Abstract
Pancreatic cancer is an almost universally lethal disease. Research over the last two decades has shown that pancreatic cancer is fundamentally a genetic disease, caused by inherited germline and acquired somatic mutations in cancer-associated genes. Multiple alterations in genes that are important in pancreatic cancer progression have been identified, including tumor suppressor genes, oncogenes, and genome maintenance genes. Furthermore, the identification of noninvasive precursor lesions of pancreatic adenocarcinoma has led to the formulation of a multi-step progression model of pancreatic cancer and the subsequent identification of early and late genetic alterations culminating in invasive cancer. In addition, an increased understanding of the molecular basis of the disease has facilitated the identification of new drug targets enabling rational drug design. The elucidation of genetic alterations in combination with the development of high-throughput sensitive techniques should lead to the discovery of effective biomarkers for early detection of this malignancy. This review focuses mainly on the current knowledge about the molecular insights of the pathogenesis of pancreatic ductal adenocarcinoma.
Key Words: Pancreatic cancer, Precursor lesion, Early detection, Mouse models, Genetics
Epidemiology
Jan-Bart M. Koorstra and Steven R. Hustinx contributed equally to this work.
Pancreatic cancer is a disease with a dismal outlook. In the United States approximately 33,000 patients are diagnosed with pancreatic cancer annually, and nearly an equal number will die from the disease, representing the fourth most common cause of cancer-related mortality. Men and women have an approximately equal risk [1]. Worldwide, pancreatic cancer causes an estimated 213,000 deaths each year [2]. For all stages combined, the 1-year survival rate is around 20%, and the overall 5-year survival rate is less than 5%, despite even the most aggressive therapies currently available [1].
A number of risk factors have been identified [3]. Pancreatic cancer is predominantly a disease of the elderly. Pancreatic cancer is rare before the age of 40, and the median age at diagnosis is 73 years. Cigarette smoking is by far the leading preventable cause of pancreatic cancer [4]. Cigarette smoking doubles the risk of pancreatic cancer (relative risk = 2) [3]. Other risk factors include diets high in meats and fat, low serum folate levels, obesity, long-standing diabetes mellitus, and chronic pancreatitis [3,5,6,7]. Approximately 10% of patients demonstrate a familial predisposition for pancreatic cancer, and a subset of these patients harbor germline mutations in BRCA2, p16/CDKN2A, PRSS1, STK11/LKB1, or the DNA mismatch repair genes (see further discussion below). In the vast majority of patients with familial risk, however, the underlying genetic predisposition remains unknown.
Complete surgical resection remains the only curative treatment. Studies from high-volume centers with optimal staging report up to a 15–20% 5-year survival rate in patients undergoing surgical resection [8, 9]. The mortality rate is so high because pancreatic cancer usually only produces symptoms when it has already metastasized, and because there are no sensitive and specific tools to detect the disease at an earlier stage. Although multiple histological subtypes of pancreatic cancer have been described, the most common and deadliest form is pancreatic ductal adenocarcinoma [10]. Novel approaches to the management of patients with this aggressive disease are urgently needed.
Research over the last two decades has shown that pancreatic cancer is fundamentally a genetic disease, caused by inherited germline and acquired somatic mutations in cancer-associated genes. A compendium of alterations in tumor suppressor genes, oncogenes, and genome-maintenance genes that are important in pancreatic cancer progression have now been identified (fig. 1). This review focuses mainly on the molecular insights on pancreatic ductal adenocarcinoma and its precursor lesions, including insights gained through experimental models of pancreatic carcinogenesis.
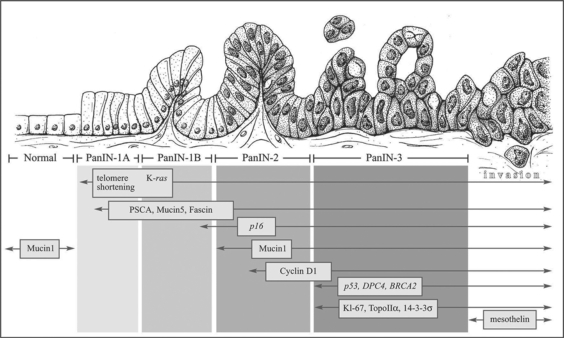
Fig. 1.
Progression model of pancreatic ductal adenocarcinoma from normal (left) to carcinoma (right). The histological progression is associated with the accumulation of specific genetic alterations. Reprinted with permission from Maitra et al. [164].
Precursor Lesions of Pancreatic Cancer
Prior to a discussion on molecular genetics of pancreatic cancer, we will briefly discuss the current state of knowledge on precursor lesions of the pancreas. This is essential from the context of separating ‘early’ genetic changes (i.e. those associated with tumor initiation) from ‘late’ abnormalities (i.e. those associated with tumor progression). A recent review in Pancreatology has extensively discussed the histology and genetics of pancreatic cancer precursors [11]; therefore, we will only discuss these in fleeting detail. Briefly, pancreatic intraepithelial neoplasias (PanINs) are classified into a four tier classification, including PanIN−1A, −1B, −2, −3, reflecting a progressive increase in histologic grade culminating in invasive neoplasia (fig. 2). The lowest grade PanIN lesions can be flat (1A) or papillary (1B), but are characterized by absence of nuclear atypia and retained nuclear polarity. PanIN-2 lesions have micropapillary features with evidence of nuclear atypia and infrequent mitoses, while PanIN-3 lesions (a.k.a. carcinoma in situ) demonstrate widespread loss of polarity, nuclear atypia, and frequent mitoses. In addition to microscopic PanIN lesions, there are now recognized macroscopic (cystic) precursor lesions of pancreatic adenocarcinoma – including intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms. Akin to PanINs, the cystic precursor lesions also demonstrate a multistep histological and genetic progression to invasive neoplasia. Since IPMNs and mucinous cystic neoplasms can be detected by radiologic scans, they represent an opportunity to diagnose invasive pancreatic cancer before it can develop [11].
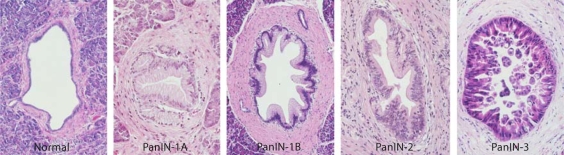
Fig. 2.
Consecutive PanIN lesions with progressive histological changes from normal to PanIN-3. Reproduced with permission from http://pathology.jhu.edu/pancreas_panin.
Tumor Suppressor Genes
Tumor suppressor genes are genes that promote tumor growth when inactivated. Tumor suppressor genes are recessive, i.e. the two copies need to be mutated for loss of function, and they can be inactivated by a variety of mechanisms. First, by an intragenic mutation in one allele (copy of a gene) coupled with loss of the second allele; second, through a deletion of both alleles (homozygous deletion), and third, by hypermethylation of the promoter of the gene-silencing gene expression. In sporadic cancers these alterations are both somatic mutations acquired during life, while patients with inherited forms of cancer inherit one mutant allele in the germline while the second allele is somatically mutated in the cancer cells.
The p16INK4A/CDKN2A gene, located on the short arm of chromosome 9 (9p), is one of the most frequently inactivated tumor suppressor genes in pancreatic cancer [12](table 1. Remarkably, virtually all pancreatic carcinomas have loss of p16INK4A/CDKN2A function, in 40% of pancreatic cancer through homozygous deletion, in 40% by an intragenic mutation coupled with loss of the second allele, and in 15% by hypermethylation of the p16INK4A/CDKN2A gene promoter [12, 13]. The protein p16 belongs to the cyclin-dependent kinase (CDK) inhibitor family and functions to prevent the phosphorylation of Rb-1 by CDKs, and cyclin D-Cdk4 and cyclin D-Cdk6 complexes, which act as cell-cycle regulators [14, 15]. Loss of p16INK4A/CDKN2A results in inappropriate phosphorylation of Rb-1, thereby facilitating progression of the cell cycle through the G1/S transition [16]. Thus, the p16/Rb pathway is inactivated in virtually all pancreatic cancers, leading to an inappropriate progression through the G1 phase of the cell cycle. Of note, in a small group of patients, inherited mutations of the p16INK4A/CDKN2A gene cause the familial atypical multiple mole melanoma (FAMM) syndrome, which is associated with an increased risk of developing melanoma and an increased risk of developing pancreatic cancer [17, 18]. Particularly, the p16 Leiden deletion, a 19-bp deletion, is associated with an increased pancreatic cancer risk [19].
Table 1.
Frequency of selected tumor suppressor genes, oncogenes and genome maintenance genes
| Gene mutations | Incidence in pancreatic adenocarcinoma, % |
|---|---|
| p16 | 80–95 |
| p53 | 50–75 |
| DPC4 | 45–55 |
| K-RAS | 75–90 |
| BRAF | 5–10 (estimated) |
| hMLH1, hMSH2 | 4 |
| BRCA2 | 7–10 |
In addition, the homozygous deletions, which inactivate p16, can encompass adjacent genes, including the MTAP, IFNA1 and IFNB1 genes [20, 21]. The MTAP gene is located approximately 100 kilobases telomeric to the p16INK4A/CDKN2A gene on chromosome 9p21, and is frequently contained in the p16INK4A/CDKN2A homozygous deletions. As a result, MTAP function is completely lost in approximately 30% of pancreatic adenocarcinomas. This is a potentially promising finding, because it may have therapeutic implications [22]. The product of the MTAP gene, the enzyme methylthioadenosine phosphorylase plays an important role in the synthesis of adenosine [23]. Chemotherapeutic agents, such as L-alanosine, a purine biosynthesis inhibitor, have been developed, to specifically target the selective loss of MTAP function in cancers, implicating that it might be effective against one third of the adenocarcinomas of the pancreas [22, 23].
