Abstract
The Neuroscience Information Framework (NIF), developed for the NIH Blueprint for Neuroscience Research and available at http://nif.nih.gov and http://neurogateway.org, is built upon a set of coordinated terminology components enabling data and web-resource description and selection. Core NIF terminologies use a straightforward syntax designed for ease of use and for navigation by familiar web interfaces, and readily exportable to aid development of relational-model databases for neuroscience data sharing. Datasets, data analysis tools, web resources, and other entities are characterized by multiple descriptors, each addressing core concepts, including data type, acquisition technique, neuroanatomy, and cell class. Terms for each concept are organized in a tree structure, providing is-a and has-a relations. Broad general terms near each root span the category or concept and spawn more detailed entries for specificity. Related but distinct concepts (e.g., brain area and depth) are specified by separate trees, for easier navigation than would be required by graph representation. Semantics enabling NIF data discovery were selected at one or more workshops by investigators expert in particular systems (vision, olfaction, behavioral neuroscience, neurodevelopment), brain areas (cerebellum, thalamus, hippocampus), preparations (molluscs, fly), diseases (neurodegenerative disease), or techniques (microscopy, computation and modeling, neurogenetics). Workshop-derived integrated term lists are available Open Source at http://brainml.org; a complete list of participants is at http://brainml.org/workshops.
Keywords: Neurodatabases, Data sharing, Terminologies, Portals
The Evolution of Scientific Information and the Neuroscience Information Framework
We introduce the core enabling terminologies for the Neuroscience Information Framework (NIF), and view the NIF itself, in the context of access to scientific information. At the dawn of science, information was disseminated via individual letters to a small number of other researchers. Printing technology enabled letters to be collected, assembled in journals, and distributed more widely. Although today an increasingly dominant mode of publication is paperless, with text and illustrations delivered via Net protocols, these are largely still as PDF or other page images. Access to this textual material, accompanied by graphical or photographic illustrations, remains conventional, with textual Google or PubMed searches that match exact tokens in publications complementing text-based indexes.
Scientific information is evolving beyond this literature page model. New media include video and 3-D via the Web, and increasingly databases deliver actual datasets, supplementing figures. Beyond neurodatabases, neuroscience web resources include knowledge bases, atlases of structure, expression, and function, genetic/genomic and material resources, and tool and modeling sites for processing, analysis, or simulation of brain data. Such sites span multiple biological scales, techniques, and data models and are often targeted towards communities of neuroscientists that use specific conventions and terminologies (Gardner et al. 2008; Koslow and Hirsch 2004).
With support from the NIH Neuroscience Blueprint Institutes and Centers, we have developed a new initiative for integrating access to and use of web resources. This Neuroscience Information Framework, accessible via http://nif.nih.gov, http://neurogateway.org, and other sites to be announced (Gardner et al. 2008) provides access to data, tools, and materials (as well as text) across scales, methods, and preparations.
Enabling Terminologies for the Neuroscience Information Framework
Framework Core Terminology Is Designed to Span—and Unify—Scales, Domains, and Uses
The NIF consortium wished to avoid a ‘Tower of Babel’ problem in which development was delayed by the many different ways neuroscientists use to describe the same thing. Humans readily map terms to the concepts they describe, although scope and meaning are often imprecise or ambiguous, but automated methods need the precision provided by terminologies, ontologies, or context-based methods. Moreover, the breadth of neuroscience is such that no single view of neuroscience, and therefore no individual terminology, is sufficient. To serve all neuroscience, we set as a design goal that the Neuroscience Information Framework respect and recognize query semantics serving multiple views of the neuroscience ecosystem (Gardner et al. 2008).
Controlled-Vocabulary Metadata Aid Access to Data or Findings
A goal was to develop terminology to serve the proliferation of web-accessible data and publications, enabling users to specify in a consistent manner important features of these data. Controlled vocabularies (CV) available for both data description by submitters and queries by those searching for relevant data avoid lexical mismatch and false negatives. For both submitters and searchers, it is of use to have a comprehensive set of terms that can be selected from, and to have such terms (semantics) arranged in an informative, useful, and intuitive structure (syntax). It is also a design goal that the semantics serve the needs of multiple communities within neuroscience. To be accurate, the terms must be those used by the neuroscience community or communities generating or recording such data. To be general, they should also be understood by investigators who work with different but related systems, preparations, or techniques, and relatable to broader areas of neuroscience (Gardner et al. 2001a, b). One early such effort, which inspired our work, was the CV keywords developed for the Society for Neuroscience (SfN) by B. Grafstein to aid classification and discovery of abstracts at the Society’s Annual Meeting.
The SfN has been an enabling partner throughout development of NIFv1, the initial version of the NIF. NIFv1 terminology development was aided by the Terminology/Ontology Subcommittee of the Society for Neuroscience’s Neuroinformatics Committee; the Subcommittee included G. Ascoli, J.G. Bjaalie, D. Gardner (Chair), G. Jacobs, and M.E. Martone. The initial charge to the subcommittee was to identify several areas spanning preparations and techniques, to convene experts to establish consensus for terms and for expansion, and to use the results as a template to expand the terminology to more areas of neuroscience. Projected uses of these prototerminology efforts were to enhance search terms for the SfN’s Neuroscience Database Gateway (predecessor to and now a component of the NIF), and to enhance keywords for the Society’s journal J. Neurosci. A longer-term goal, of moving towards an interoperable terminology/ontology for neuroscience, was acknowledged from the start. The SfN supported early workshops in this integrated terminology effort.
NIF terminology development builds on and goes beyond this core vocabulary in the NIF Standardized (NIFSTD) semantic framework, which implements e.g. lexical variants, described in this volume by Bug et al. (2008).
