T-cell prolymphocytic leukemia is a rare aggressive lymphoproliferative disease with a mature T-cell phenotype and characteristic genomic rearrangements of 14q11. This study identifies numerous new potential target genes in common breakpoints, deletions and regions of acquired uniparental disomy.
Keywords: T-cell prolymphocytic leukemia, SNP array, uniparental disomy, copy number change
Abstract
Background
T-cell prolymphocytic leukemia is a rare aggressive lymphoproliferative disease with a mature T-cell phenotype and characteristic genomic lesions such as inv(14)(q11q34), t(14;14)(q11;q32) or t(X;14)(q28;q11), mutation of the ATM gene on chromosome 11 and secondary alterations such as deletions of chromosome 8p and duplications of 8q.
Design and Methods
We analyzed malignant cells from 18 patients with T-cell prolymphocytic leukemia using high density 250K single nucleotide polymorphism arrays and molecular allelokaryotyping to refine understanding of known alterations and identify new target genes.
Results
Our analyses revealed that characteristic disruptions of chromosome 14 are frequently unbalanced. In the commonly deleted region on chromosome 11, we found recurrent microdeletions targeting the microRNA 34b/c and the transcription factors ETS1 and FLI1. On chromosome 8, we identified genes such as PLEKHA2, NBS1, NOV and MYST3 to be involved in breakpoints. New recurrent alterations were identified on chromosomes 5p, 12p, 13q, 17 and 22 with a common region of acquired uniparental disomy in four samples on chromosome 17q. Single nucleotide polymorphism array results were confirmed by direct sequencing and quantitative real-time polymerase chain reaction.
Conclusions
The first high density single nucleotide polymorphism array allelokaryotyping of T-cell prolymphocytic leukemia genomes added substantial new details about established alterations in this disease and moreover identified numerous new potential target genes in common breakpoints, deletions and regions of acquired uniparental disomy.
Introduction
T-cell prolymphocytic leukemia (T-PLL) is a rare lymphoproliferative disease with a mature T-cell phenotype. The median age at presentation is 63 years.1 Its clinical course is generally aggressive with a poor response to chemotherapy and median survival times ranging from 5 months to 2 years in patients receiving therapies containing alemtuzumab.2,3
T-PLL has several characteristic and recurring molecular lesions. These include an inversion or translocation of chromosome 14: inv(14)(q11q34) or t(14;14) (q11;q32), which lead to juxtaposition of the T-cell receptor (TCR) α/δ enhancer regions to the T-cell leukemia 1 (TCL1) locus causing deregulated expression of oncogenes located in this region.4 An alternative translocation associated with T-PLL is the t(X;14)(q28;q11) juxtaposing the TCR α/δ to the MTCP1 gene.5 Other common molecular abnormalities in T-PLL are deletions on chromosome 11 involving the ataxia-telangiectasia mutated (ATM) gene, which has been shown to be mutated in patients with T-PLL, and common chromosomal gains of 8q and losses of 8p.6–11
The TCL1 family of oncogenes enhances proliferation and survival in several lymphocytic malignancies by binding and augmenting activation of AKT12 and inhibiting activation induced cell death via impairment of the PKCθ and ERK pathways.13 This is also reflected by the clinical observation of hyperproliferative subsets of T-PLL with high levels of expression TCL1.14 Mutations in the ATM gene are known to be the cause of the rare autosomal recessive disorder ataxia-telangiectasia, which is characterized by cerebellar degeneration, immunodeficiency and increased risk of cancer.15 ATM plays a prominent role in the recognition and repair of DNA double strand breaks16,17 and the frequent disruption of this gene in T-PLL may be an explanation for the genomic instability observed in this disease. The common genomic abnormalities observed on chromosome 8 have not yet yielded any specific target genes, but the breakpoints occurring on chromosome 8 in T-PLL cluster to two regions which contain the fibroblast growth factor receptor-1 gene (FGFR1) and the MOZ gene, suggesting them as possible candidate genes.9
