Abstract
Mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS), one of the most common mitochondrial multisystemic diseases, is most commonly associated with an A-to-G transition at nucleotide position 3243 (A3243G) in mitochondrial DNA. We studied 34 individuals harboring the A3243G mutation for up to 7 years; 17 had the full MELAS phenotype and 17 who were classified as “carrier relatives” because they were either asymptomatic or had some symptoms suggestive of mitochondrial disease but no seizures or strokes. Using the sensitive real-time polymerase chain reaction to quantify the A3234G mutation, we confirmed that the percent mutation decreases progressively in DNA isolated from blood: the average percent decrease was 0.5% per year for fully symptomatic patients and 0.2% per year for oligosymptomatic carrier relatives. We also correlated mutant loads with functional status estimated by the Karnofsky score: even though the mutation load decreases, the level of functioning worsens in fully symptomatic patients, whereas the level of functioning of carrier relatives remains largely unchanged. This study suggests that A3243G mutant load in DNA isolated from blood is neither useful for prognosis nor for functional assessment.
Keywords: mitochondrial DNA mutation, MELAS, A3243G, heteroplasmy, mutant load
INTRODUCTION
Mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) is one of the most common clinically defined multisystemic diseases associated with mitochondrial DNA (mtDNA) mutations. Molecular genetic studies have associated the MELAS phenotype with more than 10 mutations in different mitochondrial tRNA or protein-coding genes [DiMauro and Schon, 2003; Kirby et al., 2004; Liolitsa et al., 2003; Ravn et al., 2001]. More than 80% of patients with MELAS harbor the A3243G mutation in the mitochondrial tRNALeu(UUR) gene [Goto et al., 1990] and the prevalence of this mutation has been estimated at 1 in 6000 [Majamaa et al., 1998] to 1 in 260 individuals [Manwaring et al., 2007].
Individuals with mtDNA point mutations carry both mutant and wild-type mtDNA within each cell, a condition called heteroplasmy [DiMauro and Moraes, 1993; Wallace, 1992]. In vitro studies in cultured skin fibroblasts have shown that the A3243G mutation impairs mitochondrial respiratory chain and protein synthesis when a certain threshold (about 85%) of mutant mtDNA is exceeded [Chomyn et al., 1991; King et al., 1992; Kobayashi et al., 1991]. Since the level of mutant mtDNA varies both among individuals and in different organs and tissues of single individuals, it is thought that the load of mutant mtDNA is in part responsible for the varied clinical expression of mtDNA defects in general and of the A3243G MELAS mutation in particular [Ciafaloni et al., 1991; Holt et al., 1990; Macmillan et al., 1993]. However, several studies have found only a weak correlation between mutation load in blood and clinical phenotype [Campos et al., 1995; Kobayashi et al., 1992; Liou et al., 1994; Martinuzzi et al., 1992; Shiraiwa et al., 1993]. In contrast, a good correlation between frequency of typical clinical features and level of mutant mtDNA is seen in muscle [Chinnery et al., 1997; Jeppesen et al., 2006].
Serial measurements in the same subject have shown that the percentage of the A3243G mutation in blood decreases as the patient gets older, but the underlying mechanisms remain unclear [Pyle et al., 2007; Rahman et al., 2001; t Hart et al., 1994].
We studied the changes in heteroplasmy in a large cohort of patients over a relatively long period of time, using the sensitive real-time polymerase chain reaction (PCR) approach to quantify the A3234G mutation. This technique can detect mutations at a frequency of less than 0.1% [Singh et al., 2006]. We also correlated mutant loads with functional status in both fully symptomatic patients and in carrier relatives.
MATERIALS AND METHODS
Patients
We studied 34 individuals harboring the A3243G mutation for up to seven years. Of these, 17 had the full MELAS phenotype because they had experienced focal central nervous system (CNS) events, either seizures, strokes, or both. The other 17 were classified as “carrier relatives” because they were either asymptomatic or had some symptoms suggestive of mitochondrial disease but no seizures or strokes. Of these, five were completely asymptomatic, five had gastrointestinal problems, five had exercise intolerance, four had hearing loss, and four had migraine headaches.
