Abstract
Brh2 plays a central role in the homologous recombination system of Ustilago maydis mediating delivery of Rad51 to single-stranded DNA. Here we report that Brh2 can pair the displaced strand of a D-loop with a complementary single-stranded DNA to form a duplexed, or double D-loop. The reaction emulates the second-end capture step envisioned in models of DNA double-strand break repair. This second-end capture reaction promoted by Brh2 proceeds efficiently when performed in the presence of Rad51 under conditions that block annealing by Rad52, or when the second single-stranded DNA substrate is replaced by double-stranded DNA. In a coupled reaction that requires extension of the D-loop more than two hundred nucleotides by DNA synthesis in order to reveal a complementary region, Brh2 was also able to promote second-end capture and thus model a synthesis-dependent strand-annealing mechanism.
Keywords: BRCA2, Rad51, Rad52, D-loops, SDSA, recombination, DNA repair
INTRODUCTION
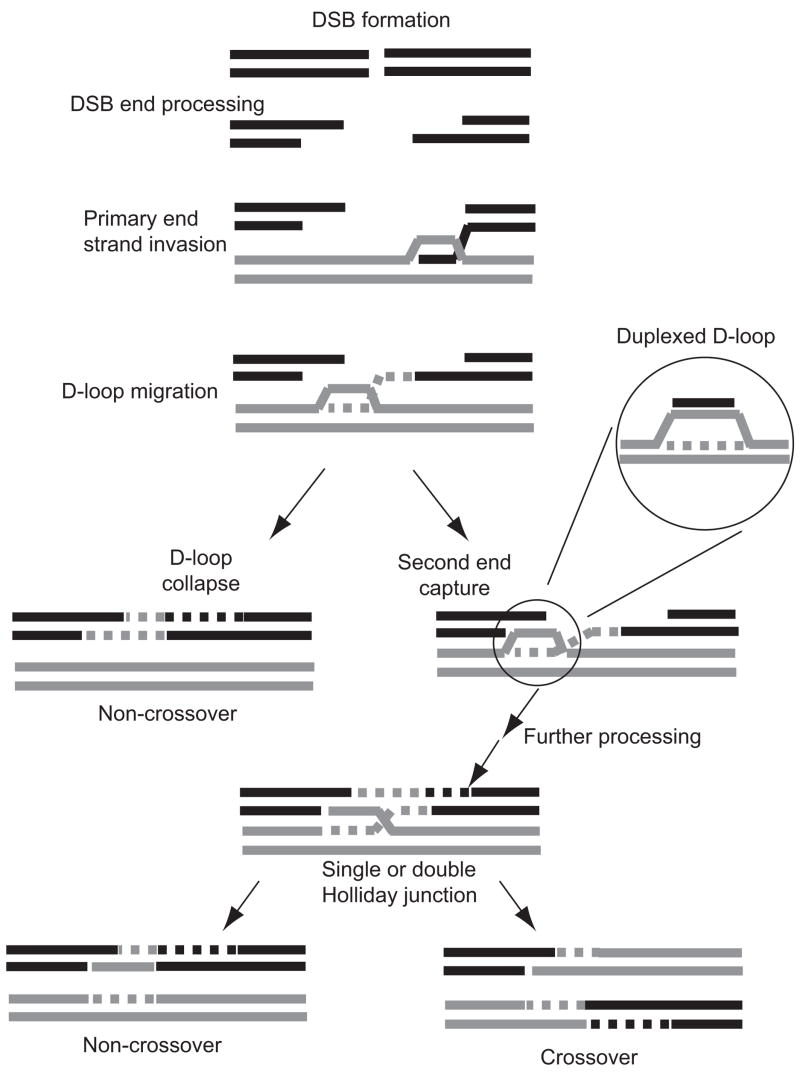
Homologous recombination plays important and even essential roles in the mitotic and meiotic cell cycles of most eukaryotes (Neale and Keeney, 2006; San Filippo et al., 2008; Symington, 2002). The DNA double-strand-break (DSB) repair model of Szostak and colleagues (Szostak et al., 1983), based on earlier conceptions by Holliday (Holliday, 1964) and Meselson and Radding (Meselson and Radding, 1975) to account for the molecular mechanism of recombination, was designed to account for the strong association between crossing over and gene conversion in meiosis, while variations of it were proposed to explain the lack of associated crossing over during mitosis and to account for the unorthodox disposition of newly synthesized DNA. Typically these variants feature asymmetric processing of the ends after DSB introduction (Haber et al., 2004). One end is resected to reveal a protruding 3′ single-stranded (ss) tail that then invades a homologous duplex forming a D-loop (Hunter and Kleckner, 2001). Driven by DNA synthesis primed from the invading 3′ end, the D-loop migrates or is enlarged to a position where there is overlap in sequence with the second end of the DSB, retrieving the information lost at the site of the DSB (Fig. 1). So positioned, the two apposing DNA ends can now be reunited. The second end can be captured by the newly synthesized invading strand if this strand is released by collapse of the D-loop (Belmaaza and Chartrand, 1994; Ferguson and Holloman, 1996; Gloor et al., 1991), or if the end of the invading strand is extruded from the D-loop (Allers and Lichten, 2001) (not shown in the figure). Alternatively the second end can be captured if it anneals to the displaced strand of the D-loop. The mode of reuniting both ends of a DSB is a key determinant of whether repair is to a crossover or noncrossover outcome (Haber et al., 2004).
Figure 1.
Second-end capture in DSB repair based on the migrating D-loop version (Ferguson and Holloman, 1996) of synthesis dependent strand annealing. DSB is resected to reveal a protruding 3′-single-strand tail, which invades a homologous duplex to form a D-loop. With DNA synthesis to extend the invading strand the D-loop migrates until there is overlap in sequence with the second end of the DSB. If the D-loop collapses releases invading strand it can anneal with complementary strand on the second end of the DSB to reunite the broken ends. Alternatively the displaced strand of the d-loop can pair with the second end. The D-loop annealed with complementary single strand is magnified to show in isolation the duplexed D-loop structure. With additional DNA synthesis and ligation a Holliday junction-containing intermediate forms. Resolution can result in crossover or non-crossover outcomes.
D-loop formation is the salient step in homologous pairing between duplex and homologous ss DNA. Pairing is catalyzed by the Rad51-nucleoprotein filament, which is formed by Rad51 polymerizing on the 3′ ss tail left after resection of the primary end of the DSB. The ability of Rad51 to assemble into filaments is controlled by a mediator. In budding yeast and related ascomycete fungi the primary mediator is Rad52, which is crucial for all modes of recombination (Sung and Klein, 2006). Paradoxically, Rad52 is conserved in vertebrates, but does not appear to play an important role in recombination (de Vries et al., 2005; Rijkers et al., 1998; Stark et al., 2004; Yamaguchi-Iwai et al., 1998). In the basidiomycete fungus Ustilago maydis a similar situation prevails in that Rad52 is conserved, but has no obvious function in recombination or repair (Kojic et al., 2008). In metazoans and U. maydis, the Rad51 mediator is a representative of the BRCA2 class of proteins, the founding member being the product of a hereditary breast cancer predisposition gene in human (Thorslund and West, 2007). Brh2, the BRCA2 homolog from U. maydis (Kojic et al., 2002), mediates filament assembly by recognizing a ss/ds DNA junction and nucleating Rad51 on the protruding 3′ ss tail coated with the single-strand DNA binding protein (RPA) (Yang et al., 2005). A simple model incorporating findings with Brh2 (Yang et al., 2005) as well as with a laboratory construct modeling the human BRCA2 protein (San Filippo et al., 2006) would be that Rad51 is physically clasped by elements in the mediator resembling a structural motif at the Rad51 polymerization interface and is directed to the ss/ds junction through DNA-binding function inherent in the mediator (Shivji and Venkitaraman, 2004; Thorslund and West, 2007).
