Abstract
Background & Aims
The peroxisome proliferator-activated receptor-γ (PPARG) is a nuclear receptor that regulates expression of mediators of lipid metabolism and the inflammatory response. There is controversy over the pro- or anti-oncogenic effects of PPARG and little is known about how its expression correlates with prognosis in patients with colon cancer.
Methods
Among 470 colorectal cancer patients (stages I-IV) identified in 2 independent prospective cohorts, PPARG expression was detected in 102 tumors (22%) by immunohistochemistry. Cox proportional hazard models computed hazard ratios (HRs) of colorectal cancer-specific and overall mortalities, unadjusted and adjusted for patient characteristics and tumor molecular features, including cyclooxygenase-2 (COX-2), fatty acid synthase (FASN), KRAS, BRAF, PIK3CA, p53, p21, β-catenin, LINE-1 hypomethylation, microsatellite instability (MSI) and the CpG island methylation phenotype (CIMP).
Results
Compared to patients with PPARG-negative tumors, patients with PPARG-positive tumors had significantly lower overall mortality, determined by Kaplan-Meier analysis (p=0.0047), univariate Cox regression (HR 0.55; 95% confidence interval [CI], 0.37-0.84; p=0.0053) and multivariate analysis (adjusted HR 0.43; 95% CI, 0.27-0.69; p=0.0004). Patients with PPARG-positive tumors experienced a lower colorectal cancer-specific mortality (HR 0.65; 95% CI, 0.40-1.07; p=0.092), which became significant in multivariate analysis (adjusted HR 0.44; 95% CI, 0.25-0.79; p=0.0054). The relationship between PPARG and lower mortality did not appear to be significantly modified by tumor stage or the other clinical and molecular variables examined (all Pinteraction>0.05).
Conclusions
Tumor expression of PPARG is independently associated with longer survival of patients. PPARG expression appears to mark an indolent subset of colorectal cancers.
INTRODUCTION
PPARG (the official symbol for peroxisome proliferator-activated receptor-γ) is a member of the nuclear hormone receptor PPAR superfamily.1, 2 Ligands for PPARG include naturally occurring fatty acids and the thiazolidinedione (TZD) class of antidiabetic drugs. PPARG plays an important role in adipose cell differentiation, modulation of metabolism and inflammatory response, and cellular apoptosis.1, 3-5 PPARG interacts with and/or regulates multiple signaling pathways, including those related to p53, p21, BCL2, NF-kappa-β, STAT, cyclin D1 and cyclooxygenase-2 (COX-2).1-7 Nonetheless, with regard to the role of PPARG in cancer, debate has still continued as to whether PPARG is pro-oncogenic or anti-neoplastic.2, 8-13 In fact, the effect of PPARG is likely multifaceted and tissue-specific. Experimental studies have suggested the role of PPARG in cell cycle regulation and cellular differentiation in colonic epithelium, supporting its anti-neoplastic effect.8, 9, 13, 14 PPARG expression has been examined in human colon cancer tissue.15-17 Although two previous studies (N=86 15 and N=99 16) did not demonstrate a prognostic value of tumoral PPARG status, these studies were limited by the small sample sizes. Thus, clinical significance of PPARG expression in human colorectal cancer remains uncertain.
We therefore examined the prognostic role of PPARG expression in a large number (N=470) of stage I-IV colorectal cancer patients identified in two independent, prospective cohort studies. Since we concurrently assessed other related molecular variables [including fatty acid synthase (FASN), COX-2, p53, p21, KRAS, BRAF, PIK3CA, LINE-1 hypomethylation, microsatellite instability (MSI), and the CpG island methylator phenotype (CIMP)], we could evaluate the independent effect of PPARG on patient survival after controlling for these molecular events. In particular, it is important to control for the effect of MSI, CIMP and LINE-1 hypomethylation (and related molecular events such as KRAS, BRAF and PIK3CA mutations), because these molecular characteristics refect genomic and epigenomic status of cancer cells, and have been related with patient survival in colon cancer.18-25
MATERIALS AND METHODS
Study Population
We utilized the databases of two independent prospective cohort studies; the Nurses' Health Study (N = 121,700 women followed since 1976),26, 27 and the Health Professionals Follow-up Study (N = 51,500 men followed since 1986).27 Every 2 years, participants have been sent follow-up questionnaires to update information on potential risk factors and to identify newly diagnosed cancer and other diseases in themselves and their first degree relatives. We calculated body mass index (BMI, kg/m2), using self-reported height from the baseline questionnaire and weight from the biennial questionnaire that immediately preceded the diagnosis of colorectal cancer. In validation studies in both cohorts, self-reported anthropometric measures were well correlated with measurements by trained technicians (r >0.96).28 On each biennial follow-up questionnaire, participants were asked whether they had a diagnosis of colorectal cancer during the previous 2 years. When a participant (or next of kin for decedents) reported colorectal cancer, we sought permission to obtain medical records. Study physicians, while blinded to exposure data, reviewed all records related to colorectal cancer, and recorded AJCC (American Joint Committee on Cancer) tumor stage and tumor location. For nonresponders, we searched the National Death Index to discover deaths and ascertain any diagnosis of colorectal cancer that contributed to death or was a secondary diagnosis. Approximately 96% of all incident colorectal cancer cases were identified through these methods. We collected paraffin-embedded tissue blocks from hospitals where patients underwent tumor resections.27 Tissue sections from all colorectal cancer cases were reviewed and confirmed by a pathologist (S.O.). We excluded cases preoperatively treated with radiation and/or chemotherapy. Tumor grade was categorized as high (≤50% glandular area) or low (>50% glandular area). Based on availability of tissue samples, we included a total of 470 stage I-IV colorectal cancer cases (180 from the men's cohort and 290 from the women's cohort) diagnosed up to 2002. We utilized the well-established colorectal cancer tissue database with long-term follow-up data. PPARG has not been examined in our colorectal cancers. Moreover, our rich tissue database readily enabled us to control for confounding by any of the clinical and tumoral molecular characteristics in survival analyses, and to assess independent effect of PPARG expression on patient outcome after controlling for possible confounders. It is analogous to a novel study that utilizes well-known cancer cell lines or well-characterized animal cancer models. Written informed consent was obtained from all study subjects. This study was approved by the Human Subjects Committees at Brigham and Women's Hospital and the Harvard School of Public Health.
