Abstract
We analyzed the dynamics of an influenza A/Albany/1/98 (H3N2) viral infection, using a set of mathematical models highlighting the differences between in vivo and in vitro infection. For example, we found that including virion loss due to cell entry was critical for the in vitro model but not for the in vivo model. Experiments were performed on influenza virus-infected MDCK cells in vitro inside a hollow-fiber (HF) system, which was used to continuously deliver the drug amantadine. The HF system captures the dynamics of an influenza infection, and is a controlled environment for producing experimental data which lend themselves well to mathematical modeling. The parameter estimates obtained from fitting our mathematical models to the HF experimental data are consistent with those obtained earlier for a primary infection in a human model. We found that influenza A/Albany/1/98 (H3N2) virions under normal experimental conditions at 37°C rapidly lose infectivity with a half-life of ~ 6.6 ± 0.2 h, and that the lifespan of productively infected MDCK cells is ~ 13 h. Finally, using our models we estimated that the maximum efficacy of amantadine in blocking viral infection is ~ 74%, and showed that this low maximum efficacy is likely due to the rapid development of drug resistance.
Keywords: drug, resistance, hollow-fiber, mathematical modeling, infectious diseases
1 Introduction
Influenza A has become a growing concern for health authorities worldwide. The annual cost of influenza illness and the threat of an imminent pandemic makes it all the more necessary to revisit the treatment options currently available. In this paper, we report on a series of experiments performed in vitro inside a hollow-fiber (HF) system. Madin-Darby canine kidney (MDCK) cells were infected with influenza A/Albany/1/98 (H3N2) while subjected to a constant concentration of the drug amantadine.
Until recently, adamantane drugs, such as amantadine and rimantadine, were the primary prophylaxis and treatment method used to prevent and control influenza infection. When used prophylactically, the effiectiveness of adamantanes against sensitive strains of influenza A is between 80% and 90% (Bright et al., 2006). When used as treatment it can reduce the duration of illness by 1.5 days if administered within 48 h of symptom onset (Bright et al., 2006). Unfortunately, resistance to adamantanes emerges rapidly during treatment and resistant variants show no evidence of fitness impairment and are readily transmissible (Deyde et al., 2007). This process, further amplified by the massive use of adamantanes, has led to wide-spread resistance among circulating influenza strains worldwide with 90.5% and 15.5% resistance prevalence among H3N2 and H1N1 strains, respectively (Deyde et al., 2007). However, clade 2 H5N1 viruses from Africa and parts of Eurasia are still susceptible to adamantanes (Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Influenza A (H5N1) Virus, 2008). In addition, the efficacy of combination therapy, including adamantanes and neuraminidase inhibitors, is an active subject of research (see Ilyushina et al., 2006; Gubareva et al., 2000). Thus, in these contexts, understanding the dynamics of adamantane treatment will be important to improve its effect.
In this paper, we use mathematical models to study the effect of drug treatment on the kinetics on influenza within the HF system. We analyzed the results of in vitro influenza infection of MDCK cells in the presence of a constant concentration of amantadine. To this end, we use variations of the ordinary differential equation (ODE) models proposed by Baccam et al. (2006) and via nonlinear parameter estimation methods parametrized the kinetics of influenza dynamics and the effects of drug dosage on the infection in the HF system. Clearly, many features of in vivo infection, such as the effects of innate and adaptive immune responses, mucociliary clearance and virion transport, differ from those found in vitro. The models that we develop highlight some of these differences and the changes in modeling strategy needed to accommodate them.
2 Materials and methods
2.1 Cells and viruses
MDCK cells (ATCC CCL-34) were obtained from the American Type Culture Collection and maintained in minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 1% sodium pyruvate, 1% MEM nonessential amino acids, and 1% penicillin-streptomycin solution. A/Albany/1/98 was obtained from the Clinical Microbiology Laboratory at the Albany Medical Center Hospital. Amantadine was purchased from Fluka, Switzerland.
2.2 Hollow-fiber system
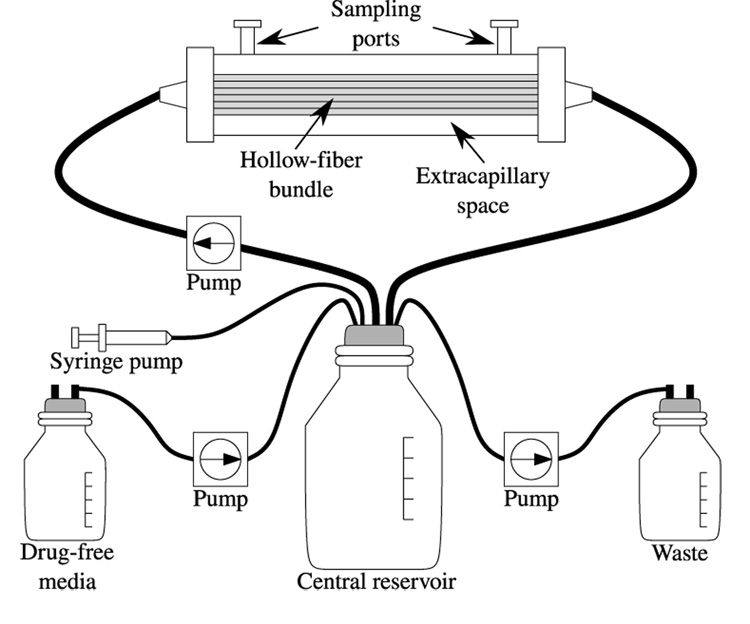
In vitro hollow fiber (HF) models for influenza infection (see Figure 1) were adapted from Bilello et al. (1994) with the following modifications. Each hollow fiber (HF) cartridge was charged with 108 MDCK cells in virus growth medium (MEM + 0.2% BSA + 2 µg/mL of TPCK-treated trypsin + 1% penicillin-streptomycin solution) and the infection was initiated by adding 100 cells pre-infected with virus. Drug delivery is controlled by a computer-driven syringe pump attached to the central reservoir, which permits the precise delivery of drug. For simplicity, the experiments described here were performed at a constant drug concentration over the full duration of the experiment. Thus, only the central reservoir was used, which contained the drug at steady-state concentration. The infection takes place in the extra-capillary space (ECS): the 15 mL of HF reactor volume not occupied by the capillary bundles. Two ports located on top of the HF reactor are used to deposit or remove materials from the ECS. At times t = 22 h, 28 h, 46 h, 72 h, 96 h, and 144 h, 3 mL of the 15 mL ECS solution are harvested for analysis. Cells and virus are obtained by mixing the uid in the ECS using the two syringes attached to the HF ports, sampling the ECS solution from one port, mixing the fluid in the ECS again, sampling the ECS solution from the opposite port and mixing a third time and then sampling the ECS from the first port. Approximately 1 mL is removed each time for a total of 3 mL. After sampling, medium is drawn into the ECS from the central reservoir so that the 15 mL are restored.
Fig. 1. Diagram of the hollow-fiber system.
The HF system allows virus-infected cells to produce high titers of cell-free virus within the HF bundle. The extracapillary space (ECS) is separated from the medium coming from the central reservoir with pore sizes that are large enough to allow nutrients, small compounds, and cellular metabolites to traverse in and out of this ECS but too small for viruses and virus-infected cells to leave the ECS. The ECS can be sampled through the sampling ports to determine the number of virus-infected cells and the amount of cell-free virus in the HF unit over time.
2.3 Plaque assays
To quantitate the amount of infectious virus taken from the hollow fiber at each time point, the number of plaque forming units (PFU) present in the clarified medium was determined by plaque assay as described by Sidwell and Smee (2000). Serial 10-fold dilutions of clarified virus supernatant were made in virus growth medium. The MDCK cell monolayers were washed twice with virus growth medium, and 0.5 mL of each virus dilution was added to the monolayers in triplicate. After a 1-h adsorption period at 37°C under an atmosphere of 5% CO2, the inoculum was removed and 5 mL of virus growth medium supplemented with 1% DEAE dextran and 0.3% agar was added to each plate. After 2 days of incubation at 37°C under an atmosphere of 5% CO2, the agar was removed, the monolayers were stained with 0.1% crystal violet in 20% ethanol, and the number of PFU were counted visually.