Biochemical studies with Brh2 have revealed capabilities that suggest functions beyond mediating Rad51-filament formation. These include the capacity to anneal complementary single-strands of DNA coated with RPA (Mazloum et al., 2007), an ability to promote strand exchange with oligonucleotide substrates, and activity in promoting D-loop formation with supercoiled plasmid DNA and a homologous single-stranded fragment (Mazloum et al., 2008). In this regard Brh2 resembles Rad52, which has been established as an annealing protein with oligonucleotide strand exchange and D-loop forming activities (Bi et al., 2004; Kagawa et al., 2001; Mortensen et al., 1996; Wu et al., 2008). Evidence supporting a role for Rad52 in second-end capture has been provided by biochemical studies with the use of purified proteins from yeast (Sugiyama et al., 2006) or human (Bugreev et al., 2007; McIlwraith and West, 2008) and by physical analysis of molecular intermediates formed during meiotic recombination in yeast (Lao et al., 2008). This critical function played by Rad52 in yeast, however, could be the exception rather than the rule. Although Rad51-dependent repair of DSBs by homologous recombination occurs universally throughout the eukaryotic domain of life, Rad52 does not. It is absent from green plants, from insects such as Anopheles and Drosophila, from nematodes such as Caenorhabditis, and from certain protists (Ramesh et al., 2005), all of which appear to have a BRCA2 homolog.
In view of the dominant function of Brh2 in the recombination system of U. maydis and concomitantly minor role of Rad52 (Kojic et al., 2002; Kojic et al., 2008), we were curious to learn whether the second-end capture step in repair of DSBs could be promoted by Brh2. Here we have investigated this reaction.
RESULTS
Experimental system
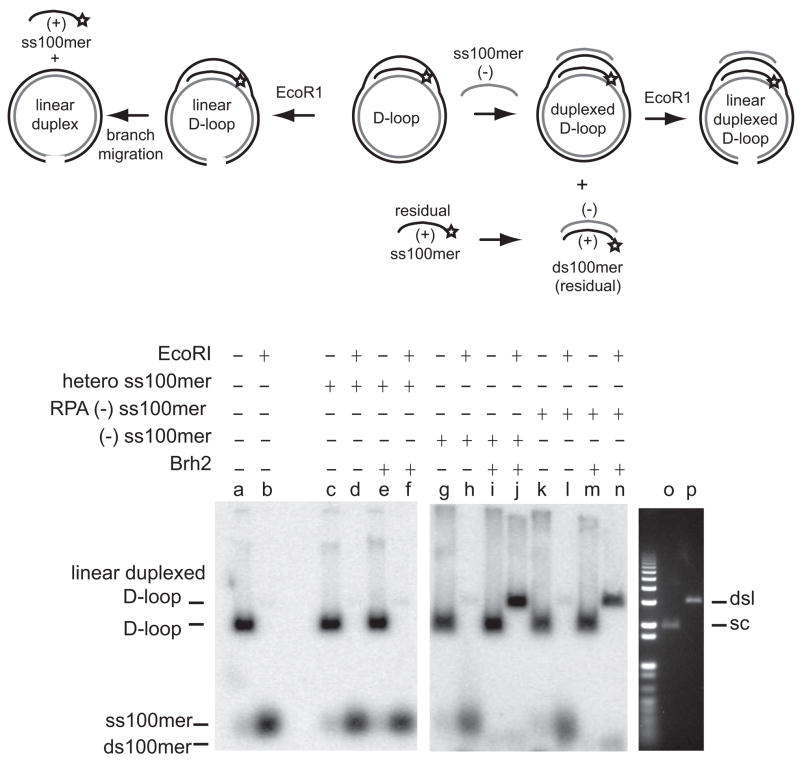
The question we posed was whether Brh2 could promote second-end capture using duplexed D-loop formation to model the reaction. A duplexed D-loop or double D-loop is a four-stranded joint molecule formed by annealing of a complementary strand to the displaced strand of a D-loop (Fig. 1). The protocol devised for determination involved stepwise addition of components to a reaction mix and was based on the procedure developed by Mazin and coworkers (Bugreev et al., 2007). In the first step, D-loop formation was initiated using superhelical plasmid DNA and 32P-labeled homologous ss oligonucleotide (+ strand) as substrates. As is well established, uptake of 32P-labeled homologous ss oligomer by plasmid DNA to form D-loops can be monitored by gel electrophoresis and is evident by the appearance in autoradiographs or phosphorimages of a radiolabeled band with mobility slower than free oligonucleotide and corresponding to that of the plasmid DNA [e.g., see (Mazloum et al., 2008)]. The diagnostic test showing that these joint molecules consist of plasmid DNA base-paired with a third strand 32P-labeled oligonucleotide in the form of a D-loop is their disappearance after endonucleolytic cleavage of the plasmid DNA at a site removed from the region homologous to the oligonucleotide sequence (Fig. 2 schematic). The joint molecules rapidly dissociate due to liberation of the oligonucleotide strand by branch migration once topological constraint in the plasmid DNA is removed (Radding et al., 1977). In the second step, an unlabeled, complementary ss100mer oligonucleotide (− strand) is added to a mixture containing the already-in-progress D-loop reaction. Annealing of the second ss oligomer to the displaced strand of the D-loop eliminates unpaired single-stranded DNA and so stabilizes the structure protecting the 32P-labeled strand from branch migration. The diagnostic test for duplexed D-loops is the stable association of the 32P-labeled strand with plasmid DNA after endonucleolytic cleavage of the plasmid DNA and the concomitant shift in the mobility of radiolabeled band representing the D-loop DNA to a mobility corresponding to linear plasmid DNA (Fig. 2 schematic).
Figure 2.