Karnofsky Score
We used this established instrument to evaluate daily living functional abilities. The scale rates performance in activities of daily living and independence of an individual on a semi-quantitative scale. For example, someone able to work without complaints and without evidence of disease is given a maximum score of 100. At the other end of the spectrum, someone who is moribund or with a rapidly progressing fatal process is given a score of 10. Within this range of 0-100, the investigators assign a score in increments of 10. Scores between 80 and 100 indicate ability to carry on normal activities, such as school or work; scores between 50 and 70 indicate lack of ability to carry on normal activity; and scores between 0 and 40 indicate a severe condition in which the subject cannot care for self and may be institutionalized or near death. The instrument has a descriptive statement next to each numeric level, e.g. a score of 50 describes someone who is moderately disabled, dependent, and who requires considerable assistance and frequent care [Karnofsky, 1949].
DNA Isolation
Total DNA was isolated from peripheral blood leukocytes of patients harboring the MELAS A3234G mutation using the Promega Wizard Genomic DNA purification kit (Promega Corporation, Madison, WI). The concentration of each DNA sample was measured spectrophotometrically, DNA was diluted in water to a concentration of 30 ng/μl and stored at -20°C before use.
Quantification of A3243G Heteroplasmy
Real-time PCR Technique
Real-time PCR was utilized to calculate the level of A-to-G mutation as described by Singh et al., [Singh et al., 2006]. Briefly, primers and MGB probes encompassing nucleotide 3243 in mtDNA were designed using Assay-by-Design software and were obtained from Applied Biosystems (Foster City, CA). The MGB probes were specific for A (wild-type) and G (mutant) nucleotide.
PCR was carried out using an ABI Prism 7000 sequence detection system and 96-well MicroAmp optical plates (Applied Biosystems). The reaction mixture contained 10 μl of Taqman Universal PCR MasterMix (Applied Biosystems), 0.36 μmol/L of each primer, 0.08 μmol/L of each probe, and 60 ng of DNA in a total volume of 25 μL. Real-time PCR conditions were one cycle of 2 minutes at 50°C and 10 minutes at 95°C, followed by 40 cycles of denaturation for 15 seconds at 95°C and annealing/extension for 60 seconds at 60°C. The fluorescent signal intensities were recorded and analyzed during PCR using the SDS (ver.1.0) software (Applied Biosystems).
Quantification of wild-type and mutant mtDNA was done by calculating the difference between wild-type and mutant threshold cycles (ΔΔCt). The results were normalized to a known mixture of 50% heteroplasmy. The ratio of mutant to wild-type DNA was calculated using the equation 2-ΔΔCt assuming the amplification efficiencies are the same for wild-type and mutant and close to 100% [Livak and Schmittgen, 2001; Singh et al., 2006]. Samples were analyzed in triplicates.
Statistical analyses
The percent heteroplasmy and the Karnofsky scores were calculated among fully symptomatic patients and among their relatives who were carriers of the A3243G mutation. We use a linear mixed model to predict the trend of heteroplasmy over the follow-up year, while taking within subject longitudinal correlation into account. i.e. we assume Yi,j = β0 + β1ti,j + αi + ei,j, where Yi,j is the jth heteroplasmy of the ith subject measured at the follow up year ti,j, αi is the random effect of subject, and ei,j is the error term. The model is fitted separately for fully symptomatic patients and 3243 carrier relatives, using RMLE estimation approach [Laird and Ware, 1982]. The significance of the slope β1 will be tested based on the corresponding Wald statistics.
