Abstract
Inactivation of mammalian telomerase leads to telomere attrition, eventually culminating in uncapped telomeres, which elicit a DNA damage response and cell cycle arrest or death. In some instances, telomerase modulation evokes a response not obviously attributable to changes in telomere length. One such example is the suppression of the DNA damage response (DDR) and changes in histone modification that occur upon repression of the telomerase reverse transcriptase, TERT, in human primary cells [1]. Here, we evaluate the contribution of TERT to the DDR in murine Tert−/− cells without critically shortened telomeres. We treated mTert−/− embryonic stem (ES) cells and murine embryonic fibroblasts (MEFs) with etoposide and irradiation, and assessed the status of p53pS15, 53BP1, ATMpS1981, SMC1pS957, and γH2AX by indirect immunofluorescence or western blotting. In four independently derived mTert−/− ES cell lines, there was no significant difference in the induction of γH2AX, 53BP1 foci formation, or the phosphorylation of ATM targets (ATM, SMC1, p53) between wildtype and mTert−/− ES cells and MEFs. A slight difference in post-translational modification of histones H3 and H4 was observed in a subset of mTert−/− ES cells, however this difference was reflected in the cellular levels of H3 and H4. Thus, in contrast to previous studies in human cells, the absence of Tert does not overtly affect the ATM-dependent response to DNA damage in murine cells.
Introduction
Telomerase, a unique cellular reverse transcriptase present in most eukaryotes, plays a crucial role in the replenishment of eroded telomeres, which occur as a by-product of the replication of chromosome termini [2,3]. Purified human telomerase consists of a telomerase reverse transcriptase (TERT), an integral RNA component (TR), and an associated subunit dyskerin [4,5]. TERT reverse transcribes a simple G-rich hexanucleotide sequence (TTAGGG in mammals) onto the 3′ end of chromosomes using TR as an internal RNA template [6]. The accessibility of telomerase to the telomere is regulated; in the budding yeast Saccharomyces cerevisiae, telomerase recruitment occurs in late S and G2 phases of the cell cycle [4]. In addition to a role in modulating access to the telomere, several telomere-associated proteins serve to protect chromosome ends from inappropriate recognition as a DNA break; these include Cdc13/Stn1/Ten1 in S. cerevisiae, and the shelterin complex in mammals (TRF1, TRF2, TIN2, TPP1, POT1, and Rap1) [4,7–9]. TRF1 acts to repress ATM-dependent signaling, and POT1 represses an ATR-dependent DNA damage response at the telomere [10,11]. In the ciliate Tetrahymena thermophila, POT1 also appears to play a role in chromosome end protection similar to humans [12]. In Arabdopsis thaliana POT1 plays a slightly different role, and appears to coordinate length regulation via an association with telomerase [13,14]. Even in the absence of environmental or replication-induced DNA damage, a transient activation of the DNA damage response at the telomere in G2 appears necessary to permit access of telomerase and telomere-processing activities essential to end protection [15,16].
In most cell types without telomerase activity, telomere erosion eventually results in ‘critically short’ termini that elicit a DNA damage response and permit end-to-end fusions [17]. The latent induction of the DDR after sufficient telomere erosion is highly conserved from yeast to humans [18–25]. The definition of a ‘critically short’ telomere is likely cell-context dependent. In primary human cells, measurement of the XpYp and 17p telomeres reveals that the majority of chromosome ends contain between 0–12.8 telomeric DNA repeats at senescence; rarely, fused ends had lost more than a kilobase of terminal DNA in a manner consistent with previously documented ‘telomere rapid deletion’ events [26–30]. The hallmarks of a damaged telomere (whether via uncapping or telomere erosion) include activation of p53-, ATM- and ATR-dependent targets, and recruitment of phosphorylated H2AX (γH2AX) and 53BP1 [31–34]. Similar to mammalian cells, in S. cerevisiae critically shortened telomeres coincide with the onset of a DDR and increased genome instability, including gross chromosomal rearrangements [35–39]. The concomitant genomic instability that arises in the presence of damaged telomeres has been suggested to be a driving force during human tumorigenesis [40,41].
Despite unequivocal evidence for the physiological function of telomerase in chromosome end maintenance, other potential roles for telomerase have emerged [42]. In S. cerevisiae, overexpression of the genes encoding the telomerase RNA, TLC1, or telomerase reverse transcriptase, EST2, suppress the temperature or damage-induced sensitivity of rad50Δ, yku80Δ, xrs2Δ, and mre11ts strains [43,44]. In yku80Δ cells, the suppression of temperature sensitivity by EST2 or TLC1 did not overtly affect overall telomere length [44]. In mice, overexpression of Tert in the skin leads to reversible neoplastic changes, increased wound healing, and stimulation of hair growth [45–48]. In neuronal cells, TERT (but not telomerase RNA) overproduction protects cells from stress-induced apoptosis [49–51] and, conversely, early generation (G1) mTert−/− MEFs are sensitive to apoptosis after treatment with staurosporine or N-methyl-D-aspartic acid [52]. TERT induction also leads to changes in cellular proliferation and expression of growth-promoting factors in primary human cells [53], and stimulates the tumorigenic potential of cells that already possess a telomerase-independent means of telomere length maintenance [54]. Since TERT is normally expressed at very low levels in primary cells, it remains unclear whether the phenotypes associated with TERT overexpression reflect a physiological role related to telomere maintenance. For example, the hair overgrowth in mTert transgenic animals is unaffected in a background lacking the telomerase RNA (mTerc−/−) [45]. In contrast, the dermal hyperplasia associated with mTert overexpression is attenuated in an mTerc−/− background, despite the absence of overt differences in telomere length [55].