NIFv1 Syntax I: Arranging Terms in Hierarchies Enables Both Broad and Specific Queries and Aids Database Development
Framework core terminologies are primarily a data description language for neuroscience, designed to specify and/or select particular data or findings. Based on this goal, we have selected a straightforward syntax designed for ease of use and for navigation by familiar web interfaces. Datasets, web resources, neuroinformatic software tools, or other entities are characterized by multiple descriptors, each addressing core concepts (e.g., data type, acquisition technique, cell type, and anatomy). Terms, like the keywords that accompany papers or abstracts, are organized in categories, each of which specifies a concept and includes a range of values. These include region or cell class of interest, neurobiological process, relevant disease, the type of data, or the technique by which the data were acquired.
Within a focused domain of neuroscience, it is important to make distinctions between similar locations, cell types, and data records. However, from outside each specialized domain, the distinction between e.g. the cortical areas AITd and AITv may be less relevant than specifying more general terms, such as AIT, or visual/multisensory, or even temporal cortex. For this reason, we arrange the terms describing each neuroscience concept in a tree or hierarchy. The tree structure allows selection of terms at the appropriate level of specificity for both description and search, with broad general terms near each root spawning more detailed entries. Each tree has at its root a set of general terms that broadly span the concept or description; more specific terms derive or branch from these.
Such trees encapsulate is-a and has-a relationships; neuroanatomical representations are largely has-a whereas techniques and data types are primarily is-a. Hierarchies also allow expansion and evolution without rendering prior entries obsolete, provided—as we intend—that the set of top-level terms for each slot span the full range of choices, and new terms are added under former leaf elements.
Recognizing the difficulty of attempting to fit terms relating distinct concepts into a single tree, we specify multiple trees, one for each concept or category. For example, one such tree includes brain areas, organized along the neuraxis. Additional trees specify e.g. depth or layer as a part of a location in the brain.
The use of multiple trees rather than a graph representation provides easier navigation for users. The simplicity of tree structures was selected for an additional purpose, to aid adoption of our neuroscientist-generated terms as seed metadata by other projects designing and developing new Web databases for additional neuroscience datasets, preparations, or techniques.
Gardner et al. (2005) noted that the use of controlled vocabulary and the context provide by the HAV representation enhance the utility and interoperability of metadata, substituting for the natural-language textual context missing from simple CV term lists. As each term is associated with a specific tree that encapsulates related concepts or entities, a text token such as ‘AIP’ can be both a brain area and a protein, and the word ‘grasp’ can be used both as a gene product and a motor action without confusion. Our work acknowledges and benefits from multiple similar organized CV efforts in both related and more general areas of biomedical science (Ashburner et al. 2000; Bota and Arbib 2004; Cimino 1998, 2000; Friedman et al. 1999; Goddard et al. 2001; Greer et al. 2002; Lindberg et al. 1993).
NIFv1 Syntax II: Detectors and Selectors Specify Web Resources and Contents
Framework terminology efforts are designed towards two important classes of descriptors. One set characterizes the focus of Web-accessible neuroscience resources. The other provides a data-description language enabling searches of individual resources (or a span of resources) for datasets, findings, techniques, tools, or materials of interest.
As a result of these variations in usage, we have found it useful to distinguish between detectors: general terms that specify the domain and contents of a database or other resource (tool repository, analytic engine, etc.) and selectors: query terms that allow specifying desired datasets. We recognize that there are additional, perhaps resource-specific, sets of metadata descriptors, less useful for search. These can include ‘analytical’ or ‘technical’ metadata such as filter settings or classifiers of local significance or useful for audit trails, such as experimenter, date, or local dataset index.
Broad Detector Terms Aid Description and NIF Integration of Disparate Web Resources
The Framework is being designed to offer access to a broad spectrum of Web-accessible resources. Fundamental to the orderly and efficient parsing of queries are terminologies describing such Web resources across multiple dimensions of knowledge or classification. To aid description and characterization of such resources, and to facilitate precise controlled-vocabulary queries, the project derived a list of detectors as neuroscience-aware descriptors of content and focus for the hundreds of resources in the proto-Framework at neurogateway.org. This process distilled a controlled vocabulary for inventoried web resource content from free-text descriptions that were provided by members of the Framework team and colleagues, and subsequently arranged in trees that describe each of several characteristic axes. These terms specify one or more of:
Resource description,
Neurobiological focus or disease and functional context,
Brain structure,
Organism,
Data type, or
Technique.
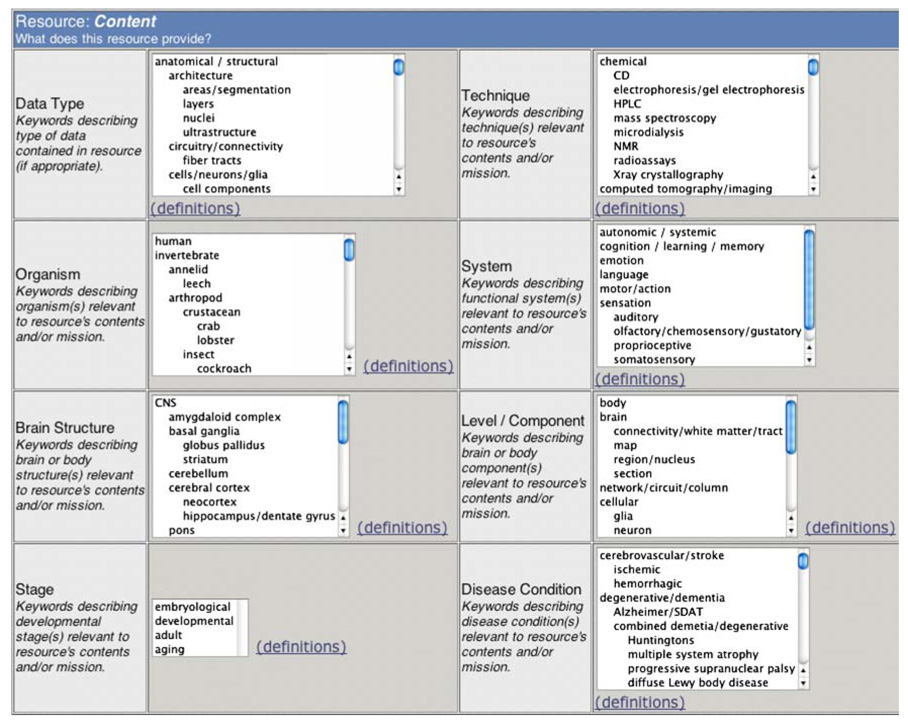
Figure 1 shows how this detector terminology, and the detector query screen, was utilized for resource characterization on the proto-Framework site at http://neurogateway.org. The full NIFv1 detector vocabulary may be accessed at: http://brainml.org/viewVocabulary.do?versionID=782
Fig. 1.
