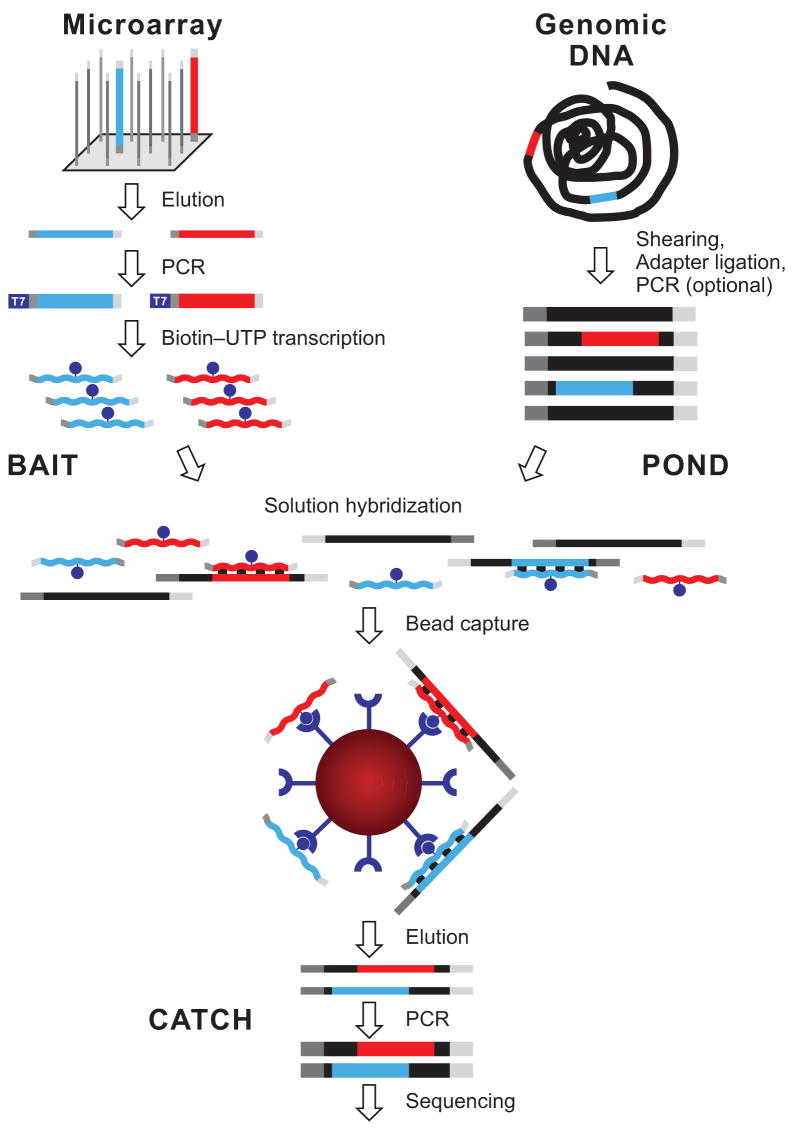
We developed a method for capturing sequencing targets that combines the flexibility and economy of oligonucleotide synthesis on a microarray with the favorable kinetics of hybridization in solution (Fig. 1). A complex pool of ultra-long 200-mer oligonucleotides is synthesized in parallel on an Agilent microarray and then cleaved from the array. Each oligonucleotide consists of a target-specific 170-mer sequence flanked by 15 bases of a universal primer sequence on each side to allow PCR amplification. After the initial PCR, a T7 promoter is added in a second round of PCR. We then use in vitro transcription in the presence of biotin-UTP to generate a single-stranded RNA hybridization “bait” for “fishing” targets of interest out of a “pond” of randomly sheared, adapter-ligated and PCR-amplified total human DNA. The hybridization is driven by the vast excess of RNA baits that cannot self-anneal. The “catch” is pulled-down with streptavidin-coated magnetic beads, PCR-amplified with universal primers, and analyzed on a “next-generation” sequencing instrument. The method allows preparation of large amounts of bait from a single oligonucleotide array synthesis that can be quality control tested, stored in aliquots and used repeatedly over the course of a large-scale targeted sequencing project.
Figure 1.
Overview of hybrid selection method. Illustrated are steps involved in the preparation of a complex pool of biotinylated RNA capture probes (“bait”; top left), whole-genome fragment input library (“pond”; top right) and hybrid-selected enriched output library (“catch”; bottom). Two sequencing targets and their respective baits are shown in red and blue. Thin and thick lines represent single and double strands, respectively. Universal adapter sequences are grey. The excess of single-stranded non-self-complementary RNA (wavy lines) drives the hybridization. See main text and Methods for details.
Capturing and sequencing exon targets
For a pilot study, we selected a set of 1,900 human genes randomly chosen to ensure unbiased sampling regardless of length, repeat content or base composition. We designed 22,000 “bait” sequences of 170 bases in length, targeting all 15,565 protein-coding exons of these genes. The baits were tiled without overlap or gaps such that the entire coding sequence was covered (see Methods). This simple design minimizes the number of synthetic oligonucleotides required; for 75% of all coding exons in the human genome, a single oligonucleotide would be sufficient. As the median size of protein-coding exons is only 120 bp16, many baits extend beyond their target exon. Our test baits for catching exons constituted 3.7 Mb, and the targeted exons comprised 2.5 Mb (67%).
Our “pond” consisted of genomic DNA, derived from a human cell line (Coriell GM15510), that had been randomly sheared, ligated to standard Illumina sequencing adapters, size-selected to 200–350 bp (mean insert size ~250 bp), and PCR-amplified for 12 cycles. We hybridized 500 ng of this whole-genome fragment library with 500 ng biotinylated RNA bait, PCR-amplified the hybrid-selected DNA and generated 36-base sequencing reads off the Illumina adapter sequence at the ends of each fragment. We obtained 85 Mb of sequence that aligned uniquely to the human genome; 76 Mb was on or within 500 bp of a bait.