In searches for new T-PLL specific target genes, recent studies have employed techniques such as comparative genomic hybridization (CGH) and 50K single nucleotide polymorphism (SNP) arrays combined with gene expression analysis. These studies have described several differentially regulated genes possibly due to gene dosage effects18 and CDKN1B haploinsufficiency as a new pathogenic mechanism in T-PLL.19
Recently, SNP arrays with a higher resolution (250,000 SNPs interrogated per array) have been developed for whole genome mapping.20 The analysis of genomic DNA with SNP arrays provides two different types of information. One is a data set comprising the intensity data of all SNPs. Since the human genome is diploid, the intensity values are raised to two after normalization, which represents the normal expression of SNPs on somatic chromosomes. A homozygous deletion results in an expression value of zero and a heterozygous deletion in an expression value of one. Amplifications result in expression values of three or higher integer copy numbers. Apart from copy number data, the method also yields a genotype data set which contains the SNP calls of either AA, AB or BB standing for the alleles of the SNPs. This, combined with the copy number data, which allows the detection of acquired uniparental disomy (UPD), which represents allelic imbalance when one allele is deleted and the other one is duplicated or amplified leading to regions with homozygous SNP calls but a copy number of two or higher. These regions typically contain a mutant tumor suppressor gene or oncogene with loss of their normal allele. Use of these high density SNP arrays in combination with a new computational calculation algorithm termed molecular allelokaryotyping21 allows robust and detailed detection of the described alterations without a need for paired normal DNA samples. In the current study, we used this new interrogational power to assess the genomes of 18 T-PLL samples and thereby identify more precisely common submicroscopic genomic lesions and breakpoints and detect novel common genomic lesions and acquired UPD as potential new pathogenic factors in T-PLL.
Design and Methods
Patients and samples
We studied 18 cases of T-PLL (TP followed by the case number). Samples TP-04, −21, −22, −25, −28, −34, −35, −37, −41, −43, −56 and −57 were obtained from the Institut Curie, Centre de Recherche (Paris, France) and are identical to the samples used in the study by Le Toriellec et al.19 Samples TP-651 and −799 came from the Department of Haematology, University Hospitals Leicester (United Kingdom) and samples TP-166, −168, −170 and −172 were from the Department of Hematology and Oncology, School of Medicine, University of Tokyo (Japan). The acquisition and analysis of patients’ DNA samples was conducted with the approval of the local ethical committees of the respective institutions.
The diagnosis of T-PLL was established according to the World Health Organization (WHO) classification of hematopoietic and lymphoid tumors. T-PLL genomic DNA was isolated from residual frozen mononuclear cells from leukemic peripheral blood taken at the time of the initial diagnosis. DNA was extracted using a NucleosSpin Tissue kit (Macherey-Nagel, Hoerdt, France). Paired normal DNA was isolated from Epstein-Barr-virus-transformed lymphoblastoid cell lines, which were generated from frozen blood samples of the corresponding patients. All patients had major lymphocytosis. One case (TP56) arose in an individual with ataxia telangiectasia.
High density single nucleotide polymorphism-array analysis
High quality genomic DNA from the 18 T-PLL cases was processed according to the genomic mapping 250K NspI protocol and hybridized to 250K NspI SNP arrays using the GeneChip Fluidics station 400 and GeneChip scanner 3000 (Affymetrix, Santa Clara, CA, USA) as described previously.21,22 Data analysis of deletions, amplifications and UPD was carried out using the CNAG software with non-matched references, as previously described.21,22 Size, position and location of genes were identified with the UCSC Genome Browser http://genome.ucsc.edu/ and the Ensemble Genome Browser http://www.ensembl.org/.