The proto-Framework catalog at http://neurogateway.org includes a broad set of detector controlled vocabulary terms that specify resources’ scope and focus, here shown in an early version exposing segments of each of eight controlled-vocabulary detector trees
We list below a sample of this detector terminology: the resource type itself. This characterizes resources by what they provide: databases deliver data, portals deliver links, atlases deliver anatomically- or spatially-organized data, knowledge bases deliver derived, generalized or canonical descriptions, and organization-supported portals deliver neuroscience-related information grouped by subject, disease, company, or institution:
data resource (neuroscience data or findings)
database (datasets)
atlas (spatially-organized data)
knowledge base (findings/knowledge derived from data)
clinical knowledge base (diagnosis/treatment)
bibliographic resource (library/publisher or literature access)
software resource (software for acquisition, analysis, display or modeling)
data acquisition software
data processing/analysis/archiving software
software for time-series analysis (nonimage)
software for spatial/image analysis
software for sequence analysis
software for pathway analysis
visualization software
3D/4D visualization
modeling/simulation software
research supplies (access to materials)
instrumentation
organism
cell line/tissue
reagent/chemical
portal (access to people, places, or sites)
lab or department
organization or institution
wide-area portal (links to external neuroscience web sites)
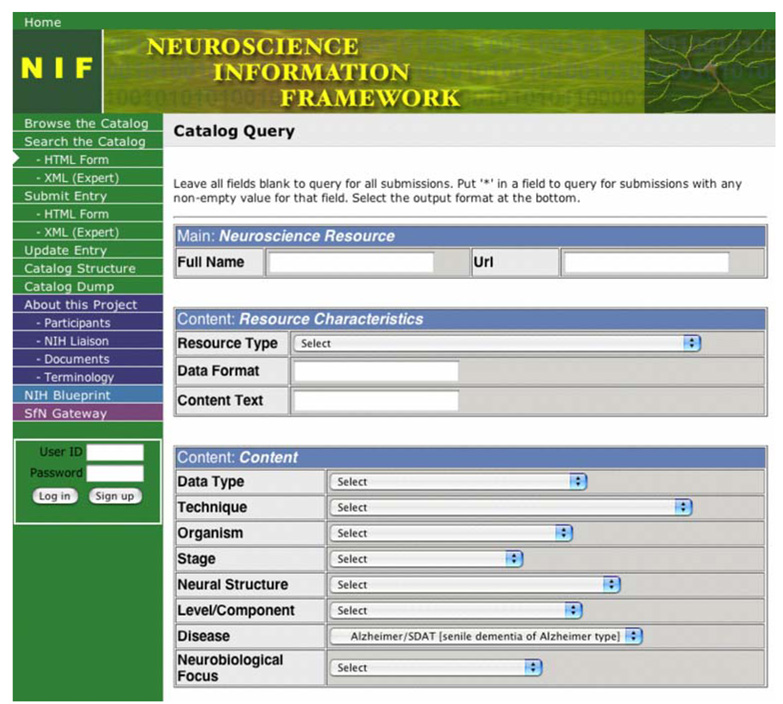
Figure 2 shows a sample search for Neurodatabase.org resources relevant to a specific disease type.
Fig. 2.
NIF Detector Terms Search the Neuroscience Web. Neurogateway.org, a NIF prototype resource provides access to hundreds of neuroscience Web resources. From possible detector search terms for data type, technique, organism, and others, the example shows search for a specific disease type using selected NIF terminology. The same underlying terminologies seen in Fig. 1 are here shown in an alternate drop-down menu format, emphasizing that the content is adaptable to multiple presentation schemas
Selector Terms Allow General or Specific Searches for Relevant Datasets or Other Resource Contents
A major Framework role is access to data and information provided by the increasing number of Web databases, tool sites, and others. In addition to the detector terminology above, useful for characterizing resources, a much larger set of selectors, again arranged in multiple hierarchies, are needed to specify and distinguish among individual datasets, tools, and findings. In a major section below, we detail the semantic complexity of these selectors and give examples of community-consensus terms derived from a series of expert terminology workshops.
Even with such broad development of specific selector terms, we emphasize that there remains a need for detectors that selector terms can not themselves serve. A major reason is that broad focus of individual resources is often implicit, and not specified in selector terms. For instance, all or most of the data in the Framework-accessible fMRIDC Web resource (http://fmridc.org; Van Horn and Gazzaniga 2005) is in fact fMRI data, so this is unlikely to appear as a selector term used to distinguish one dataset from another. This reinforces the need for a set of detector terms that are not explicit selector (search) terms, but characterize the specialization, technique, disease, or area of concentration.