RESULTS
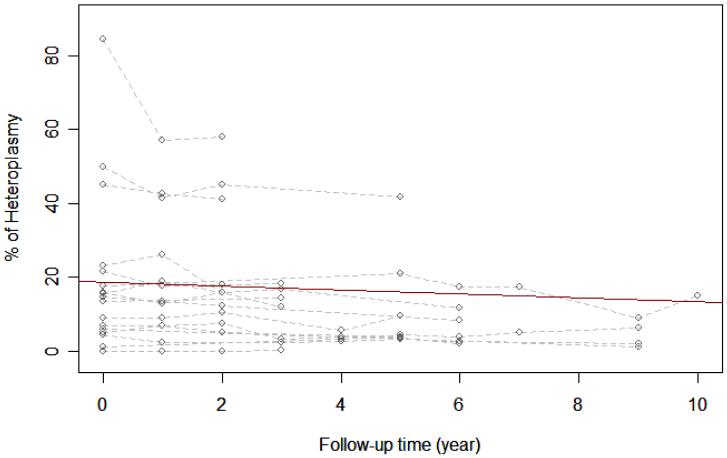
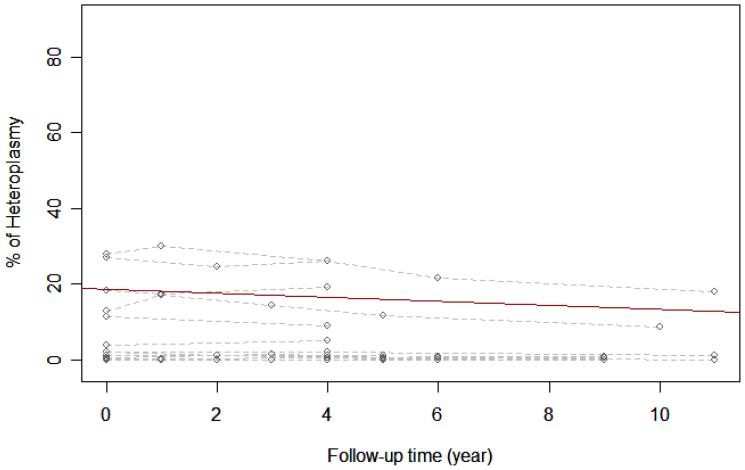
Real-time PCR was performed in DNA samples obtained from blood of 34 individuals carrying the A3234G mutation; 17 were fully symptomatic patients and 17 were their carrier relatives. Data obtained for these two groups are shown in Figure 1. Statistical analysis of data from fully symptomatic MELAS patients shows that the average percentage of heteroplasmy decreases over time (estimated slope is -0.534 with p-value of 0.0085), i.e. the mutation load in blood decreases on average by 0.534% with every follow-up year. The same statistical analysis on the carrier relatives showed that the percentage of heteroplasmy also decreases over time, 0.215% per year (estimated slope of -0.215 with p-value of 0.0011). Even though the mutation load decreases in the 2 to 7 year follow-up, the level of functioning (estimated by the Karnofsky score) of fully symptomatic patients worsens, median= -10 (Table I). However, the level of functioning of carrier relatives remains largely unchanged throughout the 2 to 6 year follow-up for this group, median=0.
Figure 1.
A. Change in heteroplasmy among fully symptomatic MELAS patients.
B. Change in heteroplasmy among carrier relatives.
The dashed lines are individual heteroplasmy paths from 17 fully symptomatic MELAS patients (Figure 1A) or 17 carrier relatives (Figure 1B). The circles are the actual measured heteroplasmy levels. The solid line indicates how the percentage of heteroplasmy changes with follow-up year on average and is estimated from a linear mixed model.
The coefficient of determination is calculated based on Nagelkerke, NJD [1991]. Coefficient of determination R2 = 0.86 (Figure 1A) and 0.91 (Figure 1B)
Table I.