Validation of acquired uniparental disomy and genomic copy number change
For confirmation of genomic copy number changes, quantitative real-time polymerase chain reaction (PCR) was performed on the genomic DNA from the hybridized T-PLL samples and from matched normal DNA from the same patients according to the calculation method described by Weksberg et al.23 Thereby, we confirmed the deletion of the FOXP1 gene on chromosome 3 in two samples and used a random region on chromosome 2p21 as a reference. Detection of acquired UPD was validated by PCR of genomic DNA and subsequent direct sequencing of SNPs in a region of acquired UPD versus a heterozygous region in sample TP28 on chromosome 17 and compared to direct sequencing of SNPs in the corresponding matched normal sample. All primer sequences are available on request.
Results
Copy number analysis
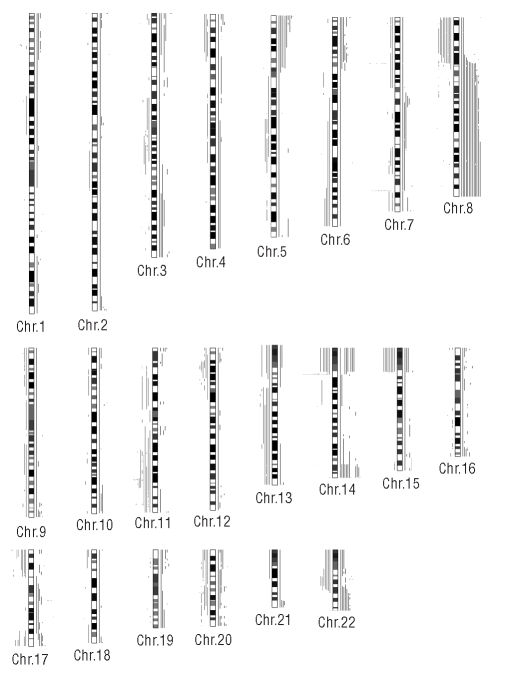
As expected from previous studies summarizing genomic lesions in T-PLL,18,24 we found a great abundance of copy number alterations present in the 18 T-PLL samples. In an initial step, we evaluated all losses and gains of genomic material detected by the allelokaryotyping software, which are depicted for each chromosome in Figure 1. We excluded alterations, which were determined to be due to background noise of SNP signals or copy number polymorphisms as recorded in data bases of the UCSC genome browser. The results of this analysis are documented in Online Supplementary Table S1. The data re-confirmed characteristic genomic lesions described in T-PLL. Moreover, a variety of other alterations containing interesting putative target genes were detected. In the first systematic measure, we sought to review the above mentioned characteristic lesions on chromosomes 14, 11 and 8.
Figure 1.
Overview of gains and losses detected by the CNAG software. Lines to the right of the cytobands document gains. Lines to the left of the cytobands represent losses. Each line represents one sample.
Chromosome 14
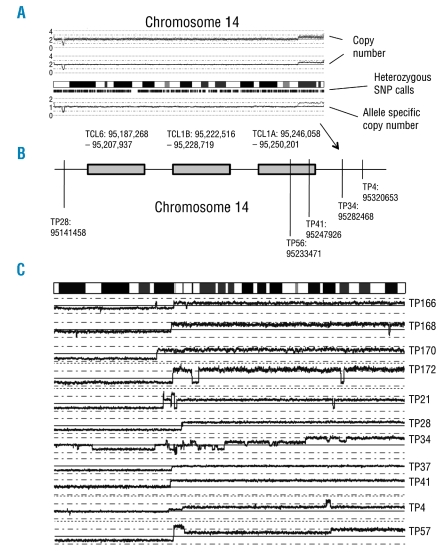
The characteristic inversion inv(14)(q11q34) or translocation t(14;14)(q11;q32) in T-PLL should not be detected by SNP array as no copy number changes occur by these balanced alterations. Interestingly, however, several copy number alterations involving the TCL oncogenes were detectable. Sample TP22 displayed a heterozygous deletion in region chr14:95130522–95211348 which partly covers T-cell leukemia/lymphoma 6 isoform TCL6a3. Furthermore, five samples had duplications of the telomeric end of chromosome 14 (example, Figure 2A) and a putative breakpoint within the TCL oncogenes or in their direct vicinity, distal to them, suggesting that the alterations affecting this site are unbalanced in these samples. The breakpoints leading to duplication or amplification in the respective samples are schematically displayed in Figure 2B, which shows that they lie in typical regions also described for the known translocations.4 An analysis of the alternate translocation t(X;14)(q28;q11) on chromosome X revealed no unbalanced lesions.