Inhibition of telomerase, on the other hand, frequently results in latent phenotypes consistent with a role in telomere integrity. For example, mice lacking one or both copies of mTert or mTerc are initially normal, however progressive telomere attrition eventually leads to end-to-end fusions, chromosome instability, infertility and loss of cell viability in various tissues [56–61]. Similarly, haploinsufficiency of human TERT or TR leads to aplastic anemia and dykeratosis congenita, which are associated with bone marrow failure and short telomeres [57,62–71]. Although inhibition of hTR in some human cancer cell lines elicits an immediate apoptotic response without detectable changes in telomere integrity, one study suggests these effects appear dependent upon the telomere-elongation activity of telomerase [72,73]. In addition to the examples mentioned above, another recent study suggests a role of TERT in the DNA damage response independent of measurable effects on telomere maintenance [1]. Targeted degradation of hTERT mRNA results in a decreased DDR after treatment with irradiation and etoposide in two human fibroblast cell lines, without a noticeable effect upon telomere length [1]. In addition, the suppression of hTERT also induces changes in nucleosomal packaging, including post-translational modification of histones H3 and H4, suggesting that hTERT itself might modulate the DDR at telomeres via effects upon chromatin remodeling [1]. In mice, however, loss of the telomerase RNA component does not sensitize animals or cells to ionizing irradiation [74]. These results prompted us to test whether the loss of Tert in mice might specifically affect the ATM-dependent response to exogenous DNA damage.
Results and Discussion
To avoid the complication of critically shortened telomeres on induction of the DDR, we analyzed the DDR in MEFs and ES cells lacking mTert that had not undergone significant telomere erosion, as judged by a robust telomeric DNA signal at chromosome ends, and the lack of detectable genome instability or chromosome end-to-end fusions [61,75,76].
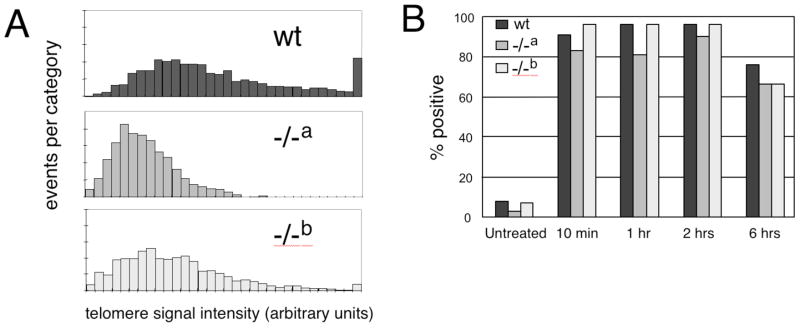
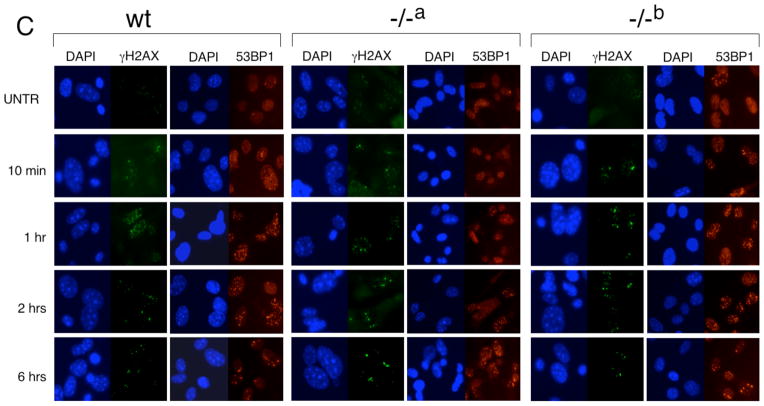
For the analysis of MEFs, we chose two independently derived mTert−/− MEF lines at early passage (mTert−/− a and mTert−/− b, passage 2–5) whose telomere lengths were slightly shorter than an mTert wildtype cell line, but did not exhibit an increase in genetic instability or end-to-end fusions relative to wildtype cells (Fig. 1A and data not shown). We tested whether mTert−/− MEFs exhibited any difference in the DDR upon treatment with 15 μg/mL etoposide (a topoisomerase II inhibitor) or 3 Gy gamma irradiation (Fig. 1B, C). At every timepoint analyzed after irradiation, both mTert−/− cell lines exhibited a similar induction of ATMpS1981 (data not shown), p53BP1, and γH2AX foci compared with wildtype cells (Fig. 1C). Quantification of the 53BP1 foci revealed no difference in the percentage of cells containing five or more foci between genotypes (Fig. 1B, and data not shown). Similar observations were noted at other doses of irradiation and etoposide (data not shown). In human and murine cells, γH2AX is enriched at short telomeres [31,32,34,77], and this recruitment is suppressed by ectopic hTERT expression [31]. However, we also observed no apparent difference in the overlap of γH2AX at telomeres in mTert−/− MEFs compared with wildtype MEFs (data not shown). Thus, we conclude that the absence of mTert in MEFs without critically shortened telomeres does not alter the extent of induction, or temporal regulation, of foci containing 53BP1 or γH2AX in response to two different types of DNA damage.