Brh2 promotes duplexed D-loop formation. The schematic depicts the course of reaction. Superhelical plasmid pBluescriipt II DNA forms D-loops by taking up 32P-labeled ss100mer (with star). Duplex D-loop formation proceeds with annealing of ss100mer (−strand) to the displaced strand of the D-loop. Residual ss100mer (−strand) anneals with ss100mer (+ strand) to form ds100mer. D-loop DNA cut with EcoRI dissociates by branch migration while duplexed D-loop DNA remains stable. A. In reaction mixes containing 32P-labeled ss100mer (+strand) and plasmid DNA, D-loop formation was initiated using 400 nM Brh2 or 750 nM Rad51. After D-loops were formed, ss100mer (−strand) was added, then mixtures were split and DNA was digested with EcoRI. B. Reactions were performed with additions as shown. Heterologous ss100mer was based on pUC19 DNA sequence. Standards (lanes o and p) show mobilities of uncut supercoiled (sc) and EcoRI-cut double-strand linear (dsl) plasmid DNA on a parallel gel after staining with ethidium bromide. C. Reaction mixes contained 32P-labeled ss100mer (+strand) precoated with RPA as in B, plasmid DNA, and protein as indicated in each panel. Rad52 (lanes a–j) a, b- no protein; c, d-80 nM; e, f-160 nM; g, h-400 nM; I, j-800 nM. Brh2 (lanes k–t) k, l-no protein; m, n-80 nM; o, p-160 nM; q, r-400 nM; s, t-800 nM. Rad51 (lanes u–d′) u, v-no protein, w, x-80 nM; y, z-160 nM; a′, b′-400 nM; c′, d′-800 nM. The results from at least 3 independent determinations were quantified and expressed as the fraction of linear duplexed D-loop relative to the fraction of D-loop (DD-loop/D-loop) at each respective protein concentration. Error bars indicate standard deviations. D. Reactions were performed as above except that Rad52 (left panel) and Brh2 (right panel) were held constant at 160 nM and 400 nM, respectively, and Rad51 was varied as indicated. Rad52 (lanes a–j), Brh2 (lanes k–t) a, b, k, l- no Rad51; c, d, m, n-80 nM Rad51; e, f, o, p-160 nM Rad51; g, h, q, r-400 nM Rad51; I, j, s, t-800 nM Rad51. The results from at least 3 independent determinations were quantified as in C.
Brh2 promotes formation of duplexed D-loops
Using pBluescript II plasmid DNA and homologous 32P-labeled 100mer oligonucleotide (+ strand) as substrates under conditions predetermined to minimize spontaneous annealing, we observed that duplexed D-loops were formed after addition of complementary (−strand) 100mer to D-loop reactions already initiated by Brh2 (Fig. 2A, lanes c, d). The yield of duplexed D-loops appeared to be almost quantitative (relative to single D-loops) based on the intensity of the product band remaining after cleavage of D-loops with EcoRI (Fig. 2A, compare lanes c and d). In contrast little duplexed D-loop product was formed in reactions with Rad51 alone under conditions in which the level of D-loop formation achieved was roughly equivalent to that in the Brh2 reaction (Fig. 2A, lanes e, f). The high preference of Brh2 for binding to D-loops noted previously (Mazloum et al., 2007) could be a key factor in directing annealing of the incoming second strand with the displaced strand of D-loop.
In controls we observed that D-loops formed by Brh2 dissociated by branch migration after cleavage of the plasmid DNA with EcoRI as expected (Fig. 2B, lanes e, f). No change in yield of duplexed D-loops in reaction promoted by Brh2 was noted when the 100mer (−strand) was precoated with RPA to reduce the rate of spontaneous annealing (Fig. 2B, compare lanes i, j with k, l). When a heterologous oligonucleotide was substituted for the second strand (100mer −strand), no band indicative of duplexed D-loop product was formed (Fig. 2B, lanes m, n). These results show that Brh2 can promote annealing of the displaced strand of a D-loop with a complementary single strand, thus modeling second-end capture.
Since Rad52 from yeast and human has been documented to promote second-end capture (Bugreev et al., 2007; McIlwraith and West, 2008; Sugiyama et al., 2006), we evaluated the U. maydis Rad52 for this activity. In agreement with these studies, we found that U. maydis Rad52 was also capable of promoting formation of duplexed D-loops to about the same extent as Brh2, although at the highest concentration tested (800 nM) D-loop formation was poor (Fig. 2C). A background level of duplexed D-loops formation was evident with Rad51, possibly due to inherent annealing activity (Kim et al., 2002), but this duplexed D-loop-forming activity was inefficient by comparison with Rad52 and Brh2 as can be seen in the graph (Fig. 2C) as the fraction of duplexed D-loop compared to the fraction of D-loop (DD-loop/D-loop).
It has been reported that Rad51 blocks Rad52-mediated annealing and the proposal made that this attenuation in annealing activity represents an important means for regulating the outcome of DSB repair events (Wu et al., 2008). In line with these findings we observed that by increasing the amount of Rad51 in reactions with Rad52, the overall level of D-loops increased but the proportional yield of duplexed D-loop product decreased suggesting Rad51 inhibits the annealing activity of Rad52 (Fig. 2D). D-loop formation was greatly stimulated when Rad51 was added to reactions containing Brh2 (compare Fig. 2D lane k with lanes m, o, q, s) demonstrating dependence of reaction on Rad51. We note that maximal D-loop formation promoted by U. maydis Rad51 with these substrates was determined to require a molar ratio of Rad51 protomer to ssDNA nucleotide of about 1:1 probably due to poor binding of Rad51 to the oligonucleotide substrate (Mazloum et al., 2008). In contrast to the situation with Rad52, duplexed D-loop formation promoted by Brh2 was not subject to inhibition by Rad51. These results show that Brh2 has an ability to promote annealing in the presence of Rad51 that is distinguished from Rad52.
D-loop DNA as substrate in second-end capture
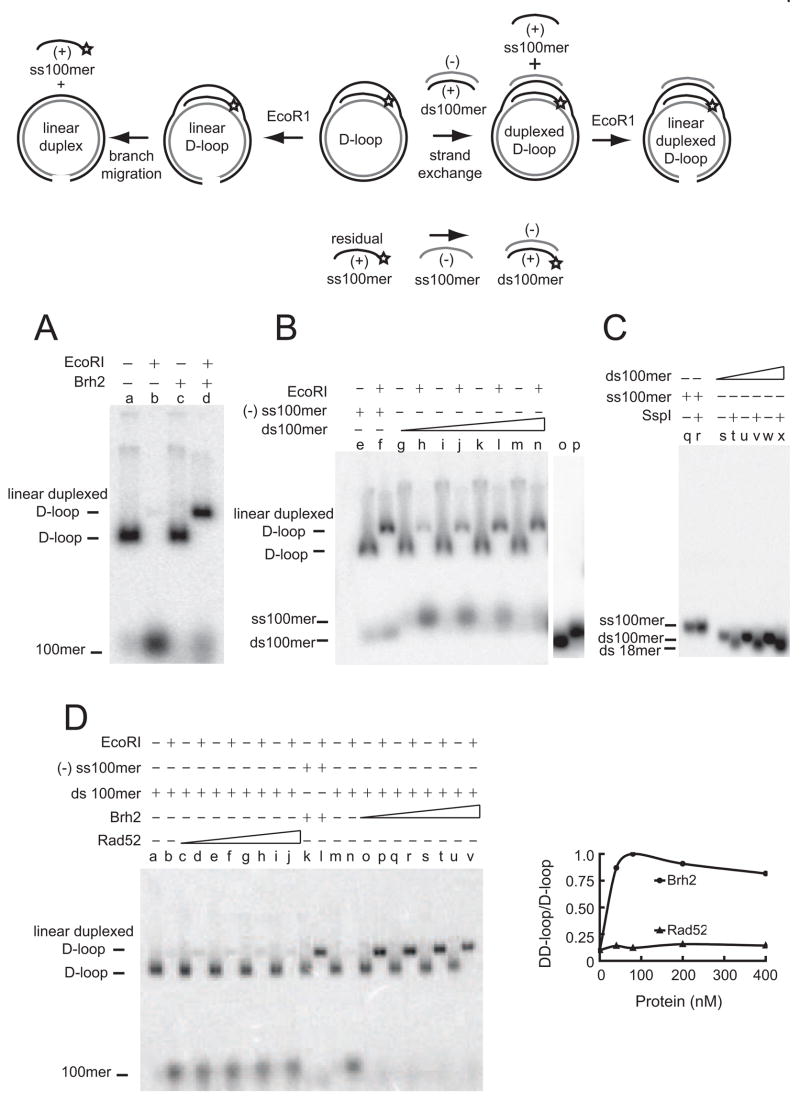
To explore the capabilities of Brh2 in promoting formation of duplexed D-loops in more detail, we used purified D-loop DNA as a substrate in reactions (Fig. 3, schematic). The D-loop DNA was prepared as above using pBluescript II plasmid DNA and homologous 32P-labeled 100mer oligonucleotide (+ strand) in reactions catalyzed by a combination of Rad51 and Brh2, then purified after deproteinization by sucrose gradient centrifugation to remove unincorporated 32P-labeled 100mer. The purified D-loops dissociated almost quantitatively when digested with EcoRI (Fig. 3 lanes a, b), in the presence or absence of heterologous ss DNA, with or without Brh2 as evident by the shift in 32P-label from the mobility of the linearized plasmid DNA to the faster moving free ss100mer (Fig. 3, lanes c–f). These controls verify the authenticity of the D-loops and show that neither heterologous ss DNA nor Brh2 interferes with cleavage of plasmid DNA by EcoRI, or that branch migration is impeded by these constituents. When complementary (−strand) 100mer was mixed with purified D-loops Brh2 efficiently promoted formation of duplexed D-loop product in the presence of absence of RPA (Fig. 3, lanes g–n). Little product formed without addition of Brh2 (Fig. 3, lanes h, l). These results add further support that Brh2 can promote annealing of the displaced strand of a D-loop with a complementary single strand.