Figure 2.
Breakpoints of unbalanced translocations involving the TCL1 locus and breakpoints on chromosome 8. (A) A view of chromosome 14 in sample TP34 showing a duplication of the chromosomal region 14q32.13 – 14q32.33 harboring a breakpoint in the TCL1 locus. Similar unbalanced translocations were detected in four other samples, and their breakpoints in relation to the three putative oncogenes TCL6, TCL1B and TCL1A are displayed in the lower panel (B). (C) T-PLL commonly displays a loss of chromosome 8p and gain of 8q. The regions of chromosomal breakpoints leading to this imbalance, as analyzed by high density SNP arrays are displayed. The images show that the breakages are frequently of highly complex nature, containing multiple changes of copy number and breakpoints in individual samples.
Chromosome 8
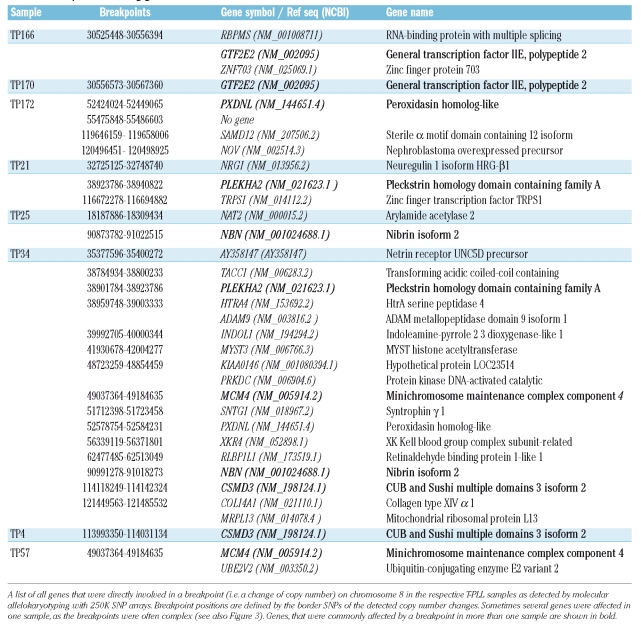
The high density genomic mapping carried out in our study revealed broad heterozygous deletions of chromosome 8p in nine samples (50%) and duplications or amplification of chromosome 8q in 13 samples (72%). The detailed mapping of the breakpoints leading to these imbalances revealed that they were scattered over a broad region on chromosome 8p and often displayed highly complex copy number alterations with numerous breakpoints (Figure 2C). In search of genes in the commonly affected regions, we analyzed all chromosomal breaks in all samples on chromosome 8 also including alterations on chromosome 8q. Although an unambiguous common breakpoint could not be identified and most breakpoints were located in regions, that did not contain any genes, some breaks were notable because they were positioned directly within genes and recurred in at least two samples. Breakpoints, that were intra-genic are summarized in Table 1. Genes commonly affected by breaks on chromosome 8 were general transcription factor IIE polypeptide 2 (GTF2E2) (two samples), peroxidasin homolog-like (PXDNL) (two samples), pleckstrin homology domain containing family A (PLEKHA2) (two samples), nibrin isoform 2 (NBN) (two samples), CUB and Sushi multiple domains 3 isoform 2 (CSMD3) (two samples) and minichromosome maintenance complex component 4 (MCM4). Furthermore, in single cases, interesting target genes such as nephroblastoma overexpressed precursor (NOV) or MYST histone acetyltransferase monocytic (MYST3) were involved directly in chromosomal breakpoints detected by SNP array.
Table 1.
Breakpoints involving genes on chromosome 8.