Fig. 1. The DNA damage response in two independently derived mTert−/− MEF cell lines.
(A) Quantitative telomeric FISH (Q-FISH) analysis of wild-type (black), mTert−/− cell line “a” (grey), and mTert−/− cell line “b” (light grey) MEFs, at passage 3 after isolation. Each mTert−/− MEF line was derived from an embryo from different parents (see Materials and Methods). Y-axis, events per category (where each ‘event’ is a metaphase telomere), and x-axis, telomere signal intensity in arbitrary fluorescence units. Each histogram represents the accumulation of data for 1600 chromosome ends (400 metaphase chromosomes). For consistency, all samples in the same diagram were prepared and analyzed in the same experiment, along with a standardized control sample from a wildtype C/57Bl6 animal to monitor reproducibility between experiments. (B) Quantification of the percentage of cells in which >5 distinct 53BP1 foci were observed (see Materials and Methods), in each genotype, and as shown in panel C. At least 100 cells were counted for each sample. (C) Indirect immunofluorescence of cells treated with 3 Gy irradiation (genotype as indicated as above), at passage 5 after isolation. From left to right under each genotype: DAPI (blue) and γH2AX (green), DAPI (blue) and 53BP1 (red). The time at which cells were fixed post-irradiation is indicated at left.
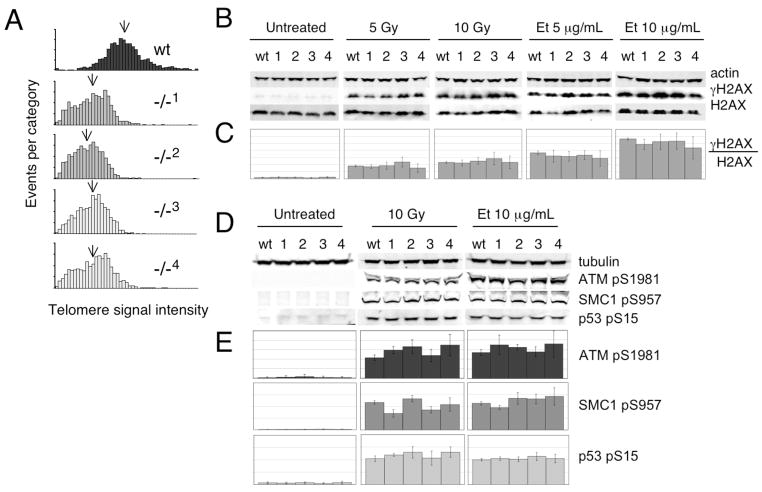
Given that the response to DNA damage may differ between cell types, we also examined four mTert−/− ES cell lines for their response to exogenous DNA damaging agents. Two ES cell lines heterozygous for mTert were independently generated via integration of an mTert disruption cassette [75,76]. From each heterozygous line (A, B), two nullizygous mTert lines were then selected via incubation at increased concentrations of G418 (cell lines 1, 2 derived from line A, and lines 3, 4 derived from line B) [75,76]. Although each mTert−/− ES cell line exhibited slightly different telomere length distributions (and all were shorter than wildtype ES cells), all lines contained <0.5% telomere signal-free ends and one or zero end-to-end fusions per metaphase preparation (Fig. 2A, data not shown). In addition, the cell growth and cell cycle profiles (by FACS analysis of asynchronous populations) were indistinguishable for all five genotypes (wt, and mTert−/− lines 1–4, data not shown). Cell lines heterozygous for mTert exhibited no differences in the DDR relative to wildtype ES cells (data not shown). Quantitative analysis of four mTert−/− ES cell lines showed a robust induction of γH2AX after treatment with irradiation or etoposide, and no significant difference compared to wildtype cells (Fig. 2B and 2C, p>0.2 see Materials and Methods for statistical methods). Indirect immunofluorescence of γH2AX in mTert−/− ES cells after irradiation and etoposide treatment was also performed (but not quantified due to poor foci resolution in ES cells) and no overt differences were noted compared with wildtype ES cells (data not shown). Consistent with these findings, we observed no significant difference in the ability to phosphorylate three other ATM targets in response to DNA damage: ATM (pS1981), SMC1 (pS957), and p53 (pS15) (Fig. 2C). We also noted no difference in the response of mTert−/− cell lines to hydroxyurea, or in the phosphorylation of another ATM target, Chk1 (pS317), upon treatment with hydroxyurea, etoposide, or irradiation (data not shown).