Mutation of the p53 gene on chromosome 17p is the most common somatic alteration in human cancer. The p53 protein plays a central role in modulating cellular responses to cytotoxic stress by contributing to both cell-cycle arrest and programmed cell death. Loss of p53 function during carcinogenesis can lead to inappropriate cell growth, increased cell survival, and genetic instability [24]. In pancreatic cancer, the p53 tumor suppressor gene is inactivated in 50–75% of the cases and occurs predominantly through single allelic loss coupled with an intragenic mutation of the second allele [25]. The loss of p53 means that two critical controls of cell number (cell division and cell death) are deregulated in the majority of pancreatic cancers. Of interest, 14–3-3σ, a p53-regulated gene, plays a role in signal transduction, apoptosis, stress response and cytoskeletal organization [26]. 14–3-3σis transcribed in response to DNA damage and in a number of cancers it is an important mediator of p53-induced G2 arrest [27].
In addition, p53-induced growth arrest is also achieved by transactivation of p21. p53binding to DNA stimulates production of the protein p21, which negatively regulates the complex consisting of cyclin D and the cell division-stimulating protein cyclin-dependent-kinase-2 [28], thereby preventing the cell from progressing from G1-S phase. This mechanism allows time for repair to damaged DNA. If p53 mutated, it is not able to bind DNA, so p21 is not made available and abnormal growth can occur. Cell lines which lack wild-type p53 show a reduced or complete absence of p21[29]. Loss of p21 activity has been observed in approximately 30–60% of pancreatic tumor specimens [30,31,32]. Pancreatic cell lines and pancreatic tumors show a correlation between active p53 and p21[33].
As stated, p53 loss is a ‘double threat’, because it results in both loss of cell cycle checkpoints, as well as deregulation of programmed cell death (i.e. apoptosis). It is now known that p53-induced apoptosis is mediated by activation of genes involved in the apoptotic pathway, for example genes such as PUMA (p53-upregulated modulator of apoptosis) and Noxa. PUMA and Noxa are activated in a p53-dependent manner following DNA damage. Once activated, they bind to Bcl-2, localize to the mitochondria to induce cytochrome c release, and activate the induction of programmed cell death [34,35,36].
Finally, the micro-RNA miR-34a deserves mention (miRNAs in general are discussed later): miR-34a is a direct transcriptional target of p53. MiR-34a activation can recapitulate elements of p53 activity, including induction of cell-cycle arrest and promotion of apoptosis, and loss of miR-34a can impair p53-mediated cell death [37, 38]. Chang et al. [39] showed that reduced expression of miR-34a is a very frequent feature of pancreatic cancer cells.
DPC4 (Smad4) is a tumor suppressor gene on chromosome 18q and is one of the most commonly inactivated genes in pancreatic ductal adenocarcinoma, detected in approximately 55% of the cases. Inactivation occurs either through homozygous deletion, in approximately 30%, or loss of one allele coupled with an intragenic mutation in the second allele in approximately 25% [40,41,42]. The transcription factor SMAD4 is an important regulator of the transforming growth factor-β (TGF-β) signaling pathway [43]. Upon receptor activation, SMAD proteins become phosphorylated and heterodimerize with Smad4 to transmit upstream signals to the nucleus and transactivate transcription of specific target genes [44]. Loss of SMAD4/DPC4 interferes with the intracellular signaling cascades downstream from TGF-β and activin, resulting in decreased growth inhibition via loss of proapoptotic signaling or inappropriate G1/S transition [43, 45]. The SMAD4 gene is remarkable for two reasons. First, inactivation of the DPC4 gene is relatively specific to pancreatic cancer, although it occurs with low incidence in other cancers, such as colon, breast, and ovarian or biliary tract carcinomas [46, 47]. Secondly, immunohistochemical labeling for Smad4 protein expression mirrors DPC4/SMAD4 gene status in pancreatic cancers with rare exceptions [42]. Inactivation of DPC4/SMAD4 is uncommon in nonductal neoplasms of the pancreas [10], and is rare in most extrapancreatic malignancies [10, 46]. Therefore, immunolabeling for loss of Smad4 is a convenient ancillary diagnostic marker in clinical specimens, including suspected metastases from an occult pancreatic primary.
Many other tumor suppressor genes that are targeted at low frequency in pancreatic cancer (<10%) deserve mentioning. Mutations in the LKB1/STK11 gene are the cause of the autosomal-dominant inherited Peutz-Jeghers syndrome. Patients with Peutz-Jeghers syndrome have an increased risk of pancreatic cancer and it is conceivable that LKB1 acts as tumor suppressor gene in pancreatic cancer as well [48, 49]. Intragenic mutations and homozygous deletions of the MKK4 gene occur in a small percentage of pancreatic cancers [50]. The MKK4 gene encodes for a component of a stress-activated protein kinase cascade and has a function in apoptosis and growth control. Furthermore, MKK4 is preferentially inactivated in subsets of pancreatic cancer metastases, suggesting that the protein product may function as a metastasis suppressor [51]. Other less frequently affected tumor suppressor genes include the TGF-β/activin signaling pathway receptors such as the TGF-β type I receptor (TGFβR1; ALK5; chromosome 9q), TGFβR2 (chromosome 3p), ACVR1β (ALK4; chromosome 12q) [52] and ACVR2 (chromosome 2q) [53, 54]. The TGFβR1 ALK5 forms a heterodimer with the TGF-β type II receptor (TGFβR2) to mediate signaling of TGF-β ligands. A downstream component of this pathway includes DPC4 (SMAD4). Signaling initiatedafter binding of TGF-β-related ligands to their cognate receptors leads to heteromerization and nuclear translocation of the Smad proteins and the transcriptional activation of target genes [55, 56]. TGF-β is a pleiotropic factor that regulates cell proliferation, angiogenesis, metastasis, and immune suppression. The involvement of the TGF-β pathway has been established in cancers of many organs including the breast, lung, colon and pancreas. TGF-β signaling is frequently attenuated in pancreatic cancer because of alterations in the components of the pathway [57, 58].
Oncogenes
Oncogenes are genes that contribute to oncogenesis when mutationally activated. In contrast to tumor suppressor genes they act in a dominant fashion, i.e. mutation of one copy of the gene suffices for activation. Oncogenes can be activated through a variety of mechanisms, including point mutations within the gene and amplification of the gene itself. A growing number of oncogenes have been identified that are targeted in pancreatic cancer.
The most common activating point mutation involves the KRAS2 oncogene, on chromosome 12p, in over 90% of pancreatic ductal adenocarcinomas [59, 60](table 1. This is the highest fraction of K-ras alteration found in any human tumor type. Frequent mutation sites involve codons 12, 13 and 61, but in pancreatic ductal cancers the majority occur in codon 12. The KRAS gene product mediates signals from growth factor receptors and other signal inputs. Mutation of KRAS results in a constitutive gain of function, because the RAS protein remains trapped in the activated state even in the absence of growth factor signals, which leads to proliferation, suppressed apoptosis and cell survival.
The RAS family proteins encode small GTP-binding cytoplasmic proteins [44]. The constitutively active RAS intrinsically binds to GTP and confers uncontrolled stimulatory signals to downstream cascades including Ras effectors. Activated KRAS engages multiple effector pathways, notably the RAF-mitogen-activated protein kinase, phosphoinositide-3-kinase (PI3K) and RalGDS pathways.
Mutant KRAS has been extensively investigated as a marker of pancreatic cancer because mutations are basically entirely limited to one codon, can be readily detected using molecular assays and are present in approximately 90% of pancreatic ductal adenocarcinomas. Unfortunately, KRAS mutations are not specific for invasive pancreatic cancer and they occur in patients with chronic pancreatitis, in individuals who smoke, and in situ neoplasias from patients without pancreatic cancer [61, 62].
The BRAF gene on chromosome 7q is a member of the RAS-RAF-MEK-ERK-MAP kinase pathway, and is mutated in one-third of the pancreatic cancers with wild-type (normal) KRAS[63]. BRAF, a serine/threonine kinase located immediately downstream in RAS signaling, is a frequent mutational target in several cell lines and nonpancreatic primary cancers including 66% of melanomas and 10% of colorectal carcinomas [64, 65]. Interestingly, KRAS and BRAF mutations are mutually exclusive and tumors with mutant forms of one of these 2 genes invariably retain wild-type copies of the other. The requirement of oncogenic KRAS or BRAF pathway-related signal transduction appears to be critically important for most instances of pancreatic ductal carcinogenesis.
The PI3K-kinase-AKT pathway is a key effector of RAS-dependent transformation of many cell types and plays a role in cell survival, cell proliferation and other growth-related processes [66]. Activated PI3K results in phosphorylated phosphatidylinositides (PIP3), a step inhibited by product of the tumor suppressor gene, PTEN. PIP3 in turn phosphorylates and activates AKT[29]. Recently activating mutations of PIK3CA, the gene encoding PI3K, have been reported in a subset of pancreatic cancer precursors, specifically in IPMNs [67]. Even in the absence of mutations, the PI3K/AKT pathway is constitutively active in the majority of pancreatic cancers [68]. This might be due to the aberrant expression of their natural antagonist PTEN[69]. Although PTEN is not mutated in pancreatic cancers, the reduction of its expression may give pancreatic cancer cells an additional growth advantage [70]. Furthermore, amplification or activation of AKT2 kinase, a major target of the PI3K complex, occurs in up to 60% of pancreatic cancers [71,72,73,74], supporting the participation of an activated PI3K-AKT axis in this disease.