Chromosome 11
In our study, 12 samples (67%) displayed heterozygous deletions on chromosome 11q, all including the ATM gene. Deletions either affected broad regions of chromosome 11q (seven samples) or very discrete deletions specifically targeting the ATM locus (five samples). Interestingly, several small defined regions other than the ATM deletions were also identified on chromosome 11q.
Sample TP4 displayed a 700 kb heterozygous microdeletion on chromosome 11q33.1 (110891599–111596604), which has its proximal breakpoint in the direct vicinity of two micro RNA, hsa-mir-34b and hsa-mir 34c. This micro RNA locus was affected either by chromosomal breakage in the direct vicinity or heterozygous deletion in eight samples (44%). Another region affected by small, confined lesions in several samples contains two members of the ETS family of transcription factors, v-ets erythroblastosis virus E26 oncogene (ETS1) and Friend leukemia virus integration 1 (FLI1). These two genes were contained in heterozygous deletions in seven samples (39%).
Homozygous deletions
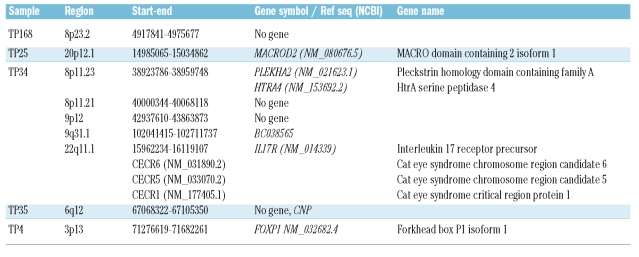
Homozygous deletions were abundantly detected on chromosomes 14 and 7 in the T-cell receptor loci. These deletions have to be understood as physiological as part of the T-cell receptor rearrangements and were, therefore, excluded from the analysis. Apart from these, scattered homozygous deletions were detected in single samples and are summarized in Table 2. Most of these deletions were single events, not recurring in other samples except for the circumscribed deletion of the transcription factor forkhead box P1 isoform 1 (FOXP1), which was additionally heterozygously deleted in two other samples. Due to this targeted deletion of FOXP1, we performed a mutation analysis of this gene by directly sequencing all exons in the samples affected by a deletion. However, this yielded no mutations.
Table 2.
Homozygous deletions: a list of all homozygous deletions detected in our data set and the genes affected.
Novel common lesions and acquired uniparental disomy
Besides the common lesions already described, our data showed a cumulation of noteworthy genomic alterations in other loci, which have not yet been described in T-PLL. As shown in Figure 1 and documented in Table 1, the allelokaryotyping software detected ten duplications and amplifications on chromosome 5p. Here, one lesion stood out to be common in three samples (TP172, TP28 and TP56). These three samples displayed a common breakpoint clustering on chromosome 5p within the direct vicinity of the gene dynein axonemal heavy chain 5 (DNAH5). Potentially, this region may represent a common fusion partner to cancer-relevant genes in these samples.
Another hot-spot of alterations was on chromosome 12p, a location long known to be a region for frequent chromosomal rearrangements in hematologic malignancies.25 Six samples featured heterozygous deletions and four samples had stretches of duplication in this region.
The smallest commonly deleted region encompassed 1.8 Mbp (chr12:14746099–16521376) and contained 16 genes. This suggests that besides haploinsufficiency of CDKN1B, another tumor suppressor gene may be involved.19 Of note, two samples (TP166, TP34) harbored breakpoints directly upstream of the promiscuous fusion partner ETV6, which is also heterozygously deleted in these samples. This could be an indicator that ETV6 may be the target of these two deletions. As reported for other hematologic malignancies, the presence of fusions involving the ETV6 gene is often associated with a deletion or lack of expression of the other ETV6 allele.26,27
A common heterozygous deletion on chromosome 13 occurred in six samples (33%) measuring 1.5 Mbp (chr13: 48879065–50392988). This region is commonly deleted in chronic lymphocytic leukemia and contains two microRNA (miR-15a and miR-16-1),28 which regulate a significant number of cancer-related genes.29
Chromosome 17 often has genomic alterations in cancer. In total, chromosome 17 was affected by deletion, amplification or acquired UPD in 12 samples. Regions of chromosome 17p were heterozygously deleted in seven cases and affected by an acquired UPD in sample TP41, which featured tetrasomy of most of its chromosomes but showed a copy number of two with deletion of one allele on chromosome 17p. Therefore, eight samples (44%) showed loss of heterozygosity of various lengths on chromosome 17p and in five of these samples, the tumor suppressor p53 was contained in the affected regions.