NIFv1 Semantics: Neuroscientist-Derived Term Sets
Core NIF Terminologies Were Derived by the Neuroscience Community at a Series of Expert Workshops
To aid precise specification and adoption of selector terms, and to aid future neuroinformatic projects in developing compatible data description schemes, the project has used as its major methodology a series of neuroscience terminology workshops. At each by-invitation workshop, experts in a selected domain of neuroscience were brought together for plenary, intensive exchanges toward developing sets of useful and clear selector terminology to describe each of several aspects of experiments, the data they produce, and the analyses and insights that derive from them.
Areas covered span real objects including anatomy and cell types, but participants recognized that anatomy is only one of several necessary components. Others included data types, methods, preparations and protocols, acquisition techniques, post-acquisition data processing, models, diseases, paradigms, and hypotheses. Participants were urged to keep in mind as they identified the concepts and entities important to each area that the terms developed should only be those that investigators working in the field can readily determine and supply, and that the community is willing to accept. We asked that this terminology not only aid the target domain, but also bridge methods and findings with data and knowledge in complementary areas, or gained using complementary techniques. Aiding participation (and adoption), it was stressed that all terminologies, like the rest of the NIFv1 deliverables, will be made available freely Open Source in a non-proprietary manner for universal adoption.
Workshops on invertebrate identified neurons, visual neuroscience I and II, hippocampus I and II, and non-pyramidal cortical neurons were carried out under SfN auspices, funded under private grants and prior NIMH contracts. The Framework added computational neuroscience and modeling, cerebellum, human neuroimaging, microscopy and neuronal ultrastructure, molluscan neurobiology, olfaction: receptors and systems, neurogenetics, neurodegenerative disease, neurodevelopment, thalamus, behavioral neuroscience, and Drosophila.
A complete list of participants is at http://brainml.org/workshops. Many participants agreed to aid future e-mail-based sessions for orderly evolution of terminologies. Post-workshop, each set of trees was edited and the majority of terms integrated in the NIFv1 core terminology; many terms were deferred for incorporation into later versions. NIFv1 trees formed the core of the NIFSTD terminologies described by Bug et al. (2008).
Workshops with Specialized Modalities
The workshop on nonpyramidal neurons was primarily a self-generated effort of several neuroscience communities that came together to codify a multi-dimensional classification scheme. (Ascoli et al. 2008). A community-approved terminology for classifying cortical neurons was thus a joint goal of this ‘Petilla nomenclature project’ (named after the meeting site at Cajal’s birthplace), directed by R. Yuste and Framework Project Director G. Ascoli. Framework project members G. Ascoli, W. Bug, D. Gardner, M.E. Martone, and G.M. Shepherd derived from parts of the Petilla nomenclature and other sources a tree with cells classified along one axis (largely morphological), with plans to have the other dimensions or schemes (e.g., molecular or physiological) represented as attributes potentially modifying terms anywhere in the basic tree.
The neuroimaging workshop was primarily devoted to spurring a collegial effort that resulted in the generous donation of several existing vocabularies and initiation of plans for sontinued cooperative development. Several classes of terms from the computational neuroscience and modeling workshop were reserved pending additional development of the complementary NeuroML (Goddard et al. 2001; Crook et al. 2007) language; these will be included in the forthcoming BrainML08 terminology, along with a tripartite scheme for representing experimental manipulations and protocols.
Multidimensional Selector Controlled Vocabulary
Central to our effort developing ‘selector’ terminology to enable individual datasets (or analytic methods, or publications) to be categorized and located via searches are vocabularies targeted towards datasets. Our scheme parses neurobiological data by three basic sets of terms, and two modifiers. These describe:
what: the neurobiological data type that is recorded or presented,
why: the neurobiological function or disease that the data relate to, and
how: the technique(s) used to acquire or derive the data.
The two modifiers are:
form: an optional modifier if data are presented as an image or a time series, and
origin: an attribute specifying how the data originated, whether from experiment or observation, simulation, or meta-analysis.
These distinct sets of terms are designed to specify the type and significance of data while avoiding the combinatorial explosion that a single tree of terms would require. Note that the terms focus on the neurobiological processes reported by the data and its significance without describing the format in which the data are presented. Similarly, we do not distinguish among closely related measures with similar neurobiological significance, such as currents vs. conductances. Many techniques listed implicitly provide such information. For example, data types include ‘blood oxygenation’ under ‘functional-imaged activity’ whereas fMRI (the technique used for data acquisition) is separately listed under techniques.