Measurement of Mortality
Patients were observed until death or June 2006, whichever came first. Ascertainment of deaths included reporting by the family or postal authorities. In addition, the names of persistent nonresponders were searched in the National Death Index. The cause of death was assigned by physicians blinded to information on lifestyle exposures and molecular changes in colorectal cancer. In rare patients who died as a result of colorectal cancer not previously reported, we obtained medical records with permission from next of kin. More than 98% of deaths in the cohorts were identified by these methods.
DNA Extraction, Pyrosequencing of KRAS, BRAF and PIK3CA, and Microsatellite Instability (MSI) Analysis
Genomic DNA from paraffin-embedded tissue was extracted, and whole genome amplification was performed.29 PCR and Pyrosequencing targeted for KRAS codons 12 and 13, BRAF codon 600 and PIK3CA exons 9 and 20 were performed as previously described.30 MSI status was determined using a microsatellite marker panel consisting of D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67 and D18S487 (i.e., 10-marker panel).29 MSI-high was defined as the presence of instability in ≥30% of the markers, MSI-low as the presence of instability in <30% of the markers, and microsatellite stability (MSS) as no unstable marker.
Real-Time PCR (MethyLight) to Determine CIMP (CpG Island Methylator Phenotype) Status
Sodium bisulfite treatment on tumor DNA and subsequent real-time PCR (MethyLight)31 assays were validated and performed.29 We quantified promoter methylation in 8 CIMP-specific genes (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1).29, 32, 33 CIMP-high was defined as ≥6/8 methylated promoters using the 8-marker CIMP panel, CIMP-low/0 as 0 to 5 methylated promoters, according to the previously established criteria.29
Pyrosequencing to Measure LINE-1 Methylation
In order to accurately quantify relatively high LINE-1 methylation levels, we utilized Pyrosequencing.24 LINE-1 methylation level measured by Pyrosequencing has been shown to correlate well with overall 5-methylcytosine level (i.e., global DNA methylation level) in tumor cells.34
Immunohistochemistry for PPARG, cyclin D1, p53, p21, p27, β-catenin, COX-2 and FASN
Tissue microarrays (TMAs) were constructed as previously described.27 Methods of immunohistochemical procedures and interpretation were previously described as follows: cyclin D1,35 β-catenin,36 p21, p27, p53,37, 38 fatty acid synthase (FASN),39 and COX-2.27
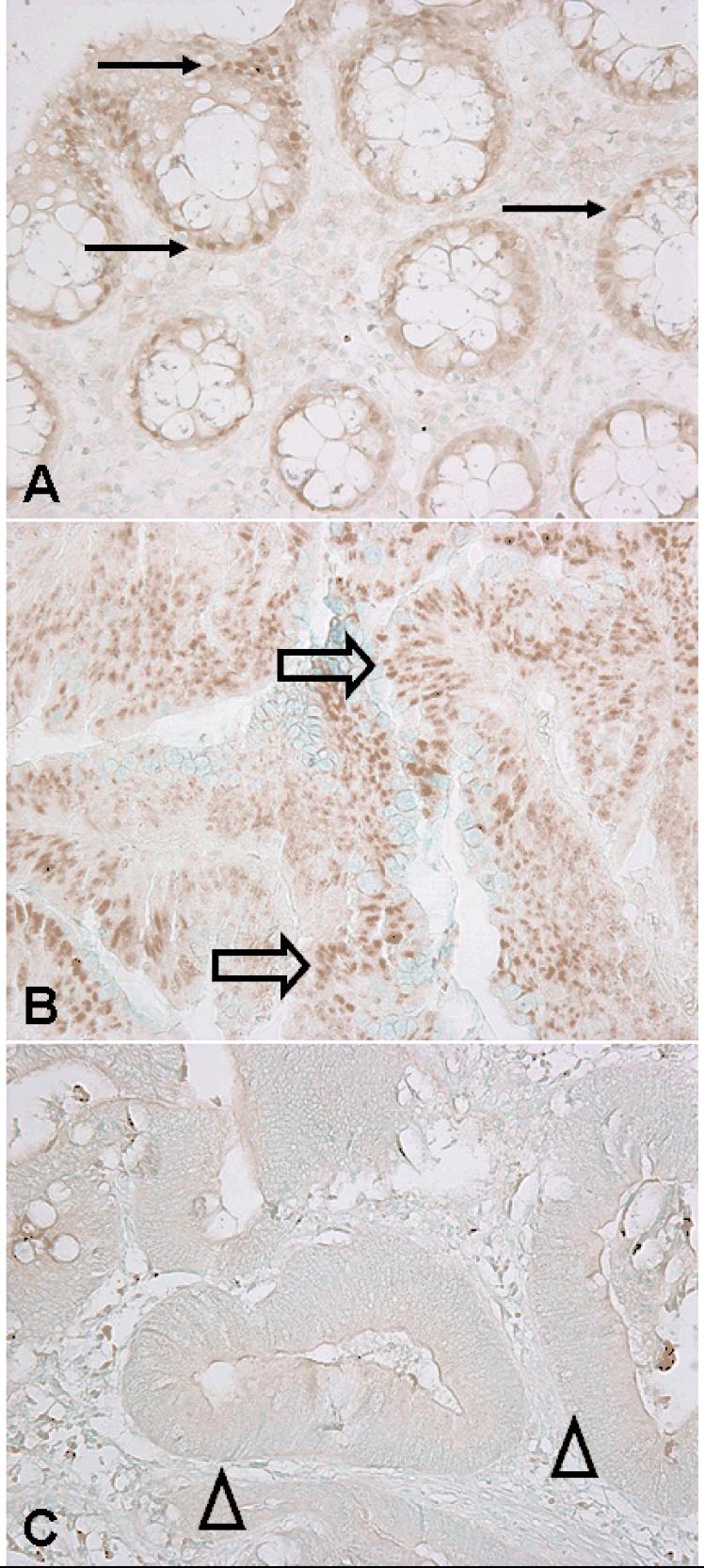
For PPARG immunohistochemistry (Figure 1), antigen retrieval was performed, and deparaffinized tissue sections in Target Retrieval Solution (pH 9.0, Dako, Glostrup, Denmark) were treated by a microwave for 5 min in a pressure cooker. Tissue sections were incubated with 10% normal goat serum (Vector Laboratories, Burlingame, CA) in phosphate-buffered saline (30 min). Primary antibody against PPARG (mouse monoclonal anti-PPARG, 1:50 dilution; Santa Cruz Biotechnology, Santa Cruz, CA) was applied, and the slides were maintained for 2 hours at room temperature. Next, we applied Envision System HRP labeled polymer anti mouse (Dako) for 30 min, followed by visualizing signal with diaminobenzidine (5 min) and methyl-green counterstain. PPARG positivity was defined as the presence of at least weak nuclear staining in ≥20% of tumor cells or moderate/strong staining in any fraction of tumor cells. Either absent staining or weak staining in <20% of tumor cells was interpreted as negative. Appropriate positive and negative controls were included in each run of immunohistochemistry. All immunohistochemically-stained slides for PPARG were interpreted by a pathologist (K.S.) unaware of other data. A random sample of 145 tumors were re-examined by a second pathologist (Y.B.) unaware of other data. The concordance between the two observers was 0.92 (κ=0.58, p<0.0001), indicating good to substantial agreement.