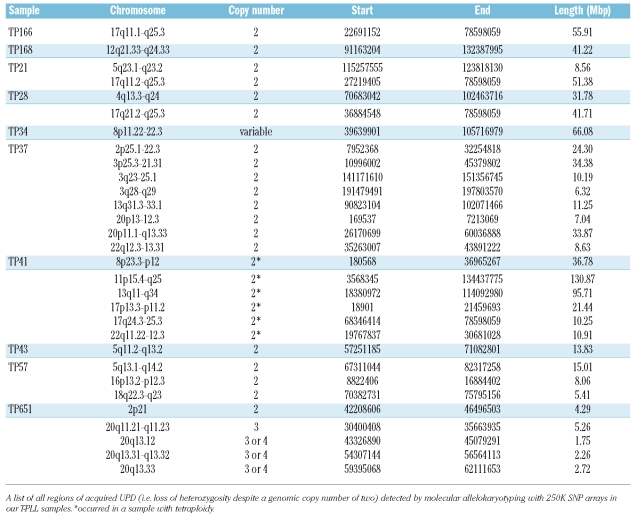
Chromosome 17q was affected by heterozygous deletions in five samples (TP22, TP34, TP35, TP4 and TP799) and acquired UPD in four samples (TP21, TP28, TP166 and TP41) and therefore exhibited loss of heterozygosity in nine cases (50%). The smallest commonly deleted region spanned 4.87 Mbp (chr17: 73729578–78599918) and contained potential target genes such as baculoviral IAP repeat containing protein 5 (BIRC5) and suppressor of cytokine signaling 3 (SOCS3). Of special interest is the recurrent acquired UPD or allelic imbalance in this region in four samples. In total, 30 regions of acquired UPD were detected in all samples (Table 3); among these, the acquired UPD observed on chromosome 17q was the only recurring acquired UPD lesion.
Table 3.
Regions of acquired uniparental disomy or allelic imbalance.
Of note, the starting point of acquired UPD of one sample (TP28) lies in the 17q21.2 region which contains the signal transducer and activator of transcription genes STAT5 A/B and STAT3, which are key factors in malignant transformation.30
Like chromosome 17, chromosome 22 was also commonly affected by loss of heterozygosity either by heterozygous deletion or acquired UPD. Seven T-PLL samples had heterozygous deletions of chromosome 22, with the smallest common lesion measuring 7.4 Mbp (chr.22: 22180211–29584212); two further samples (TP37 and TP41) displayed acquired UPD in the region, so that nine samples (50%) displayed loss of heterozygosity on chromosome 22, making this another interesting site for screening for mutated tumor suppressor genes.