Fig. 2. Induction of γH2AX after DNA damage in wildtype and mTert−/− ES cells.
(A) Q-FISH analysis of wild-type (black), two mTert−/− ES cell lines derived from one mTert+/− founder ES cell line A (1, 2; dark grey), and two mTert−/− ES cell lines derived from an independently derived mTert+/− founder ES cell line B (3, 4; light grey). Each histogram represents the accumulation of data for 1600 chromosome ends (400 metaphase chromosomes). The mean telomere signal intensity is indicated with an arrow. Y-axis, events per category, and x-axis, telomere signal intensity in arbitrary fluorescence units. (B) The same ES cell lines as in Fig. 2A were exposed to the indicated treatment, and the lysates examined by western blotting for the levels of alpha-actin (upper panel), γH2AX (middle panel), and H2AX (lower panel). One representative western blot of at least n=4 replicates is shown. (C) Quantitative analysis of n=4–7 replicates for each treatment and genotype as in panel B, as described in Materials and Methods. On the y-axis, each horizontal line marks 0.2 arbitrary units; maximum y-axes values are kept constant within a particular treatment. (D) The same ES cell lines as in A were examined for the induction of ATMpS1981 (2nd panel), SMC1pS957 (3rd panel) and p53pS15 (lower panel) after treatment with etoposide and irradiation, and normalized to gamma-tubulin (top panel) in a manner similar to Fig. 2B. One representative western blot of n=3–5 is shown. The total cellular levels of p53 remained unaltered before or after DNA damage in all genotypes (data not shown). (E) Quantitative analysis of n=3–5 replicates for each treatment and genotype as in panel D. On the y-axis, each horizontal line marks 2 arbitrary units; maximum y-axes values are kept constant within a particular treatment.
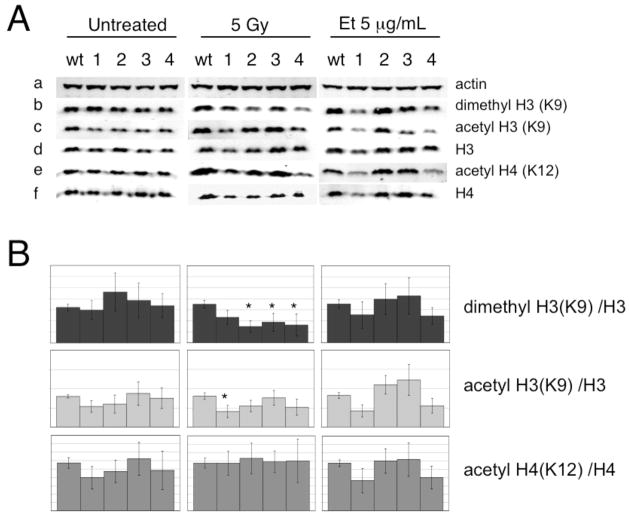
In human fibroblasts expressing shRNAs against hTERT, the suppression of an ATM-dependent response was accompanied by alterations in certain histone post-translation modifications, such as a decrease in dimethylated H3K9 (dmH3K9) and acetylated H4 (AcH4K12), and an increase in acetylated H3 (AcH3K9) [1]. Upon normalizing to total H3 and H4 levels, we observed no consistent difference between mTert−/− ES cell lines in H3 or H4 post-translational modifications relative to wildtype cells (Fig 2D) (at 5 Gy irradiation, mTert−/− line 1 exhibited a slight reduction in acetylated H3K9, p<0.05, and mTert−/− lines 2–4 a slight reduction in dimethylated H3K9, p<0.05). Thus, we cannot conclude with any statistical confidence that there exists a difference in the relative induction of these histone modifications in response to DNA damage.
In conclusion, we do not find a role of murine Tert in the ATM-dependent response of fibroblasts and embryonic stem cells to exogenous DNA damage. Our data is in agreement with Wong et al., who observed an enhanced mortality in mTerc-deficient animals after exposure to irradiation only when telomeres became critically short, and not in early mTerc−/− generations [74]. By analogy, it is possible the recently reported sensitivity of early generations of mTert-deficient MEFs to staurosporine and NMDA may not be the direct result of increased sensitivity to reactive oxygen species released during apoptosis [52]. The complete disruption of mTert does not mimic the effects upon the DDR observed in human cells expressing shRNA against hTERT [1]. This difference could be reflected in the two approaches: shRNA and gene disruption each has potential disadvantages, namely non-specificity and adaptation, respectively. It is also plausible to suppose a species-specific difference in the DDR upon telomerase suppression between murine cells and human cells. Differences in the response between murine and human cells to telomere dysfunction have been previously described [78]. More generally, murine embryonic stem cells also do not possess a canonical G1 checkpoint and are inherently hypersensitive to DNA damage [79–82]. Thus, the attenuation of the ATM-dependent DDR in the absence of TERT appears unconserved across mammalian species [1].