2.4 Parameter fitting
The models that we develop consist of systems of ordinary differential equations. These equations were solved numerically using Octave 2.1.73 (Eaton et al., 2007) and its lsode function, which uses Gear’s method if the equations are stiff, or the Adam’s method (predictor-corrector) if they are not. The delay differential equation model (which we refer to as the delay model) was solved using Matlab’s dde23 function in Octave.
The values of the models’ parameters were estimated by fitting log10(V ) predicted by numerically solving the model to log10 of the experimental viral titer. The experimental viral titer considered at each time point represents the geometric average of the three samples collected at that time point. In addition, note that log10(V ) rather than V was used for the fits because the measurement errors for viral titer are thought to follow a log-normal distribution. The fit was performed using the Octave 2.1.73 (Eaton et al., 2007) leasqr function, which is an implementation of the Levenberg-Marquardt nonlinear regression method (Seber and Wild, 1989). The 95% confidence interval (95% CI) provided for each parameter estimate corresponds to the 25th and 975th instances of 1,000 ordered bootstrap replicates (Efron and Tibshirani, 1986).
To quantify the quality of each fit, we computed the sum of squared residuals (SSR) between the experimental viral titer and the models’ results. In order to compare models with different numbers of parameters (e.g. the simple vs eclipse model or the simple model with 6 free parameters vs the simple model with 5 free parameter and 1 fixed parameter), we also computed the small-sample (second order) Akaike’s “An Information Criterion” (AICC) for each fit using
which when applied to our case of least square fits, can be re-written as
where K = Npar+1, Npar is the number of parameters of the fitted model and the +1 term arises because we estimate the variance σ2, by SSR=Npts, where Npts is the number of data points fitted by the model, and SSR is the sum of squared residuals (Burnham and Anderson, 2002). The model with the lowest AICC is considered to be the better model given the experimental data it is approximating.
3 Results
3.1 Rate of loss of viral infectivity
In our experiments, viral titer is determined from plaque assays, which yield the concentration of infectious virions (pfu/mL). Between sampling times virions cannot leave the extracellular space of the HF. Thus, the loss of infectious virions can only come from the physical degradation of virion integrity, loss of virions through cell entry, or the loss of virion infectivity such that they no longer form plaques when used in plaque assays. The cell-independent loss of virion infectivity was determined by means of the following simple experiment. Virions were incubated in the same medium and under the same conditions that exist in the HF system but in the absence of any cells. Viral titer was determined at various times by performing plaque forming assays on the virus collected from the container. This scenario can be described by
which integrates to V (t) = V0 e−ct, where V (t) and V0 are the infectious influenza virus titers at time t and time 0, respectively, and c is the first order rate constant characterizing the loss of viral infectivity we wish to determine. A linear regression was performed to fit
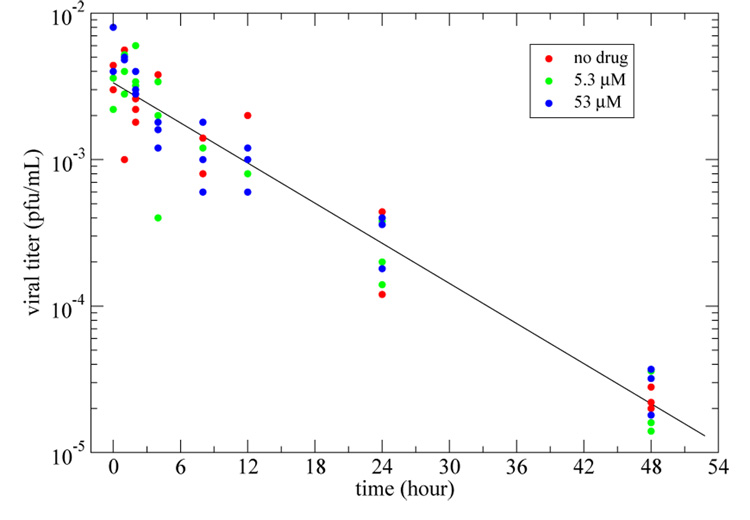
to the plaque assay triplicates from three different experiments: (i) in the absence of amantadine; (ii) with amantadine at a concentration of 5.3 µM; and (iii) with amantadine at a concentration of 53 µM. Since the presence of amantadine did not seem to affect the rate of loss of infectivity (see Figure 2), data from all three experiments were combined into a single set. The linear least square regression performed on the combined data set yielded a rate of loss of infectivity of c = 0.105 ± 0.003 h−1, corresponding to a half-life of 6.6 ± 0.2 h for infectious influenza A/Albany/1/98 (H3N2) virions at 37°C. Viral infectivity decays should be taken into consideration in the context of infection experiments. For example, if a viral sample is not used quickly, the actual multiplicity of infection (MOI) may not be the desired MOI.
Fig. 2. Rate of loss of infectivity of influenza virions.
A linear least square regression yielded a rate of loss of infectivity (slope of the best-fit line) of c = 0.105 ± 0.003 h−1. Since the presence of amantadine in various concentrations (red, green, blue) did not appear to affect the rate of loss of infectivity, data from all three experiments were combined as a single set for fitting.
3.2 Preliminary models
3.2.1 Simple model
The model
| (1) |
| (2) |
| (3) |
is a modified version of the simple infection model proposed in Baccam et al. (2006) where a term for the effect of amantadine has been added. T is the density of available target cells, I the density of infected cells secreting virions, V is the infectious viral titer as detected by plaque assays.
The parameters are such that β is the rate at which virions infect the available target cells, δ is the rate of loss of productively infected cells, p is the production rate of infectious virions by productively infected cells, and c is the rate of loss of infectious virions.
The drug used in the experiments, amantadine, is an adamantane. Adamantanes act as antiviral agents by blocking the ion channel activity of influenza virus’ matrix M2 protein (Takeda et al., 2002; Schnell and Chou, 2008; Stouffer et al., 2008). Virions, which bind to cell surface sialic acid receptors, are typically brought into a cell through the endocytic pathway. The endocytic vesicles are acidified and influenza virus’ M2 protein allows the interior of the virus to also acidify. In the presence of adamantanes, the virion interior does not acidify and this prevents viral uncoating. This, in turn, hinders the virions’ ability to replicate and successfully infect new cells (Takeda et al., 2002; Schnell and Chou, 2008; Stouffer et al., 2008). For this reason, we chose to model amantadine as affecting β, the rate at which virions successfully infect new cells. For this purpose, we introduce a parameter for drug efficacy, ϵ(t), which depends on the drug concentration, D(t). The pharmacodynamic effect of a drug is frequently modeled using some form of the Emax model (Holford and Sheiner, 1981). Here, we use
| (4) |
where D(t) is the drug concentration at time t, ϵmax is the maximum effect of the drug such that 0 < ϵmax ≤ 1, IC50 is the concentration of drug necessary to inhibit the response by 50%, and the parameter n, called the Hill coefficient, controls the steepness of the sigmoidal function. In preliminary fits, we determined that n = 1 gives a reasonable fit to the drug effect and thus we fixed n to be 1. Also, in the experiments reported here, D(t) = D is held constant such that ϵ(t) = ϵ is also constant over time.
3.2.2 Eclipse model
A more biologically accurate model should include an eclipse phase (Baccam et al., 2006). The eclipse phase is the period of time that elapses between the entry of the virus into the target cell and the release of virions produced by that newly infected cell. The eclipse model is
| (5) |
| (6) |
| (7) |
| (8) |
where ϵ is as in (4). T, I, and V are as in (1)–(3), and E is the density of infected cells in the eclipse phase, which have not yet begun releasing virions. The parameters are also as in (1)–(4) with the addition of parameter k which is the rate at which newly infected cells move out of the eclipse phase and start releasing virions. In other words, 1/k is the mean time elapsed between the successful infection of the cell and the initial release of virions produced by that cell.