A third downstream pathway activated trough RAS is the RalGDS pathway. RalGDS is one of several known Ras-regulated guanine-nucleotide exchange factors, or GEFs, that function by activating Ral A and Ral B GTPases [75]. Recently, RAL A was shown to be activated in a variety of pancreatic cancers, and knockdown of RAL A suppressed tumorigenicity of RAS-transformed human cells [76]. In the same studies, knockdown of RAL B had no effect on tumor initiation, but suppressed tumor progression (i.e. metastases), suggesting divergent roles for the two RAL proteins in the context of pancreatic neoplasia. Whether or not these signaling moieties can be utilized as therapeutic targets remains to be determined.
The mammalian Hedgehog family of secreted signaling proteins – comprised of Sonic, Indian, and Desert Hedgehog (Shh, Ihh, and Dhh) – regulates the growth and patterning of many organs, including the pancreas, during embryogenesis [77]. The Hedgehog pathway is under negative regulation by the Patched (PTC) tumor suppressor protein that inactivates the Smoothed (SMO) protein. The Hedgehog ligands engage the PTC transmembrane protein, disrupting the inhibition of SMO and thereby enabling signaling transduction to the GLI family of transcriptional regulators [78]. Loss of PTC, activating mutations in SMO and overexpression of GLI and Hedgehog proteins are associated with a variety of cancers [79]. Activation of the Hedgehog pathway has been implicated in both the initiation of pancreatic ductal neoplasia and in the maintenance of advanced cancers [80]. The expression of the Hedgehog ligands, the transcriptional target gene PTC, and the essential pathway component SMO is undetectable in normal human pancreatic ducts. In contrast, a relative increase in the expression of these proteins is observed during pancreatic ductal tumorigenesis [78, 81, 82]. Moreover, it has been confirmed that the Hedgehog pathway plays a role in metastases. Inhibition of Hedgehog signaling has been shown to reduce the incidence of systemic metastasis in pancreatic adenocarcinoma xenografts [83]. Recently, Ji et al. [84] showed that there is a cross-talk between oncogenic KRAS and the Hedgehog signaling pathway in pancreatic cancer cell lines. Their studies suggest that oncogenic KRAS through the RAF/MEK/MAPK pathway suppresses GLI1 protein degradation and consequently plays an important role in activating Hedgehog signaling pathway in the absence of additional Hedgehog ligand during pancreatic tumorigenesis.
The Notch signaling pathway is another pathway which is important in directing cell fate and cell proliferation during embryonic development. Later in life, the Notch signaling pathway plays a critical role in maintaining the balance among cell proliferation, differentiation, and apoptosis [85]. In mammals, this signaling pathway involves interaction of the membrane-bound Notch receptors (Notch 1–4) and Notch ligands (Delta-like, and Jagged) on adjacent cells [85, 86]. The function of Notch signaling in tumorigenesis can be either oncogenic or antiproliferative, and the function is context dependent. In a limited number of tumor types, including human hepatocellular carcinoma and small cell lung cancer, Notch signaling is antiproliferative rather than oncogenic. However, most of the studies show an opposite effect of Notch in many human cancers including pancreatic cancer [87]. In the normal adult pancreas, Notch and its ligands are expressed at low levels. Interestingly, aberrant expression of its ligands, expression of mutant Notch1 oncoprotein, and abnormal expression of transcription targets of Notch signaling can be observed in early stages of pancreatic tumorigenesis as well as in invasive pancreatic cancer [88].
Several other oncogenes that are targeted in pancreatic cancer by amplifications deserve mentioning. First, the AKT2 gene on chromosome 19q is a downstream effector of the PI3K/AKT pathway, and is amplified in 10–15% of pancreatic cancers [73, 89]. AKT2 can be activated by stimuli such as platelet-derived growth factor, basic fibroblast growth factor, and insulin through the PI3K/AKT pathway, suggesting this pathway's importance in this tumor type [72]. Secondly, the AIB1 gene on chromosome 20q is amplified in approximately 60% of pancreatic cancers [90]. The nuclear receptor coactivator amplified in breast cancer 1 (AIB1/SRC-3) belongs to the p160/steroid receptor coactivator family (SRC)[91]. AIB1 amplification and/or overexpression is not only detected in hormone-sensitive tumors, such as breast, prostate and ovarian, but it is also found in nonsteroid-targeted tumors such as pancreatic cancer, colorectal carcinoma and hepatocellular carcinoma [92]. Thirdly, the MYB gene on chromosome 6q is amplified in 10% of pancreatic carcinomas [93]. Abnormalities in the locus of the human MYB gene have been observed in several human cancers. In a majority of these tumors, these abnormalities seem to be accompanied by an amplification of the MYB gene followed by enhanced transcription [94].
Genome Maintenance Genes
Genome maintenance genes are those that function to identify and repair damage to DNA. When a genome maintenance gene is inactivated, DNA damage is not repaired efficiently and DNA mutations accumulate. If these mutations occur in cancer-associated genes they can contribute to tumorigenesis [90]. Although gross chromosomal abnormalities are frequent in pancreatic ductal adenocarcinomas, genetic instability also occurs through DNA mismatch repair defects [95]. The DNA mismatch repair genes hMLH1 and hMSH2 are examples of genome maintenance genes targeted in pancreatic cancer [96]. When one of these genes is inactivated, DNA changes occur leading to ‘microsatellite instability’ (MSI). MSI is associated with poor differentiation, lack of KRAS2 and p53 mutations, and germline mutations of this gene are associated with the human nonpolyposis colorectal cancer syndrome (HNPCC) [96,97,98]. Approximately 4% of pancreatic cancers have MSI and these cancers have a specific microscopic appearance called ‘medullary type’, which includes a syncytial growth pattern, pushing borders and lymphocytic infiltrate [96].
The causative genes of Fanconi anemia, FANCC and FANCG, alsoplay a role in pancreatic tumorigenesis [99]. Fanconi anemia is a hereditary cancer susceptibility disorder, with the occurrence of hematologic abnormalities or acute myelogenous leukemia at an early stage, usually leading to death before the age of 20. Patients who survive into adulthood often develop solid tumors [99]. The BRCA2 gene represents Fanconi complementation group D1 and is thought to aid DNA strand and interstrand crosslinking repair. BRCA2 has therefore been categorized as genome maintenance gene rather than a standard tumor suppressor. In ductal pancreatic cancers 7–10% harbor an inactivating intragenic inherited mutation of one copy of the BRCA2 gene, accompanied by loss of heterozygosity [100, 101]. Of interest, it has been shown that the presence of BRCA2/Fanconi anemia gene mutations in pancreatic cancer may make them particularly sensitive to chemotherapeutic agents that cause DNA crosslinks such as Mitomycin C, because these cancers are unable to repair DNA interstrand crosslinks [102].
Growth Factors
Several of the genes known to be overexpressed in pancreatic cancer include growth factors and their receptors. Growth factors are the proteins that control cell differentiation and proliferation. Disturbances in growth inhibition and an abundance of growth-promoting factors give cancer cells a distinct growth advantage, which clinically results in rapid tumor progression. The epidermal growth factor receptor (EGFR) is overexpressed and plays a distinct role in pancreatic cancer. The four receptors of the EGF family are membrane-spanning glycoproteins composed of an amino terminal extracellular ligand-binding domain, a hydrophobic transmembrane region and a cytoplasmic domain that contains both the tyrosine kinase domain as well as the receptor [103]. The classical EGF receptor is also known as HER1 or ErbB-1. The remaining three receptors are designated HER-2/Neu (ErbB-2), HER-3 (ErbB-3), and HER-4 (ErbB-4). HER-2/neu overexpression is most prominent in well-differentiated ductal adenocarcinoma, as well as in the early-stage precursor lesions, and appears to correlate with the grade of dysplasia in the precursor lesions [104, 105]. In pancreatic cancer, HER-2/neu amplification has been observed with a variable incidence of 10–60% [106, 107]. In addition, increased levels of fibroblast growth factor (FGF), FGF-receptor, insulin-like growth factor I (IGF-I), IGF-I receptor, nerve growth factor, and vascular endothelial growth factor (VEGF) are also reported in pancreatic cancer [108, 109].
Tumor growth requires accompanying expansion of the host vasculature with tumor progression, which is often correlated with vascular density. VEGF is the best-characterized inducer of tumor angiogenesis. Interestingly, Delta-like ligand 4 (Dll4), a Notch ligand, is dynamically regulated by VEGF [110]. Several studies demonstrated that Dll4 may act downstream of VEGF as a ‘brake’ on VEGF-mediated angiogenic sprouting [111]. Dll4, a transmembrane ligand for the Notch family of receptors, is induced by VEGF as a negative feedback regulator and acts to prevent overexuberant angiogenic sprouting [112].