Figure 3.
Duplexed D-loop formation with RPA coated DNA. The schematic depicts the endpoint of reactions in which purified D-loop DNA was used as substrate. D-loop DNA was prepared with pBluescript II plasmid DNA and 32P-labeled ss100mer (+ strand), deproteinized and purified by sucrose gradient centrifugation (lane a). After preincubating purified D-loop DNA with 400 nM Brh2 for 5 min, reactions were initiated by addition of ss100mer, incubated further as indicated and digested with EcoRI for analysis. Heterologous ss100mer was based on pUC19 DNA sequence. Mobility standards for supercoiled and double-strand linear plasmid (lanes o, p) are shown in the ethidium stained gel.
Brh2 promotes capture of a second-end in the absence of resection
Processing the ends of DNA after introduction of DSBs to reveal 3′-protruding tails (Sun et al., 1991; White and Haber, 1990) is thought to be prerequisite for recombinational repair because single-stranded DNA tails are considered to be necessary for Rad51 loading and filament formation, strand invasion of a homologous sequence, and securing the apposing DNA ends to bridge the DSB (Haber et al., 2004). Tail lengths have been determined by direct physical measurement (Sun et al., 1991; White and Haber, 1990) and can be inferred to some extent from conversion tract lengths of restriction site markers as the invading strand contributes to heteroduplex DNA formation (Sun et al., 1991; Sweetser et al., 1994). A few hundred residues is a length that investigators in the field would probably take as a reasonable average, but depending on the particular system under analysis the lengths may vary considerably (Palmer et al., 2003; Paques and Haber, 1999). Furthermore, the tail lengths on either side of a DSB may not necessarily be the same due to asymmetry in how the ends are processed (Neale et al., 2005). This could result from inherent asymmetry built into the mechanism and could also be due to additional factors such as differences in chromatin structure on either side of a DSB. In the extreme case in which the second apposing end of a DSB is not resected, we were curious whether it was still possible for that end to be captured by Brh2.
Since Brh2 is capable of promoting strand invasion and exchange to a limited extent (Mazloum et al., 2007; Mazloum et al., 2008) we tested whether Brh2 could promote the pairing between purified D-loops and ds100mer, again, using the assay of duplexed D-loop formation (Fig. 4, schematic). In this case formation of duplexed D-loop product would necessarily be accomplished by strand exchange between the looped out or displaced strand of the D-loop and the strand of the ds100mer with identical sequence. D-loop DNA prepared as above with 32P-labeled (+ strand) 100mer was incubated with unlabeled ds100mer. Duplexed D-loop product was efficiently formed implying that the complementary strand (− strand) from the ds100mer had been taken up with concomitant displacement of the unlabeled (+) strand (Fig. 4A).
Figure 4.
Pairing of D-loops and double-stranded DNA. The schematic depicts the course of reaction in which purified D-loop DNA and ds100mer were used as substrates. A. Reactions (15 μl) contained 1 nM D-loop DNA prepared with pBluescript II plasmid DNA and 32P-labeled ss100mer (+ strand) that was purified by sucrose gradient centrifugation (lane a), and 1 nM unlabeled ds100mer and 100 nM Brh2 (lanes c, d). B. Reactions contained 1 nM D-loop DNA with 32P-labeled ss100mer (+ strand), 100 nM Brh2, and either 1 nM ss100mer (− strand) (lanes e, f), or a ds100mer in increasing concentrations of 0.5 (lanes g, h), 1 (lanes i, j), 2 (lanes k, l), and 3 nM (lanes m, n) respectively. In the panel to the right, ds100mer (lane o) and ss100mer (+ strand) (lane p) are shown as mobility standards. C. Mixes contained 500 units per ml SspI and 0.5 nM 32P-labeled ss100mer (− strand) (lanes q, r), or 0.5 (lanes s, t), 1 (lanes u, v), or 3 nM (lanes w, x) ds100mer prepared with 32P-labeled ss100mer (− strand), as indicated. D. Mixes contained purified D-loop DNA prepared from pBluescript II plasmid DNA and 32P-labeled ss100mer (+ strand) and either Rad52 (left panel) or Brh2 (right panel). Reactions were started by addition of unlabeled ds100mer as above. Rad52 (lanes a–j) a, b, m, n-no protein; c, d, o, p-80 nM; e, f, q, r-160 nM; g, h, s, t-400 nM; i, j, u, v-800 nM.
Several lines of evidence support the notion that formation of the duplexed D-loop proceeds by strand exchange with the double stranded oligonucleotide. It was shown previously that Brh2 prepared according to our protocol is free of contaminating exonuclease activity that might nibble the ends of the ds100mer making a single stranded tail available for annealing (Mazloum et al., 2007). The ds100mer was prepared by annealing equimolar amounts of complementary ss100mer strand, then purifying the annealed product after electrophoresis in a polyacrylamide gel. In any event, we considered the possibility that the duplexed D-loop product might be formed by annealing of free (− strand) ss100mer contaminating the ds100mer preparation with the looped out strand of the D-loop. Arguing against this possibility was the following observation. There was a trace of unincorporated 32P-labeled (+ strand) ss100mer present in the D-loop DNA preparation after purification by sucrose gradient centrifugation. This 32P-labeled (+ strand) ss100mer could anneal to form ds100mer upon addition of (− strand) ss100mer as evidenced by the fast migrating band in gel electrophoresis. However, no annealing of the unincorporated 32P-labeled (+ strand) 100mer to duplex form was evident in samples to which the ds100mer was added suggesting that there was no free (− strand) ss100mer in the preparation (Fig. 4B). We also examined the ds100mer for the presence of free (− strand) ss100mer by testing for any DNA remaining uncut after digestion with a double-strand specific endonuclease (SspI restriction endonuclease cuts once in the 100mer sequence). ds100mer was prepared by our standard protocol but the (−strand) ss100mer was 32P-labeled. After digestion with SspI and gel electrophoresis, analysis indicated that all the 32P-labeled (− strand) was in the form of ds100mer and that all of the DNA was cleaved by SspI to yield a ds18mer product (Fig. 4C). These controls show that there is no free (− strand) ss100mer present in the substrate. We conclude that Brh2 can promote pairing between the displaced strand of a D-loop and a double-stranded DNA oligonucleotide.