3.2.3 Delay model
Another way to include an eclipse phase is to introduce it as a delay in the simple model such that
| (9) |
| (10) |
| (11) |
where ϵ is as in (4). T, I, and V are as in (1)–(3), and τ is the delay between viral entry into a cell and the start of virus production by that cell. In other words, once a virion has entered a cell the latter is removed from the target population immediately, but only enters the productively infected cell population after a time τ. During the eclipse phase the cell does not contribute to the dynamics of the simulation. We also neglect cell death during the eclipse phase consistent with our neglecting death of target cells during this brief infection. We propose this model as an alternative to the eclipse model above, where cells infected by a virion enter the eclipse compartment and remain there on average for time 1/k, with some cells becoming infectious (migrating to the I compartment) almost immediately and others taking as much as an infinite amount of time to become infected. While this fixed delay model does not provide the variability in the transition times that one would expect with any biological system, it does prevent unrealistically short or long transitions times. Models with a distributed delay have been used to model HIV infection (Mittler et al., 1998) and could also be used here, but involve adding extra parameters.
3.2.4 Basic reproductive number
In order to quantitate the ability of influenza to spread in the HF system, we computed the basic reproductive number, R0, using the parameters estimated by fitting our model to the viral titer data. The basic reproductive number represents the average number of cells that will become infected as a result of introducing a single infected cell into a population of fully susceptible cells. For all three of our models, it is given by
| (12) |
where T0 is the density of target cells available at time t = 0. Note that p/δ is the average number of virions produced by an infected cell over its lifespan, and βT0/c is the average number of cells infected per virion. Including an eclipse phase does not alter this formula as infected cells only produce virus at rate p after the eclipse phase and during their average time of viral production 1/δ.
3.2.5 Estimation of key parameters of influenza dynamics
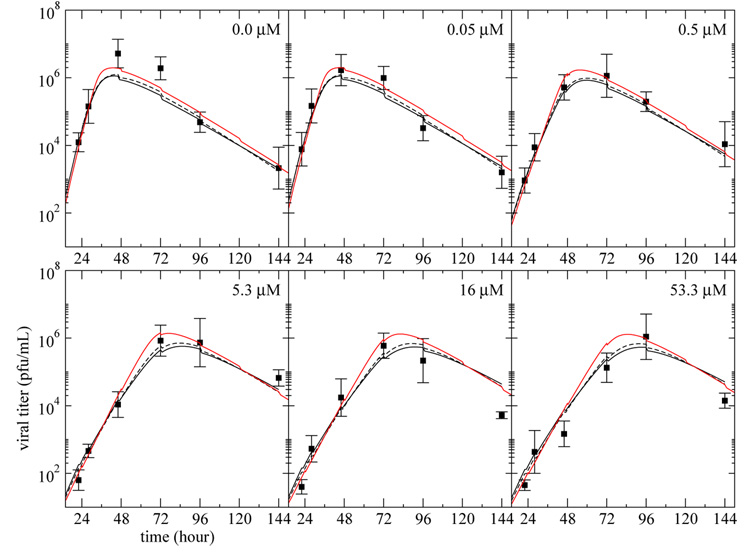
We used the simple, eclipse, and delay models to fit the viral titer data over time. To this end, each model was fitted simultaneously to data from 6 different continuous-infusion experiments each done at a different fixed drug concentration, D. In our fitting only D was allowed to vary between experiments. In Table 1, we present the best fit parameters obtained, and Figure 3 shows the fits of the models to the experimental results.
Table 1.
Parameter estimates for the simple, eclipse, and delay models
| Param. | Simple model | (95% CI) | Eclipse model | (95% CI) | Delay model | (95% CI) |
|---|---|---|---|---|---|---|
| β (mL · h−1) | 7.5 × 10−7 | (4.2–14)×10−7 | 23 × 10−7 | (6.3–53)×10−7 | 56 × 10−7 | (9.7–160)×10−7 |
| k or τ (h−1 or h) | – | – | 0.17 | (0.071–62) | 4.8 | (1.2–6.0) |
| δ (h−1) | 0.066 | (0.049–0.084) | 0.075 | (0.057–0.18) | 0.070 | (0.055–0.086) |
| p (h−1) | 0.064 | (0.036–0.105) | 0.084 | (0.049–0.23) | 0.098 | (0.056–0.15) |
| IC50 (µM) | 0.33 | (0.15–0.84) | 0.26 | (0.13–0.60) | 0.16 | (0.068–0.45) |
| ϵmax | 0.67 | (0.61–0.73) | 0.82 | (0.68–0.86) | 0.92 | (0.75–0.96) |
| 1/k or τ (h) | — | — | 6.0 | (0.016–14) | 4.8 | (1.2–6.0) |
| 1/δ (h) | 15 | (12–21) | 13 | (5.4–17) | 14 | (12–18) |
| p/δ | 0.97 | (0.68–1.4) | 1.1 | (0.78–1.6) | 1.4 | (0.92–1.9) |
| R0 | 46 | (33–69) | 170 | (44–290) | 491 | (75–1300) |
| SSR | 4.9 | (2.4–6.6) | 4.1 | (1.9–5.4) | 4.2 | (1.9–5.3) |
| AICC | −57 | (36 pts, 5 par) | −60 | (36 pts, 6 par) | −60 | (36 pts, 6 par) |
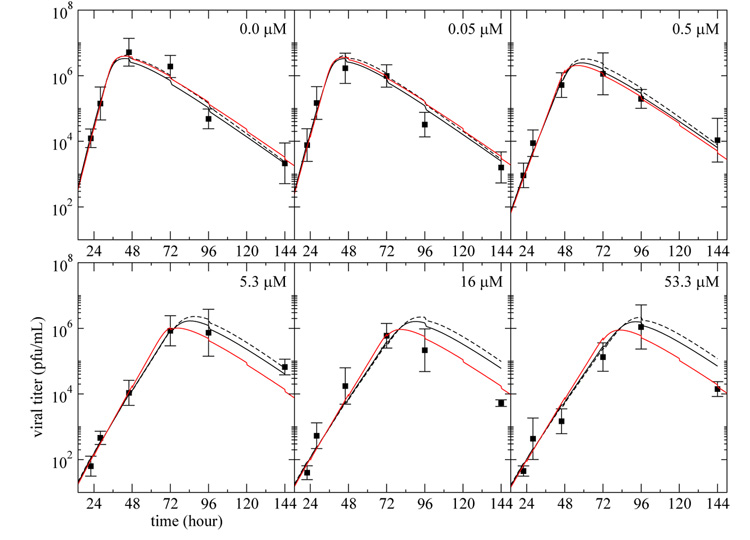
Fig. 3. Infectious viral titer predicted by the three models.
The curves represent the best fit of the infectious viral titer, V , predicted by the simple model (solid), (1)–(3), eclipse model (dashed), (5)–(8), and delay model (red), (9)–(11), to the plaque assay data (square) from experiments performed on MDCK cells in the HF system at various drug dosages. The vertical bars on the experimental points indicate the standard deviation of the experimental measurements done in triplicate. Each panel corresponds to a different drug concentration — indicated in the top right corner — maintained constant over the full duration of the experiment. All the data were fitted simultaneously and the parameter values obtained are given in Table 1.
Since infection in our experiments is initiated with 108 uninfected cells mixed with 100 infected cells deposited into the 15 mL HF reactor, the initial conditions used are T0 = 6.7 × 106 cell · mL−1, E0 = 0, I0 = 6.7 cell · mL−1, and V0 = 0. In addition, at each sampling time, we reduce T, E, I, and V by 20% because 3 mL of the 15 mL of the HF extracellular fluid are harvested for sampling. Below (see Viral Loss and Production) we will examine the effect of assuming that the cells adhere to the fiber and thus are not removed as readily as virions. Additionally, we fixed c = 0.105 h−1.