Of the specifically captured 76 Mb of sequence, 49 Mb (65%) lay directly on a bait. The proportion of this sequence directly within the exons (36 Mb total) closely matched the proportion of exonic sequence within the bait. Overall, 58% and 42% of the 85 Mb uniquely aligning human sequence mapped to baits and exons, respectively.
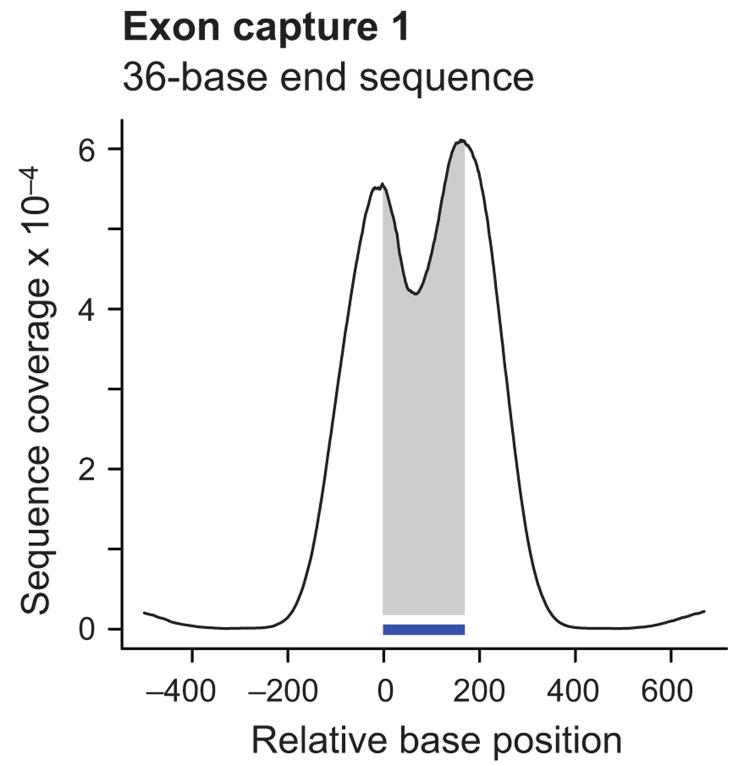
The high stringency of hybridization selects for fragments that contain a substantial portion of the bait sequence. As a result, fragments with both ends near or outside the ends of the bait sequence are overrepresented relative to fragments that have less overlap and thus end near the middle. Merely end-sequencing the fragments with short 36-base reads therefore leads to elevated coverage near the end of the baits, with many reads falling outside the target, and a pronounced dip in coverage in the center. This effect is evident in the cumulative coverage profile representing 7,052 free-standing single-bait targets (Fig. 2a).
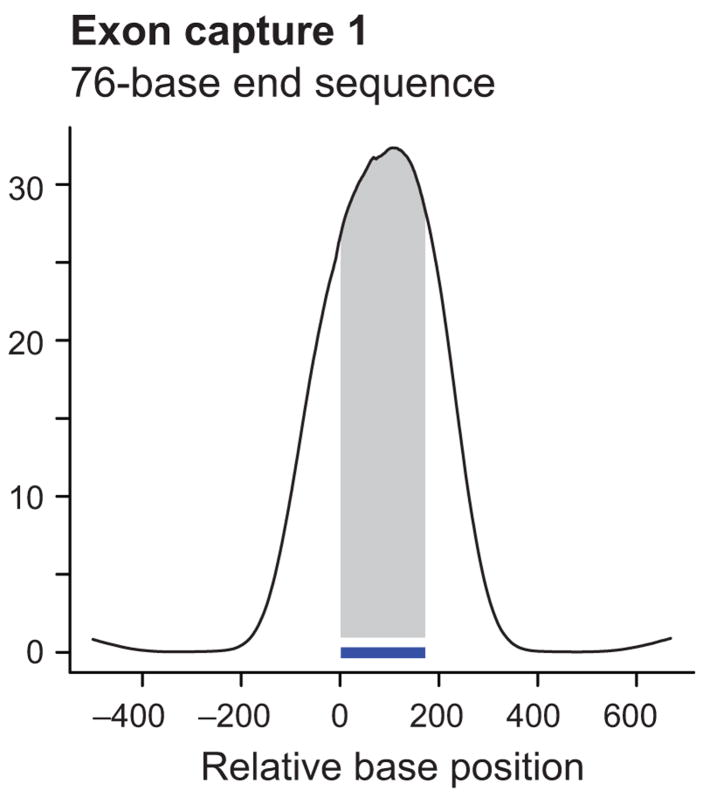
Figure 2.
Coverage profiles of exon targets by end sequencing and shotgun sequencing. Shown are cumulative coverage profiles that sum the per-base sequencing coverage along 7,052 single-bait target exons. Only free-standing baits that were not within 500 bases of another one were included in this analysis. End sequencing of exon capture 1 with 36-base reads (a) produced a bimodal profile with high sequence coverage near and slightly beyond the ends of the 170-base baits (indicated by the horizontal bar). Shotgun sequencing of capture 2 from a different pond library (containing fragments with generic rather than Illumina-specific adapters) with 36-base reads after concatenating and re-shearing (b) gave more coverage on bait (shaded area) than near bait. Re-sequencing of capture 1 with 76-base end reads (c) had a similar effect, although the peak was slightly wider and the on-bait fraction of the peak area slightly less. Note that the scale on the Y-axis and hence the absolute peak height is different in each case. The different scales reflect the different numbers of sequenced bases which is much lower for GA-I lanes (a, b) than for a GA-II lane (c).