Since Rad52 shares with Brh2 the ability to promote formation of D-loops and duplexed D-loops with a single-strand-DNA donor, we investigated whether it was also capable of forming duplexed D-loops with a double-strand donor. Here we used the same experimental design as above with purified D-loop DNA containing 32P-labeled (+ strand) 100mer and unlabeled ds100mer as substrates. Brh2 promoted formation of duplexed D-loops as readily with ds100mer as with ss100mer (compare Fig 4D, lanes k, l with o–v), but Rad52 had little activity in forming duplexed D-loops with ds100mer substrate (Fig. 4D, lanes c–j). Controls showed that the ds100mer did not inhibit cleavage of the D-loop DNA by EcoRI (Fig. 4D, lane b) nor was it contaminated by free ss100mer (−strand) (Fig. 4D, lane n). These results show that Brh2 has an ability to promote homologous pairing of the displaced strand of a D-loop with a duplex DNA molecule.
Brh2 promotes DNA synthesis-dependent second-end capture
In a biochemical investigation aimed at identifying DNA polymerase activities in mammalian cells capable of promoting synthesis from a model recombination intermediate, it was found that the translesion polymerase Pol eta could extend synthesis from a synthetic oligonucleotide-based D-loop (McIlwraith et al., 2005). Deletion of the gene for Pol eta in chicken DT40 cells was found to reduced the frequency of DSB-induced gene conversion (Kawamoto et al., 2005). On the other hand genetic studies in S. cerevisiae support an alternative view that the replicative polymerase Pol delta is preferentially recruited for repair synthesis during induced homologous recombination, while Pol eta does not appear to affect gene conversion (Maloisel et al., 2008). We were interested to know whether Brh2 would be capable of promoting second-end capture by pairing a remotely situated oligomer with the displaced strand of a D-loop that was formed as a consequence of extension by DNA synthesis (Fig 5, top schematic). In the absence of any direct knowledge regarding which DNA polymerase might be responsible for extending D-loops in U. maydis, we used E. coli DNA polymerase I Klenow fragment (3′-5′ exo−) (hereafter, Pol) as a representative generic enzyme for catalyzing DNA synthesis.
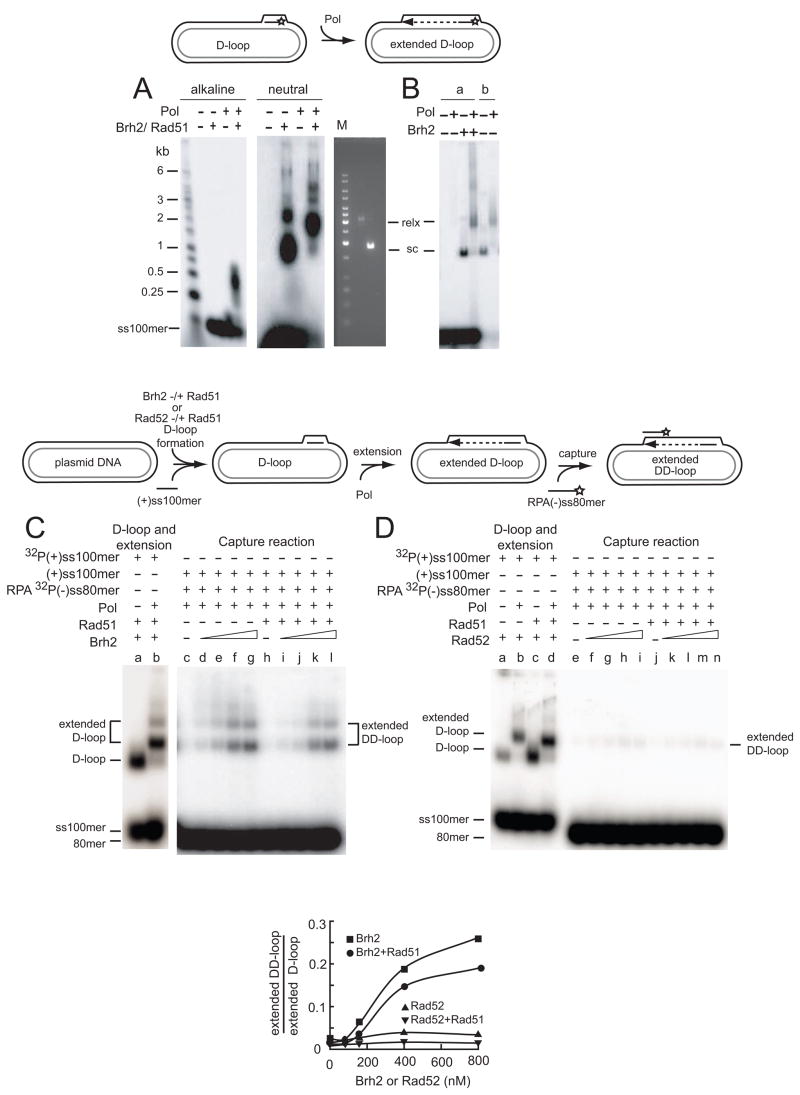
Figure 5.
Brh2-promoted duplexed D-loop formation coupled with DNA synthesis. The schematic depicts use of the third (invading) strand in a D-loop as a primer for DNA synthesis. A. D-loops were formed in standard reactions optimized for yield in which 6 nM 32P-labeled (with star) ss100mer (+ strand) had been preincubated with 750 nM Rad51 in the presence of 2 mM ATP, 2 mM CaCl2 and 18 nM pBluescript plasmid DNA had been pre-mixed with 400 nM Brh2. After D-loop formation, deoxynucleoside triphosphates and DNA polymerase (Pol) were added. Following incubation for 30 min the mixtures were split, deproteinized and electrophoresed under neutral (left panel) or denaturing conditions (30 mM NaOH, middle panel). A gel run in parallel with plasmid DNA in supercoiled (sc) or relaxed form (relx) using wheat germ topoisomerase (Promega) was stained with ethidium bromide. B. In the lanes indicated under a, D-loop reactions were performed with Brh2 as above, but without Rad51, followed by addition of DNA polymerase. In the lanes indicated under b, preformed purified D-loop DNA free of unincorporated 32P-labeled ss100mer was used in reaction with DNA polymerase. C. The schematic depicts the stepwise protocol for D-loop formation and DNA-synthesis-coupled second-end capture. D-loop formation controls were performed with Brh2 (400 nM) and Rad51 (750 nM) with or without Pol (lanes a, b) as in A using 32P-labeled ss100mer (+ strand) to gauge the level of D-loop formation. In the second end capture determinations with increasing levels of Brh2 in the presence or absence of 750 nM Rad51 (lanes c–l) D-loop formation was initiated in the same way, but with the use of unlabeled ss100mer (+ strand). After the extension period with DNA polymerase, 32P-ss80mer (− strand) coated with RPA was added and mixtures incubated an additional 30 min. Brh2 was added as follows: lanes c, h-no Brh2; lanes d, i-80 nM; lanes e, j-160 nM; lanes f, k-400 nM; lanes g, l-800 nM. Product formation was quantified by comparison of the extended duplexed D-loop product against the standard of extended D-loops (lane b) and expressed graphically below as extended DD-loop/extended D-loop. D. Reactions were performed as in C except that Rad52 was used instead of Brh2. In the controls (lanes a–d) D-loop formation by Rad52 (200 nM) and Rad51 (750 nM) and extension by Pol were gauged using 32P-labeled ss100mer (+ strand). Second-end capture was performed as above with increasing Rad52 as follows: lanes e, j-no Rad52; lanes f, k-80 nM; lanes g, l-160 nM; lanes h, m-400 nM; lanes I, n-800 nM. Product formation was quantified as in C by comparison of the extended duplexed D-loop product against the standard of extended D-loops minus or plus Rad51 (lane b or d, respectively) and expressed graphically below.