The fitted models are in good agreement with the experimental data, and the AICC values indicate that the eclipse and delay models are slightly better supported by the experimental data than the simple model. Unfortunately, the parameter estimates suggest that over its lifespan, an infected cell will produce p/δ = 0.97 (95% confidence interval: 0.68–1.4) infectious virions with the simple model, 1.1 (0.78–1.6) with the eclipse model, or 1.4 (0.92–1.9) with the delay model. If an infected cell did indeed produce less than one infectious virus over its infected lifespan, the infection could not be sustained. Strangely, however, due to the initially large density of target cells (T0 = 6.7 × 106 cells/mL) and the small clearance rate of virions, the basic reproductive numbers computed using (12) from the parameter estimates are much greater than one and much greater than the average number of infectious virions produced. But in order to be meaningful, the basic reproductive number should be smaller than the number of virions produced by a cell over its infected lifespan: at best, each infectious virion will infect only one cell. This is a clear indication that the models are not properly capturing the mechanisms involved in the infection process, or that our parameter estimation procedure is not providing accurate estimates.
3.3 Incorporating loss of virions due to cell entry
In models of in vivo viral infection virus loss due to entry into target cells is frequently neglected as this process is a minor mechanism of virion loss compared with processes such as phagocytosis and mucociliary clearance. An important consequence of the value obtained for the virion clearance rate is that in our models, the rate at which a virion is lost due to target cell entry, βT, is not negligible compared to the rate at which a virion loses infectivity, c. As per our parameter estimates, the rate of loss of infectious virus due to loss of infectivity is c = 0.105 h−1 and the rate of loss of virus due to cell entry at time t = 0, is βT0 = 5.0 h−1. Thus, loss of virions through cell entry cannot be neglected and a term needs to be added in all three models to incorporate the effect of virus entry into target cells, namely
| (13) |
where γ is the rate at which infectious virions are lost due to target cell entry. Note that this new formulation of the model will affect the calculation of the basic reproductive number, which should now be computed as
| (14) |
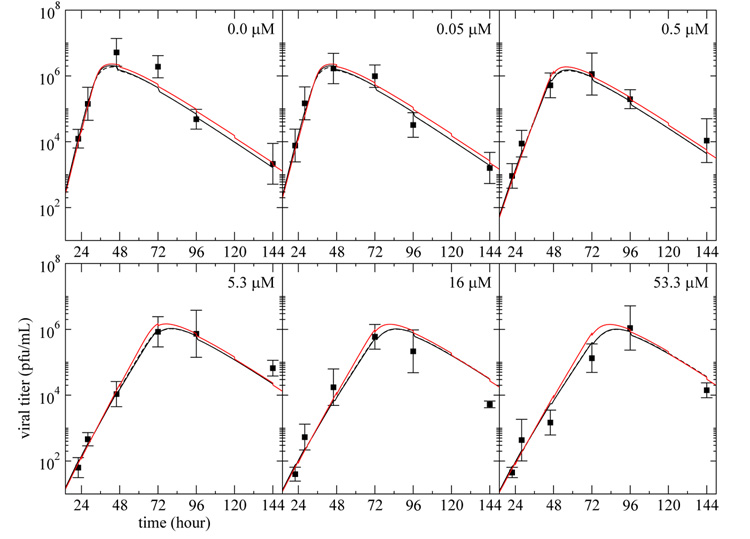
We repeated the fits to the three models where equations (3), (8), and (11) were replaced with (13), and the viral clearance rate was still held fixed at c = 0.105 h−1. The best-fit parameter estimates are presented in Table 2. The fit of the models to the experimental data are presented in Figure 4, and the predicted evolution of the target and infected cell populations over the course of the infection using the simple model are presented in Figure 5.
Table 2.
Parameter estimates for the three models with virion entry into cells
| Param. | Simple model | (95% CI) | Eclipse model | (95% CI) | Delay model | (95% CI) |
|---|---|---|---|---|---|---|
| β (mL · h−1) | 8.7 × 10−7 | (5.1–16)×10−7 | 18 × 10−7 | (7.1–54)×10−7 | 9.6 × 10−7 | (6.1–19)×10−7 |
| k or τ (h−1 or h) | – | – | 0.31 | (0.082–23) | 0.22 | (0.0028–1.3) |
| δ (h−1) | 0.076 | (0.063–0.093) | 0.076 | (0.063–0.14) | 0.075 | (0.062–0.090) |
| p (h−1) | 0.13 | (0.075–0.24) | 0.12 | (0.072–0.28) | 0.12 | (0.082–0.20) |
| γ (mL · h−1) | 1.3 × 10−7 | (0.37–3.3)×10−7 | 0.61 × 10−7 | (0.072–2.2)×10−7 | 1.1 × 10−7 | (0.38–2.3)×10−7 |
| IC50 (µM) | 0.40 | (0.23–0.78) | 0.30 | (0.17–0.68) | 0.38 | (0.21–0.76) |
| ϵmax | 0.56 | (0.51–0.64) | 0.74 | (0.55–0.83) | 0.60 | (0.54–0.71) |
| 1/k or τ (h) | — | — | 3.2 | (0.041–12) | 0.22 | (0.0028–1.3) |
| 1/δ (h) | 13 | (11–16) | 13 | (6.9–16) | 13 | (11–16) |
| p/δ | 1.7 | (1.1–2.8) | 1.6 | (1.0–2.5) | 1.7 | (1.2–2.4) |
| β/γ | 6.9 | (2.9–21) | 29 | (5.1–330) | 8.8 | (5.0–26) |
| R0 | 11 | (7.5–19) | 36 | (10–120) | 13 | (9.5–25) |
| SSR | 3.6 | (1.7–4.5) | 3.7 | (1.7–4.5) | 3.6 | (1.7–4.7) |
| AICC | −65 | (36 pts, 6 par) | −61 | (36 pts, 7 par) | −61 | (36 pts, 7 par) |
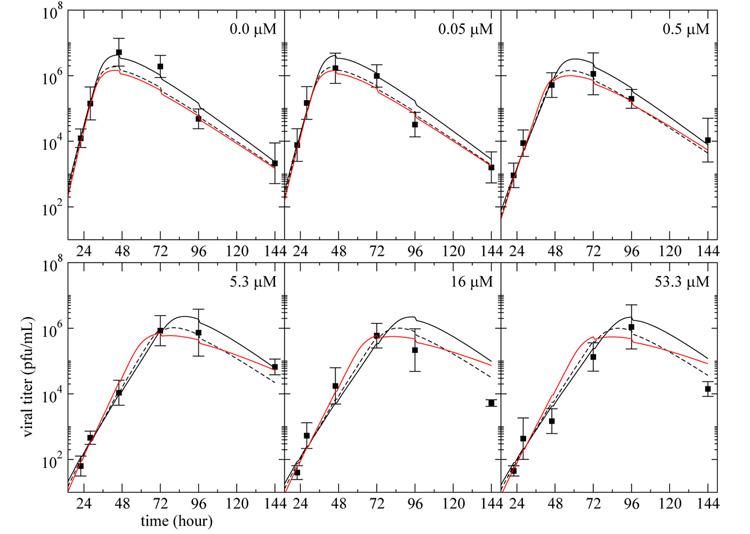
Fig. 4. Infectious viral titer predicted by the three models with virion entry into cells.
The curves represent the best fit of the infectious viral titer, V , predicted by the simple model (solid), eclipse model (dashed), and delay model (red), modified to include loss of virus due to cell entry as in Eqn. (13), to the plaque assay data (square) from experiments performed on MDCK cells in the HF system at various drug dosages. Each panel corresponds to a different drug dosage experiment where the drug is maintained at a constant concentration — indicated in the top right corner — over the full duration of the experiment. All the data were fitted simultaneously and the parameter values obtained are given in Table 2.
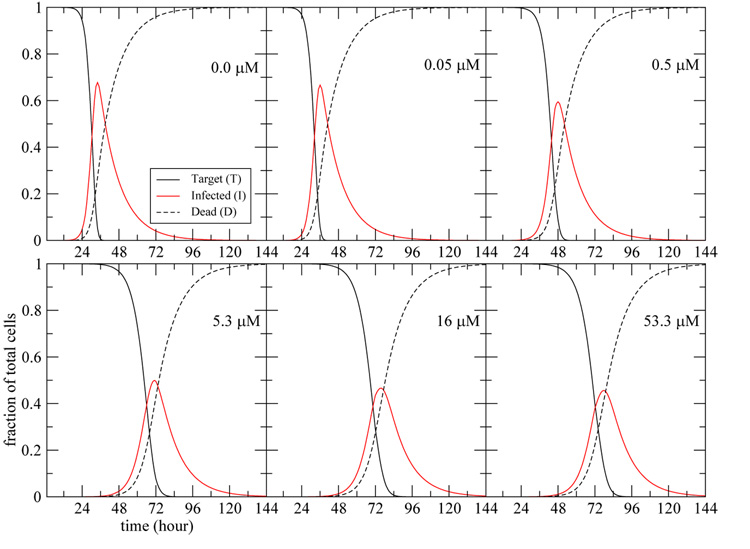
Fig. 5. Prediction of the evolution of the cell populations over time.