Telomere Shortening
Defective telomeres may be the major cause of the chromosomal instability observed in many cancers and in the vast majority of pancreatic cancers [113]. Telomeres are structures at the end of linear chromosomes that normally function to protect the terminal sequences and prevent the ends of chromosomes from joining aberrantly [114, 115]. Telomeres serve as protective ‘caps’ and are composed of short repeated DNA sequences and associated proteins. It appears that telomeres become abnormally short very early in the development of pancreatic neoplasia [114]. These shortened telomeres can presumably lead to the abnormal fusion of chromosome ends and in this fashion to chromosome instability, promoting further neoplastic progression in these cells [90]. Such a chromosome fusion leads to so-called anaphase bridges during mitosis [116]. These anaphase bridges frequently break during cellular replication, generating unstable chromosome ends that are subject to abnormal fusion events and subsequent chromosomal rearrangements [117]. This process, called breakage-fusion-bridge cycles, has been observed in pancreatic cancers and is believed to be one of the major causes underlying loss of function of tumor suppressor genes and the gain of function of oncogenes as described earlier [90]. In most instances, cells harboring this degree of genomic instability are eliminated through activation of p53. However, chromosomal rearrangements likely persist in cells with p53 mutations, and these cells will then quickly accrue further genomic alterations [118]. Thus telomere dysfunction and p53 loss cooperate to promote the development of carcinomas in multiple tissues [79]. Chromosomal instability provides a tumor with the genetic diversity to overcome certain barriers in carcinogenesis. However, ultimately, chromosomal instability might prove counterproductive to tumor growth, which may explain why neoplasms seem to acquire mechanisms to elongate their telomeres at later stages in the development of a malignancy, often through the reactivation of the enzyme telomerase, or through alternate lengthening of telomeres [119].
Familial Pancreatic Cancer
In the majority of cases, cancer is a multifactorial disorder in which genetic and environmental factors interact to initiate carcinogenesis. However, in a minority, the disease follows a familial pattern of transmission, suggesting a hereditary cancer syndrome. Characterization of the genetic mutations segregating in such families has helped to elucidate the molecular events that underlie tumorigenesis in the more common multifactorial form of the disease. Elucidation of the mechanisms of hereditary colorectal cancer and breast/ovarian cancer syndromes represents some of the greatest triumphs of the last century in the field of cancer genetics.
It has been estimated that 10% of pancreatic cancers have a familial basis [120, 121]. Having a first-degree relative with pancreatic cancer doubles the risk of developing pancreatic cancer [122], and the risk increases with increasing numbers of affected relatives [123]. Segregation analyses have suggested that an autosomal dominant pattern of inheritance is the most parsimonious genetic model for this increased risk [124], but the gene responsible for the familial aggregation of pancreatic cancer in the majority of cases has not yet been identified [125]. In different countries familial pancreatic cancer registries have been established to investigate the epidemiology and genetic background of these families, and to organize the screening programs for high-risk relatives and for follow-up. The largest such registry, the National Familial Pancreas Tumor Registry, is located at the Johns Hopkins Medical Institutions, Baltimore, Md., USA(http://pathology2.jhu.edu/pancreas/nfptr.cfm)[125].
To date, at least five hereditary disorders that significantly increase the risk of pancreatic cancer have been described. These include familial breast/ovarian cancer syndrome (caused by inherited mutations in the BRCA2 gene), the FAMM syndrome (caused by germline mutations in the p16 gene), the Peutz-Jeghers syndrome (caused by inherited mutations in the STK11/LKB1 gene), hereditary pancreatitis (caused by germline mutations in the PRSS1 gene), and hereditary HNPCC caused by mutations in hMLH1 or hMSH2.
Familial breast/ovarian cancer syndrome is associated with an increased risk of breast cancer in men and women, and a subset of these families also harbor an increased risk for pancreatic cancer [126]. Germline mutations of the BRCA2 gene, residing on 13q12–13, are identified in 4–17% of familial pancreatic cancer, with a particular propensity for occurring in families of Ashkenazi Jewish heritage [100, 127]. As mentioned earlier, the protein product of the BRCA2 gene has been shown to interact with protein products of several of the Fanconi anemia genes and to function in the repair of double-strand DNA breaks [99].
The FAMM syndrome is an autosomal dominant disorder characterized by the familial occurrence of multiple melanocytic naevi, atypical naevi, and an increased risk of both melanoma and pancreatic cancer [128, 129]. FAMM can be caused by germline mutations in the p16/CDKN2A gene on chromosome 9p. The carriers of the germline p16-Leiden mutation have an estimated risk of 17% to develop pancreatic cancer by the age of 75 years [19, 130].
The Peutz-Jeghers syndrome is a rare, autosomal dominant condition characterized by the development of hamartomatous gastrointestinal polyps, mucocutaneous pigmentation and high lifetime risk of developing cancer, affecting both gastrointestinal and extragastrointestinal sites. The lifetime risk of developing pancreatic cancer is approximately 36% [131]. In 50% of families the pathogenesis is caused by germline mutations occurring in the STK11/LKB1 gene [48, 132].
Hereditary pancreatitis is characterized by the familial occurrence of pancreatitis with an early age of onset [133]. Germline mutations in the PRSS1 gene cause an autosomal dominant form of the disease, whereas germline mutations in SPINK1 lead to an autosomal recessive pattern of inheritance. An estimated 40% of patients with familial pancreatitis will develop pancreatic cancer by the age of 70 years [134].
HNPCC has an autosomal dominant pattern of inheritance, it affects approximately 1 in 200 persons and is associated with multiple forms of cancer, most importantly colorectal, but also gastric, endometrial, and pancreatic cancer [135]. As discussed before, HNPCC is caused by mutations in one of the DNA mismatch repair genes. The group of individuals with a known predisposing familial syndrome, and with a history of familial pancreatic cancer would be among the first to benefit from screening tests for early detection of pancreatic cancer.
Mouse Models of Pancreatic Cancer
Although the pancreas was the first organ where transgenesis was attempted over two decades ago [136], the development of a mouse model that faithfully recapitulates the multistep progression of human pancreatic adenocarcinoma has been elusive. In 2003, Hingorani et al. [137] developed a mouse model of pancreatic neoplasia by conditional mis-expression of mutant KRAS in the pancreas from its endogenous promoter. The bitransgenic mice express a ‘knock-in’ KrasG12D upon Cre-mediated recombination and removal of a lox-STOP-lox allele within the Pdx1 expression domain. Pdx1 is a transcription factor that is expressed in the developing pancreas and foregut, restricting mutant KRAS expression to these organs. The Pdx1-Cre, lox-STOP-lox-KrasG12D mice develop the entire histologic compendium of murine PanIN (mPanIN) lesions observed in the cognate human disease, and a subset of mice develop invasive pancreatic carcinomas as well. Subsequent models have utilized additional cooperating mutations with Kras (for example, an oncogenic Trp53R172H allele or biallelic deletions of INK4a/Arf) – these compound transgenic mice develop metastatic pancreatic cancers with near-universal penetrance, and represent biologically relevant models of advanced pancreatic cancer in humans [138,139,140].
Several important lessons have been learnt from these newly developed mouse models of pancreatic cancer. First, these studies indicate the likely absolute requirement of mutant Kras in order to initiate pancreatic neoplasia along the mPanIN pathway, which might also explain the extremely high frequency of KRAS abnormalities in human PanIN lesions and pancreatic cancer [141]. Thus, misexpression of other oncogenes by themselves results in pancreatic ‘cancer’ in mice (for example, aberrant expression of the Hedgehog transcription factor GLI2) [82], but it is only upon coexpression with mutant Kras thatthese mice develop cancers preceded by mPanINs. Second, the expression of mutant Kras from its endogenous promoter appears to be a prerequisite as well, since earlier models of transgenic Kras expression have resulted in cancers of acinar histogenesis without mPanIN formation [142]. Third, these mouse models have helped elucidate some insights into the putative cell-of-origin of pancreatic cancer. For example, recent studies by Guerra and colleagues have demonstrated that mPanINs and adenocarcinomas can be reproduced in the pancreas of adult mice by conditional misexpression of mutant Kras to the elastase-expressing acinar/centroacinar compartment [143]; the one caveat is that the mature acinar/centroacinar compartment appears to be resistant to the oncogenic transformation unless accompanied by an ongoing injurious stimulus (i.e. chronic pancreatitis). These studies provide remarkable experimental reiteration to the long-standing epidemiological associations between chronic pancreatitis and an increased incidence of pancreatic cancer [3]. They also underscore the possibility that the moniker of ‘ductal’ adenocarcinoma might not reflect the true histogenesis of these cancers, at least in the context of murine pancreatic neoplasia. Fourth, and not the least, the development of these models have provided an unprecedented opportunity to explore preclinical diagnostic and therapeutic strategies in autochthonous models not afforded by short-term xenograft studies. For example, the cancers developing in these mice recapitulate not only the morphology of the cognate human disease, but also many of the oncogenic signaling pathways like EGFR, Notch and Hedgehog[137, 140]. Small molecule inhibitors targeted against these pathways can now be tested in the transgenic models prior to clinical trials. There is little doubt that the development of these models has fulfilled a critical lacuna on the field of pancreatic cancer research.
Molecular Biomarkers and Therapy
The gene expression patterns in pancreatic cancer have been studied using multiple platforms. A decade ago, gene expression was studied through analysis of the product of one gene at a time. Currently, gene expression patterns can be studied using technologies that assay nearly the entire genomesimultaneously. Examples of such technologies that have been applied to pancreatic cancer include serial analysis of gene expression, cDNA arrays and oligonucleotide arrays [144,145,146,147]. The protein products of differentially expressed genes have proven useful as diagnostic markers in tissue biopsies, as serum markers, and as therapeutic targets. For example, prostate stem cell antigen and mesothelin were identified to be overexpressed in the majority of pancreatic cancers by serial analysis of gene expression, and immunolabeling for these two proteins can be used to aid in the interpretation of challenging pancreatic biopsies [148, 149]. Similarly, osteopontin was identified as overexpressed in pancreatic carcinoma using oligonucleotide microarrays, and serum osteopontin levels have a sensitivity of 80% and a specificity of 97% for pancreatic cancer [150].