To improve coverage in the middle, we replaced end sequencing of the catch with shotgun sequencing of the catch. Specifically, we changed the Illumina adapter on the whole-genome fragment library to a generic sequencing-platform independent adapter and amplified the catch with PCR primers carrying a NotI site at their 5′ ends. NotI-digestion of the PCR product generates sticky ends and facilitates concatenation by co-ligation for subsequent re-shearing and shotgun sequencing of the hybrid selected DNA. This modification to the protocol shifted the coverage to the middle (Fig. 2b). About 90 of 102 Mb unique human sequence (88%) aligned within 500 bases of a bait. The proportion of bait sequence in the specific catch (90 Mb) rose from 65% to 77% (69 Mb; 51 Mb thereof on exon). The fraction of bait and exon sequence in the uniquely aligning human Illumina sequence was 67% and 50%, respectively.
Although shearing the catch improved the proportion of bait sequence, the process adds an additional round of library construction with associated costs, amplification steps, and potential biases. It also generates reads containing uninformative adapter sequence as a by-product. During the course of these experiments, it became possible to increase the sequence read length on the Illumina platform. We reasoned that simply increasing the read length would also increase coverage in the middle and thus obviate the need for shotgun-library construction. Indeed, we performed end sequencing of the very same catch that had produced the bimodal coverage profile shown in Figure 2a, this time running 76-base instead of 36-base reads on an Illumina GA-II instrument. The longer reads resulted in a unimodal, center-weighted cumulative coverage profile (Fig. 2c). This lane generated 492 Mb of sequence that aligned uniquely to the genome, of which 445 Mb were on or near a bait. Of the specifically captured sequence, 321 Mb (72%) was directly on the bait itself and 235 Mb (53%) was contained within the exons. About 65% of the unique human sequence was on bait; 48% was on exons proper. The average coverage of bases was 86-fold within baits and 94-fold within coding exons.
Specificity
The percentage of the uniquely aligning human sequence that falls on or near a bait (e.g., 445/492 = 90% for the 76-base end reads) provides an upper bound for estimating the specificity of hybrid selection. In this experiment, 358 Mb (42%) of the 851 Mb of raw sequence did not align uniquely to the human genome (Table 1) and were not considered. By comparison, typically ~55% of raw bases in whole-genome-sequencing lanes do not align uniquely. The raw bases likely contain hybrid-selected human sequence that is not unique. The lower bound, assuming that all discarded sequence represented repetitive human background sequence rather than low-quality reads, was 445/851 = 52%. To obtain a more precise number, we aligned the raw reads again to the human genome, this time allowing multiple placements, and determined the fraction of all human alignable sequence that lay on or within 500 bp of a bait. Based on this calculation, our best estimate for the specificity of this catch was 82%. Of note, the specifically captured sequence included near-target hits that were not on exons proper. The percentage of uniquely aligning Illumina sequence that actually lay on coding sequence, i.e., the upper bound of the overall specificity of targeted exon sequencing, was 48% in this experiment. Table 1 shows a detailed breakdown of raw and uniquely aligned Illumina sequences and measures of specificity for the three targeted exon-sequencing experiments.
Table 1.
Detailed breakdown of Illumina sequences generated from exon catches
| Length and kind of Illumina sequencing reads | 36-base GA-I end sequences | 36-base GA-I shotgun sequences | 76-base GA-II end sequences |
|---|---|---|---|
| Aggregate length of targeta | 2.5 Mb | 2.5 Mb | 2.5 Mb |
| Aggregate length of baits | 3.7 Mb | 3.7 Mb | 3.7 Mb |
|
| |||
| Total raw unfiltered sequence | 152 Mb | 219 Mbb | 851 Mb |
| Raw sequence not aligned uniquely to genomec | 67 Mb | 116 Mb | 358 Mb |
| Uniquely aligned human sequence | 85 Mb | 102 Mb | 492 Mb |
|
| |||
| Uniquely aligned sequence on target | 36 Mb | 51 Mb | 235 Mb |
| Uniquely aligned sequence near targetd | 40 Mb | 38 Mb | 210 Mb |
| Uniquely aligned sequence on or near target | 76 Mb | 90 Mb | 445 Mb |
|
| |||
| Fraction of uniquely aligned sequence on or near targete | 89% | 88% | 90% |
| Fraction of raw bases uniquely aligned on or near targetf | 50% | 41%g | 52% |
| Fraction of uniquely aligned bases on targeth | 42% | 50% | 48% |
Protein-coding exon sequence only
Each unit of concatenated catch contains 44–46 bases (~18%) of generic adapter sequence. Therefore, ~18% (39 Mb) of the 219 Mb is not of human origin.
All raw sequence that fails to align uniquely to the human reference genome including low-quality sequence
Outside but within 500 bp of a target exon
Upper bound for estimating the specificity of hybrid selection
Lower bound for estimating the specificity of hybrid selection
The denominator (219 Mb) includes ~39 Mb of sequence from the generic adapters. Excluding these 39 Mb, the lower bound for the estimated specificity of this catch is 90/180 = 50%.
Upper bound for the overall specificity of targeted exon sequencing
Regional capture and sequencing
Next, we designed and tested a pool of 170-mer baits for targeted sequencing of four genomic regions ranging from 0.22 to 0.75 Mb in size (Supplementary Table 2 online). The combined span of the regions was 1.68 Mb. The target regions included a large portion of ENCODE region ENr113 as well as the genes IGF2BP2, CDKN2A and B, and CDKAL1. For a pilot experiment, we designed non-overlapping 170-mers that largely excluded repetitive sequence (allowing no more than 40 bases of repetitive sequence in each). The baits totaled 0.75 Mb in length, while the remaining 0.93 Mb was not covered owing to repetitive sequence content. We fished in a pond containing 350–500 bp fragments of DNA from the human cell line GM15510. The catch was analyzed with the shotgun sequencing approach above, with 36-base reads. The experiment preceded the development of the 76-base reads.