Materials and Methods
Cell Lines and Telomere Length Analysis
Murine embryonic fibroblasts (MEF; C57BL/6J mTert−/− and wild-type) were derived according to standard protocols from 13.5-day-old embryos whose parents were G10-mTert+/− (i.e. after 10 generations of breeding between +/− and +/+) [83]. The wildtype and mTert−/− MEFs used in this study were not immortalized (e.g. were used between passages 3–5). Telomere length was measured using quantitative fluorescence in situ hybridization (Q-FISH) (Fig. 1A) [84,85]. Embryonic stem cells (wildtype, mTert+/−, mTert−/−) have been previously described and characterized [75,76]. In this study, mTert−/− ES cells were used between passages 25–30, at which time >99% chromosome ends possessed detectable telomeric DNA as measured by Q-FISH (Fig. 2A and data not shown).
DNA Damage
For environmental damage, cells were irradiated with 3, 5, 8 or 10 Gy and incubated for the indicated times at 37°C (1 hr, unless otherwise indicated). For replicative damage, cells were treated with hydroxyurea (2.5 mM) or etoposide (indicated dose) for 1 hour at 37°C. For western analysis, cells were lysed in radio-immunoprecipitation assay (RIPA) buffer (7 M urea, 2 M thiourea, 1% C7BzO, 40 mM Tris-HCl pH 7.5, 1 mM Na3VO4, 10 mM NaF) and 50 μg (histones) or 100 μg (p53-pS15, SMC1-pS957, ATM-pS1981) total protein was analyzed in each sample. After resolution on a 16% (histones), 4–12% (p53-pS15, SMC1-pS957) or 3–8% (ATMpS1981) w/v acrylamide gel (Invitrogen) and transfer to PVDF membrane (Invitrogen), blots were incubated with two or more of the following primary antibodies in TBST (Tris-buffered saline containing 0.1% Tween-20): rabbit anti-actin (Sigma-Aldrich), murine anti-γ-tubulin (Sigma-Aldrich), rabbit anti-H2AX (Novus), mouse anti-γH2AX, rabbit anti-dimethyl H3 (K9), rabbit anti-acetyl H3 (K9), rabbit anti-acetyl H4 (K12), murine anti-H3, and rabbit anti-H4 (Upstate Biotech), rabbit anti-p53 (Santa Cruz Biotechnology), rabbit anti-p53-pS15, mouse anti-SMC1-pS957 (Cell Signaling) and mouse anti-ATMpS1981 (Rockland Immunochemical Inc.). Immunoblots were incubated with a 1:15,000 dilution of IRDye680-conjugated anti-rabbit IgG and/or IRDye800CW-conjugated anti-mouse IgG for 1 hour at room temperature, then scanned and quantified on a Licor Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, Nebraska).
Image Quantification and Statistics
For each set of treatment conditions exhibited in Figures 2 and 3, numerous replicates were carried out for all five respective genotypes (n=4–7). Individual sample values were normalized via the ratio difference between the actin (or tubulin) average (across all 5 genotypes) and raw intensity value in each lane. Between blots, sample values were similarly normalized against the wildtype ‘normalized’ values (hence the slightly lower standard deviation attributed to all wildtype samples). To calculate statistical significance, a one-way ordinary ANOVA (across all 5 cell lines) was performed on the ratio of each histone modification (H2AX, H3, or H4) relative to total protein levels for each treatment condition. Except where noted in the text, the ANOVA analysis failed to detect a significant difference between mTert−/− lines and wild-type cells. ANOVA is preferable to a student’s t-test because it takes into account the unequal variation across all five genotypes, and reduces the likelihood of statistical errors when comparing many different treatment groups.
Fig. 3.
(A) The levels of di-methylated H3K9 (panel b), acetylated H3K9 (panel c), were normalized to actin (panel a) and total H3 (panel d), and the levels of acetylated H4K12 (panel e), normalized to actin and total H4 (panel f) after the indicated treatments in wildtype and four mTert−/− ES cell lines. One representative western blot (of n=4–7) is shown. (B) Quantitative analysis of the histone modifications, a representative blot of which is shown in panel A. The asterisks indicate treatment conditions that demonstrated a statistically significant decrease in modified histone H3 (normalized to total H3) in mTert−/− ES cell lines compared to wildtype cells (p<0.05).