Recently, micro-RNAs (miRNAs), a novel class of 18–23 nucleotide noncoding RNAs, have gained attention as another family of molecules involved in cancer development. Current evidence has illustrated that miRNAs are misexpressed in various human cancers, and further indicates that miRNAs can function as tumor suppressors (‘TSGmiRs’) or oncogenes (‘oncomiRs’) [151, 152]. Upon binding to their target RNAs, miRNAs cause posttranscriptional gene silencing by either cleaving the target mRNA or by inhibiting the translation process [153].
As several studies have highlighted, miRNA expression is deregulated in pancreatic cancer. A miRNA signature of pancreatic cancer has been elucidated, and it includes the upregulation of miR-21, miR-155, miR-221 and miR-222 [154, 155]. Moreover, Chang et al. [39] found that miR-34a is frequently lost in pancreatic cancer cell lines. These studies demonstrate that miRNAs may become useful biomarkers for pancreatic cancer diagnostics. In addition, these aberrantly expressed miRNAs might be useful as potential therapeutic targets, with the recent availability of in vivomiRNA knockdown strategies (‘antagomirs’) [156].
The revolution in our understanding of the genetics of cancer and the exploration of gene expression on a large scale has brought with it the hope that novel therapies can be developed specifically exploiting the genetic deletions and resultant absolute biochemical deficiencies present in pancreatic cancer. Two promising examples of therapies using a specific biochemical difference, including mitomycin C for pancreatic cancers harboring BRCA2 gene mutations and L-alanosine, a purine biosynthesis inhibitor, for pancreatic cancers with loss of MTAP function were already mentioned above.
The downregulation of Notch signaling could also be a novel therapeutic approach for pancreatic cancer. Numerous studies have proposed inhibition of Notch signaling as a strategy for cancer treatment, such as with the pharmacological block of γ-secretase enzyme with small molecule inhibitors, which has a striking antineoplastic effect in Notch expressing transformed cells in vitro and in xenograft models [157]. Inhibitors of γ-secretase prevent the second ligand-induced proteolytic cleavage of the Notch receptor, thereby blocking the Notch signaling pathway. Importantly, in pancreatic cancer cells it has been shown that downregulation of Notch1 inhibits cell growth and induces apoptosis [87]. In other compartments of the gastrointestinal tract, notably the colorectum and the esophagus, regression of tumorigenesis is observed after chemical inhibition of Notch [158, 159].
Furthermore, developmental signaling pathways, like the Hedgehog signaling pathway, have emerged as therapeutic targets in pancreatic cancers [160]. This pathway is aberrantly activated in the majority of pancreatic ductal adenocarcinomas [78]. Drugs such as cyclopamine which specifically inhibit the Hedgehog pathway have been shown to be effective in xenograft models of human pancreatic cancer in treated mice [81]. Interestingly, the realization of cross-talk between RAS/MAPK and Hedgehog signaling pathways in pancreatic carcinomas also suggest that targeting the RAS and Hedgehog pathways synergistically may represent a new therapeutic strategy [84]. Additionally, there are a few promising agents on the therapeutic horizon, being tested in clinical trials, like bevacizumab, the monoclonal antibody against VEGF, which targets tumor vascularization andcetuximab, the monoclonal antibody against the EGFR [161]. Of note, trastuzumab (Herceptin®) is a humanized monoclonal antibody that acts on the HER2/neu (erbB2) receptor, a member of the EGFR family, and shows profound beneficial results with breast cancer patients whose tumors overexpress this receptor [103]. Whether trastuzumab will be as effective a form of treatment in pancreatic cancer as it appears to be in breast cancer, is currently the focus of several studies [162, 163].
Future Perspectives
Intensive research over the last two decades has shown that pancreatic cancer is fundamentally a genetic disease, caused by inherited germline and/or acquired somatic mutations in cancer-associated genes. It has uncovered multiple alterations in many genes that are important in pancreatic cancer progression. In addition, an increased understanding of the molecular basis of the disease has provided the identification of new drug targets enabling rational drug design, and facilitated the production of animal models of the disease on which such therapies can be tested.
Pancreatic ductal adenocarcinoma is nevertheless still one of the most lethal cancers of all human malignancies. The poor prognosis and late presentation of pancreatic cancer patients emphasize the importance of early detection, which is the sine qua non for the fight against pancreatic cancer. It is hoped for the future that the understanding of genetic alterations in combination with the development of high-throughput sensitive techniques will lead to the rapid discovery of an effective biomarker.
Acknowledgements
This study was supported by the Sol Goldman Pancreatic Cancer Research Center, the Michael Rolfe Foundation for Pancreatic Cancer Research, NIH R01CA113669 and NIH P50CA062924.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(suppl 8):S4–S66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 3.Lowenfels AB, Maisonneuve P. Epidemiology and risk factors for pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:197–209. doi: 10.1016/j.bpg.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Villeneuve PJ, Johnson KC, Hanley AJG, Mao Y. Alcohol, tobacco and coffee consumption and the risk of pancreatic cancer: results from the Canadian Enhanced Surveillance System case-control project. Eur J Cancer Prev. 2000;9:49–58. doi: 10.1097/00008469-200002000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Michaud DS, Liu SM, Giovannucci E, Willett WC, Colditz GA, Fuchs CS. Dietary sugar, glycemic load, and pancreatic cancer risk in a prospective study. J Natl Cancer Inst. 2002;94:1293–1300. doi: 10.1093/jnci/94.17.1293. [DOI] [PubMed] [Google Scholar]
- 6.Michaud DS, Giovannucci E, Willett WC, Colditz GA, Stampfer MJ, Fuchs CS. Physical activity, obesity, height, and the risk of pancreatic cancer. JAMA. 2001;286:921–929. doi: 10.1001/jama.286.8.921. [DOI] [PubMed] [Google Scholar]
- 7.Everhart J, Wright D. Diabetes mellitus as a risk factor for pancreatic cancer – a meta-analysis. JAMA. 1995;273:1605–1609. [PubMed] [Google Scholar]
- 8.Sohn TA, Yeo CJ, Cameron JL, Koniaris L, Kaushal S, Abrams RA, Sauter PK, Coleman J, Hruban RH, Lillemoe KD. Resected adenocarcinoma of the pancreas-616 patients: results, outcomes, and prognostic indicators. J Gastrointest Surg. 2000;4:567–579. doi: 10.1016/s1091-255x(00)80105-5. [DOI] [PubMed] [Google Scholar]
- 9.Winter JM, Cameron JL, Campbell KA, Arnold MA, Chang DC, Coleman J, Hodgin MB, Sauter PK, Hruban RH, Riall TS, Schulick RD, Choti MA, Lillemoe KD, Yeo CJ. 1423 pancreaticoduodenectomies for pancreatic cancer: a single-institution experience. J Gastrointest Surg. 2006;10:1199–1210. doi: 10.1016/j.gassur.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 10.Hruban RH, Klimstra DS, Pitman MB. Tumors of the pancreas. Washington: Armed Forces Institute of Pathology; 2006. Atlas of Tumor Pathology. [Google Scholar]
- 11.Singh M, Maitra A. Precursor lesions of pancreatic cancer: molecular pathology and clinical implications. Pancreatology. 2007;7:9–19. doi: 10.1159/000101873. [DOI] [PubMed] [Google Scholar]
- 12.Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 13.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W, Baylin SB, Kern SE, Herman JG. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 14.Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 15.Liggett WH, Jr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998;16:1197–1206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- 16.Sellers WR, Rodgers JW, Kaelin WG., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartsch DK, Kress R, Sina-Frey M, Grutzmann R, Gerdes B, Pilarsky C, Heise JW, Schulte KM, Colombo-Benkmann M, Schleicher C, Witzigmann H, Pridohl O, Ghadimi MB, Horstmann O, von Bernstorff W, Jochimsen L, Schmidt J, Eisold S, Estevez-Schwarz L, Hahn SA, Schulmann K, Bock W, Gress TM, Zugel N, Breitschaft K, Prenzel K, Messmann H, Endlicher E, Schneider M, Ziegler A, Schmiegel W, Schafer H, Rothmund M, Rieder H. Prevalence of familial pancreatic cancer in Germany. Int J Cancer. 2004;110:902–906. doi: 10.1002/ijc.20210. [DOI] [PubMed] [Google Scholar]
- 18.de vos tot Nederveen Cappel WH, Lagendijk MA, Lamers CB, Morreau H, Vasen HF. Surveillance for familial pancreatic cancer. Scand J Gastroenterol Suppl. 2003;329:94–99. doi: 10.1080/00855920310002762. [DOI] [PubMed] [Google Scholar]
- 19.de vos tot Nederveen Cappel WH, Offerhaus GJ, van Puijenbroek M, Caspers E, Gruis NA, De Snoo FA, Lamers CB, Griffioen G, Bergman W, Vasen HF, Morreau H. Pancreatic carcinoma in carriers of a specific 19 base pair deletion of CDKN2A/p16 (p16-Leiden) Clin Cancer Res. 2003;9:3598–3605. [PubMed] [Google Scholar]