D-loop DNA formed with plasmid DNA and homologous ss100mer in reactions with Brh2 and Rad51 was first investigated to determine the extent to which the invading single strand could serve as a primer for DNA synthesis. Under the experimental conditions we employed for agraose gel electrophoresis the D-loops formed with 32P-labeled ss100mer and pBluescript II plasmid DNA migrated with nearly the same mobility as control supercoiled plasmid DNA. Since the plasmid DNA prepared by our procedure contains 30–35 superhelical turns as determined by 2-D electrophoresis in gels containing chloroquine (data not shown), it would be expected that those molecules should have the capacity to accommodate a completely base-paired third strand of up to about 350 residues in length. In such case nearly all superhelical turns would be removed and the D-loops would run with a mobility of completely relaxed DNA during gel electrophoresis.
Under conditions previously optimized for D-loop formation in which Rad51 was pre-incubated with 32P-labeled ss100mer then mixed with pre-formed complexes of Brh2 and plasmid DNA, the D-loops ran as expected with mobility equivalent to control superhelical DNA (Fig. 5A). However, after addition of Pol and all four deoxynucleoside triphosphates (dNTPs) the mobility of the D-loop was quantitatively reduced to that of topoisomerase-relaxed plasmid DNA (Fig. 5A, middle panel), consistent with the notion that the ss100mer invading strand was serving as a primer for DNA synthesis and that synthesis proceeded to the extent that all remaining negative superhelical turns in the plasmid were removed. When the length of the 32P-labeled strand was analyzed under denaturing conditions, the bulk of the label appeared to migrate between standards of 250 and 500 residues (Fig. 5A, left panel) indicating that indeed the ss100mers in the D-loops had been elongated to approximately the extent anticipated based on complete unwinding of the superhelical plasmid DNA. D-loops prepared with Brh2 alone or deproteinized and purified by sucrose gradient centrifugation were similarly capable of being extended by DNA synthesis based on shift in mobility of the 32P-labeled product band from the position of superhelical to relaxed plasmid DNA (Fig. 5B).
Having established that D-loops formed by Brh2 with or without Rad51 could be elongated by DNA synthesis, we tested whether a second ss oligomer downstream from the D-loop sequence formed during initial strand invasion could be captured. For this experiment we used unlabeled ss100mer (+ strand) as the invading single strand for D-loop formation and a 32P-labeled ss80mer (− strand) lying 200 residues away from the 3′-end of the 100mer sequence as the second-end target (Fig. 5C,D schematic). Capture of the second strand in this case would depend on DNA synthesis to extend the D-loop through the 200 residue intervening sequence and past the downstream locus spanning the target sequence, i.e., 280 residues.
In reactions with Brh2, product with mobility corresponding to that of relaxed plasmid DNA was formed that was dependent on addition of Pol (Fig. 5C, lanes d–g). No product was formed without Brh2 or with Rad51 alone (Fig. 5C, lane c, h), but product was formed in the combined presence of Brh2 and Rad51 (Fig. 5C, lanes i–l). The results were quantified as the fraction of product, or extended duplexed D-loops, generated with respect to a standard of extended D-loops formed with 32P-labeled ss100mer (+ strand) and elongated by DNA synthesis (Fig. 5C lane b) and is denoted graphically as extended DD-loop per extended D-loop. By comparison Rad52 was reduced in ability to promote second-end capture in the DNA synthesis-dependent reactions (Fig. 5D, lanes e–n), even though it was active in D-loop formation with or without Rad51 and did not interfere with DNA synthesis as judged by relaxation of the plasmid DNA template (Fig. 5D, lanes a–d). The poor activity of Rad52 in the synthesis-dependent reaction contrasts with the relatively high activity observed in the non-synthesis dependent duplexed D-loop reaction noted above (Fig. 2C). In pairing reactions Rad52, like Rad51, is strongly influenced by the order of addition of DNA substrates (Kagawa et al., 2001). Possibly, the ability of Rad52 to anneal DNA is different when it is presented with static single-stranded DNA substrates as opposed to the dynamic situation in which the DNA structure changes from duplex to single-stranded during the course of reaction as a consequence of DNA synthesis.
Two caveats should be kept in mind regarding this analysis and its presentation. First, it is to be emphasized that in the experimental determinations (Fig. 5C,D--“capture reaction” panels) the extended D-loop is primed for DNA synthesis with an unlabeled ss100mer (+strand). Formation of the extended duplexed D-loop product becomes evident only upon annealing of the 32P-labeled ss80mer (− strand). The standard of extended D-loop used for comparison (Fig. 5C, D--“D-loop and extension” panels) was prepared in a different reaction in which synthesis was primed from 32P-labeled ss100mer (+ strand) in a D-loop. Thus, there is no direct or absolute measure for how much D-loop DNA is converted into the final product of extended duplexed D-loop, only a relative level. Second, in contrast to the simpler case of Brh2-promoted duplexed D-loop formation (Fig. 2D) in which total product formed is stimulated by Rad51, it appears that Rad51 becomes inhibitory in the DNA synthesis-dependent reactions (Fig. 5C), although the actual level of inhibition is not clear due to the lack of direct measurement as above. The basis for the difference between annealing a complementary strand to the displaced strand of a D-loop after formation compared to annealing after extension of the D-loop by over 200 nucleotides is not known, but most likely reflects a change in nucleoprotein structure under the conditions for DNA synthesis. In the presence of Mg2+, which is added as a cofactor for DNA polymerase, Rad51 inactivates itself (Bugreev and Mazin, 2004) and forms a dead-end complex on DNA. Addition of Rad54, which serves to sweep Rad51 from double-stranded DNA (Solinger et al., 2002), might well relieve the inhibition of synthesis-dependent second-end capture.