Immunofluorescence
MEFs (mTert−/− and wild-type, at passage 5) were grown on chamber slides and fixed with 100% v/v ice-cold methanol for 10 minutes at −20°C. Fixed cells were incubated with primary antibodies overnight at 4°C, followed by the appropriate fluorochrome-conjugated secondary antibody for 1–2 hours at room temperature and mounted in Vectashield with DAPI (Vector Laboratories, Inc.). Images were captured on a Leica microscope with Volocity acquisition software (Improvision Inc.). Cells were scored positive if they displayed 5 or more discrete foci of 53BP1. At least 100 cells were analyzed for each treatment. Primary antibodies used were rabbit anti-53BP1 (Cell Signaling, at 1/1000 dilution) and mouse anti-γH2AX (Upstate Biotechnology, at 1/4000 dilution), and secondary antibodies included Alexa Fluor 546 goat anti-rabbit IgG and Alexa Fluor 488 donkey anti-mouse IgG (Invitrogen, at 1/1000 dilution).
Acknowledgments
We thank Denis Bouchard, Peter Cheng, Gordon Duncan, Alastair Kerr, Yie Liu, Svetlana Makovets, Lisa Martin, and David Sealey for technical or statistical advice, Dick Hill for access to the OCI cell irradiator, and Elizabeth Blackburn, Dan Durocher, Titia de Lange, Mike Tyers, and members of the Harrington lab for critical discussion and comments on the manuscript. L.A.H. gratefully acknowledges the support of the Campbell Family Institute for Breast Cancer Research, and grants from the NIH (RO1 AG02398) and the Howard Hughes Medical Institute International Scholar Award Program (HHMI 55005945).
Abbreviations
- TERT
telomerase reverse transcriptase
- MEF
murine embryonic fibroblast
- ES
embryonic stem
- DDR
DNA damage response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Masutomi K, Possemato R, Wong JM, Currier JL, Tothova Z, Manola JB, Ganesan S, Lansdorp PM, Collins K, Hahn WC. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc Natl Acad Sci U S A. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price C, Jacob NK. Engineering the end: DNA processing at human telomeres. Mol Cell. 2005;18:147–148. doi: 10.1016/j.molcel.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 3.Greider CW. Telomeres and senescence: the history, the experiment, the future. Curr Biol. 1998;8:R178–181. doi: 10.1016/s0960-9822(98)70105-8. [DOI] [PubMed] [Google Scholar]
- 4.Hug N, Lingner J. Telomere length homeostasis. Chromosoma. 2006 doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 5.Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 6.Kelleher C, Teixeira MT, Forstemann K, Lingner J. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem Sci. 2002;27:572–579. doi: 10.1016/s0968-0004(02)02206-5. [DOI] [PubMed] [Google Scholar]
- 7.Bertuch AA, Lundblad V. The maintenance and masking of chromosome termini. Curr Opin Cell Biol. 2006;18:247–253. doi: 10.1016/j.ceb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 9.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 10.Churikov D, Wei C, Price CM. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol Cell Biol. 2006;26:6971–6982. doi: 10.1128/MCB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 12.Jacob NK, Lescasse R, Linger BR, Price CM. Tetrahymena POT1a Regulates Telomere Length and Prevents Activation of a Cell Cycle Checkpoint. Mol Cell Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shakirov EV, Surovtseva YV, Osbun N, Shippen DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surovtseva YV, Shakirov EV, Vespa L, Osbun N, Song X, Shippen DE. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. Embo J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 16.Frank CJ, Hyde M, Greider CW. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell. 2006;24:423–432. doi: 10.1016/j.molcel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Harrington L. Those dam-aged telomeres! Curr Opin Genet Dev. 2004;14:22–28. doi: 10.1016/j.gde.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 19.von Zglinicki T, Saretzki G, Ladhoff J, d’Adda di Fagagna F, Jackson SP. Human cell senescence as a DNA damage response. Mech Ageing Dev. 2005;126:111–117. doi: 10.1016/j.mad.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 20.d’Adda di Fagagna F, Teo SH, Jackson SP. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 21.Reaper PM, di Fagagna F, Jackson SP. Activation of the DNA damage response by telomere attrition: a passage to cellular senescence. Cell Cycle. 2004;3:543–546. [PubMed] [Google Scholar]
- 22.Chan SW, Blackburn EH. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- 23.Hackett JA, Greider CW. Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene. 2002;21:619–626. doi: 10.1038/sj.onc.1205061. [DOI] [PubMed] [Google Scholar]
- 24.Lydall D, Whitehall S. Chromatin and the DNA damage response. DNA Repair (Amst) 2005;4:1195–1207. doi: 10.1016/j.dnarep.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Shay JW, Wright WE. Telomeres are double-strand DNA breaks hidden from DNA damage responses. Mol Cell. 2004;14:420–421. doi: 10.1016/s1097-2765(04)00269-2. [DOI] [PubMed] [Google Scholar]