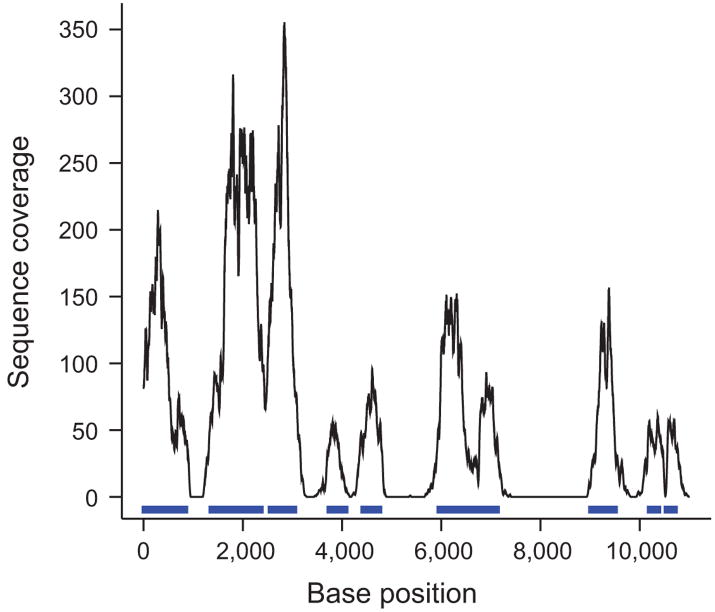
We generated one lane of Illumina GA-I sequence, yielding 191 Mb that aligned uniquely to the human reference sequence. Of this sequence, 179 Mb (94%) fell within the four targeted genome segments. About 164 Mb were on bait whereas 15 Mb aligned uniquely within the 0.95 Mb that were not covered by baits. Essentially all unique sequence within the bait-free zones was within 500 bp of a bait sequence, suggesting that it was “fish” rather than “weed”. A typical coverage profile along 11 kb is shown in Figure 3. As expected, the coverage was not uniform and had peaks at unique segments that were represented in the bait pool and deep valleys or holes at mostly repetitive regions outside the baits. The average depth of coverage for the 0.75 million genome bases covered by bait in the four target regions was 221.
Figure 3.
Sequence coverage along a contiguous target. Shown is base-by-base sequence coverage along a typical 11-kb segment (chr4:118635000-118646000) out of 1.7 Mb. Sequence corresponding to bait is marked in blue. Segments that had more than 40 repeat-masked bases per 170-base window were not targeted by baits and received little or no coverage with sequencing reads aligning uniquely to the genome except directly adjacent to a bait.
Evenness of coverage
Uniformity of capture, along with specificity, is the main determinant for the efficiency and practical utility of any bulk enrichment method for targeted sequencing. The larger the differences in relative abundance, the deeper one has to sequence to cover the underrepresented targets. We sought to display the data in a form that is independent of the absolute quantity of sequence (Fig. 4). Specifically, we normalized the coverage of each base to the mean coverage observed across the entire set of targets. This allows comparison of results from experiments with widely differing sequence yields, different template preparation methods or different sequencing instruments.
Figure 4.
Normalized coverage-distribution plots. Shown is the fraction of bait-covered bases in the genome achieving coverage with uniquely aligned sequence equal or greater than the normalized coverage indicated on the X-axis. The absolute per base coverage was divided by the mean coverage of all bait positions (18 in a; 221 in b). The curve for the shotgun-sequenced exon capture (a) is steeper than the curve for the regional capture (b) indicating a less uniform representation of sequencing targets in the exon catch. Dashed lines point to the fraction of bases achieving at least half or one fifth the mean coverage.
The two graphs in Figure 4 show the fraction of bases contained within a bait at or above a given normalized coverage level; the normalized coverage was obtained by dividing the observed coverage by the mean coverage, which was 18 for the shotgun-sequenced exon capture (Fig. 4a) and 221 for the regional capture (Fig. 4b).
In the exon-capture experiment, more than 60% of the bases within baits achieved at least half the mean coverage, and almost 80% received at least one fifth. Twelve percent had no coverage in this particular sequencing lane. The normalized coverage-distribution plot for targeted regional sequencing is considerably flatter, indicating even better capture uniformity: 80% of the bases within baits received at least half the mean coverage; 86% received at least one fifth; 5% were not covered in this experiment.
We attribute the differences in performance mainly to the fact that exon targets are generally short and isolated and often targeted by a single capture oligonucleotide (with few additional ones to choose from without widening the segment covered by bait). In contrast, the regional capture benefits from synergistic effects between adjacent baits, i.e., an overhanging genome fragment caught by one bait contributing to the coverage underneath neighboring ones. The slightly longer DNA fragments used in this experiment (350–500 bases compared to 200–350 bases for exon capture) may have contributed to this effect. Additional coverage-distribution data, including graphs that were truncated at a normalized coverage of 5 instead of 1 to show the tail of the distribution, are available in Supplementary Figures 1 and 2 online.
Effects of base composition
Separating the exon-capture baits into five categories based on their GC content revealed a systematic difference in coverage – with targets having GC content in the range of 50–60% receiving the highest coverage and those with very high (70–80%) or very low (30–40%) GC content getting the least coverage (see Supplementary Fig. 3 online). The effects of base composition most likely reflect genuine systematic differences in hybridization behavior. However, it is also conceivable that GC bias at other steps in the process contribute to this effect. For example, we know from microarray assays that PCR can deplete oligonucleotide sequences with extreme base compositions up to ~5-fold (data not shown). In addition, bias at the oligonucleotide-synthesis step may play a role. PCR amplification of the catch and sequencing itself is also known to introduce bias19,20.