Figure 1.
PPARG expression in normal colonic mucosa and colorectal cancer.
A. Normal colonic epithelial cells with nuclear PPARG expression (arrows). B. Colorectal cancer cells with nuclear PPARG expression in (block arrows). C. Colorectal cancer cells with no nuclear PPARG expression (arrowheads).
Statistical Analysis
We used stage-matched, conditional Cox proportional hazard models to calculate hazard ratios (HRs) of death according to tumoral PPARG status, adjusted for age, sex, year of diagnosis, BMI, family history of colorectal cancer in any first degree relative, tumor location, stage, grade, MSI, CIMP, LINE-1, KRAS, BRAF, PIK3CA, β-catenin, p53, p21, p27, cyclin D1, FASN and COX-2. In addition, we also performed Cox regression analysis to assess the unadjusted, main effect of PPARG expression on mortality. For analyses of colorectal cancer-specific mortality, death as a result of colorectal cancer was the primary end point and deaths as a result of other causes were censored. The proportionality of hazards assumption was satisfied by evaluating time-dependent variables, which were the cross-product of the PPARG variable and survival time (p=0.62 for colon cancer-specific mortality; p=0.58 for overall mortality). To adjust for potential confounding, age, year of diagnosis and LINE-1 methylation were used as continuous variables, and all of the other covariates were used as categorical variables. Tumor stage (I, IIA, IIB, IIIA, IIIB, IIIC, IV) was used as a matching variable. We dichotomized family history (present vs. absent), BMI (<30 kg/m2 vs. ≥30 kg/m2), tumor location (rectum vs. colon), grade (high vs. low), CIMP (high vs. low/0), MSI (high vs. low/MSS), p53, p21, p27, cyclin D1, β-catenin, KRAS, PIK3CA, BRAF, FASN and COX-2. For cases with missing information in other covariates [including BMI (2.8% missing), tumor location (0.9% missing), MSI (0.4% missing), KRAS (0.2% missing), BRAF (2.8% missing), β-catenin (7.4% missing), p53 (0.6% missing), p21 (2.1% missing), p27 (4.5% missing) and cyclin D1 (4.7% missing)], we included those cases in a majority category, in order to minimize the number of “missing” indicator variables and maximize the efficiency of multivariate Cox regression analyses. We confirmed that excluding cases with missing information in any of the covariates did not substantially alter results (data not shown).
An interaction was assessed by including the cross product of the PPARG variable and another variable of interest in a multivariate Cox model, and the likelihood ratio test was performed. P values were conservatively interpreted, considering multiple hypotheses testing. To assess an interaction of PPARG and stage, we dichotomized tumor stage (I-II vs. III-IV) as well as treated stage as a linear ordinal variable (I to IV).
The Kaplan-Meier method was used to describe the distribution of colon cancer-specific and overall survival time, and the log-rank test was performed. The chi square test was used to examine an association of PPARG with any of the categorical variables. The t-test assuming unequal variances was performed to compare mean age and mean LINE-1 methylation level. All analyses used SAS version 9.1 (SAS Institute, Cary, NC) and all p values were two-sided.
RESULTS
PPARG expression in colorectal cancer and normal mucosa
Among 470 patients with stage I-IV colorectal cancer, PPARG expression was observed in 102 (22%) tumors by immunohistochemistry (Figure 1). We assessed clinical and molecular characteristics of colorectal cancers according to tumoral PPARG status, to assess potential confounders (Table 1). Compared to PPARG-negative cases, PPARG-positive cases were more likely diagnosed in 1995 or after, slightly more like to be located in rectum, and less likely to have a family history of colorectal cancer.
Table 1.