We present two sample trees. The first lists techniques:
chemical
separations
gel/electrophoresis
Southern
northern
western
chromatography/HPLC [high-pressure liquid chromatography]
centrifugation
spectroscopy
mass spectrometry [MS]
circular dichroism [CD]
absorbency/absorbance/fluorescence
microdialysis
NMR [nuclear magnetic resonance]
calorimetry/microcalorimetry
radioassay
Xray crystallography
computer tomography/imaging
CAT [computer axial tomography]
MRI [magnetic resonance imaging]
structural
diffusion/diffusion tensor imaging [DTI]
manganese enhanced
functional [fMRI]
spectrographic [sMRI]
PET [positron emission tomography]
SPECT [single photon emission computed tomography]
electrode-based
extracellular
single electrode
tetrode
electrode array
sharp electrode array
flat/flexible electrode array
intracellular /whole-cell/clamp
voltage-clamp
patch
amperometric
pH
ion sensitive (non-H ion)
macroelectrode
cuff/suction
field/surface
EEG [electroencephalography]
electron microscopy [EM]
SEM [scanning electron microscopy]
secondary electron microscopy
x-ray microscopy
back scattered electron microscopy
TEM [transmission electron microscopy]
high voltage electron microscopy/HVEM
intermediate voltage electron microscopy/IVEM
energy filtering/EFTEM
electron diffraction camera
STEM [scanning transmission electron microscopy]
genomic/proteomic assays
expression chip/microarray
in situ hybridization
FISH [fluorescent in situ hybridization]
RNA in situ
sequence
BLAST [basic local alignment search tool]
homology-based search
CGH [comparative genome hybridization]
co-immunoprecipitation
chromatin IPCH
chip on chip
transcript quantitation
SAGE
Solexa
PCR
quantitative
real time
structure comparison
probes/markers
histological staining [to become protocol in BrainML08]
Golgi/silver stains
immunocytochemistry
immunohistochemistry
myelin
nissl
organelle/subcellular markers
cell death markers
nuclear markers
caspases
labeling [to become protocol in BrainML08]
radiolabeling
conformational stain
reporter assays/dyes
voltage indicator dyes
Ca indicator dyes
fluorescent probes
fluro-J
genetically coded
enzymatic function reporter(s)
transcription reporter(s)
fate mapping
genetic
lineage tracing
birth dating
thymidine
deoxyuridine derivatives/brdu
bioactive molecules [to become protocol in BrainML08]
physiological manipulators
cell activation
cell inactivation
RNAi
viral vectors
chemical genetics
light microscopy / optical imaging
bright field
dark field
intrinsic
fluorescence/chemiluminescence/phosphorescence
multi-photon
confocal
FRET [fluorescence resonance energy transfer]
Nomarski/DIC [differential interference contrast]
phase
polarization
spectroscopy
ratio imaging
stereology
TIRM [total internal reflection microscopy]
MEG [magnetoencephalography]
mechanical sensors
displacement transducer
force transducer / optical tweezers
audio/acoustic
spectral
sound pressure level detector [SPL]
scanning probe / AFM [atomic force microscopy]
ultrasound imaging
physiological monitors
EKG [electrocardiograph]
polygraph/polysomnograph
spirometer/respirometer
thermal monitors
reports/narratives
clinical reports
clinical rating scales [use standard nomenclature]
Barthel stroke ratings
updrs [Parkinsons]
uhdrs scale
Neuropsychological testing
subject reports
videography and photography
time-lapse
videomicroscopy
Other trees specify the structure from which the data were obtained, the level of examination, and the cell type. This neuroanatomy terminology reflects extensive refinement in our thalamus workshop, co-chaired by E.G. Jones and building on work of prior workshops, functional cortical parcellation of Felleman and Van Essen (1991), and NeuroNames (Bowden and Dubach 2003), with partial rationalization by D. Bowden and by E.P. Gardner. In this scheme, we place many neural structures in a single tree, organized along a primary rostral to caudal (or superior to inferior) neuraxis. As the brain is three-dimensional, other conceptual axes are needed for second physical axis, layering or depth. Terms that are important but which supplement the tree structure, such as ‘ipsilateral’ or ‘contralateral’ are indicated as attributes modifying the tree-selected term or level. Consistent with contemporary usage, terms freely mix Latin (or Greek) derived terms with English. As example, we provide an excerpt of the primary neuroanatomy tree, using the thalamus to illustrate the overall tree structure and the level of detail for many structures; ellipses (…) mark the remaining 75% of the tree not shown here:
CNS [central nervous system]
brain
telencephalon
. . .
diencephalon
pretectum
thalamus
epithalamus
paraventricular nucleus
anterior paraventricular nucleus
posterior paraventricular nucleus
habenular nucleus
medial habenular nucleus (Hm)
lateral habenular nucleus (Hl)
parvocellular subnucleus (Hlpc)
magnocellular subnucleus (Hlmc)
pineal/Conerium/Epiphysis
dorsal thalamus
principal nuclei
anterior group
anterodorsal nucleus (AD)
anteroventral nucleus (AV)
anteromedial nucleus (AM)/interanteromedial nuc (IAV)
lateral dorsal nucleus (LD)
medial group
parataenial nucleus (Pt)
medioventral nucleus (MV)/reuniens nucleus
mediodorsal nucleus (MD)
magnocellular (medial) division (MDmc)
parvocellular (central) division (MDpc)
multiform (lateral) div (MDmf)/paralaminar div
densocellular/paralamellar division
submedial nucleus (Sm)
ventral group
ventral anterior nucleus (VA)
principal division (VAp)
magnocellular division (VAmc)
ventral lateral nucleus (VL)
ventral lateral anterior nucleus (VLa)
ventral lateral posterior nucleus (VLp)
ventral lateral posterior nucleus dorsal (VLpd)
ventral lateral posterior nucleus ventral
(VLpv/VIM)
ventral posterior complex (VP)
ventral posterior lateral nucleus (VPL)
VPL nucleus anterior (VPLa/VPS) [Anterodorsal
shell]
VPL nucleus posterior (VPLp) [Central core]
ventral posterior medial nucleus (VPM)
ventral posterior inferior nucleus (VPI)
parvocellular division of VP complex (VPMpc/VPpc)
[Basal ventral medial nucleus(VMb)]
ventral medial nucleus (VM)
lateral posterior/pulvinar complex
lateral posterior nucleus (LP)
pulvinar nuclei (Pl)
anterior pulvinar nucleus (Pla)
inferior pulvinar nucleus (Pli)
lateral pulvinar nucleus (Pll)
medial pulvinar nucleus (Plm)
posterior group
posterior nucleus (Po)
posterior medial nucleus (Pom)
posterior lateral nucleus (Pol)
posterior intermediate nucleus (Poi)
posterior intralaminar nucleus (Pil)
[for Limitans/suprageniculate nucleus (L/SG) see IL]
medial geniculate complex
ventral nucleus (MGv)
dorsal nucleus (MGd)
anterior-dorsal division (MGad)
posterior-dorsal division (MGpd)
medial (magnocellular) nucleus (MGmc) [IL-like zone]
lateral geniculate nucleus
dorsal lateral geniculate (LGd)
magnocellular layers
parvocellular layers
koniocellular layers
medial interlaminar nucleus
A Layers
C Layers
[for ventral lateral geniculate see ventral thalamus]
intralaminar nuclei (IL)
anterior intralaminar group
central/midline/rhomboid nucleus (Rh)/(Ce)
central medial nucleus (CeM)
central lateral nucleus (CL)
paracentral nucleus
central or midline group [midline ext. of Pt, Rh, CL and CeM
nuclei]
intermediodorsal nucleus [rat]
posterior intralaminar group
centre median nucleus (CM)
parafascicular nucleus (Pf)
subparafascicular nucleus (SPf)
limitans/suprageniculate nucleus (L/SG)
limitans division (L)
suprageniculate division (SG)
ventral thalamus/(Prethalamus)
reticular nucleus (R)
zona incerta (ZI)
nucleus of the field of Forel (FF)
ventral lateral geniculate complex/pregeniculate nucleus (Prg)
pars principalis
medial division
dorsal cap
intergeniculate leaflet (IGL)
subthalamic nucleus
hypothalamus
hypophysis
third ventricle
. . .