Reproducibility
To assess the reproducibility of targeted exon sequencing we compared the results from independent technical replicates. Specifically, we performed two separate hybrid selections with ~250 bp fragments prepared from the same source DNA (NA15510) and generated one lane of Illumina shotgun sequence each. As shown in Figure 5a, the ratio of the mean normalized sequence coverage for individual exons in the two experiments was distributed closely around 1, indicating much less experiment-to-experiment than target-to-target variability. Base-by-base coverage profiles for individual exons were remarkably similar between the two technical replicates (purple and teal lines in Fig. 5b), consistent with the notion that variability in coverage is by and large systematic rather than stochastic. The coverage profile along the same exon in a different source DNA (NA11994) followed a similar pattern (black line in Fig. 5b). Additional data that demonstrate the sample-to-sample consistency of targeted sequencing of whole-genome amplified DNA samples can be found in Supplementary Figure 4 online.
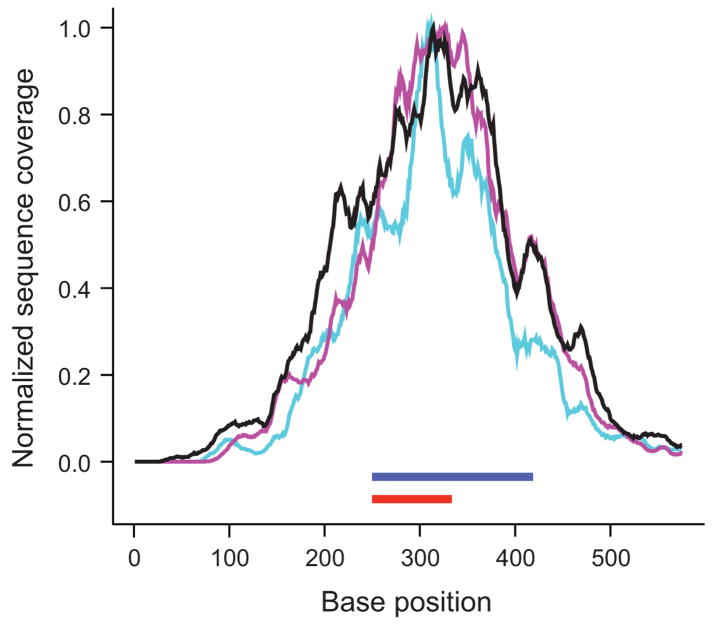
Figure 5.
Reproducibility of hybrid selection. For each exon (n = 15,565), the ratio of the mean coverage in two independent hybrid selection experiments performed on the same source DNA (NA15510) was plotted over its mean coverage in one experiment (a). Coverage was normalized to adjust for the different number of sequencing reads. The average ratio (black line) is close to 1. Standard deviations are indicated by purple lines. The graph on the right (b) shows base-by-base sequence coverage along one target in three independent hybrid selections, two of them performed on NA15510 (purple and teal lines) and one on NA11994 source DNA (black). Note the similiarities at this fine resolution of the three profiles which were normalized to the same height. The position of target exon (ENSE00000968562) and bait is indicated by red and blue bars, respectively.
The number of exon positions where we called a high-confidence genotype (see Methods) in the two technical replicates was 1,586,379 and 1,578,975, respectively, i.e., ~64% of the 2.5 Mb of targeted exon sequence. A total of 1,459,172 nucleotide positions were called in both. Of these, only 14 disagreed, indicating an overall discordance rate of ~10−5 which is consistent with our threshold for genotype calls (LOD ≥5). The excellent reproducibility permits sequencing of essentially the same subset of the genome in different experiments. It also allows accurate predictions of target coverage at a given number of total sequencing reads. According to a normalized coverage distribution plot for exon as opposed to bait sequence (Supplementary Fig. 1a online), quadrupling the number of sequenced bases would increase the fraction of exon sequence called at high confidence to >80%. This can be easily achieved by longer reads and higher cluster densities on a newer Illumina GA-II instrument. Indeed, a single lane of 76-base end-sequencing reads provided high-confidence genotypes for 89% (~2.2 Mb) of the targeted exon space.
Accuracy of SNP detection
To assess the accuracy of SNP detection, we fished for exons in three different human samples (Coriell NA11830, NA11992 and NA11994) that had been previously genotyped for the International HapMap project. With one lane of Illumina GA-I sequence for each sample, we were able to call 7,712 sequencing-based genotypes in coding exons for direct comparison with previously obtained genotypes. Each cell line had about 3,850 genotypes in HapMap within our target exons, of which ~22% were heterozygous. As expected, the detection sensitivity of 67% (7,712 high-confidence genotype calls for 11,544 HapMap genotypes) closely matched the percentage of exon bases scanned with high confidence (64%) in these particular GA-I sequencing lanes.
The discordance rate at high-confidence sites was low (0.6%) and close to the estimated error rate of HapMap genotypes21. Of note, the HapMap discordancy for the very same loci in whole-genome Illumina sequencing experiments was essentially the same (0.6%). Hence, there is no evidence that the hybrid selection process per se compromises the accuracy.
To resolve a representative subset of the discrepancies we genotyped two DNA samples (NA11830 and NA11992) on the Sequenom platform. A list of all 44 discordant genotypes plus 22 Sequenom genotypes is shown in Supplementary Table 1 online. In 19 of 22 informative cases (86%), the Sequenom assay confirmed the sequencing-based result. Three cases were bona fide hybrid-selection sequencing errors that missed the non-reference allele at heterozygous positions. Bias against the non-reference allele may be due to preferential capture of the reference allele present in the capture probes, to preferential alignment against the reference genome or both.
Overall, the two alleles at heterozygous loci were represented almost equally on average. Based on 1,722 heterozygous SNP calls, the fraction of reads supporting the reference allele had a mean of 0.53 and a standard deviation of 0.12. The nearly balanced recovery of both alleles increases the power to detect heterozygotes. Consequently, the sensitivity to detect SNPs is mainly limited by sequence coverage rather than by systematic or stochastic allelic bias or drop-out effects.