- 20.Chen ZH, Zhang H, Savarese TM. Gene deletion chemoselectivity: codeletion of the genes for p16(INK4), methylthioadenosine phosphorylase, and the alpha- and beta-interferons in human pancreatic cell carcinoma lines and its implications for chemotherapy. Cancer Res. 1996;56:1083–1090. [PubMed] [Google Scholar]
- 21.Zhang H, Chen ZH, Savarese TM. Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-alpha1, interferon-beta1, and other 9p21 markers in human malignant cell lines. Cancer Genet Cytogenet. 1996;86:22–28. doi: 10.1016/0165-4608(95)00157-3. [DOI] [PubMed] [Google Scholar]
- 22.Hustinx SR, Hruban RH, Leoni LM, Iacobuzio-Donahue C, Cameron JL, Yeo CJ, Brown PN, Argani P, Ashfaq R, Fukushima N, Goggins M, Kern SE, Maitra A. Homozygous deletion of the MTAP gene in invasive adenocarcinoma of the pancreas and in periampullary cancer: a potential new target for therapy. Cancer Biol Ther. 2005;4:83–86. doi: 10.4161/cbt.4.1.1380. [DOI] [PubMed] [Google Scholar]
- 23.Illei PB, Rusch VW, Zakowski MF, Ladanyi M. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin Cancer Res. 2003;9:2108–2113. [PubMed] [Google Scholar]
- 24.Kirsch DG, Kastan MB. Tumor-suppressor p53: implications for tumor development and prognosis. J Clin Oncol. 1998;16:3158–3168. doi: 10.1200/JCO.1998.16.9.3158. [DOI] [PubMed] [Google Scholar]
- 25.Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, Kern SE. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025–3033. [PubMed] [Google Scholar]
- 26.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 27.Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3 Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 28.Weiss RH, Marshall D, Howard L, Corbacho AM, Cheung AT, Sawai ET. Suppression of breast cancer growth and angiogenesis by an antisense oligodeoxynucleotide to p21 (Waf1/Cip1) Cancer Lett. 2003;189:39–48. doi: 10.1016/s0304-3835(02)00495-0. [DOI] [PubMed] [Google Scholar]
- 29.Doucas H, Garcea G, Neal CP, Manson MM, Berry DP. Chemoprevention of pancreatic cancer: a review of the molecular pathways involved, and evidence for the potential for chemoprevention. Pancreatology. 2006;6:429–439. doi: 10.1159/000094560. [DOI] [PubMed] [Google Scholar]
- 30.Ahrendt SA, Brown HM, Komorowski RA, Zhu YR, Wilson SD, Erickson BA, Ritch PS, Pitt HA, Demeure MJ. p21(WAF1) expression is associated with improved survival after adjuvant chemoradiation for pancreatic cancer. Surgery. 2000;128:520–528. doi: 10.1067/msy.2000.108052. [DOI] [PubMed] [Google Scholar]
- 31.Song MM, Nio Y, Dong M, Tamura K, Furuse K, Tian YL, He SG, Shen K. Comparison of K-ras point mutations at codon 12 and p21 expression in pancreatic cancer between Japanese and Chinese patients. J Surg Oncol. 2000;75:176–185. doi: 10.1002/1096-9098(200011)75:3<176::aid-jso5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 32.Garcea G, Neal CP, Pattenden CJ, Steward WP, Berry DP. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur J Cancer. 2005;41:2213–2236. doi: 10.1016/j.ejca.2005.04.044. [DOI] [PubMed] [Google Scholar]
- 33.Harada N, Gansauge S, Gansauge F, Gause H, Shimoyama S, Imaizumi T, Mattfeld T, Schoenberg MH, Beger HG. Nuclear accumulation of p53 correlates significantly with clinical features and inversely with the expression of the cyclin-dependent kinase inhibitor p21(WAF1/CIP1) in pancreatic cancer. Br J Cancer. 1997;76:299–305. doi: 10.1038/bjc.1997.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 35.Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A. p53- and drug-induced apoptotic responses mediated by BH3-only proteins Puma and Noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 36.Yu J, Zhang L. The transcriptional targets of p53 in apoptosis control. Biochem Biophys Res Commun. 2005;331:851–858. doi: 10.1016/j.bbrc.2005.03.189. [DOI] [PubMed] [Google Scholar]
- 37.He L, He XY, Lim LP, De Stanchina E, Xuan ZY, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen CF, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel Sequencing – miR-34a is a p53 target that induces apoptosis and G(1)-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 39.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 41.Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, Kern SE, Hruban RH. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–2006. [PubMed] [Google Scholar]
- 42.Wilentz RE, Su GH, Dai JL, Sparks AB, Argani P, Sohn TA, Yeo CJ, Kern SE, Hruban RH. Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas : a new marker of DPC4 inactivation. Am J Pathol. 2000;156:37–43. doi: 10.1016/S0002-9440(10)64703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 44.Schneider G, Schmid RM. Genetic alterations in pancreatic carcinoma. Mol Cancer. 2003;2:15. doi: 10.1186/1476-4598-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 46.Schutte M, Hruban RH, Hedrick L, Cho KR, Nadasdy GM, Weinstein CL, Bova GS, Isaacs WB, Cairns P, Nawroz H, Sidransky D, Casero RA, Jr, Meltzer PS, Hahn SA, Kern SE. DPC4 gene in various tumor types. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 47.Hahn SA, Bartsch D, Schroers A, Galehdari H, Becker M, Ramaswamy A, Schwarte-Waldhoff I, Maschek H, Schmiegel W. Mutations of the DPC4/Smad4 gene in biliary tract carcinoma. Cancer Res. 1998;58:1124–1126. [PubMed] [Google Scholar]
- 48.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Höglund P, Järvinen H, Kristo P, Pelin K, Ridanpää M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 49.Hemminki A, Tomlinson I, Markie D, Järvinen H, Sistonen P, Björkqvist AM, Knuutila S, Salovaara R, Bodmer W, Shibata D, de la Chapelle A, Aaltonen LA. Localization of a susceptibility locus for Peutz-Jeghers syndrome to 19p using comparative genomic hybridization and targeted linkage analysis. Nat Genet. 1997;15:87–90. doi: 10.1038/ng0197-87. [DOI] [PubMed] [Google Scholar]
- 50.Su GH, Hilgers W, Shekher MC, Tang DJ, Yeo CJ, Hruban RH, Kern SE. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 1998;58:2339–2342. [PubMed] [Google Scholar]
- 51.Xin W, Yun KJ, Ricci F, Zahurak M, Qiu W, Su GH, Yeo CJ, Hruban RH, Kern SE, Iacobuzio-Donahue CA. MAP2K4/MKK4 expression in pancreatic cancer: genetic validation of immunohistochemistry and relationship to disease course. Clin Cancer Res. 2004;10:8516–8520. doi: 10.1158/1078-0432.CCR-04-0885. [DOI] [PubMed] [Google Scholar]
- 52.Su GH, Bansal R, Murphy KM, Montgomery E, Yeo CJ, Hruban RH, Kern SE. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc Natl Acad Sci USA. 2001;98:3254–3257. doi: 10.1073/pnas.051484398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK, Yeo CJ, Hruban RH, Kern SE. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63:994–999. [PubMed] [Google Scholar]
- 54.Hruban RH, Iacobuzio-Donahue C, Wilentz RE, Goggins M, Kern SE. Molecular pathology of pancreatic cancer. Cancer J. 2001;7:251–258. [PubMed] [Google Scholar]
- 55.Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58:5329–5332. [PubMed] [Google Scholar]
- 56.Heldin CH, Miyazono K, tenDijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 57.Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005;23:2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 58.Gaspar NJ, Li LY, Kapoun AM, Medicherla S, Reddy M, Li G, O'Young G, Quon D, Henson M, Damm DL, Muiru GT, Murphy A, Higgins LS, Chakravarty S, Wong DH. Inhibition of transforming growth factor beta signaling reduces pancreatic adenocarcinoma growth and invasiveness. Mol Pharmacol. 2007;72:152–161. doi: 10.1124/mol.106.029025. [DOI] [PubMed] [Google Scholar]
- 59.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 60.Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, Kensler TW, Bose KK, Cameron JL, Bos JL. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–554. [PMC free article] [PubMed] [Google Scholar]
- 61.Tada M, Omata M, Kawai S, Saisho H, Ohto M, Saiki RK, Sninsky JJ. Detection of Ras gene mutations in pancreatic juice and peripheral blood of patients with pancreatic adenocarcinoma. Cancer Res. 1993;53:2472–2474. [PubMed] [Google Scholar]
- 62.Jimeno A, Hidalgo M. Molecular biomarkers: their increasing role in the diagnosis, characterization, and therapy guidance in pancreatic cancer. Mol Cancer Ther. 2006;5:787–796. doi: 10.1158/1535-7163.MCT-06-0005. [DOI] [PubMed] [Google Scholar]
- 63.Calhoun ES, Jones JB, Ashfaq R, Adsay V, Baker SJ, Valentine V, Hempen PM, Hilgers W, Yeo CJ, Hruban RH, Kern SE. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol. 2003;163:1255–1260. doi: 10.1016/S0002-9440(10)63485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 65.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 66.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 67.Schonleben F, Qiu WL, Ciau NT, Ho DJ, Li XJ, Allendorf JD, Remotti HE, Su GH. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin Cancer Res. 2006;12:3851–3855. doi: 10.1158/1078-0432.CCR-06-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reichert M, Saur D, Hamacher R, Schmid RM, Schneider G. Phosphoinositide-3-kinase signaling controls S-phase kinase-associated protein 2 transcription via E2F1 in pancreatic ductal adenocarcinoma cells. Cancer Res. 2007;67:4149–4156. doi: 10.1158/0008-5472.CAN-06-4484. [DOI] [PubMed] [Google Scholar]