Changes in Heteroplasmy and Karnofsky scores
| Change in Heteroplasmy | Change in Karnofsky Scale | Follow-up years | ||||
|---|---|---|---|---|---|---|
| Median | Range | Median | Range | Median | Range | |
| Fully Symptomatic (n=17) | -4 | -26 to +6 | -10 | -40 to +20 | 4 | 2 to 7 |
| Carrier Relatives (n=17) | 0 | -8 to +0.5 | 0 | -20 to 0 | 3 | 2 to 6 |
DISCUSSION
We analyzed the proportion of mutant genomes in DNA isolated from leukocytes in 34 patients carrying the MELAS A3243G mutation for up to 10 years and also studied the correlation between changes in heteroplasmy and levels of functioning.
Our finding of decreased mutant load in DNA isolated from leukocytes is consistent with previous reports [Pyle et al., 2007; Rahman et al., 2001; Hart et al., 1994]. In a series of eleven A3243G subjects, Pyle et al., reported that the percentage of mutant mtDNA in blood decreased progressively with an overall mean decrease of 0.6% per year (standard deviation 0.5) [Pyle et al., 2007]. In our study, we found that fully symptomatic patients experience an average 0.5% decrease (standard error 0.2%, p-value = 0.0085) in the proportion of mutant mtDNA per year whereas the decrease for their oligosymptomatic carrier relatives is 0.2% (standard error 0.06 %, p-value= 0.0011). The underlying mechanism for this decrease remains incompletely understood, but it may be associated with random segregation of mutant and wild-type mtDNA into daughter cells resulting in the loss of mutant mtDNA to the advantage of wild-type DNA in rapidly dividing tissues. On the other hand, slower dividing, postmitotic tissues, such as skeletal muscle, brain, and endocrine organs, may show accumulating levels of mutant mtDNA and are often clinically affected [Sue et al., 1998].
Our data did not show a correlation between mutant load in leukocyte DNA and a measure of clinical impairment [Karnofsky activity of daily living score]. We observed that, despite the fact that mutant load decreased, there was a progressive decrease in Karnofsky scores (median= -10) in fully symptomatic MELAS patients (Table I). While Karnofsky scores in relatives carrying the A3243G mutation appeared to remain the same over the period of follow-up, the difference between patients affected with MELAS and their relatives was not statistically significant (p value 0.1951).
Studies based on mutation analysis in muscle tissue had found a genotypephenotype correlation with regards to mutation load [Chinnery et al., 1997; Jeppesen et al., 2006; Macmillan et al., 1993]. This suggests that leukocyte DNA is not an ideal source for studies of mutation load, and that mutant load in DNA isolated from blood is neither useful for prognosis nor for functional assessment. While the most accurate way to study phenotype-genotype correlations longitudinally might be to assess heteroplasmy using real-time PCR in serial muscle biopsies, repeat muscle biopsies are obviously not feasible. Several studies have shown that mtDNA mutation loads are greater in urinary sediment than in blood [McDonnell et al., 2004; Shanske et al., 2004] and more closely resemble mutant loads in muscle. Thus, real-time PCR using DNA from urinary sediments from patients through the years may provide a better correlation between changes in heteroplasmy and functional levels in individuals harboring the 3243G mutation.
ACKNOWLEDGMENTS
This work has been supported by NIH Grant HD32062 and the Marriott Mitochondrial Disorders Clinical Research Fund (MMD-CRF).