Clinical and molecular characteristics according to PPARG status in colorectal cancer
| Clinical or molecular feature | No. of cases | PPARG |
|
|---|---|---|---|
| Negative | Positive | ||
| Total N | 470 | 368 | 102 |
| Sex | |||
| Male (HPFS) | 180 (38%) | 148 (40%) | 32 (31%) |
| Female (NHS) | 290 (62%) | 220 (60%) | 70 (69%) |
| Mean age ± SD | 66.0 ± 8.6 | 65.8 ± 8.9 | 66.5 ± 7.6 |
| Body mass index (BMI, kg/m2) | |||
| <30 | 200 (44%) | 159 (45%) | 41 (41%) |
| 25-30 | 174 (38%) | 130 (36%) | 44 (44%) |
| ≥30 | 83 (18%) | 68 (19%) | 15 (15%) |
| Family history of colorectal cancer | |||
| Absent | 353 (75%) | 270 (73%) | 83 (81%) |
| Present | 117 (25%) | 98 (27%) | 19 (19%) |
| Year of diagnosis | |||
| Prior to 1995 | 193 (41%) | 167 (45%) | 26 (25%) |
| 1995 to 2002 | 277 (59%) | 201 (55%) | 76 (75%) |
| Tumor location | |||
| Right (cecum to transverse colon) | 230 (49%) | 176 (48%) | 54 (54%) |
| Left colon (splenic flexure to sigmoid colon) | 150 (32%) | 128 (35%) | 22 (22%) |
| Rectum | 86 (18%) | 62 (17%) | 24 (24%) |
| Tumor stage | |||
| I | 106 (23%) | 79 (21%) | 27 (26%) |
| II | 163 (35%) | 130 (35%) | 33 (32%) |
| III | 140 (30%) | 110 (30%) | 30 (29%) |
| IV | 61 (13%) | 49 (13%) | 12 (12%) |
| Tumor grade | |||
| Low | 428 (91%) | 333 (90%) | 95 (93%) |
| High | 42 (8.9%) | 35 (9.5%) | 7 (6.9%) |
| MSI | |||
| MSI-low/MSS | 385 (82%) | 299 (82%) | 86 (84%) |
| MSI-high | 83 (18%) | 67 (18%) | 16 (16%) |
| CIMP | |||
| CIMP-low/0 | 395 (84%) | 309 (84%) | 86 (84%) |
| CIMP-high | 75 (16%) | 59 (16%) | 16 (16%) |
| Mean LINE-1 methylation (%) | 60.3 ± 9.4 | 60.1 ± 9.7 | 61.1 ± 8.2 |
| BRAF mutation | |||
| (-) | 392 (86%) | 307 (86%) | 85 (84%) |
| (+) | 65 (14%) | 49 (14%) | 16 (16%) |
| KRAS mutation | |||
| (-) | 295 (63%) | 231 (62%) | 64 (63%) |
| (+) | 174 (37%) | 136 (37%) | 38 (37%) |
| PIK3CA mutation | |||
| (-) | 363 (86%) | 277 (85%) | 86 (88%) |
| (+) | 60 (14%) | 48 (15%) | 12 (12%) |
| β-catenin* | |||
| Inactive (score 0-2) | 274 (63%) | 211 (63%) | 63 (64%) |
| Active (score 3-5) | 161 (37%) | 126 (37%) | 35 (36%) |
| p53 expression | |||
| (-) | 285 (61%) | 226 (62%) | 59 (59%) |
| (+) | 182 (39%) | 141 (38%) | 41 (41%) |
| p21 (CDKN1A) | |||
| Expressed | 86 (19%) | 70 (19%) | 16 (16%) |
| Lost | 374 (81%) | 291 (81%) | 83 (84%) |
| p27 (CDKN1B) | |||
| Nuclear expression | 94 (21%) | 72 (20%) | 22 (23%) |
| Cytoplasmic expression or loss of expression | 355 (79%) | 280 (80%) | 75 (77%) |
| Cyclin D1 expression | |||
| (-) | 133 (30%) | 109 (31%) | 24 (24%) |
| (+) | 315 (70%) | 241 (69%) | 74 (76%) |
| Fatty acid synthase (FASN) expression | |||
| (-) | 322 (83%) | 249 (86%) | 73 (77%) |
| (+) | 64 (17%) | 42 (14%) | 22 (23%) |
| Cyclooxygenase-2 (COX-2) expression | |||
| (-) | 82 (17%) | 68 (18%) | 14 (14%) |
| (+) | 388 (83%) | 300 (82%) | 88 (86%) |
(%) indicates the proportion of tumors with a specific clinical or molecular feature in PPARG(-) (or PPARG+) tumors.
β-catenin score was calculated as previously described.36
CIMP, CpG island methylator phenotype; HPFS, Health Professionals Follow-up Study; LINE-1, long interspersed nucleotide element-1; MSI, microsatellite instability; MSS, microsatellite stable; NHS, Nurses' Health Study; SD, standard deviation.
In 264 cases for which we could evaluate PPARG expression in normal colonic mucosa, 134 cases (51%) show at least weak positivity. Tumoral PPARG positivity was significantly more common in cases with PPARG-positive normal mucosa (26%=35/134) than those with PPARG-negative normal mucosa (13%=17/130; p=0.008). This phenomenon could be a result of field effect. An alternative possibility was the presence of some poor quality specimens that showed false negative in either tumor or normal mucosa or both, driving PPARG staining in tumor and normal mucosa towards a concordant pattern. PPARG expression in normal mucosa was not significantly related with patient survival (data not shown).