Discussion
The Neuroscience Information Framework is built upon a set of coordinated terminology components enabling data and web-resource description and selection. The NIFv1 core terminologies described here form a data description language to specify and select particular neuroscience data or findings, not a true ontology. Its purpose is to provide a set of usable terms in a hierarchy so that investigators recording from, assaying, or otherwise sampling an area or a function of the nervous system can have a set of terms that encompass areas of current and likely future interest. Additional development of ontologies for the NIF is described in the accompanying Bug et al. (2008).
The NIFv1 data description language satisfies the following design goals:
It incorporates current usage by those who are not expert in specific areas, such as neuroanatomy, but is informed by the understanding of those who are. Thus the electrophysiologist, the neuroimager, or the molecular biologist need a context in which to place commonly-used descriptive terms in their fields. There is inevitably a tension between common usage of terms such as “pons” and “Broca’s area” and precise definitions, but we recognize that some terms will be used imprecisely and some ambiguously.
As different techniques yield, and different experimenters seek, more or less precision of location in the nervous system, the syntax allows for variable specificity. For the purposes of data description, terms are included that describe both broad areas (“parietal cortex” and “lumbar spinal cord”) and very specific locations. These terms are arranged in a tree hierarchy, with the most specific terms the leaves and the most general at the root.
Because a researcher looking for data relevant to a question does not know the degree of specificity used to describe a dataset placed in a database, or a finding in the literature, searches using general terms find as well more specific ones located on finer branches. As noted above, it would be possible to implement this terminology using graphs rather than trees, allowing multiple inheritance, but this is difficult for casual users to navigate and therefore awkward for the neuroscience community.
In the development of these terminologies, we have recognized that no single scheme can completely encompass the wide range of disparate data types, preparations, or techniques seen in contemporary neuroscience, let alone in likely future development. In particular, we have tried to develop a scheme that can intelligently record and relate what may be similar areas in principal model animals and perhaps aid integrated knowledge of nervous system function. A unified list enables description of and thereby access to data across scales and preparations, one of our contracted goals from the NIH. The alternative to this comprehensive scheme would be a distinct and precise atlas or neuranatomy for each species; these are of course available for many model animals but to represent each in a NIF-compatible form is beyond the limited scope of this project.
The results of multiple workshops have been integrated in the terminology being developed for the NIF and are also made freely available via Open Source for universal adoption. In this terminology, we have specified many descriptors, and arranged the terms useful to each in hierarchic trees. These terminologies are designed to satisfy such immediate NIF-related goals as identifying the concepts and entities important to specific areas of neuroscience, including data and experimental techniques as well as neurons and preparations. Longer-term goals include stimulating further community adoption of these terms to aid additional development of neuroinformatic resources (Gardner et al. 2003; Kennedy 2004, 2006; Koslow and Hirsch 2004; Liu and Ascoli 2007), and future efforts linking findings obtained in specific areas or preparations, or using particular techniques that yield specific data types, to related or relatable data of different types.
Our current development may therefore be thought of as an index for a book that is still being written. Completeness—defined depending on the level of detail to which each investigator can go or wishes to go—is unattainable, and this is why we our syntax represents more specific terms as branches of more general ones. If a very detailed term is not (yet) in the tree, the next level up encompasses it.
Increasingly, we believe that ontologies or knowledge bases for neuroscience are only one aspect of the wider problem of representing knowledge by metadata in other fields that directly impact real contemporary data in the neurosciences. One obvious need is for terms that bridge to, and interoperate with, conventional sequence and structure bioinformatics. For an example, consider what is needed to classify the different patch clamp data (or action potential shape or spike train patterns) resulting from manipulations that include changes in promoters, gene sequence, allelic selection, post-translational modification, alterations in protein phosphatases, and more, all of which need to be encoded in appropriate metadata in order to make sense of the data. Companion development of the NIFSTD semantic framework is designed toward this goal (Bug et al. 2008).
Complementary NIFv1 Terminology Components
Although the core NIFv1 terminologies here described do not form an ontology, these terms should inform such development, and as noted above, workshop terms are being integrated with parallel NIF-derived and integrated ontology and terminology components to form NIFSTD (Bug et al. 2008). Similarly, these terms are presented only as defined by context in trees and via common usage; we expect that extensions to this work will provide precise definitions as well. Another NIFv1 terminology project is Caltech’s Textpresso, which parses and extracts terms from a large contemporary neuroscience corpus (Müller et al. 2008). As related in this issue by Marenco et al. (2008), mediators will be able to take OWL-based and purely XML-based schemes and rationalize them probabilistically.