It is not clear what is the basis for the second product band with slower mobility, but we think it is probably a dimer structure that forms between two plasmid molecules when the invading strand elongated by DNA synthesis forms a D-loop in common with a second plasmid molecule. As the invading strand is extended by DNA synthesis to the point where the topological constraint of the plasmid molecule limits further synthesis, possibly the 5′-end of the invading strand becomes dissociated and is taken up by a second plasmid molecule. Experimentation to clarify the nature of this second product band is underway. These findings show that Brh2 can promote annealing of a downstream, second strand with the displaced strand in a D-loop formed initially by coupled Brh2/Rad51-promoted strand invasion at a distant locus, then extended by DNA synthesis.
We conclude that Brh2 can promote pairing of the displaced strand in a D-loop extended by DNA synthesis with a downstream homologous ssDNA, thus modeling second-end capture in repair of a DSB.
DISCUSSION
Accumulating evidence has implicated Rad52 as the enabling factor of the recombinational machinery that promotes second strand capture. In biochemical studies with purified yeast or human proteins, Rad52 has been shown to catalyze single-strand annealing of tailed molecules with the displaced strand of a D-loop intermediate (Bugreev et al., 2007; McIlwraith and West, 2008; Sugiyama et al., 2006). In Saccharomyces cerevisiae there is strong physical evidence supporting this notion (Lao et al., 2008). Analysis of molecular intermediates formed during recombination demonstrated that joint molecules representing nascent strand invasion intermediates accumulate in mutants devoid of Rad52. However, formation of subsequent double Holliday junction intermediates was defective strongly implying that Rad52 is required for progression from the single-end strand invasion joint molecule intermediate to the post-invasion crossover precursor formed by second-end capture. As the activity responsible for transition to second-end capture was localized to the N-terminal DNA-interaction domain of Rad52, it was inferred that its inherent strand annealing function was likely the activity promoting reunion of both DSB ends.
The first conclusion from the present study is that Brh2 can promote intermolecular pairing between the displaced strand of a D-loop and a complementary strand in the presence of Rad51, a condition that attenuates the annealing activity of Rad52 (Wu et al., 2008). The complementary strand can be a free single strand, in which case it can become paired with the displaced strand of the D-loop through the annealing activity inherent in Brh2. Alternatively, if the complementary strand is present in a duplex structure, it can swap partners and become paired with the D-loop displaced strand through the strand exchange activity inherent in Brh2. One can imagine that with the advent of a flap endonuclease to trim or polish any protruding or displaced single strand resulting from strand exchange, capture of the second end could be enabled by Brh2 and would require minimal resectioning to prepare the end. The second important conclusion from this work is that Brh2 can function in a coupled reaction with DNA polymerase to promote pairing between the displaced strand of a D-loop extended by new synthesis and a complementary strand. These reactions emulate the second-end capture step envisioned in models for repair of DSBs (Haber et al., 2004) and raise the possibility that Brh2 serves to perform an additional role in recombination that is functionally distinct and temporally removed from the Rad51-mediator activity.
Studies with the purified U. maydis Rad52 protein have shown it is imbued with potent strand annealing activity as in the case of the yeast protein (Mazloum et al., 2007). Nevertheless, genetic evidence for a role of Rad52 in second-end capture is lacking in U. maydis. rad52 deletion mutants have few discernible defects in DNA repair, recombination, or meiosis, contrary to expectations for a function essential in reuniting the two ends of a broken DNA molecule (Kojic et al., 2008). On the other hand brh2 mutants are extremely deficient in all of these processes (Kojic et al., 2002). An additional role for Brh2 in a process downstream of mediating Rad51-filament formation would not be inconsistent with the genetic findings. Separating the mediator function from second-end capture function, however, is a difficult genetic challenge. Studies are now underway aimed at physical characterization of the DNA strands processed after introduction of a single DSB so that protein trafficking events on both ends of a single DSB can be monitored.
It is instructive to compare and contrast the findings presented here with the published studies on Rad52-promoted second-end capture using reconstituted in vitro systems from yeast (Sugiyama et al., 2006) or human ((Bugreev et al., 2007; McIlwraith and West, 2008). All three of those systems vary in experimental design and implementation, but share the common strategy of stepwise addition of Rad52 to reactions containing previously formed D-loops or joint molecules. The intent in those investigations was to isolate the second-end capture step from the D-loop formation step. In present study, however, a different design was followed. Here Brh2 (or Rad52) and Rad51 were mixed at the onset to emulate the situation in vivo where presumably these proteins are in physical contact when D-loop formation initiates. The different ordering of addition of the components could well account for certain discrepancies, such as in the requirement for Rad52 in second-end capture following Rad51-promoted D-loop formation as observed by the Mazin laboratory (Bugreev et al., 2007) using human proteins, versus the inhibitory effect of Rad51 on Rad52-promoted second-end capture as noted here with the U. maydis system. In the human system developed by the West laboratory, Rad52 was shown to promote second-end capture with pre-formed D-loop DNA created by annealing oligonucleotides, but no Rad51 was present. It would be interesting to learn how the human system responds with Rad51 and Rad52 added together. In both human systems but not the yeast system, second-end capture was dependent on DNA synthesis, in each case catalyzed by DNA polymerase eta, and in each case the extent of synthesis was limited to about 30 residues. As shown in the present work, second-end capture with Brh2 can be extended over a much larger span of residues.
It is fascinating to consider the biological puzzle presented by Rad52. On the one hand it is pivotal for every aspect of recombination in S. cerevisiae and apparently for other ascomycete fungi as well. Yet it is absent in green plants, flies, mosquitoes, nematodes, the protist clade represented by plasmodium, and is relegated to an apparently minor role in recombination in vertebrates and the basidiomycete fungi, if we extrapolate from U. maydis. BRCA2 homologs appear present in all these latter organisms, and are evident by BLAST analysis in other fungal phyla in which genome sequences are available such as the zygomycetes and chytrids (e.g., see http://genome.jgi-psf.org Phycomyces blakesleeanus Phybll 65076 and Batrachochytrium dendrobatidis Batde5 22020, respectively), but not in the ascomycete fungi. Therefore, it seems logical to conclude that BRCA2 was lost in the course of evolution of the ascomycota and that somehow Rad52 took over the role of BRCA2. That the Rad52 paralog Rad59, which is absent in U. maydis and higher eukaryotes, can help alleviate the inhibitory effect of Rad51 on Rad52-promoted annealing (Wu et al., 2008), analogous to the activity of Brh2 in promoting second-end capture in the presence of Rad51, suggests additional compensating functions might have evolved to fill in for the loss of BRCA2. The role that Rad52 plays in the non-ascomycete fungi and in other eukaryotic kingdoms awaits discovery.
MATERIALS AND METHODS
Reagents
Brh2 in complex with Dss1, Rad51, Rad52, and RPA proteins were purified after overproduction in E. coli as described previously (Mazloum et al., 2007). Oligonucleotides were synthesized by Integrated DNA Technologies and purified by electrophoresis in 8% acrylamide gels. Oligonucleotide sequences are based on plasmid pBluescript II SK+ DNA nucleotides 2–101 (ss100mer), 2701–2780 (ss80mer), or pUC19 DNA ss80mer (as heterologous control), respectively as previously described (Mazloum et al., 2007). ds100mer was prepared by thermal annealing of complementary ss100mers from mixes containing equimolar concentrations, then purifying the ds product by electrophoresis in acrylamide gel as above. Plasmid pBluescript II SK+ DNA was purified without an alkaline denaturation step as described (Mazloum et al., 2008). D-loop DNA used a substrate was prepared in reactions with both Brh2 and Rad51 to maximize yield (Mazloum et al., 2008). It was then purified after removal of protein by extraction with phenol followed by sucrose gradient centrifugation. All DNA concentrations are expressed as moles of molecules rather than nucleotide, unless indicated otherwise. Oligonucleotides were 5′-end-labeled using [γ-32P]ATP and T4 polynucleotide kinase.