PPARG expression and prognosis in colorectal cancer
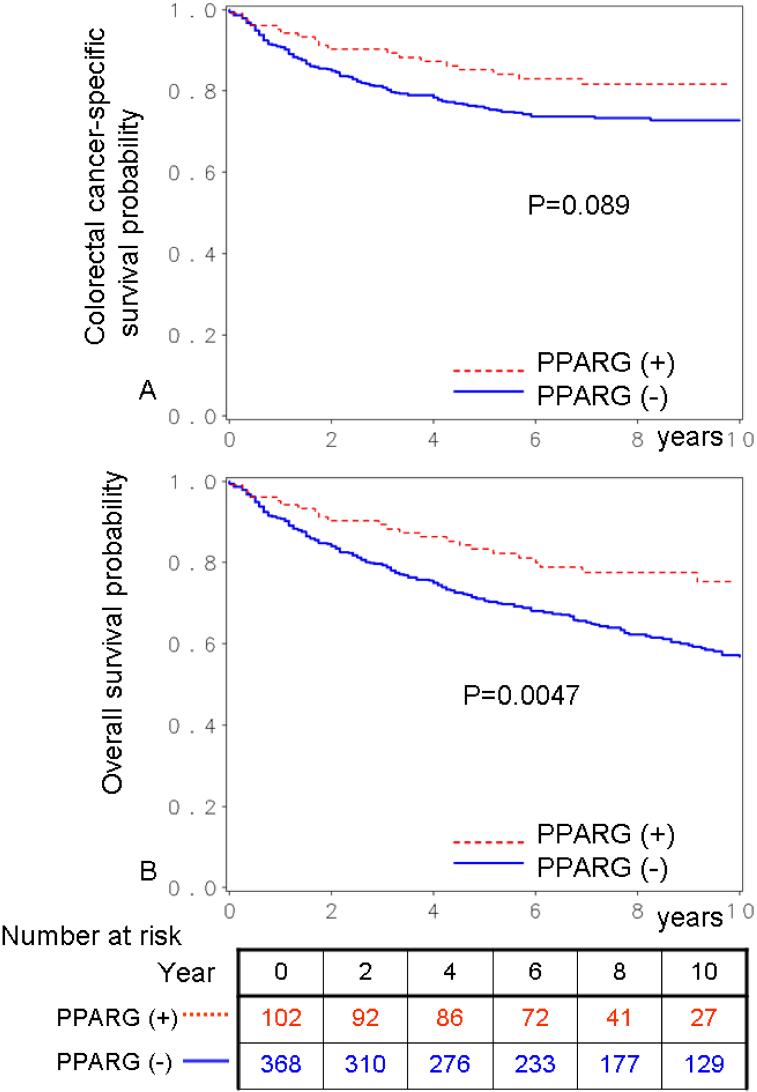
During follow-up, there were 199 deaths, including 118 colorectal cancer-specific deaths. We assessed the influence of PPARG expression on patient survival. In Kaplan-Meier analysis, PPARG-positive patients experienced significantly longer overall survival (log-rank p=0.0047) (Figure 2). Five-year overall survival was 83% in PPARG-positive patients and 71% in PPARG-negative patients. In univariate Cox regression analysis, compared to patients with PPARG-negative tumors, those with PPARG-positive tumors experienced a significantly lower overall mortality [HR 0.55; 95% confidence interval (CI), 0.37-0.84; p=0.0053] (Table 2). In the multivariate Cox model adjusting for potential predictors of patient outcome, PPARG positivity was associated with a significantly lower overall mortality (multivariate HR 0.43; 95% CI, 0.27-0.69; p=0.0004).
Figure 2.
Kaplan-Meier survival curves in colorectal cancer according to PPARG status. A. Colorectal cancer-specific survival. B. Overall survival.
Table 2.
PPARG expression in colorectal cancer and patient mortality
| Total N | Colorectal cancer-specific mortality | Overall mortality | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Deaths / person-years | Univariate HR (95% CI) | Stage-matched HR (95% CI) | Multivariate HR (95% CI) | Deaths / person-years | Univariate HR (95% CI) | Stage-matched HR (95% CI) | Multivariate HR (95% CI) | ||
| PPARG(-) | 368 (78%) | 99/2982 | 1 (referent) | 1 (referent) | 1 (referent) | 173/2982 | 1 (referent) | 1 (referent) | 1 (referent) |
| PPARG(+) | 102 (22%) | 19/789 | 0.65 (0.40-1.07) | 0.54 (0.33-0.90) | 0.44 (0.25-0.79) | 26/789 | 0.55 (0.37-0.84) | 0.52 (0.34-0.79) | 0.43 (0.27-0.69) |
| P value | 0.092 | 0.017 | 0.0054 | 0.0053 | 0.0025 | 0.0004 | |||
The multivariate, stage-matched conditional Cox regression model included age, year of diagnosis, sex, family history of colorectal cancer, body mass index (BMI), tumor location, stage, grade, KRAS, BRAF, PIK3CA, p53, p21, p27, cyclin D1, β-catenin, COX-2, FASN, LINE-1 methylation, microsatellite instability (MSI), and CpG island methylator phenotype (CIMP).
CI, confidence interval; HR, hazard ratio.
In the analysis using colorectal cancer-specific mortality as the end point, PPARG-positive cases experienced a similar reduction of mortality in univariate and multivariate analyses (multivariate HR 0.44; 95% CI, 0.25-0.79; p=0.0054). The greater effect of PPARG on cancer-specific mortality in multivariate analysis than univariate analysis was simply due to the effect of adjusting for tumor stage. When we simply adjusted for tumor stage, HR for colorectal cancer specific mortality was 0.54 (95% CI, 0.33-0.90; p=0.017) (Table 2). No other major confounder was present.