- 26.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lustig AJ. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet. 2003;4:916–923. doi: 10.1038/nrg1207. [DOI] [PubMed] [Google Scholar]
- 29.Li B, Lustig AJ. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 30.Williams B, Bhattacharyya MK, Lustig AJ. Mre 11 p nuclease activity is dispensable for telomeric rapid deletion. DNA Repair (Amst) 2005;4:994–1005. doi: 10.1016/j.dnarep.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Meier A, Fiegler H, Munoz P, Ellis P, Rigler D, Langford C, Blasco MA, Carter N, Jackson SP. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. Embo J. 2007;26:2707–2718. doi: 10.1038/sj.emboj.7601719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 33.Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004;18:2108–2119. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 35.IJpma AS, Greider CW. Short Telomeres Induce a DNA Damage Response in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:987–1001. doi: 10.1091/mbc.02-04-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hackett J, Greider CW. End resection initiates genomic instability in the absence of telomerase. Mol Biol Cell. 2003 doi: 10.1128/MCB.23.23.8450-8461.2003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- 38.Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 39.Pennaneach V, Putnam CD, Kolodner RD. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol. 2006;59:1357–1368. doi: 10.1111/j.1365-2958.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- 40.Feldser DM, Hackett JA, Greider CW. Telomere dysfunction and the initiation of genome instability. Nat Rev Cancer. 2003;3:623–627. doi: 10.1038/nrc1142. [DOI] [PubMed] [Google Scholar]
- 41.De Lange T. Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- 42.Shay J, Wright WE. Does Telomerase Moonlight? Scientist. 2005;18:18–19. [Google Scholar]
- 43.Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase) Genetics. 2002;160:49–62. doi: 10.1093/genetics/160.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teo SH, Jackson SP. Telomerase subunit overexpression suppresses telomere-specific checkpoint activation in the yeast yku80 mutant. EMBO Rep. 2001;2:197–202. doi: 10.1093/embo-reports/kve038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 47.Artandi SE, Alson S, Tietze MK, Sharpless NE, Ye S, Greenberg RA, Castrillon DH, Horner JW, Weiler SR, Carrasco RD, DePinho RA. Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc Natl Acad Sci U S A. 2002;99:8191–8196. doi: 10.1073/pnas.112515399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez-Suarez E, Geserick C, Flores JM, Blasco MA. Antagonistic effects of telomerase on cancer and aging in K5-mTert transgenic mice. Oncogene. 2005;24:2256–2270. doi: 10.1038/sj.onc.1208413. [DOI] [PubMed] [Google Scholar]
- 49.Fu W, Killen M, Culmsee C, Dhar S, Pandita TK, Mattson MP. The catalytic subunit of telomerase is expressed in developing brain neurons and serves a cell survival-promoting function. J Mol Neurosci. 2000;14:3–15. doi: 10.1385/JMN:14:1-2:003. [DOI] [PubMed] [Google Scholar]
- 50.Lu C, Fu W, Mattson MP. Telomerase protects developing neurons against DNA damage-induced cell death. Brain Res Dev Brain Res. 2001;131:167–171. doi: 10.1016/s0165-3806(01)00237-1. [DOI] [PubMed] [Google Scholar]
- 51.Kang HJ, Choi YS, Hong SB, Kim KW, Woo RS, Won SJ, Kim EJ, Jeon HK, Jo SY, Kim TK, Bachoo R, Reynolds IJ, Gwag BJ, Lee HW. Ectopic expression of the catalytic subunit of telomerase protects against brain injury resulting from ischemia and NMDA-induced neurotoxicity. J Neurosci. 2004;24:1280–1287. doi: 10.1523/JNEUROSCI.4082-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J, Sung YH, Cheong C, Choi YS, Jeon HK, Sun W, Hahn WC, Ishikawa F, Lee HW. TERT promotes cellular and organismal survival independently of telomerase activity. Oncogene. 2008;27:3754–3760. doi: 10.1038/sj.onc.1211037. [DOI] [PubMed] [Google Scholar]
- 53.Smith LL, Coller HA, Roberts JM. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 54.Stewart SA, Hahn WC, O’Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, Popescu NC, Weinberg RA. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc Natl Acad Sci U S A. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cayuela ML, Flores JM, Blasco MA. The telomerase RNA component Terc is required for the tumour-promoting effects of Tert overexpression. EMBO Rep. 2005;6:268–274. doi: 10.1038/sj.embor.7400359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheong C, Hong KU, Lee HW. Mouse models for telomere and telomerase biology. Exp Mol Med. 2003;35:141–153. doi: 10.1038/emm.2003.20. [DOI] [PubMed] [Google Scholar]
- 57.Greider CW. Telomerase RNA levels limit the telomere length equilibrium. Cold Spring Harb Symp Quant Biol. 2006;71:225–229. doi: 10.1101/sqb.2006.71.063. [DOI] [PubMed] [Google Scholar]
- 58.Harrington L. Making the most of a little: dosage effects in eukaryotic telomere length maintenance. Chromosome Res. 2005 doi: 10.1007/s10577-005-0994-5. in press. [DOI] [PubMed] [Google Scholar]