The populations of target, infected, and dead cells over the course of the infection are predicted using the simple model modified to include loss of virus due to cell entry as in Eqn. (13). The dead cells, D, are defined as dD/dt = δI, and the fraction of total cells are computed by dividing T, I, or D by (T + I + D) at each time point. Each panel corresponds to a different drug dosage experiment where the drug is maintained at a constant concentration — indicated in the top right corner — over the full duration of the experiment. The parameter values used are given in Table 2.
The fits obtained when including loss of virus due to cell entry are better (smaller AICC) than those obtained previously, and the simple model is now statistically better supported by the experimental data than the eclipse and delay model.
It is interesting to compare the parameter estimates obtained here for MDCK cells infected with influenza A/Albany/1/98 (H3N2) in the HF system to those obtained in Baccam et al. (2006) where similar models were used to analyze experimental data from human volunteers infected with influenza A/Hong Kong/123/77 (H1N1) in the absence of drug (ϵ = 0). Table 3 presents the results side-by-side for the two systems.
Table 3.
Comparison of parameter values for human and MDCK cells in a hollow-fiber system
| Parameters | |||||||
|---|---|---|---|---|---|---|---|
| System | Model | tEa | (95% CI) | 1/δb | (95% CI) | R0c | (95% CI) |
| Human d | simple | — | — | 6.0 h | (3.9–9.2 h) | 11 | (6.6–19) |
| eclipse | 6.0 h | (4.6–7.9 h) | 4.6 h | (2.8–7.5 h) | 22 | (10–46) | |
| HFe | simple | — | — | 13 h | (11–16 h) | 11 | (7.5–19) |
| eclipse | 3.2 h | (0.041–12 h) | 13 h | (6.9–16 h) | 36 | (10–120) | |
| delay | 0.22 h | (0.0028–1.3 h) | 13 h | (11–16 h) | 13 h | (9.5–25) | |
Length of the eclipse phase where tE = 1/k (eclipse) or τ (delay).
Length of time over which infected cells produce virions.
Basic reproductive number.
Averaged results for infection of 6 human volunteers with inuenza A/HK/123/77 (H1N1) from Baccam et al. (2006).
Results from the hollow-fiber (HF) system as per Table 2.
For human infection, we found the length of the eclipse phase, 1/k, to be about 6.0 h, almost twice the 3.2 h found here for the MDCK HF system. This difference, however, is not statistically significant due to the wide confidence interval on both parameter estimates. Both values are also in rough agreement with values reported elsewhere (Bocharov and Romanyukha, 1994; Möhler et al., 2005).
Fitting the data from human infection, Baccam et al. (2006) estimated the length of time over which infected cells produce virions, 1/δ, to be about 4.6 h to 6.0 h, about half of the 13 h found here for the MDCK HF system. The smaller productively infected cell lifespan found for in vivo infection in humans may reflect the active killing of infected cells by the immune system. It could also be due to differences between MDCK cells used in our experiments and human lung epithelial cells. Also, note that in Baccam et al. (2006), we used models that neglected the loss of virions due to cell entry as non-specific and immune-mediated clearance of infectious virions in vivo would also be taking place.
The fits to the experimental viral titer allowed us to obtain new estimates for the number of infectious virions a cell is expected to produce over its lifespan. We found p/δ = 1.7 (1.1–2.8) virions from the simple model, 1.6 (1.0–2.5) from the eclipse model, or 1.7 (1.2–2.4) in the delay model. These estimates are greater than one for all models, but are still low. It is important to note that in the plaque assay, one only measures infectious virions. However, we expect that the majority of viral particles produced by an infected cell are non-infectious. To test this, in another set of experiments, performed on MDCK cells in the HF system, we assayed both the infectious viral titer (measured through plaque assays) and the total viral titer (measured through PCR assays). We found that infectious virions made up only about 1 in 10,000 virions (data not shown). Thus, assuming that an infected cell will produce 1 infectious virion for each 10,000 virions produced, an infected cell is expected to produce about 17,000 virions over its lifespan. This is in agreement with values suggested elsewhere (Bocharov and Romanyukha, 1994; Möhler et al., 2005).
We also computed the basic reproductive number using (14) and the parameter estimates found for both models. We found it to be 11 (7.5–19) for the simple model, 36 (10–120) for the eclipse model, and 13 (9.5–25) for the delay model. These values compare well with the basic reproductive numbers of 11 (6.6–19) and 22 (10–46) for the human infection data analyzed using the simple and eclipse models, respectively (Baccam et al., 2006), but they are still larger than our estimate of the number of infectious virions produced by an infected cell over its lifespan. Furthermore, the rate at which infectious virions infect new cells, β, is larger than the rate at which infectious virions are lost due to cell entry, γ, suggesting that a single infectious virion can infect more than one cell. These odd results are addressed in detail below.
3.4 Viral loss and production
The influenza infection process is such that a target cell becomes productively infected after one or more virions have entered it. Thus, one would expect the rate at which virions are cleared due to entry into target cell, γ, to be either larger or at least equal to the rate at which virions infect cells, β. Unfortunately, our best parameter estimates suggest that γ is smaller than β. If we force β = γ the fit of the simple model is not as good (AICC = −51) as when is free (AICC = −65), with the model showing systematic deviations from the data. How then can this be explained?
One possible explanation is that the number of virions counted by the plaque assay underestimates the true number of infectious virions. Say the true number of infectious virions, Vreal, is such that Vreal = αV, where V is the number of infectious virions counted by the plaque assay and α > 1. If this were the case, our model with virion entry into cells would become
where V was replaced with Vreal=/α. Thus, the underestimation of the number of infectious virions would affect the rate of infection of target cells, β, and the rate of production of virus, p, such that βreal = β/α, and preal = α p. This means that the rate of infection of target cell by virions, βreal, could in fact be equal to or greater than the rate of virus entry into target cells, γ, provided that α is such that βreal = β/α ≤ γ While this would not affect the value of the basic reproductive number, it could explain why we obtained values of R0 that were greater than the number of infectious virions produced by a cell over its infected lifespan. Indeed, we would have
and if α is such that β/α ≤ γ, then R0 would indeed be smaller than (p α)/δ = preal/δ.
In order to have β/α ≤ γ, α would have to be at least 6.9 (2.9–21) with the simple model, 29 (5.1–330) with the eclipse model, or 8.8 (5.0–26) with the delay model when including virion entry into cells (see β/γ in Table 2). Assuming that exactly one infectious virus is lost every time a cell is infected implies βreal = β/α = γ. That is, α = 6.9 in the simple model, 29 in the eclipse model, and 8.8 in the delay model. Since our estimated values of p/δ are 1.7, 1.6, and 1.7 for the simple, eclipse, and delay models, respectively, such values of α imply that an infected cell is expected to produce at least (p α)/δ ~ 12 infectious virions according to the simple model or as many as 46 infectious virions according to the eclipse model, or 15 according to the delay model. These numbers are now larger than their respective basic reproductive numbers of 11, 36, and 13, as they should be. But what could account for the fact that no more than 14% (1/6.9) of infectious virions are being counted?
Several factors may be responsible for this underestimation of the true count of infectious virions. The most significant contribution likely comes from the fact that the virions sampled from the HF reactor are frozen for storage, and subsequently thawed before they are counted by plaque assay. Experimental data suggests that there is a 10-fold reduction in plaque counts after each freeze/thaw cycle: a loss of 90% of infectious virions (data not shown). Infectious virions may also be lost between the time of thawing and plaquing because the rate of loss of infectivity is high, and may be even higher depending on how carefully the virions were handled. In addition, plaques that are not well separated may be miscounted resulting in a systematic underestimation of infectious virus count.