NIFv1 terminology also acknowledges multiple parallel efforts. An informal survey conducted among NIF Team members yielded the following list of other terminology or ontology efforts in the biomedical sciences that one or more were involved in: Gene Ontology,WormBase, NeuroNames, BrainInfo, GENSAT, Gene Network, fMRIDC, BrainML, Brain Map, W3C BioONT, IUPHAR Nomenclature, Unified Medical Language System, BIRN Ontology, Ontology of Biomedical Investigation, National Center for Biomedical Ontologies, OBO Relations / Foundry, and the International Committee on Cortical Interneuron Nomenclature.
The NIF Terminologies, Like the NIF Itself, Are Designed for Evolution and Migration
In addition to the dynamic inventory of neuroscience Web resources forthcoming at http://nif.nih.gov and http://neurogateway.org, which are annotated using NIF terminologies, terminologies (and code) are available Open Source to enable any interested group, journal, or society to establish, mirror, or enhance a Framework site. An expanding Textpresso literature repository for neuroscience is available at http://textpresso.org/neuroscience and above sites. NIFv1 and later term lists will be referenceable at http://brainml.org.
NIF terminologies are expanding. Many selector terms are being enriched through term integration by later workshops. In addition to those described here, terms are being collated to produce vocabulary trees for BrainML08’s protocols and paradigms, post-acquisition data processing, and models, diseases, and hypotheses. Believing that community development of vocabularies by neuroscientists facilitates community acceptance, we have tried to construct a terminology whose utility will itself encourage neuroscientists, in the cooperative spirit of the Open Source movement, to propose additional enhancements or extensions to this work.
Exportable Metadata and Semantic Data Models Aid Database Development as well as Resource Integration
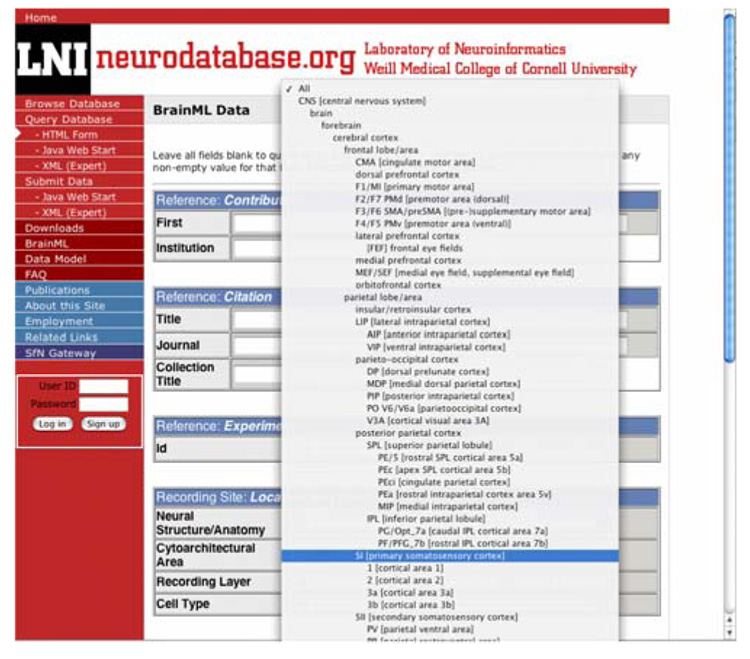
Neurodatabase.org, our Weill-Cornell Laboratory of Neuroinformatics archive for neurophysiology data, now incorporates the Open Source NIFv1 terminology for brain area and other descriptors. As noted above, the neuraxis serves as the main tree for these adoptable Open Source selector terms; other trees (not shown) serve second axes, layer, or depth. This standardizes metadata and can potentially facilitate direct database access via NIF query methods (Fig. 3)
Fig. 3.
Neurodatabase.org, the Laboratory of Neuroinformatics-developed archive of neurophysiology data, now incorporates the Open Source NIFv1 terminology for brain area and other descriptors. Exportable NIF terminology, available at http://brainml.org, standardizes metadata, aids future development of descriptors and query terms for databases, and can facilitate direct database access via NIF query screens
Information Sharing Statement
All elements of the Framework are open and Open Source. See the NIF at http://nif.nih.gov and http://neurogateway.org; terminologies are at http://brainml.org.
Acknowledgements
This project has been funded in whole or in part through the NIH Blueprint for Neuroscience Research with Federal funds from the National Institute on Drug Abuse, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN271200577531C to Weill Cornell Medical College. BrainML representation and BrainML08 development are supported by MH57153 from NIMH and computational and related metadata partially funded by MH68012 from NIMH, both to Weill Cornell Medical College. Cortical and other mammalian terminology development was aided by NS44820 from NINDS to E.P. Gardner. Early terminology meetings were funded by the Society for Neuroscience under a generous gift from Paul Allen and Jody Patton and under contract (NIH Order No. 263-MD-409125-1) from NIMH, NINDS, and NIDA. We thank the many engaged and productive participants at our expert workshops. These included five Society for Neuroscience Presidents: Michael E. Goldberg, Bernice Grafstein, Edward G. Jones, Pasco Rakic, and David Van Essen, and chairs and co-chairs who in addition to the authors included Gwen Jacobs, Maryann E. Martone, Gordon M. Shepherd, Nick Strausfeld, Jack Van Horn, Robert W. Williams, and Rafa Yuste. Additional help was provided by John H. Byrne, Holly Cline, Katherine Graubard, Ray Guillery, Takao K. Hensch, Steven S. Hsiao, Kei Ito, Harvey Karten, Robert LaMotte, Roger Lemon, Steve Lisberger, Margaret S. Livingstone, Carol A. Mason, George Paxinos, and Joseph L. Price. We had hoped to include the complete set of participants’ names as an Appendix, but at the direction of the Editors these are available only as Supplementary Material at http://brainml.org/workshops. We gratefully acknowledge the professional encouragement and cooperation received from the Society for Neuroscience and the NIF Advisory Committee: H. Akil, G. Ascoli, D. Gardner, B. Grafstein, M.E. Martone, G.M. Shepherd, P. Sternberg, D.C. Van Essen, and R. W. Williams.