DISCUSSION
We have developed a hybrid-selection method for enriching specific subsets of a genome that is flexible, scalable, and efficient. It combines the economy of oligodeoxynucleotide synthesis on an array with the favorable kinetics of RNA-driven hybridization in solution and works well for short dispersed segments and long contiguous regions alike. With further optimization, routine implementation of hybrid selection would enable deep targeted “next-generation” sequencing of thousands of exons as well as of megabase-sized candidate regions implicated by genetic screens. Hybrid-selection based targeting may be potentially useful for a variety of other applications as well, where traditional single-plex PCR is either too costly or too specific in that specific primers may fail to produce a PCR product that represents all genetic variation in the sample. Examples are enrichment of precious ancient DNA that is heavily contaminated with unwanted DNA, deep sequencing of viral populations in patient material or metagenomic analyses of environmental or medical specimens.
Previous methods for hybrid selection have used cloned DNA, such as BACs or cosmids, to create capture probes for cDNA22,23 or genomic DNA fragments24. Clone-based probes are suboptimal for several reasons. Readily available clones often contain extraneous sequences and are not easily configured into custom pools. Moreover, cDNAs are inefficient for capturing very short exons (data not shown). Instead of cloned DNA, we use pools of ultra-long custom-made oligonucleotides which are synthesized in parallel on a microarray and offer much greater flexibility. In principle, one can target any arbitrary sequence. As with all hybridization-based methods, repeat elements have to be either circumvented at the bait design stage or physically blocked during the hybridization. We currently do both. There are also fundamental limits to the power of hybridization to discriminate between close paralogues, members of gene families, pseudogenes, or segmental duplications.
We perform a simple pull-down with streptavidin-coated magnetic beads, a generic laboratory technique that does not require customized equipment. It can be performed in almost any tube or multi-well plate format, and there are numerous precedents for processing many samples in parallel. Our method is also largely independent of the sequencing platform. As shown here, it works well in combination with the Illumina platform whereby the hybrid-selected material can be either end sequenced or shotgun sequenced. Direct end sequencing with longer reads is clearly preferred as it is far less complex and requires fewer amplification steps. Our protocol can also be easily adapted for the 454 instrument (data not shown) which produces fewer but even longer reads, and, presumably, for other sequencing platforms as well.
The length of the baits allows thorough washes at high stringency to minimize contamination with non-targeted sequences that would cross-hybridize to the bait or hybridize to legitimate target fragments via the common adapter sequence. A related source of background, indirect pull-down of repetitive “passenger” DNA fragments, is suppressed by addition of C0t-1 DNA to block repeats during the hybridization.
To prepare the bait, we amplify the complex pool of synthetic oligonucleotides twice by PCR. The risk of introducing bias during the amplification is more than compensated by its advantages: first, PCR selects for full-length synthesis products; second, it helps amortizing the fixed cost of chemical oligonucleotide synthesis over a large number of DNA samples; third and most importantly perhaps, it allows storage and testing at various stages of aliquots and obviates the need for frequent chemical re-synthesis and quality control of a given set of DNA oligonucleotides.
The sensitivity is in part due to the use of single-stranded RNA as capture agent. While a 5′-biotinylated double-stranded PCR product is equally specific (data not shown), it is not as good a hybridization driver. In a hybrid selection with single-stranded RNA, each bait is present in vast (several hundred-fold) excess over its cognate target. The excess RNA drives the hybridization reaction toward completion and reduces the amount of input fragment library needed. Further, saturating the available target molecules with an excess of bait prevents all-or-none single-molecule capture events that give rise to the stochastic and skewed representation of targets and alleles in multiplex amplification14. It also helps normalizing differences in abundance and hybridization rates of individual baits to some extent.
An important parameter for capturing short and dispersed targets such as exons is fragment size. Longer fragments extend beyond their baits and thus contain more sequence that is slightly off-target. On the other hand, shearing genomic DNA to a shorter size range generates fewer fragments that are long enough to hybridize to a given bait at high stringency. By virtue of the high excess of bait, our protocol works well for fishing in whole-genome libraries with a mean insert size of ~250 bp, i.e., only slightly longer than the average protein-coding exon and minimum target size (164 and 170 bp, respectively). In contrast, microarray capture has a lower effective concentration of full-length probes, requires more input fragment library to drive the hybridization and becomes less efficient with input fragment libraries that have insert sizes much smaller than 500 bp12. Array capture is therefore better suited for longer targets, for which edge effects and target dilution by over-reaching baits or overhanging fragment ends are negligible. In fact, capturing fragments larger than the oligonucleotides is beneficial for this application as it helps extend coverage into segments next to repeats that must be excluded from the baits. Because of synergistic effects between neighboring baits, contiguous regions are less demanding targets than short isolated exons.
One advantage of hybrid selection is that long capture probes are more tolerant to polymorphisms than the shorter sequences typically used as primers for PCR or multiplex amplification. We have seen very little allelic bias and few cases of allelic drop out at SNP loci. The concordance of sequencing-base genotype calls and known HapMap genotypes was excellent (99.4%). For the majority of discrepancies that we looked at, the sequencing genotype was validated by a specific SNP-genotyping assay. We have not examined other genetic variation such as indels, translocations and inversions; the capture efficiency may be lower for such sequence variants because they differ more from the reference sequence used to design the baits.
In conclusion, the protocol described here should allow extensive sequencing of targeted loci in genomes. Still, it remains imperfect with some unevenness in selection and some gaps in coverage. Fortunately, these imperfections appear to be largely systematic and reproducible. We anticipate that additional optimization, more sophisticated bait design based on physicochemical as well as empirical rules, and comprehensive libraries of pre-designed and pre-tested oligonucleotides will enable efficient, cost-effective and routine deep resequencing of important targets and contribute to the identification of biologically and medically relevant mutations.