Unfortunately, without a good estimate of the ratio of counted infectious virions to true number of infectious virions, our estimates for the production rate of infectious virus, the infection rate of cells, and the number of virions needed to infect a cell need to be interpreted with caution. Additional experiments are needed in order to estimate α, and thus to obtain better estimates for these parameters.
3.5 Hollow fiber system sampling
At each sampling time, we reduced T, E, I, and V by 20% because 3 mL of the 15 mL of the HF extracellular fluid is harvested for sampling. But it is possible that inside the HF, uninfected cells and cells in the early stage of infection attach themselves solidly to the surface of the capillary bundle. In that case, less than 20% of the cells present in the HF system may be harvested. We explored the effect of having anywhere between 0–20% of uninfected and/or infected cells removed at each sampling time on the parameter estimates obtained using the simple model with virion entry into cells. We found that varying the percentage of cells removed at each sampling time did not have a significant effect on the parameter estimates with the exception of β/γ which varied from 6.9 when 20% of cells were removed to 29 when no cells were removed, and ϵmax which varied from 0.56 when 20% of cells were removed to 0.67 when no cells were removed.
3.6 Models without multiple infection
In the models developed above, the effect of amantadine has been modeled by assuming that the drug blocked infection and hence the loss of target cells. Thus with a perfect drug (ϵ = 1), target cells would never be lost. However, in the presence of amantadine virus still enters cells. Thus, implicit in these models is the assumption that multiple virions can enter cells and the successive entry of virions into target cells does not reduce their ability to accommodate more virions. Successive entry of infectious virions into cells is commonly called multiple infection. Here because target cells are never lost when ϵ = 1, in principle an infinite number of virions can enter. This is not very realistic (Huang et al., 2008) and thus we decided to also explore two different approaches to correct the models.
3.6.1 Removal of target cells after virion entry
This approach is equivalent to taking the opposite limiting case. Whereas before an infinite number of virions could enter a target cell, here only a single infectious virion will be allowed to enter. In other words, entry of an infectious virus removes a cell from the target cell population whether or not this cell becomes infected. This is modeled by removing the drug effect, (1−ϵ), from the target cell equations in the three models, such that
| (15) |
leaving the equations for E, I, and V as before. Indeed, since adamantanes affect the successful infection of the cell and not the rate of viral entry into the cell, the drug should only affect the rate at which target cells enter the infected compartment, not the rate at which they leave the target cell compartment.
We repeated the fit to the three models where the equation for the virus titer, V , was replaced with (13), the equation for the target cells, T, was replaced with (15), and holding the viral clearance rate fixed at c = 0.105 h−1.
The best-fit of this model occurred for k = ∞ (no eclipse phase). In order to keep an eclipse phase, we fixed k = 0.25 h−1 (1/k = 4 h) consistent with values obtained here and adopted in an earlier mathematical model for influenza A/Equi 2 (H3N8) infection in large-scale microcarrier cultures of MDCK cells (Möhler et al., 2005). The best-fit parameter estimates are presented in Table 4. Figure 6 shows the fit of the models to the viral titer data.
Table 4.
Parameter estimates for the three models with virion entry into cells and no multiple infection
| Param. | Simple model | (95% CI) | Eclipse model | (95% CI) | Delay model | (95% CI) |
|---|---|---|---|---|---|---|
| β (mL · h−1) | 5.6 × 10−7 | (3.5–9.8)×10−7 | 8.7 × 10−7 | (4.8–17)×10−7 | 6.3 × 10−7 | (4.1–12)×10−7 |
| k or τ (h−1 or h) | — | — | 0.25 | Fixed | 0.45 | (0.0013–2.7) |
| δ (h−1) | 0.080 | (0.067–0.096) | 0.086 | (0.071–0.11) | 0.078 | (0.066–0.094) |
| p (h−1) | 0.22 | (0.14–0.39) | 0.30 | (0.18–0.49) | 0.22 | (0.14–0.40) |
| γ(mL · h−1) | 1.5 × 10−7 | (0.66–3.3)×10−7 | 0.90 × 10−7 | (0.27–2.1)×10−7 | 1.2 × 10−7 | (0.37–2.4)×10−7 |
| IC50 (µM) | 0.40 | (0.25–0.76) | 0.31 | (0.17–0.58) | 0.37 | (0.18–0.67) |
| ϵmax | 0.56 | (0.51–0.62) | 0.75 | (0.70–0.80) | 0.63 | (0.54–0.82) |
| 1/k or τ (h) | — | — | 4.0 | Fixed | 0.45 | (0.0013–2.7) |
| 1/δ (h) | 12 | (10–15) | 12 | (9.3–14) | 13 | (11–15) |
| p/δ | 2.8 | (1.9–4.2) | 3.4 | (2.3–5.0) | 2.8 | (2.0–4.5) |
| β/γ | 3.7 | (1.8–8.0) | 9.6 | (4.6–26) | 5.1 | (2.7–20) |
| R0 | 9.3 | (6.9–13) | 28 | (20–46) | 13 | (8.4–42) |
| SSR | 3.2 | (1.6–3.9) | 3.5 | (1.7–4.5) | 3.3 | (1.7–3.9) |
| AICC | −69 | (36 pts, 6 par) | −66 | (36 pts, 6 par) | −65 | (36 pts, 7 par) |
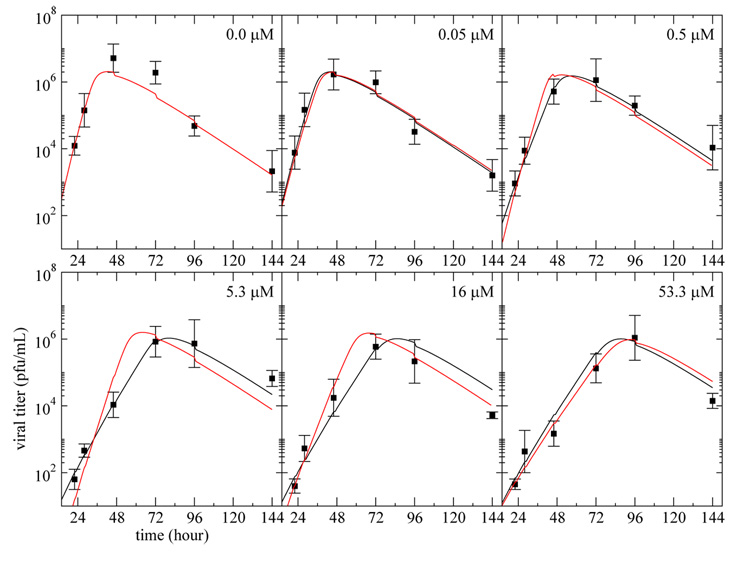
Fig. 6. Infectious viral titer predicted by the three models with virion entry into cells and no multiple infection.
The curves represent the best fit of the infectious viral titer, V , predicted by the simple model (solid), eclipse model (dashed), and delay model (red), modified to include loss of virus due to cell entry as per (13), and no multiple infection as per (15), to the plaque assay data (square) from experiments performed on MDCK cells in the HF system at various drug dosages. Each panel corresponds to a different drug dosage experiment where the drug is maintained at a constant concentration — indicated in the top right corner — over the full duration of the experiment. All the data were fitted simultaneously and the parameter values obtained are given in Table 4.
The fitted models are in good agreement with the experimental data, and the AICC values indicate that this model formulation is slightly better supported by the experimental data than the formulation allowing multiple viral entries into target cells. The parameter estimates have not been significantly affected by the change, and the parameters presented in Table 2 and Table 4 are indistinguishable given the confidence intervals.
3.6.2 Lengthening of the eclipse phase
This approach lays probably somewhere between the case where cells are removed upon entry of a single virion and the case where cells can absorb an infinite number of virions and remain susceptible to infection. This is modeled by putting the drug effect, (1 − ϵ), in front of k rather than β, such that the eclipse model becomes
| (16) |
| (17) |
| (18) |
| (19) |
We fit this model to the data holding the viral clearance rate fixed at c = 0.105 h−1. Unfortunately, convergence could not be achieved. In order to facilitate convergence, we fixed the length of the eclipse phase in the absence of drug to 1/k = 4 h, as above. The best-fit parameter estimates are presented in Table 5. Figure 7 shows the fit of the models to the viral titer data.