REFERENCES
- Campos Y, Bautista J, Gutierrez-Rivas E, Chinchon D, Cabello A, Segura D, Arenas J. Clinical heterogeneity in two pedigrees with the 3243 bp tRNA(leu(UUR)) mutation of mitochondrial DNA. Acta Neurol Scand. 1995;91:62–65. doi: 10.1111/j.1600-0404.1995.tb05845.x. [DOI] [PubMed] [Google Scholar]
- Chinnery P, Howell N, Lightowlers R, Turnbull D. The relationship between mutation load and clinical phenotypes. Brain. 1997;120:1713–1721. doi: 10.1093/brain/120.10.1713. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Meola G, Bresolin N, Lai ST, Scarlato G, Attardi G. In vitro genetic transfer of protein synthesis and respiration defects to mitochondrial DNA-less cells with myopathy-patient mitochondria. Mol Cell Biol. 1991;11:2236–2244. doi: 10.1128/mcb.11.4.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafaloni E, Ricci E, Servidei S, Shanske S, Silvestri G, Manfredi G, Schon EA, DiMauro S. Widespread tissue distribution of a tRNALeu(UUR) mutation in the mitochondrial DNA of a patient with MELAS syndrome. Neurology. 1991;41:1663–1664. doi: 10.1212/wnl.41.10.1663. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Moraes CT. Mitochondrial encephalomyopathies. Arch Neurol. 1993;50:1197–1208. doi: 10.1001/archneur.1993.00540110075008. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. New Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Petty RK, Morgan Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- Jeppesen TD, Schwartz M, Frederiksen AL, Wibrand F, Olsen DB, Vissing J. Muscle phenotype and mutation load in 51 persons with the 3243A>G mtDNA mutation. Arch Neurol. 2006;63:1701–1706. doi: 10.1001/archneur.63.12.1701. [DOI] [PubMed] [Google Scholar]
- Karnofksky DA. The clinical evaluation of chemotherapeutic agents in cancer. In: MacLeod CM, editor. Evaluation of chemotherapeutic agents. Columbia University Press; New York, NY: 1949. pp. 196–206. [Google Scholar]
- King MP, Koga Y, Davidson M, Schon EA. Defects in mitochondrial protein synthesis and respiratory chain activity segragate with the tRNALeu(UUR) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. Mol Cell Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby DM, Salemi R, Sugiana C, Ohtake A, Parry L, Bell KM, Kirk EP, Boneh A, Taylor RW, Dahl H-HM, Ryan MT, Thorburn DR. NUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J Clin Invest. 2004;114:837–845. doi: 10.1172/JCI20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Momoi MY, Tominaga K, Shimoizumi H, Nihei K, Yanagisawa M, Kagawa Y, Ohta S. Respiration-deficient cells are caused by a single point mutation in the mitochondrial tRNA-Leu(UUR) gene in mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes (MELAS) Am J Hum Genet. 1991;49:590–599. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Ichihashi K, Ohta S, Kagawa Y, Yanagisawa M, Momoi MY. The mutant imtochondrial genes in mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) were selectively amplified through generations. J Inher Metab Dis. 1992;15:803–808. doi: 10.1007/BF01800025. [DOI] [PubMed] [Google Scholar]
- Laird NM, Ware JH. Random-Effects Models for Longitudinal Data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- Liolitsa D, Rahman S, Benton S, Carr LJ, Hanna MG. Is the mitochondrial complex I ND5 gene a hot-spot for MELAS causing mutations? Ann Neurol. 2003;53(1):128–32. doi: 10.1002/ana.10435. [DOI] [PubMed] [Google Scholar]
- Liou C-W, Huang CC, Chee EC, Jong YJ, Pang CY, Lee HC, Wei YH. MELAS syndrome: correlation between clinical features and molecular genetic analysis. Acta Neurol Scand. 1994;90:354–359. doi: 10.1111/j.1600-0404.1994.tb02737.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-delta.deltaCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Macmillan C, Lach B, Shoubridge EA. Variable distribution of mutant mitochondrial DNAs (tRNALeu[3243]) in tissues of symptomatic relatives with MELAS: the role of mitotic segregation. Neurology. 1993;43:1586–1590. doi: 10.1212/wnl.43.8.1586. [DOI] [PubMed] [Google Scholar]