- 59.Hathcock KS, Hemann MT, Opperman KK, Strong MA, Greider CW, Hodes RJ. Haploinsufficiency of mTR results in defects in telomere elongation. Proc Natl Acad Sci U S A. 2002;99:3591–3596. doi: 10.1073/pnas.012549799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hao LY, Armanios M, Strong MA, Karim B, Feldser DM, Huso D, Greider CW. Short telomeres, even in the presence of telomerase, limit tissue renewal capacity. Cell. 2005;123:1121–1131. doi: 10.1016/j.cell.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 61.Erdmann N, Liu Y, Harrington L. Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc Natl Acad Sci U S A. 2004;101:6080–6085. doi: 10.1073/pnas.0401580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 63.Xin ZT, Beauchamp AD, Calado RT, Bradford JW, Regal JA, Shenoy A, Liang Y, Lansdorp PM, Young NS, Ly H. Functional characterization of natural telomerase mutations found in patients with hematologic disorders. Blood. 2007;109:524–532. doi: 10.1182/blood-2006-07-035089. [DOI] [PubMed] [Google Scholar]
- 64.Goldman F, Bouarich R, Kulkarni S, Freeman S, Du HY, Harrington L, Mason PJ, Londono-Vallejo A, Bessler M. The effect of TERC haploinsufficiency on the inheritance of telomere length. Proc Natl Acad Sci U S A. 2005;102:17119–17124. doi: 10.1073/pnas.0505318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Armanios M, Chen JL, Chang YP, Brodsky RA, Hawkins A, Griffin CA, Eshleman JR, Cohen AR, Chakravarti A, Hamosh A, Greider CW. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci U S A. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 67.Vulliamy TJ, Knight SW, Mason PJ, Dokal I. Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cells Mol Dis. 2001;27:353–357. doi: 10.1006/bcmd.2001.0389. [DOI] [PubMed] [Google Scholar]
- 68.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 69.Theimer CA, Finger LD, Trantirek L, Feigon J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc Natl Acad Sci U S A. 2003;100:449–454. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ly H, Schertzer M, Jastaniah W, Davis J, Yong SL, Ouyang Q, Blackburn EH, Parslow TG, Lansdorp PM. Identification and functional characterization of 2 variant alleles of the telomerase RNA template gene (TERC) in a patient with dyskeratosis congenita. Blood. 2005;106:1246–1252. doi: 10.1182/blood-2005-01-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamaguchi H, Baerlocher GM, Lansdorp PM, Chanock SJ, Nunez O, Sloand E, Young NS. Mutations of the human telomerase RNA gene (TERC) in aplastic anemia and myelodysplastic syndrome. Blood. 2003;102:916–918. doi: 10.1182/blood-2003-01-0335. [DOI] [PubMed] [Google Scholar]
- 72.Goldkorn A, Blackburn EH. Assembly of mutant-template telomerase RNA into catalytically active telomerase ribonucleoprotein that can act on telomeres is required for apoptosis and cell cycle arrest in human cancer cells. Cancer Res. 2006;66:5763–5771. doi: 10.1158/0008-5472.CAN-05-3782. [DOI] [PubMed] [Google Scholar]
- 73.Li S, Rosenberg JE, Donjacour AA, Botchkina IL, Hom YK, Cunha GR, Blackburn EH. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004;64:4833–4840. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- 74.Wong KK, Chang S, Weiler SR, Ganesan S, Chaudhuri J, Zhu C, Artandi SE, Rudolph KL, Gottlieb GJ, Chin L, Alt FW, DePinho RA. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat Genet. 2000;26:85–88. doi: 10.1038/79232. [DOI] [PubMed] [Google Scholar]
- 75.Liu Y, Kha H, Ungrin M, Robinson MO, Harrington L. Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc Natl Acad Sci U S A. 2002;99:3597–3602. doi: 10.1073/pnas.062549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu Y, Snow BE, Hande MP, Yeung D, Erdmann NJ, Wakeham A, Itie A, Siderovski DP, Lansdorp PM, Robinson MO, Harrington L. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr Biol. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- 77.Hao LY, Strong MA, Greider CW. Phosphorylation of H2AX at short telomeres in T cells and fibroblasts. J Biol Chem. 2004;279:45148–45154. doi: 10.1074/jbc.M403924200. [DOI] [PubMed] [Google Scholar]
- 78.Smogorzewska A, de Lange T. Different telomere damage signaling pathways in human and mouse cells. Embo J. 2002;21:4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 80.Hong Y, Stambrook PJ. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc Natl Acad Sci U S A. 2004;101:14443–14448. doi: 10.1073/pnas.0401346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 82.Van Sloun PP, Jansen JG, Weeda G, Mullenders LH, van Zeeland AA, Lohman PH, Vrieling H. The role of nucleotide excision repair in protecting embryonic stem cells from genotoxic effects of UV-induced DNA damage. Nucleic Acids Res. 1999;27:3276–3282. doi: 10.1093/nar/27.16.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2003. [Google Scholar]
- 84.Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci U S A. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]