- 69.Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL, Reddy SA. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23:8571–8580. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- 70.Ebert MP, Fei G, Schandl L, Mawrin C, Dietzmann K, Herrera P, Friess H, Gress TM, Malfertheiner P. Reduced PTEN expression in the pancreas overexpressing transforming growth factor-beta 1. Br J Cancer. 2002;86:257–262. doi: 10.1038/sj.bjc.6600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altomare DA, Tanno S, De Rienzo A, Klein-Szanto AJ, Tanno S, Skele KL, Hoffman JP, Testa JR. Frequent activation of AKT2 kinase in human pancreatic carcinomas. J Cell Biochem. 2003;88:470–476. doi: 10.1002/jcb.10287. [DOI] [PubMed] [Google Scholar]
- 72.Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, Watson DK, Testa JR. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruggeri BA, Huang L, Wood M, Cheng JQ, Testa JR. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol Carcinog. 1998;21:81–86. [PubMed] [Google Scholar]
- 74.Schlieman MG, Fahy BN, Ramsamooj R, Beckett L, Bold RJ. Incidence, mechanism and prognostic value of activated AKT in pancreas cancer. Br J Cancer. 2003;89:2110–2115. doi: 10.1038/sj.bjc.6601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 76.Lim KH, O'Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, Counter CM. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 77.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 78.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, Yajnik V, Antoniu B, McMahon M, Warshaw AL, Hebrok M. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, DePinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 80.Taipale J, Beachy PA. The Hedgehog and Wnt signaling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 81.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, Beachy PA. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 82.Pasca di Magliano M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, Karikari C, Alvarez H, Iacobuzio-Donahue C, Jimeno A, Gabrielson KL, Matsui W, Maitra A. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 85.Sjolund J, Manetopoulos C, Stockhausen MT, Axelson H. The Notch pathway in cancer: differentiation gone awry. Eur J Cancer. 2005;41:2620–2629. doi: 10.1016/j.ejca.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 86.Miele L, Golde T, Osborne B. Notch signaling in cancer. Curr Mol Med. 2006;6:905–918. doi: 10.2174/156652406779010830. [DOI] [PubMed] [Google Scholar]
- 87.Wang Z, Zhang Y, Li Y, Banerjee S, Liao J, Sarkar FH. Down-regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol Cancer Ther. 2006;5:483–493. doi: 10.1158/1535-7163.MCT-05-0299. [DOI] [PubMed] [Google Scholar]
- 88.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, Hruban RH, Ball DW, Schmid RM, Leach SD. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 89.Miwa W, Yasuda J, Murakami Y, Yashima K, Sugano K, Sekine T, Kono A, Egawa S, Yamaguchi K, Hayashizaki Y, Sekiya T. Isolation of DNA sequences amplified at chromosome 19q13.1-q13.2 including the AKT2 locus in human pancreatic cancer. Biochem Biophys Res Commun. 1996;225:968–974. doi: 10.1006/bbrc.1996.1280. [DOI] [PubMed] [Google Scholar]
- 90.Maitra A, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:211–226. doi: 10.1016/j.bpg.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 91.Henke RT, Haddad BR, Kim SE, Rone JD, Mani A, Jessup JM, Wellstein A, Maitra A, Riegel AT. Overexpression of the nuclear receptor coactivator AIB1 (SRC-3) during progression of pancreatic adenocarcinoma. Clin Cancer Res. 2004;10:6134–6142. doi: 10.1158/1078-0432.CCR-04-0561. [DOI] [PubMed] [Google Scholar]
- 92.Yan J, Tsai SY, Tsai MJ. SRC-3/AIB1:transcriptional coactivator in oncogenesis. Acta Pharmacol Sin. 2006;27:387–394. doi: 10.1111/j.1745-7254.2006.00315.x. [DOI] [PubMed] [Google Scholar]
- 93.Wallrapp C, Muller-Pillasch F, Solinas-Toldo S, Lichter P, Friess H, Buchler M, Fink T, Adler G, Gress TM. Characterization of a high copy number amplification at 6q24 in pancreatic cancer identifies c-myb as a candidate oncogene. Cancer Res. 1997;57:3135–3139. [PubMed] [Google Scholar]
- 94.Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 95.Han HJ, Yanagisawa A, Kato Y, Park JG, Nakamura Y. Genetic instability in pancreatic cancer and poorly differentiated type of gastric cancer. Cancer Res. 1993;53:5087–5089. [PubMed] [Google Scholar]
- 96.Goggins M, Offerhaus GJ, Hilgers W, Griffin CA, Shekher M, Tang D, Sohn TA, Yeo CJ, Kern SE, Hruban RH. Pancreatic adenocarcinomas with DNA replication errors (RER+) are associated with wild-type K-ras and characteristic histopathology. Poor differentiation, a syncytial growth pattern, and pushing borders suggest RER+ Am J Pathol. 1998;152:1501–1507. [PMC free article] [PubMed] [Google Scholar]
- 97.Wilentz RE, Goggins M, Redston M, Marcus VA, Adsay NV, Sohn TA, Kadkol SS, Yeo CJ, Choti M, Zahurak M, Johnson K, Tascilar M, Offerhaus GJ, Hruban RH, Kern SE. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: a newly described and characterized entity. Am J Pathol. 2000;156:1641–1651. doi: 10.1016/S0002-9440(10)65035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamamoto H, Itoh F, Nakamura H, Fukushima H, Sasaki S, Perucho M, Imai K. Genetic and clinical features of human pancreatic ductal adenocarcinomas with widespread microsatellite instability. Cancer Res. 2001;61:3139–3144. [PubMed] [Google Scholar]
- 99.van der Heijden MS, Yeo CJ, Hruban RH, Kern SE. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–2588. [PubMed] [Google Scholar]
- 100.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH, Kern SE. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 101.Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, Korte B, Gerdes B, Kress R, Ziegler A, Raeburn JA, Campra D, Grutzmann R, Rehder H, Rothmund M, Schmiegel W, Neoptolemos JP, Bartsch DK. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 102.van der Heijden MS, Brody JR, Dezentje DA, Gallmeier E, Cunningham SC, Swartz MJ, DeMarzo AM, Offerhaus GJ, Isacoff WH, Hruban RH, Kern SE. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res. 2005;11:7508–7515. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
- 103.Cohenuram M, Saif MW. Epidermal growth factor receptor inhibition strategies in pancreatic cancer: past, present and the future. JOP. 2007;8:4–15. [PubMed] [Google Scholar]
- 104.Day JD, Digiuseppe JA, Yeo C, Lai-Goldman M, Anderson SM, Goodman SN, Kern SE, Hruban RH. Immunohistochemical evaluation of HER-2/neu expression in pancreatic adenocarcinoma and pancreatic intraepithelial neoplasms. Hum Pathol. 1996;27:119–124. doi: 10.1016/s0046-8177(96)90364-0. [DOI] [PubMed] [Google Scholar]
- 105.Tomaszewska R, Okon K, Nowak K, Stachura J. HER-2/Neu expression as a progression marker in pancreatic intraepithelial neoplasia. Pol J Pathol. 1998;49:83–92. [PubMed] [Google Scholar]
- 106.Stoecklein NH, Luebke AM, Erbersdobler A, Knoefel WT, Schraut W, Verde PE, Stern F, Scheunemann P, Peiper M, Eisenberger CF, Izbicki JR, Klein CA, Hosch SB. Copy number of chromosome 17 but not HER2 amplification predicts clinical outcome of patients with pancreatic ductal adenocarcinoma. J Clin Oncol. 2004;22:4737–4745. doi: 10.1200/JCO.2004.05.142. [DOI] [PubMed] [Google Scholar]
- 107.Talar-Wojnarowska R, Malecka-Panas E. Molecular pathogenesis of pancreatic adenocarcinoma: potential clinical implications. Med Sci Monit. 2006;12:RA186. [PubMed] [Google Scholar]
- 108.Zhu ZW, Friess H, Wang L, Bogardus T, Korc M, Kleeff J, Buchler MW. Nerve growth factor exerts differential effects on the growth of human pancreatic cancer cells. Clin Cancer Res. 2001;7:105–112. [PubMed] [Google Scholar]
- 109.Korc M. Role of growth factors in pancreatic cancer. Surg Oncol Clin N Am. 1998;7:25–41. [PubMed] [Google Scholar]
- 110.Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 111.Suchting S, Freitas C, le Noble F, Benedito R, Bréant C, Duarte A, Eichmann A. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc Natl Acad Sci USA. 2007;104:3225–3230. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lobov IB, Renard RA, Papadopoulos N, Gale NW, Thurston G, Yancopoulos GD, Wiegand SJ. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci USA. 2007;104:3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gisselsson D, Pettersson L, Hoglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci USA. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Van Heek NT, Meeker AK, Kern SE, Yeo CJ, Lillemoe KD, Cameron JL, Offerhaus GJ, Hicks JL, Wilentz RE, Goggins MG, De Marzo AM, Hruban RH, Maitra A. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meeker AK, Hicks JL, Platz EA, March GE, Bennett CJ, Delannoy MJ, De Marzo AM. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res. 2002;62:6405–6409. [PubMed] [Google Scholar]