METHODS
Capture probes (“bait”)
Libraries of synthetic 200-mer oligodeoxynucleotides were obtained from Agilent Technologies Inc. The pool for exon capture consisted of 22,000 oligonucleotides of the sequence 5′-ATCGCACCAGCGTGTN170CACTGCGGCTCCTCA-3′ with N170 indicating the target-specific bait sequences. Baits were tiled along exons without gaps or overlaps starting at the “left”-most coding base in the strand of the reference genome sequence shown in the UCSC genome browser (i.e., 5′ to 3′ or 3′ to 5′ along the coding sequence, depending on the orientation of the gene) and adding additional 170-mers until all coding bases were covered. The synthetic oligonucleotides for regional capture consisted of 10,000 200-mers that targeted 4,409 distinct 170-mer sequences, of which 3,227 were represented twice (i.e., the sequence above plus its reverse complement) and 1,182 were represented thrice. For baits designed to capture a pre-defined set of targets we first choose the minimal set of unique olignonucleotides and then add additional copies (alternating between reverse complements and the original plus strands) until the maximum capacity of the synthetic oligonucleotide array (currently up to 55,000) has been reached. The PCR product and the biotinylated RNA bait is the same for forward and reverse-complemented oligonucleotides. Synthesizing plus and minus oligonucleotides for a given target may provide better redundancy at the synthesis step than synthesizing the very same sequence twice, although we have no hard evidence that reverse complementing the oligonucleotides has any measurable benefit. Complete lists of sequencing targets and oligonucleotide sequences are available as Supplementary Table 2 and Supplementary Data 1–3 online. Oligonucleotide libraries were resuspended in 100 μl TE0.1 buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0). A 4-μl aliquot was PCR-amplified in 100 μl containing 40 nmol of each dNTP, 60 pmol each of 21-mer PCR primers A (5′-CTGGGAATCGCACCAGCGTGT-3′) and B (5′-CGTGGATGAGGAGCCGCAGTG-3′), and 5 units PfuTurboCx Hotstart DNA polymerase (Stratagene). The temperature profile was 5 min. at 94°C followed by 10 to 18 cycles of 20 s at 94°C, 30 s at 55°C, 30 s at 72°C. The 212-bp PCR product was cleaned-up by ultrafiltration (Millipore Montage), preparative electrophoresis on a 4% NuSieve 3:1 agarose gel (Lonza) and QIAquick gel extraction (Qiagen). The gel-purified PCR product (100 μl) was stored at −70°C. To add a T7 promoter, a 1-μl aliquot was re-amplified in 200 μl as before, except that the forward primer was T7-A (5′-GGATTCTAATACGACTCACTATAGGGATCGCACCAGCGTGT-3′) and 12 to 15 PCR cycles were sufficient. Qiagen-purified 232-bp PCR product (1 μg) was used as template in a 100-μl MAXIscript T7 transcription (Ambion) containing 0.5 mM ATP, CTP and GTP, 0.4 mM UTP and 0.1 mM Biotin-16-UTP (Roche). After 90 min. at 37°C, the unincorporated nucleotides and the DNA template were removed by gel filtration and TURBO DNase (Ambion). The yield was typically 10–20 μg of biotinylated RNA as determined by a Quant-iT assay (Invitrogen), i.e., enough for 20–40 hybrid selections. Biotinylated RNA was stored in the presence of 1 U/μl SUPERase-In RNase inhibitor (Ambion) at −70°C.
Whole-genome fragment libraries (“pond”)
Whole-genome fragment libraries were prepared using a modification of Illumina’s genomic DNA sample preparation kit. Briefly, 3 μg of human genomic DNA (Coriell) was sheared for 4 min. on a Covaris E210 instrument set to duty cycle 5, intensity 5 and 200 cycles per burst. The mode of the resulting fragment-size distribution was ~250 bp. End repair, non-templated addition of a 3′-A, adapter ligation and reaction clean-up followed the kit protocol except that we used a generic adapter for libraries destined for shotgun sequencing after hybrid selection. This adapter consisted of oligonucleotides C (5′-TGTAACATCACAGCATCACCGCCATCAGTCxT-3′ with “x” denoting a phosphorothioate bond resistant to excision by 3′-5′ exonucleases) and D (5′-[PHOS]GACTGATGGCGCACTACGACACTACAATGT-3′). The ligation products were cleaned up (Qiagen) and size-selected on a 4% NuSieve 3:1 agarose gel followed by QIAquick gel extraction. A standard prep starting with 3 μg of genomic DNA yielded ~500 ng of size selected material with genomic inserts ranging from ~200 to ~350 bp, i.e., enough for one hybrid selection. To increase the yield we typically amplified an aliquot by 12 cycles of PCR in Phusion High-Fidelity PCR master mix with HF buffer (NEB) using Illumina PCR primers 1.1 and 2.1, or, for libraries with generic adapters, oligonucleotides C and E (5′-ACATTGTAGTGTCGTAGTGCGCCATCAGTCxT-3′) as primers. After QIAquick clean-up, if necessary, fragment libraries were concentrated in a vacuum microfuge to 250 ng per μl before hybrid selection.