Table 5.
Parameter estimates for the eclipse model with the drug affecting the length of the eclipse phase
| Param. | Eclipse model | (95% CI) |
|---|---|---|
| β (mL · h−1) | 30 × 10−7 | (13–82)×10−7 |
| k (h−1) | 0.25 | Fixed |
| δ (h−1) | 0.075 | (0.057–0.093) |
| p (h−1) | 0.093 | (0.055–0.18) |
| γ(mL · h−1) | 0.84 × 10−7 | (0.11–3.8)×10−7 |
| IC50 (µM) | 0.27 | (0.12–0.71) |
| ϵmax | 0.83 | (0.78–0.88) |
| 1/k (h) | 4.0 | Fixed |
| 1/δ (h) | 13(h) | (11–17) |
| p/δ | 1.2 | (0.86–2.0) |
| β/γ | 36 | (13–190) |
| R0 | 38 | (24–85) |
| SSR | 5.1 | (1.9–6.8) |
| AICC | −52 | (36 pts, 6 par) |
Fig. 7. Infectious viral titer predicted by the eclipse model with virion entry into cells, with no multiple infection, or with the drug affecting the length of the eclipse phase.
The curves represent the best fit of the infectious viral titer, V , predicted by the eclipse model with virion entry into cell (dashed), with no multiple infection (solid), or with the drug affecting the length of the eclipse phase (red), to the plaque assay data (square) from experiments performed on MDCK cells in the HF system at various drug dosages. Each panel corresponds to a different drug dosage experiment where the drug is maintained at a constant concentration — indicated in the top right corner — over the full duration of the experiment. All the data were fitted simultaneously and the parameter values obtained are given in Table 2 (dashed), Table 4 (solid), and Table 5 (red).
The newly fitted system is in reasonable agreement with the experimental data, but the AICC value indicates that the model formulation is actually the worst of all those proposed so far. The parameter estimates have not been significantly affected by the change, and the parameters presented in Table 2, Table 4, and Table 5 are indistinguishable given the confidence intervals. From Figure 7, we can see that the fit of the model where the drug affects the length of the eclipse phase is most similar to that of the model with viral clearance due to cell entry with multiple infection. The two models differ most at higher drug concentrations.
3.7 Effect of amantadine
Figure 4, shows that the presence of amantadine has the effect of delaying the viral titer peak rather than reducing it. This is because adamantanes only lower the rate of infection of new cells. Figure 5, shows that the presence of the drug not only delays the peak of infected cells, but also reduces the fraction of cells infected at the peak from about 68% in the absence of drug to less than 48% at the higher drug concentrations. Note, however, that the total number of cells infected during the experiment (area under the curve) remains roughly the same since essentially all cells are infected.
Fitting our models to viral titer data obtained at different amantadine drug concentrations simultaneously provides an estimate of the effect of amantadine on viral titer. We found the IC50 to be 0.40 µM (0.23–0.78 µM) with the simple model, 0.30 µM (0.17–0.68 µM) with the eclipse model, and 0.38 µM (0.21– 0.76 µM) with the delay model. These values are in good agreement with the value reported in Ilyushina et al. (2006) of 0.56 µM for amantadine in the context of an in vitro infection of MDCK cells with influenza A/Panama/2007/99 (H3N2).
We also obtained estimates of ϵmax, the maximum drug effect, of 56% (51–64%) with the simple model, 74% (55–83%) with the eclipse model, and 60% (54–71%) with the delay model. These low values for ϵmax are surprising given that in tissue cultures, amantadine blocks influenza virus yield 90–99%, depending on the virus strain (Takeda et al., 2002). This discrepancy may be due to the emergence of drug resistance under treatment. Indeed, adamantanes are known to rapidly generate drug resistance as a single amino acid mutation is sufficient to confer drug resistance to a mutant (Abed et al., 2005; Belshe et al., 1988; Bright et al., 2006; Saito et al., 2003). In order to explore this possibility, we investigated the effect of fixing ϵmax to a higher value, 95%, and setting
where τµ is the time where the virus population shifts from being composed of mostly wild-type strains with IC50 as in Table 2 to mostly drug-resistant strains with IC50 = IC50,mutant. Note that the IC50,mutant is not meant to represent the IC50 of a specific mutant strain (e.g. S31N), but rather that of the combined drug-resistant mutant population. In reality, the development of drug-resistant mutations is a stochastic process and a number of different mutants can emerge.
We used the least square fitting method to find the best estimates for a common IC50,mutant for all experiments and individual τµ for each experiment with a different drug concentration (see Table 6). In the fitting process, all other parameters were fixed and given the values presented in Table 2. The viral titer curves resulting from this modification are presented in Figure 8. One expects that at higher drug concentrations the mutant population will have greater selective advantage, and thus should overtake the wild-type population more rapidly. In fact, our results in Table 6 show that τµ is lower for higher drug concentrations.
Table 6.
Parameters for the case where mutants are taken into consideration
| Drug concentration (µM) |
τµ (h) |
IC50,mutant (µM) |
ϵmax |
|---|---|---|---|
| 0.0 | 144 | ||
| 0.05 | 120 | ||
| 0.5 | 13 | 33 | 95% |
| 5.3 | 15 | ||
| 16 | 9.2 | ||
| 53.3 | 0 |
Fig. 8. Effect of considering emergence of drug-resistance on viral titer predictions.
The best fit of the viral titer data using the simple model with virion entry into cells, with and without considering the effect of the emergence of drug-resistant mutants on ϵmax are presented. The simple model using the parameters presented in Table 2 with ϵmax = 56% (black) is compared against the simple model using ϵmax = 95% and IC50 = 0.40 µM for t ≤ τµ and IC50 = 33 µM for t > τµ with τµ as in Table 6 (red). All other parameters are as in Table 2.
We obtained a SSR of 5.5 fitting the viral titer curves to the simple model with ϵmax = 95%. While this fit is not as good as that obtained using ϵmax = 56% (SSR = 3.6), it did improve the fit at higher drug concentrations. Indeed, the fits with ϵmax = 95% are notably worse for the intermediate drug concentrations 0.5 µM and 5.3 µM, but better for the higher concentrations 16 µM and 53.3 µM. This suggests that if the mutant population could be explicitly modeled, a larger estimate for ϵmax would likely be found. Unfortunately, such an explicit model of the drug resistant mutants involves too many parameters and could not be explored with the present data. The model presented here, with τµ, is a conceptual model which shows that drug resistance could account for the low value of ϵmax. This model, with its extra parameter exhibited very slow and unreliable parameter convergence when fitted to the experimental data. For this reason, we were not able to obtain bootstrap CI for the parameter estimates.
4 Discussion
We analyzed the dynamics of an influenza A/Albany/1/98 (H3N2) viral infection of MDCK cells under various constant concentrations of amantadine using mathematical models. The experiments were conducted in vitro inside a hollow-fiber (HF) system.
We first attempted to fit the experimental data with the models used previously in Baccam et al. (2006), i.e. a simple model describing the population of uninfected and infected cells, and virions, and a more realistic eclipse-phase model where the time elapsed between the entry of the infectious virus and the release of infectious virus by the newly infected cell (the eclipse-phase) is explicitly taken into consideration. These models, which were developed for in vivo infection, assume virions are lost by a first-order process. We also considered a delay model where the eclipse-phase is included as a delay in the creation of new infected cells rather than as an additional compartment as in the eclipse-phase model.
In vitro, mucosal and immune-mediated clearance of virus cannot occur, but nonetheless we observed loss of infectious virus within the HF system. This caused us to consider the possibility that loss of viral infectivity rather than loss of viral particles could be playing a significant role. Through an independent experiment, we determined that under normal experimental conditions at 37°C, influenza A/Albany/1/98 (H3N2) virions lose infectivity rapidly with a half-life of 6.6 h. This has important implications for the design of experimental procedures as inconsistent delays before plating a viral inoculum for plaque counting could result in significant discrepancies in quantitation of viral titer.