- 116.Gisselsson D, Jonson T, Petersén A, Strömbeck B, Dal Cin P, Höglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O'Hagan RC, Chang S, Maser RS, Mohan R, Artandi SE, Chin L, DePinho RA. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- 118.Meeker AK, De Marzo AM. Recent advances in telomere biology: implications for human cancer. Curr Opin Oncol. 2004;16:32–38. doi: 10.1097/00001622-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 119.Von Hoff DD, Evans DB, Hruban RH. Pancreatic Cancer. Sudbury: Jones and Bartiett; 2005. pp. 31–43. [Google Scholar]
- 120.Klein AP, Hruban RH, Brune KA, Petersen GM, Goggins M. Familial pancreatic cancer. Cancer J. 2001;7:266–273. [PubMed] [Google Scholar]
- 121.Lynch HT, Smyrk T, Kern SE, Hruban RH, Lightdale CJ, Lemon SJ, Lynch JF, Fusaro LR, Fusaro RM, Ghadirian P. Familial pancreatic cancer: a review. Semin Oncol. 1996;23:251–275. [PubMed] [Google Scholar]
- 122.Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Arnason S, Gulcher JR, Bjornsson J, Kong A, Thorsteinsdottir U, Stefansson K. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, Offerhaus GJ, Griffin C, Cameron JL, Yeo CJ, Kern S, Hruban RH. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–2638. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 124.Klein AP, Beaty TH, Bailey-Wilson JE, Brune KA, Hruban RH, Petersen GM. Evidence for a major gene influencing risk of pancreatic cancer. Genet Epidemiol. 2002;23:133–149. doi: 10.1002/gepi.1102. [DOI] [PubMed] [Google Scholar]
- 125.Brune K, Abe T, Canto M, O'Malley L, Klein AP, Maitra A, Volkan AN, Fishman EK, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Multifocal neoplastic precursor lesions associated with lobular atrophy of the pancreas in patients having a strong family history of pancreatic cancer. Am J Surg Pathol. 2006;30:1067–1076. [PMC free article] [PubMed] [Google Scholar]
- 126.Berman DB, Costalas J, Schultz DC, Grana G, Daly M, Godwin AK. A common mutation in BRCA2 that predisposes to a variety of cancers is found in both Jewish Ashkenazi and non-Jewish individuals. Cancer Res. 1996;56:3409–3414. [PubMed] [Google Scholar]
- 127.Schutte M, da Costa LT, Hahn SA, Moskaluk C, Hoque AT, Rozenblum E, Weinstein CL, Bittner M, Meltzer PS, Trent JM. Identification by representational difference analysis of a homozygous deletion in pancreatic carcinoma that lies within the BRCA2 region. Proc Natl Acad Sci USA. 1995;92:5950–5954. doi: 10.1073/pnas.92.13.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lal G, Liu G, Schmocker B, Kaurah P, Ozcelik H, Narod SA, Redston M, Gallinger S. Inherited predisposition to pancreatic adenocarcinoma: role of family history and germ-line p16, BRCA1, and BRCA2 mutations. Cancer Res. 2000;60:409–416. [PubMed] [Google Scholar]
- 129.Parker JF, Florell SR, Alexander A, DiSario JA, Shami PJ, Leachman SA. Pancreatic carcinoma surveillance in patients with familial melanoma. Arch Dermatol. 2003;139:1019–1025. doi: 10.1001/archderm.139.8.1019. [DOI] [PubMed] [Google Scholar]
- 130.Vasen HF, Gruis NA, Frants RR, van Der Velden PA, Hille ET, Bergman W. Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden) Int J Cancer. 2000;87:809–811. [PubMed] [Google Scholar]
- 131.Giardiello FM, Brensinger JD, Tersmette AC, Goodman SN, Petersen GM, Booker SV, Cruz-Correa M, Offerhaus JA. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000;119:1447–1453. doi: 10.1053/gast.2000.20228. [DOI] [PubMed] [Google Scholar]
- 132.Lim W, Hearle N, Shah B, Murday V, Hodgson SV, Lucassen A, Eccles D, Talbot I, Neale K, Lim AG, O'Donohue J, Donaldson A, Macdonald RC, Young ID, Robinson MH, Lee PW, Stoodley BJ, Tomlinson I, Alderson D, Holbrook AG, Vyas S, Swarbrick ET, Lewis AA, Phillips RK, Houlston RS. Further observations on LKB1/STK11 status and cancer risk in Peutz-Jeghers syndrome. Br J Cancer. 2003;89:308–313. doi: 10.1038/sj.bjc.6601030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Finch MD, Howes N, Ellis I, Mountford R, Sutton R, Raraty M, Neoptolemos JP. Hereditary pancreatitis and familial pancreatic cancer. Digestion. 1997;58:564–569. doi: 10.1159/000201502. [DOI] [PubMed] [Google Scholar]
- 134.Lowenfels AB, Maisonneuve P, Dimagno EP, Elitsur Y, Gates LK, Jr, Perrault J, Whitcomb DC. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst. 1997;89:442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 135.Lynch HT, Smyrk TC, Watson P, Lanspa SJ, Lynch JF, Lynch PM, Cavalieri RJ, Boland CR. Genetics, natural history, tumor spectrum, and pathology of hereditary nonpolyposis colorectal cancer: an updated review. Gastroenterology. 1993;104:1535–1549. doi: 10.1016/0016-5085(93)90368-m. [DOI] [PubMed] [Google Scholar]
- 136.Ornitz DM, Palmiter RD, Hammer RE, Brinster RL, Swift GH, Macdonald RJ. Specific expression of an elastase human growth-hormone fusion gene in pancreatic acinar-cells of transgenic mice. Nature. 1985;313:600–602. doi: 10.1038/313600a0. [DOI] [PubMed] [Google Scholar]
- 137.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 138.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, Hanahan D, Redston MS, Chin L, DePinho RA. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 141.Deramaudt T, Rustgi AK. Mutant KRAS in the initiation of pancreatic cancer. Biochim Biophys Acta. 2005;1756:97–101. doi: 10.1016/j.bbcan.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 142.Grippo PJ, Nowlin PS, Demeure MJ, Longnecker DS, Sandgren EP. Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 2003;63:2016–2019. [PubMed] [Google Scholar]
- 143.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, Dubus P, Sandgren EP, Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 144.Hustinx SR, Cao D, Maitra A, Sato N, Martin ST, Sudhir D, Iacobuzio-Donahue C, Cameron JL, Yeo CJ, Kern SE, Goggins M, Mollenhauer J, Pandey A, Hruban RH. Differentially expressed genes in pancreatic ductal adenocarcinomas identified through serial analysis of gene expression. Cancer Biol Ther. 2004;3:1254–1261. doi: 10.4161/cbt.3.12.1238. [DOI] [PubMed] [Google Scholar]
- 145.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–8622. [PubMed] [Google Scholar]
- 146.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, Van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, Wilentz RE, Murugesan SR, Leach SD, Jaffee E, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE) Clin Cancer Res. 2001;7:3862–3868. [PubMed] [Google Scholar]
- 149.McCarthy DM, Maitra A, Argani P, Rader AE, Faigel DO, Van Heek NT, Hruban RH, Wilentz RE. Novel markers of pancreatic adenocarcinoma in fine-needle aspiration: mesothelin and prostate stem cell antigen labeling increases accuracy in cytologically borderline cases. Appl Immunohistochem Mol Morphol. 2003;11:238–243. doi: 10.1097/00129039-200309000-00006. [DOI] [PubMed] [Google Scholar]
- 150.Koopmann J, Fedarko NS, Jain A, Maitra A, Iacobuzio-Donahue C, Rahman A, Hruban RH, Yeo CJ, Goggins M. Evaluation of osteopontin as biomarker for pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2004;13:487–491. [PubMed] [Google Scholar]
- 151.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nature Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 152.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 153.Valencia-Sanchez MA, Liu JD, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 154.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 155.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 157.Kimura K, Satoh K, Kanno A, Hamada S, Hirota M, Endoh M, Masamune A, Shimosegawa T. Activation of Notch signaling in tumorigenesis of experimental pancreatic cancer induced by dimethylbenzanthracene in mice. Cancer Sci. 2007;98:155–162. doi: 10.1111/j.1349-7006.2006.00369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 159.van Es JH, Clevers H. Notch and Wnt inhibitors as potential new drugs for intestinal neoplastic disease. Trends Mol Med. 2005;11:496–502. doi: 10.1016/j.molmed.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 160.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 161.Von Hoff DD. What's new in pancreatic cancer treatment pipeline? Best Pract Res Clin Gastroenterol. 2006;20:315–326. doi: 10.1016/j.bpg.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 162.Safran H, Iannitti D, Ramanathan R, Schwartz JD, Steinhoff M, Nauman C, Hesketh P, Rathore R, Wolff R, Tantravahi U, Hughes TM, Maia C, Pasquariello T, Goldstein L, King T, Tsai JY, Kennedy T. Herceptin and gemcitabine for metastatic pancreatic cancers that overexpress HER-2/neu. Cancer Invest. 2004;22:706–712. doi: 10.1081/cnv-200032974. [DOI] [PubMed] [Google Scholar]
- 163.Kimura K, Sawada T, Komatsu M, Inoue M, Muguruma K, Nishihara T, Yamashita Y, Yamada N, Ohira M, Hirakawa K. Antitumor effect of trastuzumab for pancreatic cancer with high HER-2 expression and enhancement of effect by combined therapy with gemcitabine. Clin Cancer Res. 2006;12:4925–4932. doi: 10.1158/1078-0432.CCR-06-0544. [DOI] [PubMed] [Google Scholar]
- 164.Maitra A, Adsay NV, Argani P, Iacobuzio-Donahue C, De Marzo A, Cameron JL, Yeo CJ, Hruban RH. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16:902–912. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]