Stratified analysis of PPARG and mortality
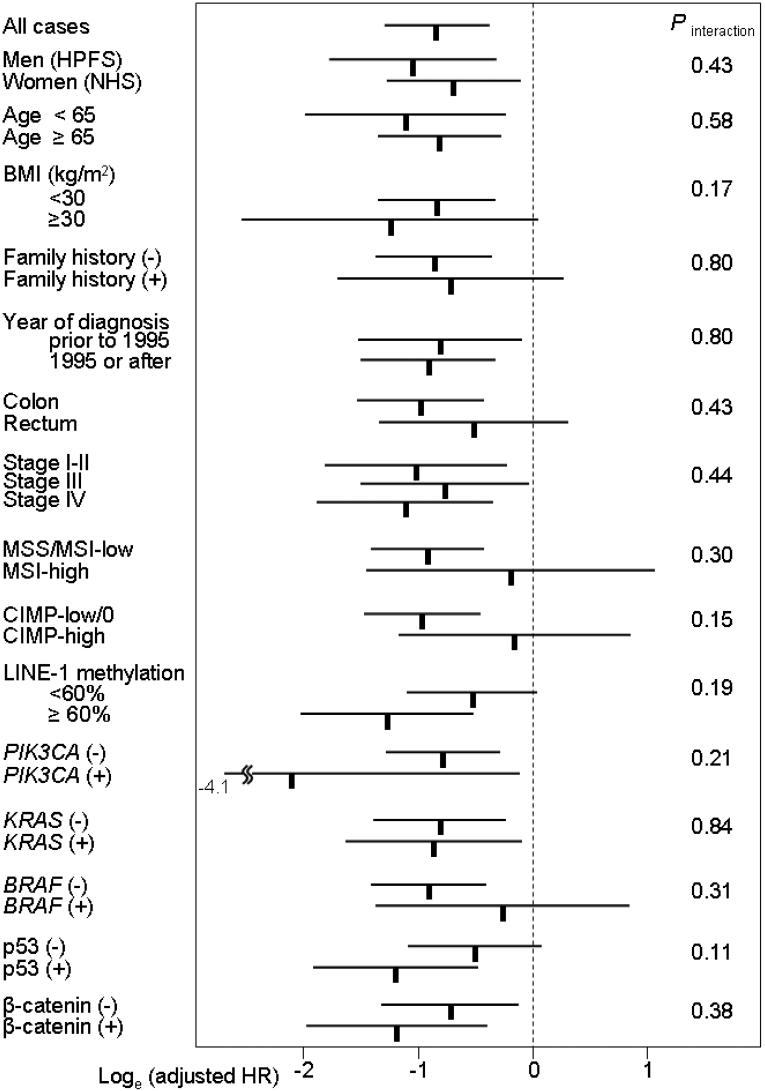
We examined the influence of PPARG positivity on overall mortality across strata of other potential predictors of patient survival (Figure 3). Accordingly, we assessed whether there was potential modifying effect (on the relation between PPARG and a low mortality) by any of the other variables examined, including sex (cohort), age, BMI, family history of colorectal cancer, year of diagnosis, tumor location, stage, grade, MSI, CIMP, LINE-1 methylation, KRAS, BRAF,PIK3CA, β-catenin, p53, p21, p27, cyclin D1, FASN and COX-2. There was no evidence for significant effect modification by any of the variables (all p for interaction >0.05). Notably, the effect of PPARG did not significantly differ between the two independent cohort studies (p for interaction = 0.43).
Figure 3.
PPARG status and overall mortality in various strata.
Loge(adjusted HRs) with 95% CI for overall mortality in PPARG+ tumors (vs. PPARG-tumors) are shown.
BMI, body mass index; CI, confidence interval; CIMP, CpG island methylator phenotype; HPFS, Health Professionals Follow-up Study; HR, hazard ratio; NHS, Nurses' Health Study.
DISCUSSION
We conducted this study to examine the relation between expression of PPARG (the official symbol for peroxisome proliferator-activated receptor-γ) and patient survival in 470 patients with stage I-IV colorectal cancer. We have shown that PPARG expression is independently associated with good prognosis in colorectal cancer. We have been able to demonstrate that this relation did not appear to be significantly modified by tumor stage or any of the other clinical features and tumoral molecular characteristics. Furthermore, our resource of a large number of colorectal cancers derived from the two independent, prospective cohort studies has enabled us to precisely estimate the frequency of colorectal cancers with PPARG expression, and provided us with robust statistics in survival analysis. Our results suggest that PPARG expression in colorectal cancer is independently associated with low mortality, and marks colorectal cancer with indolent biological behavior.
Examining molecular alterations is important in cancer research.40-46 In particular, determining status of microsatellite instability (MSI), the CpG island methylator phenotype (CIMP) and LINE-1 hypomethylation in colorectal cancer is increasingly important, because these molecular characteristics refect genomic and epigenomic status of cancer cells, and have been related with patient survival in colon cancer.19, 21, 23, 24 In addition to examining status of CIMP, MSI and LINE-1 hypomethylation, we assessed various molecular variables potentially related with PPARG and/or energy balance, including fatty acid synthase (FASN), cyclooxygenase-2 (COX-2), p53, p21, KRAS, BRAF and PIK3CA. Thus, unlike other studies, we were able to evaluate the independent effect of PPARG on patient survival after controlling for those related molecular events.
A role of PPARG in colorectal cancer has been controversial. Animal models generated by different methods show different effects of PPARG. Genetic models with APC mutations suggest that PPARG promote tumorigenesis.10-12 On the contrary, colon tumors induced by carcinogens can be suppressed by thiozolidinedione (TZD) based PPARG agonists, suggesting a tumor-suppressor role of PPARG.4, 13 A large epidemiologic study of a diabetic population suggests that thiozolidinedione usage may reduce the risk of a number of cancers including lung, colon and prostate.47 Various studies have shown the effect of PPARG ligands on normalization of cell cycle progression and cellular differentiation.8, 9, 14 It is proposed that PPARG could be a conditional tumor suppressor or conditional oncogene that modulates the tumor pathogenesis depending on cellular conditions, tissue types, or genetic background of individuals.2 Nonetheless, our current data suggest that PPARG expression may mark an indolent subset of colorectal cancers.
Excess energy balance, obesity and lack of exercise have been linked to increased risks of a variety of human cancers including colorectal cancer.48, 49 PPARG is one of potential molecules that link between energy balance, cellular metabolism and cancer pathogenesis. PPARG has been shown to play an important role in the control of gene expression linked to a variety of cellular processes.1 Activation of PPARG improves insulin sensitivity through a combination of metabolic actions, including partitioning of lipid stores and the regulation of metabolic and inflammatory mediator adipokines.1 Thus, we could hypothesize that there might be an interactive effect of PPARG expression and energy balance (or related tumoral molecular events) on tumor aggressiveness. However, we did not show any significant interaction of PPARG with patient body mass index (BMI), FASN expression or PIK3CA mutation in terms of patient survival. Nonetheless, it is still possible that energy balance may differentially influence the occurrence of colorectal cancer according to PPARG expression. We await future studies that examine the effect of energy balance on the occurrence of PPARG-positive or negative tumors.