Footnotes
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Daniel Gardner, Email: dan@med.cornell.edu, Laboratory of Neuroinformatics and Department of Physiology, Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10065, USA.
David H. Goldberg, Laboratory of Neuroinformatics and Department of Physiology, Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10065, USA
Bernice Grafstein, Laboratory of Neuroinformatics and Department of Physiology, Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10065, USA.
Adrian Robert, Laboratory of Neuroinformatics and Department of Physiology, Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10065, USA.
Esther P. Gardner, Department of Physiology and Neuroscience, NYU School of Medicine, New York, NY 10016, USA
References
- Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, et al. Petilla Interneuron Terminology Group. Petilla terminology: Nomenclature of features of GABAergic interneurons of the cerebral cortex. Nature Reviews Neuroscience. 2008;9:318–324. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: Tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Arbib MA. Integrating databases and expert systems for the analysis of brain structures: Connections, similarities, and homologies. Neuroinformatics. 2004;2:19–58. doi: 10.1385/NI:2:1:019. [DOI] [PubMed] [Google Scholar]
- Bowden DM, Dubach MF. NeuroNames 2002. Neuroinformatics. 2003;1:43–60. doi: 10.1385/NI:1:1:043. [DOI] [PubMed] [Google Scholar]
- Bug W, Ascoli GA, Grethe JS, Gupta A, Fennema-Notestine C, Laird A, et al. The NIFSTD and BIRNLex vocabularies: Building comprehensive ontologies for neuroscience. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods of Information in Medicine. 1998;37:394–403. [PMC free article] [PubMed] [Google Scholar]
- Cimino JJ. From data to knowledge through conceptoriented terminologies: experience with the Medical Entities Dictionary. Journal of the American Medical Informatics Association. 2000;7:288–297. doi: 10.1136/jamia.2000.0070288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook S, Gleeson P, Howell F, Svitak J, Silver RA. MorphML: Level 1 of the NeuroML standards for neuronal morphology data and model specification. Neuroinformatics. 2007;5:96–104. doi: 10.1007/s12021-007-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cerebral Cortex (New York, N.Y.) 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Friedman C, Hripcsak G, Shagina L, Liu H. Representing information in patient reports using natural language processing and the extensible markup language. Journal of the American Medical Informatics Association. 1999;6:76–87. doi: 10.1136/jamia.1999.0060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D, Akil H, Ascoli GA, Bowden DM, Bug W, Donohue DE, et al. The Neuroscience Information Framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D, Abato M, Knuth KH, DeBellis R, Erde SM. Dynamic publication model for neurophysiology databases. Philosophical Transactions of the Royal Society of Neuroinform London. Series B, Biological Sciences. 2001a;356:1229–1247. doi: 10.1098/rstb.2001.0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D, Abato M, Knuth KH, Robert A. Neuroinformatics for neurophysiology: The role, design, and use of databases. In: Koslow SH, Subramaniam S, editors. Databasing the brain: The role, design, and use of databases. New York: Wiley; 2005. pp. 47–67. [Google Scholar]
- Gardner D, Knuth KH, Abato M, Erde SM, White T, DeBellis R, et al. Common data model for neuroscience data and data model interchange. Journal of the American Medical Informatics Association. 2001b;8:17–31. doi: 10.1136/jamia.2001.0080017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D, Toga AW, Ascoli GA, Beatty J, Brinkley JF, Dale AM, et al. Towards effective and rewarding data sharing. Neuroinformatics. 2003;1:289–295. doi: 10.1385/NI:1:3:289. [DOI] [PubMed] [Google Scholar]
- Goddard NH, Hucka M, Howell F, Cornelis H, Shankar K, Beeman D. Towards NeuroML: Model description methods for collaborative modelling in neuroscience. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2001;356:1209–1228. doi: 10.1098/rstb.2001.0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer DS, Westbrook JD, Bourne PE. An ontology driven architecture for derived representations of macromolecular structure. Bioinformatics (Oxford, England) 2002;18:1280–1281. doi: 10.1093/bioinformatics/18.9.1280. [DOI] [PubMed] [Google Scholar]
- Kennedy DN. Barriers to the socialization of information. Neuroinformatics. 2004;2:367–368. doi: 10.1385/NI:2:4:367. [DOI] [PubMed] [Google Scholar]
- Kennedy DN. Where’s the beef? Missing data in the information age. Neuroinformatics. 2006;4:271–274. doi: 10.1385/NI:4:4:271. [DOI] [PubMed] [Google Scholar]
- Koslow SH, Hirsch MD. Celebrating a decade of neuroscience databases. Looking to the future of high-throughput data analysis, data integration, and discovery neuroscience. Neuroinformatics. 2004;2:267–270. doi: 10.1385/NI:2:3:267. [DOI] [PubMed] [Google Scholar]
- Lindberg DAB, Humphreys BL, McCray AT. The unified medical language system. Methods of Information in Medicine. 1993;32:281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ascoli GA. Value added by data sharing: Longterm potentiation of neuroscience research. Neuroinformatics. 2007;5:143–145. doi: 10.1007/s12021-007-0009-0. [DOI] [PubMed] [Google Scholar]
- Marenco L, Li Y, Martone ME, Sternberg PW, Shepherd GM, Miller PL. Issues in the design of a pilot concept-based query interface for the Neuroinformatics Information Framework. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H-M, Rangarajan A, Teal TK, Sternberg PW. Textpresso for neuroscience: searching the full text of thousands of neuroscience research papers. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn JD, Gazzaniga MS. Maximizing information content in shared and archived neuroimaging studies of human cognition. In: Koslow SH, Subramaniam S, editors. Databasing the brain: The role, design, and use of databases. New York: Wiley; 2005. pp. 449–458. [Google Scholar]