- Majamaa K, Moilanen JS, Uimonen S, Remes AM, Salmela PI, Karppa M, Majamaa-Voltti KAM, Rusanen H, Sorri M, Peuhkurinen KJ, Hassinen IE. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes: Prevalence of the mutation in an adult population. Am J hum Genet. 1998;63:447–454. doi: 10.1086/301959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manwaring N, Jones MM, Wang JJ, Rochtchina E, Howard C, Mitchell P, Sue CM. Population prevalence of the MELAS A3243G mutation. Mitochondrion. 2007;7:230–233. doi: 10.1016/j.mito.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Martinuzzi A, Bartolomei L, Carrozzo R, Mostacciuolo M, Carbonin C, Toso V, Ciafaloni E, Shanske S, DiMauro S, Angelini C. Correlation between clinical and molecular features in two MELAS families. J Neurol Sci. 1992;113:222–229. doi: 10.1016/0022-510x(92)90250-o. [DOI] [PubMed] [Google Scholar]
- McDonnell MT, Schaefer AM, Blakely EL, McFarland R, Chinnery PF, Turnbull DM, R.W. T. Noninvasive diagnosis of the 3243A>G mitochondrial DNA mutation using urinary epithelial cells. Eur J Hum Genet. 2004;12:778–781. doi: 10.1038/sj.ejhg.5201216. [DOI] [PubMed] [Google Scholar]
- Nagelkerke NJD. A note on a general definition of the coefficient of determination. Biometrica. 1991;78:691–692. [Google Scholar]
- Pyle A, Taylor RW, Durham SE, Deschauer M, Schaefer AM, Samuels DC, Chinnery PF. Depletion of mitochondrial DNA in leucocytes harbouring the 3243A>G mtDNA mutation. Am J Med Genet. 2007;44:69–74. doi: 10.1136/jmg.2006.043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Poulton J, Marchington D, Suomalainen A. Decrease of 3243 A>G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am J Hum Genet. 2001;68:238–240. doi: 10.1086/316930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravn K, Wibrand F, Juul Hansen F, Horn N, Rosenberg T, Schwartz M. An mtDNA mutation, 144453G>A, in the NADH dehydrogenase subunit 6 associated with severe MELAS syndrome. Eur J Hum Genet. 2001;9:805–809. doi: 10.1038/sj.ejhg.5200712. [DOI] [PubMed] [Google Scholar]
- Shanske S, Pancrudo J, Kaufmann P, Engelstad K, Jhung S, Lu J, Naini A, DiMauro S, De Vivo DC. Varying loads of the mitochondrial DNA A3243G mutation in different tissues: Implications for diagnosis. Am J Med Genet. 2004;130A:134–137. doi: 10.1002/ajmg.a.30220. [DOI] [PubMed] [Google Scholar]
- Shiraiwa N, Ishii A, Iwamoto H, Mizusawa H, Kagawa Y, Ohta S. Content of mutant mitochondrial DNA and organ dysfunction in a patient with a MELAS subgroup of mitochondrial encephalomyopathies. J Neurol, Sci. 1993;15:174–179. doi: 10.1016/0022-510x(93)90270-9. [DOI] [PubMed] [Google Scholar]
- Singh RB, Ellard S, Hattersley A, Harries LW. Rapid and sensitive real-time polymerase chain reaction method for detection and quantification of 3243A>G mitochondrial point mutation. J Mol Diagn. 2006;8:225–230. doi: 10.2353/jmoldx.2006.050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue CM, Crimmins D, Quigley A. Detection of MELAS point mutation 3243 in muscle, blood and hair follicles. JNeurolSci. 1998;161:36–39. doi: 10.1016/s0022-510x(98)00179-8. [DOI] [PubMed] [Google Scholar]
- t Hart LM, Lemkes HH, Heine RJ, Stolk RP, Feskens EJ, Jansen JJ, van der Does FE, Grobbe DE, Kromhout D, van den Ouweland JM. Prevalence of maternally inherited diabetes and deafness in diabetic populations in The Netherlands. Diabetologia. 1994;37:1169–1170. doi: 10.1007/BF00418385. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]