In our dataset, compared to PPARG-negative cases, PPARG-positive cases were more likely to be diagnosed in 1995 or after. This might have been due to poor antigenicity of PPARG in older specimens. However, when we examined the strata of “year of diagnosis” and a potential interaction between PPARG and “year of diagnosis”, “year of diagnosis” did not significantly alter the relation between PPARG and patient survival (p for interaction = 0.80; see Figure 3). This suggests that misdiagnosis due to poor antigenicity, if any, did not substantially alter survival analysis results. Moreover, in our multivariate Cox regression analysis, we adjusted hazard ratio (HR) for clinical and tumoral characteristics, including year of diagnosis. Thus, any potential confounding effect of year of diagnosis on patient survival was controlled in our multivariate analysis model.
In our cohorts, data on cancer treatment were limited. Nonetheless, it is unlikely that chemotherapy use differed according to tumoral PPARG status, since such data were not available to patients or treating physicians. In addition, beyond cause of mortality, data on cancer recurrences were not available in these cohorts. Nonetheless, given the median survival for metastatic colon cancer was approximately 10 to 12 months during much of the time period of this study,50 colon cancer-specific survival should be a reasonable surrogate for cancer-specific outcomes.
In summary, our large cohort study suggests that PPARG expression is independently associated with good prognosis in colorectal cancer. Our findings may have considerable clinical implications, given that PPARG has been used as a drug target. Future studies are needed to confirm this association as well as to elucidate exact mechanisms by which PPARG affects tumor behavior.
ACKNOWLEDGEMENTS
We deeply thank the Nurses' Health Study and Health Professionals Follow-up Study cohort participants who have generously agreed to provide us with biological specimens and information through responses to questionnaires; hospitals and pathology departments throughout the U.S. for providing us with tumor tissue materials; Frank Speizer, Walter Willett, Susan Hankinson, Graham Colditz, Meir Stampfer, and many other staff members who implemented and have maintained the cohort studies; and Bruce Spiegelman for helpful discussion.
Funding: The U.S. National Institute of Health (P01 CA87969, P01 CA55075, P50 CA127003, and K07 CA122826 to S.O.); the Bennett Family Fund; the Entertainment Industry Foundation National Colorectal Cancer Research Alliance (NCCRA). K.N. was supported by a fellowship grant from the Japanese Society for Promotion of Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of NCI or NIH. Funding agencies did not have any role in the design of the study; the collection, analysis, or interpretation of the data; the decision to submit the manuscript for publication; or the writing of the manuscript.
Abbreviations and the HUGO Gene Nomenclature Committee-approved official gene symbols
- AJCC
American Joint Committee on Cancer
- BMI
body mass index
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- COX-2
cyclooxygenase-2
- FASN
fatty acid synthase
- HPFS
Health Professionals Follow-up Study
- HR
hazard ratio
- LINE-1
long interspersed nucleotide element-1
- MSI
microsatellite instability
- MSS
microsatellite stable
- NHS
Nurses' Health Study
- PPARG
peroxisome proliferator-activated receptor gamma.
References
- 1.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 2.Wang YL, Miao Q. To Live or to Die: Prosurvival Activity of PPARgamma in Cancers. PPAR Res. 2008;2008:209629. doi: 10.1155/2008/209629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarraf P, Mueller E, Jones D, et al. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat Med. 1998;4:1046–52. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 4.Girnun GD, Smith WM, Drori S, et al. APC-dependent suppression of colon carcinogenesis by PPARgamma. Proc Natl Acad Sci U S A. 2002;99:13771–6. doi: 10.1073/pnas.162480299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elrod HA, Sun SY. PPARgamma and Apoptosis in Cancer. PPAR Res. 2008;2008:704165. doi: 10.1155/2008/704165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang WL, Frucht H. Activation of the PPAR pathway induces apoptosis and COX-2 inhibition in HT-29 human colon cancer cells. Carcinogenesis. 2001;22:1379–83. doi: 10.1093/carcin/22.9.1379. [DOI] [PubMed] [Google Scholar]
- 7.Subbaramaiah K, Lin DT, Hart JC, et al. Peroxisome proliferator-activated receptor gamma ligands suppress the transcriptional activation of cyclooxygenase-2. Evidence for involvement of activator protein-1 and CREB-binding protein/p300. J Biol Chem. 2001;276:12440–8. doi: 10.1074/jbc.M007237200. [DOI] [PubMed] [Google Scholar]
- 8.Burton JD, Goldenberg DM, Blumenthal RD. Potential of peroxisome proliferator-activated receptor gamma antagonist compounds as therapeutic agents for a wide range of cancer types. PPAR Res. 2008;2008:494161. doi: 10.1155/2008/494161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson EA. PPARgamma physiology and pathology in gastrointestinal epithelial cells. Mol Cells. 2007;24:167–76. [PubMed] [Google Scholar]
- 10.Lefebvre AM, Chen I, Desreumaux P, et al. Activation of the peroxisome proliferator-activated receptor gamma promotes the development of colon tumors in C57BL/6JAPCMin/+ mice. Nat Med. 1998;4:1053–7. doi: 10.1038/2036. [DOI] [PubMed] [Google Scholar]
- 11.Saez E, Tontonoz P, Nelson MC, et al. Activators of the nuclear receptor PPARgamma enhance colon polyp formation. Nat Med. 1998;4:1058–61. doi: 10.1038/2042. [DOI] [PubMed] [Google Scholar]
- 12.Yang K, Fan KH, Lamprecht SA, et al. Peroxisome proliferator-activated receptor gamma agonist troglitazone induces colon tumors in normal C57BL/6J mice and enhances colonic carcinogenesis in Apc1638 N/+ Mlh1+/- double mutant mice. Int J Cancer. 2005;116:495–9. doi: 10.1002/ijc.21018. [DOI] [PubMed] [Google Scholar]
- 13.Osawa E, Nakajima A, Wada K, et al. Peroxisome proliferator-activated receptor gamma ligands suppress colon carcinogenesis induced by azoxymethane in mice. Gastroenterology. 2003;124:361–7. doi: 10.1053/gast.2003.50067. [DOI] [PubMed] [Google Scholar]