Given the nature of the mechanisms by which infectious virions are lost within the HF system, we then considered a modified version of the standard models with one term for non-specific clearance due to loss of infectivity, and a second term for viral loss due to entry of infectious virus into uninfected cells. The clearance term due to loss of viral infectivity was fixed to the value determined in our independent experiment, thus reducing the number of parameters to be estimated. The addition of the term for loss of virions due to cell entry while fixing the value for the clearance rate due to loss of viral infectivity significantly improved the fit of the models to the experimental data. This confirmed that these more accurate models better capture the experimental reality.
Using these models, we found the lifespan of infected cells to be ~ 13 h. We were also able to compute the basic reproductive number from our parameter estimates and found it to be 11 for the simple model, 36 for the eclipse model, and 13 for the delay model. These values are in good agreement with the values of 11 (simple) and 22 (eclipse) found in Baccam et al. (2006) for an in vivo human primary infection with influenza A/HK/123/77 (H1N1).
We also obtained estimates for the parameters describing the drug effect. Our estimates for the IC50 of amantadine of 0.4 µM for the simple model, 0.3 µM for the eclipse model, and 0.38 µM for the delay model are in good agreement with the value reported in Ilyushina et al. (2006) of 0.56 µM for amantadine in the context of an in vitro infection of MDCK cells with influenza A/Panama/2007/99 (H3N2). On the other hand, we found a maximum drug efficacy for amantadine of 56%, 74%, and 60% with the simple, eclipse, and delay models, respectively. Amantadine, however, is reported to have a maximum efficacy of 90–99%, depending on the virus strain (Takeda et al., 2002). Using a simplistic model, we confirmed that the rapid emergence of drug resistance over the course of treatment with amantadine could account for the low value obtained for the maximum drug efficacy.
In conclusion, we developed a set of models of influenza infection in vitro in a hollow fiber system that allows precise control of antiviral drug concentrations. Fitting the models to data allowed us to estimate key parameters that characterize influenza A infection in vitro and the effects of amantadine treatment. We determined that the emergence of drug-resistant mutants over the course of treatment needs to be considered explicitly in order to accurately characterize the effect of adamantanes on the dynamics of an influenza viral infection. This is particularly important as understanding the process of emergence of drug-resistance under adamantane treatment could be of great help in guiding the design of new drug compounds or drug-combination therapies.
Acknowledgments
The authors wish to thank Kristine Zager for technical assistance. Portions of this work were done under the auspices of the U.S. Department of Energy under contract DE-AC52-06NA25396 and supported by the LANL/LDRD Program (RMR, ASP), a UNM/LANL Joint Science and Technology Laboratory grant and NIH grant R21-AI73607 (CAAB). In addition, support was provided by Adamas Pharmaceuticals Inc. to the Ordway Research Institute to perform the experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abed Y, Goyette N, Boivin G. Generation and characterization of recombinant inuenza A (H1N1) viruses harboring amantadine resistance mutations. Antimicrob. Agents Chemother. 2005 February;42(2):556–559. doi: 10.1128/AAC.49.2.556-559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccam P, Beauchemin C, Macken CA, Hayden FG, Perelson AS. Kinetics of inuenza A virus infection in humans. J. Virol. 2006 August;80(15):7590–7599. doi: 10.1128/JVI.01623-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshe RB, Smith MH, Hall CB, Betts R, Hay AJ. Genetic basis of resistance to rimantadine emerging during treatment of inuenza virus infection. J. Virol. 1988 May;62(5):1508–1512. doi: 10.1128/jvi.62.5.1508-1512.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilello JA, Bauer G, Dudley MN, Cole GA, Drusano GL. Effect of 2’,3’-didehydro-3’-deoxythymidine in an in vitro hollow-fiber pharmacodynamic model system correlates with results of dose-ranging clinical studies. Antimicrob. Agents Chemother. 1994 June;38(6):1386–1391. doi: 10.1128/aac.38.6.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov GA, Romanyukha AA. Mathematical model of antiviral immune response III. Inuenza A virus infection. J. Theor. Biol. 1994;167(4):323–360. doi: 10.1006/jtbi.1994.1074. [DOI] [PubMed] [Google Scholar]
- Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among inuenza A viruses isolated early during the 2005–2006 inuenza season in the United States. JAMA. 2006 February 22;295(8):891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model Selection and Multi-model Inference: A Practical Information-Theoretic Approach. 2nd Edition. New York, USA: Springer-Verlag New York Inc; 2002. [Google Scholar]
- Deyde VM, Xu X, Bright RA, Shaw M, Smith CB, Zhang Y, Shu Y, Gubareva LV, Cox NJ, Klimov AI. Surveillance of resistance to adamantanes among inuenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 2007 July 15;196(2):249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- Eaton JW, et al. GNU Octave version 2.1.73. A free open-source soft-ware for solving linear and nonlinear problems numerically, and for performing other numerical experiments using a language that is mostly compatible with Matlab. 2007 Available at: http://www.octave.org/
- Efron B, Tibshirani R. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Statistical Science. 1986 February;1(1):54–75. [Google Scholar]
- Gubareva LV, Kaiser L, Hayden FG. Inuenza virus neuraminidase inhibitors. Lancet. 2000 March 4;355(9206):827–835. doi: 10.1016/S0140-6736(99)11433-8. [DOI] [PubMed] [Google Scholar]
- Holford N, Sheiner L. Understanding the dose-effect relationship: Clinical application of pharmacokinetic-pharmacodynamic models. Clin. Pharmacokinet. 1981 November–December;6(6):429–453. doi: 10.2165/00003088-198106060-00002. [DOI] [PubMed] [Google Scholar]
- Huang I-C, Li W, Sui J, Marasco W, Choe H, Farzan M. Inuenza A virus neuraminidase limits viral superinfection. J. Virol. 2008 May;82(10):4834–4843. doi: 10.1128/JVI.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyushina NA, Bovin NV, Webster RG, Govorkova EA. Combination chemotherapy, a potential strategy for reducing the emergence of drug-resistant inuenza A variants. Antiviral Res. 2006 July;70(3):121–131. doi: 10.1016/j.antiviral.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Mittler JE, Sulzer B, Neumann AU, Perelson AS. Inuence of delayed viral production on viral dynamics in HIV-1 infected patients. Math. Biosci. 1998 September;152(2):143–163. doi: 10.1016/s0025-5564(98)10027-5. [DOI] [PubMed] [Google Scholar]
- Möohler L, Flockerzi D, Sann H, Reichl U. Mathematical model of inuenza A virus production in large-scale microcarrier culture. Biotechnol. Bioeng. 2005 April 5;90(1):46–58. doi: 10.1002/bit.20363. [DOI] [PubMed] [Google Scholar]
- Saito R, Sakai T, Sato I, Sano Y, Oshitani H, Sato M, Suzuki H. Frequency of amantadine-resistant inuenza A viruses during two seasons featuring cocirculation of H1N1 and H3N2. J. Clin. Microbiol. 2003 May;41(5):2164–2165. doi: 10.1128/JCM.41.5.2164-2165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of inuenza a virus. Nature. 2008 January 31;451(7178):591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seber GAF, Wild CJ. Nonlinear Regression. New York, USA: John Wiley & Sons, Inc; 1989. [Google Scholar]
- Sidwell RW, Smee DF. In vitro and in vivo assay systems for study of inuenza virus inhibitors. Antiviral Res. 2000 October;48(1):1–16. doi: 10.1016/s0166-3542(00)00125-x. [DOI] [PubMed] [Google Scholar]
- Stouffer AL, Acharya R, Salom D, Levine AS, Costanzo LD, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an inuenza virus proton channel. Nature. 2008 January 31;451(7178):596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. Inuenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 2002 February;76(3):1391–1399. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Inuenza A (H5N1) Virus. Update on avian inuenza A (H5N1) virus infection in humans. N Engl J Med. 2008 January 17;358(3):261–273. [Google Scholar]