D-loop and second-end capture reactions
For D-loop reactions catalyzed by Rad51, 6 nM 32P-labeled 100mer (+ strand) was preincubated with protein at 37° in a 15 μl reaction in a buffer containing 25 mM Tris-HCl, pH 7.5, 20 mM KCl, 1 mM DTT, 4 mM CaCl2, 2 mM ATP. After 15 min reactions were started by addition of plasmid DNA (18 nM unless otherwise indicated) and incubation continued for an additional 30 min. D-loop reactions promoted by Rad52 were similarly performed with preincubation of protein on ss100mer. D-loop reactions promoted by Brh2 were also performed similarly except that order of addition was changed. The preincubation step with ss100mer was not performed, but instead, Brh2 was preincubated with plasmid DNA. In reactions utilizing dual combinations of Rad51, Rad52 and Brh2, the proteins were preincubated, separately if required, under their respective optimal conditions (i.e., Rad51 and Rad52 with ssDNA first; Brh2 with plasmid DNA first) before mixing as described previously (Mazloum et al., 2008). For duplexed D-loop formation, after the 30 min incubation period to allow D-loop formation 3 nM ss100mer (− strand) coated with RPA was then added and incubation continued for an additional 15 min. ssDNA was precoated with RPA by incubation in buffer containing 1 mM MgCl2 at 7.5 moles nucleotide per mole RPA for 15 min. MgCl2 was then brought to 10 mM and EcoRI (500 units per ml) was added. After 15 min reactions were stopped by addition of 10 mM EDTA. SDS and proteinase K were added to 1.2 % and 1.7 mg/ml, respectively, and after an additional incubation for 15 min tracking dye was added and DNA products were resolved by electrophoresis on 1% agarose gels. For reactions involving DNA synthesis, D-loop formation was performed as above using ss100mer (+ strand) for 30 min. Then a mixture of all four deoxynucleoside triphosphates (125 μM each), 10 mM MgCl2 and 250 units per ml E. coli DNA polymerase Klenow fragment (3′-5′ exo−, New England BioLabs) was added (concentrations refer to the final levels in the reaction). After incubation for 15 min, extension was stopped by addition of 10 mM EDTA. When second-end capture coupled with DNA synthesis was being performed, 1 nM 32P-ss80mer (− strand) coated with RPA was added and mixtures incubated for an additional 15 min. Reactions were then stopped with SDS and proteinase K and 20 nM ss80mer (+strand). Gels were dried onto Whatman DE81 paper, exposed to phosphor storage screens (Molecular Dynamics) and processed with a Typhoon 9400 PhosphorImager (Amersham Biosciences). Quantitation was performed using ImageQuaNT software (Molecular Dynamics). In experiments involving DNA synthesis dependent second-end capture 32P-ss100mer (+ strand) and 32P-ss80mer (− strand) were prepared simultaneously and under identical conditions so that the specific activities of the labeled oligonucleotides would be the same.
Acknowledgments
We thank laboratory members Qingwen Zhou and Milorad Kojic for generous help and advice. WKH thanks Lorraine Symington, Columbia University for stimulating discussions. Support for this work was provided by grants GM42482 and GM79859 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allers T, Lichten M. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol Cell. 2001;8:225–231. doi: 10.1016/s1097-2765(01)00280-5. [DOI] [PubMed] [Google Scholar]
- Belmaaza A, Chartrand P. One-sided invasion events in homologous recombination at double-strand breaks. Mutat Res. 1994;314:199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- Bi B, Rybalchenko N, Golub EI, Radding CM. Human and yeast Rad52 proteins promote DNA strand exchange. Proc Natl Acad Sci U S A. 2004;101:9568–9572. doi: 10.1073/pnas.0403205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nat Struct Mol Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci U S A. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries FA, Zonneveld JB, van Duijn-Goedhart A, Roodbergen M, Boei J, van Buul PP, Essers J, van Steeg H, van Zeeland AA, van Benthem J, Pastink A. Inactivation of RAD52 aggravates RAD54 defects in mice but not in Schizosaccharomyces pombe. DNA Repair (Amst) 2005;4:1121–1128. doi: 10.1016/j.dnarep.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Ferguson DO, Holloman WK. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci U S A. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR. Targeted gene replacement in Drosophila via P element-induced gap repair. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- Haber JE, Ira G, Malkova A, Sugawara N. Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday’s model. Philos Trans R Soc Lond B Biol Sci. 2004;359:79–86. doi: 10.1098/rstb.2003.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. Homologous pairing promoted by the human Rad52 protein. J Biol Chem. 2001;276:35201–35208. doi: 10.1074/jbc.M104938200. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Kim HK, Morimatsu K, Norden B, Ardhammar M, Takahashi M. ADP stabilizes the human Rad51-single stranded DNA complex and promotes its DNA annealing activity. Genes Cells. 2002;7:1125–1134. doi: 10.1046/j.1365-2443.2002.00588.x. [DOI] [PubMed] [Google Scholar]
- Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- Kojic M, Mao N, Zhou Q, Lisby M, Holloman WK. Compensatory role for Rad52 during recombinational repair in Ustilago maydis. Mol Microbiol. 2008;67:1156–1168. doi: 10.1111/j.1365-2958.2008.06116.x. [DOI] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol Cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel L, Fabre F, Gangloff S. DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol Cell Biol. 2008;28:1373–1382. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazloum N, Zhou Q, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazloum N, Zhou Q, Holloman WK. D-loop formation by Brh2 protein of Ustilago maydis. Proc Natl Acad Sci U S A. 2008;105:524–529. doi: 10.1073/pnas.0707031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, West SC. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol Cell. 2008;29:510–516. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Meselson MS, Radding CM. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975;72:358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci U S A. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Schildkraut E, Lazarin R, Nguyen J, Nickoloff JA. Gene conversion tracts in Saccharomyces cerevisiae can be extremely short and highly directional. Nucleic Acids Res. 2003;31:1164–1173. doi: 10.1093/nar/gkg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding CM, Beattie KL, Holloman WK, Wiegand RC. Uptake of homologous single-stranded fragments by superhelical DNA. IV. Branch migration. J Mol Biol. 1977;116:825–839. doi: 10.1016/0022-2836(77)90273-x. [DOI] [PubMed] [Google Scholar]
- Ramesh MA, Malik SB, Logsdon JM., Jr A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Shivji MK, Venkitaraman AR. DNA recombination, chromosomal stability and carcinogenesis: insights into the role of BRCA2. DNA Repair. 2004;3:835–843. doi: 10.1016/j.dnarep.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. Embo J. 2006;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Sweetser DB, Hough H, Whelden JF, Arbuckle M, Nickoloff JA. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol Cell Biol. 1994;14:3863–3875. doi: 10.1128/mcb.14.6.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls Rad52-mediated DNA annealing. J Biol Chem. 2008;283:14883–14892. doi: 10.1074/jbc.M801097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Buerstedde JM, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]