- 14.Cesario RM, Stone J, Yen WC, et al. Differentiation and growth inhibition mediated via the RXR:PPARgamma heterodimer in colon cancer. Cancer Lett. 2006;240:225–33. doi: 10.1016/j.canlet.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Theocharis S, Giaginis C, Parasi A, et al. Expression of peroxisome proliferator-activated receptor-gamma in colon cancer: correlation with histopathological parameters, cell cycle-related molecules, and patients' survival. Dig Dis Sci. 2007;52:2305–11. doi: 10.1007/s10620-007-9794-4. [DOI] [PubMed] [Google Scholar]
- 16.Gustafsson A, Hansson E, Kressner U, et al. EP1-4 subtype, COX and PPAR gamma receptor expression in colorectal cancer in prediction of disease-specific mortality. Int J Cancer. 2007;121:232–40. doi: 10.1002/ijc.22582. [DOI] [PubMed] [Google Scholar]
- 17.Konstantinopoulos PA, Vandoros GP, Sotiropoulou-Bonikou G, et al. NF-kappaB/PPAR gamma and/or AP-1/PPAR gamma 'on/off' switches and induction of CBP in colon adenocarcinomas: correlation with COX-2 expression. Int J Colorectal Dis. 2007;22:57–68. doi: 10.1007/s00384-006-0112-y. [DOI] [PubMed] [Google Scholar]
- 18.Van Rijnsoever M, Elsaleh H, Joseph D, et al. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–903. [PubMed] [Google Scholar]
- 19.Ward RL, Cheong K, Ku SL, et al. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21:3729–36. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- 20.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–9. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 21.Popat S, Hubner R, Houlston RS. Systematic Review of Microsatellite Instability and Colorectal Cancer Prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 22.Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–6. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 23.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2008 doi: 10.1136/gut.2008.155473. in press (published online on 2 Oct 2008. doi:10.1136/gut.2008.155473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino S, Nosho K, Kirkner GJ, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colditz GA, Hankinson SE. The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5:388–96. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 27.Chan AT, Ogino S, Fuchs CS. Aspirin and the Risk of Colorectal Cancer in Relation to the Expression of COX-2. New Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 28.Rimm EB, Stampfer MJ, Colditz GA, et al. Validity of self-reported waist and hip circumferences in men and women. Epidemiology. 1990;1:466–73. doi: 10.1097/00001648-199011000-00009. [DOI] [PubMed] [Google Scholar]
- 29.Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 34.Yang AS, Estecio MR, Doshi K, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nosho K, Kawasaki T, Chan AT, et al. Cyclin D1 is frequently overexpressed in microsatellite unstable colorectal cancer, independent of CpG island methylator phenotype. Histopathology. 2008;53:588–98. doi: 10.1111/j.1365-2559.2008.03161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawasaki T, Nosho K, Ohnishi M, et al. Correlation of beta-catenin localization with cyclooxygenase-2 expression and CpG island methylator phenotype (CIMP) in colorectal cancer. Neoplasia. 2007;9:569–577. doi: 10.1593/neo.07334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino S, kawasaki T, Kirkner GJ, et al. Loss of nuclear p27 (CDKN1B/KIP1) in colorectal cancer is correlated with microsatellite instability and CIMP. Mod Pathol. 2007;20:15–22. doi: 10.1038/modpathol.3800709. [DOI] [PubMed] [Google Scholar]
- 38.Ogino S, Meyerhardt JA, Cantor M, et al. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005;11:6650–6. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]
- 39.Ogino S, Brahmandam M, Cantor M, et al. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 40.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–99. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goel A, Nagasaka T, Arnold CN, et al. The CpG Island Methylator Phenotype and Chromosomal Instability Are Inversely Correlated in Sporadic Colorectal Cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 42.Ponferrada A, Caso JR, Alou L, et al. The role of PPARgamma on restoration of colonic homeostasis after experimental stress-induced inflammation and dysfunction. Gastroenterology. 2007;132:1791–803. doi: 10.1053/j.gastro.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 43.Chung SK, Lee MG, Ryu BK, et al. Frequent alteration of XAF1 in human colorectal cancers: implication for tumor cell resistance to apoptotic stresses. Gastroenterology. 2007;132:2459–77. doi: 10.1053/j.gastro.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 44.Konishi K, Shen L, Wang S, et al. Rare CpG island methylator phenotype in ulcerative colitis-associated neoplasias. Gastroenterology. 2007;132:1254–60. doi: 10.1053/j.gastro.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 45.Shin SK, Nagasaka T, Jung BH, et al. Epigenetic and genetic alterations in netrin-1 receptors UNC5C and DCC in human colon cancer. Gastroenterology. 2007;133:1849–1857. doi: 10.1053/j.gastro.2007.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawamura YI, Toyota M, Kawashima R, et al. DNA hypermethylation contributes to incomplete synthesis of carbohydrate determinants in gastrointestinal cancer. Gastroenterology. 2008;135:142–151. e3. doi: 10.1053/j.gastro.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 47.Govindarajan R, Ratnasinghe L, Simmons DL, et al. Thiazolidinediones and the risk of lung, prostate, and colon cancer in patients with diabetes. J Clin Oncol. 2007;25:1476–81. doi: 10.1200/JCO.2006.07.2777. [DOI] [PubMed] [Google Scholar]
- 48.Chen YQ, Edwards IJ, Kridel SJ, et al. Dietary fat-gene interactions in cancer. Cancer Metastasis Rev. 2007;26:535–51. doi: 10.1007/s10555-007-9075-x. [DOI] [PubMed] [Google Scholar]
- 49.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–77. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 50.Meyerhardt JA, Giovannucci EL, Holmes MD, et al. Physical activity and survival after colorectal cancer diagnosis. J Clin Oncol. 2006;24:3527–3534. doi: 10.1200/JCO.2006.06.0855. [DOI] [PubMed] [Google Scholar]