Hybrid selection
A 7-μl mix containing 2.5 μg human C0t-1 DNA (Invitrogen), 2.5 μg salmon sperm DNA (Stratagene) and 500 ng whole genome fragment library was heated for 5 min. at 95°, held for 5 min. at 65° in a PCR machine and mixed with 13 μl prewarmed (65°C) 2X hybridization buffer (10X SSPE, 10X Denhardt’s, 10 mM EDTA and 0.2% SDS) and a 6-μl freshly prepared, prewarmed (2 min. at 65°C) mix of 500 ng biotinylated RNA and 20 U SUPERase-In. After 66 h at 65°C, the hybridization mix was added to 500 ng (50 μl) M-280 streptavidin Dynabeads (Invitrogen), that had been washed 3 times and were resuspended in 200 μl 1M NaCl, 10 mM Tris-HCl, pH 7.5, and 1 mM EDTA. After 30 min. at RT, the beads were pulled down and washed once at RT for 15 min. with 0.5 ml 1X SSC/0.1% SDS, followed by three 10-min. washes at 65°C with 0.5 ml prewarmed 0.1X SSC/0.1% SDS, resupending the beads once at each washing step. Hybrid-selected DNA was eluted with 50 μl 0.1 M NaOH. After 10 min. at RT, the beads were pulled down, the supernatant transferred to a tube containing 70 μl 1 M Tris-HCl, pH 7.5, and the neutralized DNA desalted and concentrated on a QIAquick MinElute column and eluted in 20 μl. We routinely use 500 ng of “pond” and “bait” per reaction but have seen essentially identical results in proportionally scaled-down 5- μl reactions with 100 ng each.
“Catch”processing and sequencing
For fragment libraries carrying standard Illumina adapter sequences, 4 μl of hybrid-selected material was amplified for 14 to 18 cycles in 200 μl Phusion polymerase master mix and PCR primers 1.1 and 2.1 (Illumina) and the PCR product cluster-amplified and end-sequenced for 36 or 76 cycles. Hybrid-selected material with generic adapter sequences (8 μl) was amplified in 400 μl Phusion High-Fidelity PCR master mix for 14 to 18 cycles using PCR primers F (5′-CGCTCAGCGGCCGCAGCATCACCGCCATCAGT-3′) and G (5′-CGCTCAGCGGCCGCGTCGTAGTGCGCCATCAGT-3′). Initial denaturation was 30 s at 98°C. Each cycle was 10 s at 98°C, 30 s at 55°C and 30 s at 72°C. Qiagen-purified PCR product (~1 μg) was digested with NotI (NEB), cleaned-up (Qiagen MinElute) and concatenated in a 20-μl ligation reaction with 400 U T4 DNA ligase (NEB). After 16 h at 16°C, reactions were cleaned up (Qiagen) and sonicated (Covaris). Sample preparation for Illumina sequencing followed the standard protocol except that the PCR amplification was limited to 10 cycles.
Computational methods
All coverage and SNP statistics are for single lanes of sequencing data. Illumina reads were collected from the instrument and aligned to the human genome using Broad’s in-house aligner which is the ImperfectLookupTable (ILT) of the ARACHNE2 genome assembly suite25 and available for downloading at http://www.broad.mit.edu/wga along with documentation. Briefly, a lookup table of the locations of every 12-mer in the genome was computed. For a single read, each 12-mer in the read was looked up, and all occurrences of each 12-mer were considered putative placements. Each putative placement of the read in the genome was interrogated for number of mismatches. No insertions or deletions were considered. To ensure high quality and unique placements, only reads with 4 or fewer errors and a next-best placement at least 3 errors worse were considered. Coverage at each reference position was accumulated from the unique alignments. All aligned bases were included in the basic coverage calculations. High-confidence base calls (and coverage calculations based thereon) excluded bases that failed a signal clarity filter. The filter was that the ratio of brightest dye color to next-brightest dye color had to be 2 or greater. Typically, about 80% of aligned bases passed this filter. Genotypes at each position were inferred with a straightforward Bayesian model. The likelihood of the observed data P(data|genotype) assuming each genotype at each position were computed with the assumptions that each allele is equally likely to be observed and miscalls occur with a rate of 1/1000. These genotypes were combined with a prior probability over the genotypes defined by the reference. The prior used was: P(homozygous reference) = 0.999, P(heterozygous ref/nonref) = 0.001, P(nonref) = 0.00001. This yields the posterior probability P(genotype|data). The most likely genotype was selected. The “confidence” in our call of the specific genotype was the ratio of the best to next-best theory. We used a best-to-next-best ratio of 105 (LOD score 5) as threshold for calling a high-confidence genotype. The confidence in our belief that there was a SNP (independent of the specific genotype) was the ratio of the best theory to the reference. We used a best-to-reference ratio of 105 as our minimum confidence cutoff for reporting a SNP. Genome coordinates are zero-offset and for NCBI Build 35 (hg17).
Supplementary Material
Acknowledgments
We thank the staff of the Broad Institute Genome Sequencing Platform and Genetic Analysis Platform for generating sequencing and genotyping data. This work was supported by NHGRI grant HG03067-05 (to ESL) and funds of the Broad Institute.
References
- 1.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 3.Bentley DR, et al. Accurate whole genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DR, et al. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 2008;18:1638–1642. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–66. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 8.Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 10.Dahl F, et al. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci USA. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredriksson S, et al. Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector. Nucleic Acids Res. 2007;35:e47. doi: 10.1093/nar/gkm078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodges E, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 13.Okou DT, et al. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 14.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 15.Krishnakumar S, et al. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc Natl Acad Sci USA. 2008;105:9296–9301. doi: 10.1073/pnas.0803240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clamp M, et al. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci USA. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nilsson M, et al. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 18.Hardenbol P, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 19.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quail MA, et al. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovett M, Kere J, Hinton LM. Direct selection: a method for the isolation of cDNAs encoded by large genomic regions. Proc Natl Acad Sci USA. 1991;88:9628–9632. doi: 10.1073/pnas.88.21.9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parimoo S, Patanjali SR, Shukla H, Chaplin DD, Weissman SM. cDNA selection: efficient PCR approach for the selection of cDNAs encoded in large chromosomal DNA fragments. Proc Natl Acad Sci USA. 1991;88:9623–9627. doi: 10.1073/pnas.88.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bashiardes S, et al. Direct genomic selection. Nat Methods. 2005;2:63–69. doi: 10.1038/nmeth0105-63. [DOI] [PubMed] [Google Scholar]
- 25.Jaffe DB, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.