Validation of copy number changes and acquired uniparental disomy
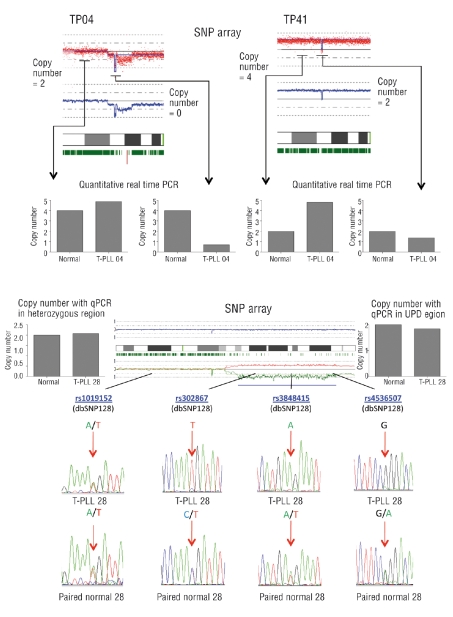
Copy number changes, loss of heterozygosity and acquired UPD detected by high density SNP arrays and molecular allelokaryotyping21 have been validated extensively by alternative methods in previous studies.27,31 In the current study, we confirmed copy number results by quantitative real-time PCR on the genomic DNA of the putatively deleted region and the adjacent region to the lesion in patients and matched normal samples in accordance to a method specifically designed to analyze genomic DNA by real-time PCR.23 We confirmed a homozygous deletion of the FOXP1 gene on chromosome 3p in sample TP4 and a heterozygous deletion of this gene in sample TP41 (Figure 3A). Acquired UPD was confirmed on chromosome 17 in sample TP28 by sequencing SNPs in the region displaying acquired UPD as compared to the adjacent heterozygous region on the same chromosome (Figure 3B).
Figure 3.
Validation of copy number analysis and acquired UPD. (A) Copy number results obtained by SNP arrays were validated by performing quantitative real-time PCR on genomic DNA of samples containing putative deletions of the FOXP1 gene. Chromosome views of chromosome 3 are displayed. In sample TP04, FOXP1 was putatively homozygously deleted (left image, FOXP1 locus = copy number 0, adjacent regions = copy number: two), while in sample TP41, FOXP1 was deleted heterozygously in a tetraploid setting (right image, FOXP1 locus = copy number: two, adjacent regions = copy number: four). Quantitative real-time PCR in the corresponding T-PLL samples and their matched normal DNA confirmed the copy number states estimated by the allelokaryotyping software. (B) Acquired UPD detected by SNP arrays was validated using sample TP28 on chromosome 17. The chromosome view of chromosome 17 in sample TP28 is depicted. Acquired UPD is present on chromosome 17q, as visualized by the divergence of the estimated allele specific copy number (red and green lines in the lower panel of the image), indicating the duplication of one allele and concomitant loss of the other allele and loss of heterozygosity, as evidenced by the abrupt absence of heterozygous SNP calls (vertical green bars directly below the cytoband image). Chromatographs of sequenced SNPs within the acquired UPD region and the adjacent heterozygous region in T-PLL DNA (TP28) and matched normal DNA (paired normal DNA) show that SNPs were homozygous in the T-PLL sample in the acquired UPD region and heterozygous in the matched normal DNA. In the adjacent region, which did not display acquired UPD, the sequenced SNPs were heterozygous in both T-PLL and matched normal samples. Quantitative real-time PCR confirmed a copy number state of two in all regions sequenced in each of the samples.
Discussion
In this study, we aimed to utilize the increased interrogational power of high density SNP arrays and molecular allelokaryotyping21 to refine the understanding of known genomic lesions and discover new ones present in the malignant cells of patients with T-PLL. With high quality genomic DNA from 18 T-PLL patients analyzed by 250K SNP arrays, this study currently represents the most detailed genomic examination of this hematologic malignancy.
In a first step of analysis, we sought to re-evaluate established molecular hallmarks of T-PLL. By doing this, we demonstrated that the well-known disruptions of chromosome 14, inv(14)(q11q34) or t(14;14) (q11;q32),4,12,13 are often unbalanced, indicating excess copies of the involved chromosome 14q fragments.
We refined knowledge of the breakpoints leading to characteristic abnormalities on chromosome 818,24,9 at a submicroscopic level and showed that they were often of a highly complex nature. Although a common breakpoint was not identified for all these cases, certain genes were recurrently and directly involved in breakpoints in several samples. PLEKHA2 is an adaptor protein with a pleckstrin homology domain involved in signaling after activation of lymphocytes.32 Nibrin isoform 2 (NBN), the Nijmegen Breakage Syndrome protein is an important member of the DNA breakage recognition and repair complex consisting of Mre11, Rad50, Nbs1 and ATM16 and was disrupted in one of the few samples that did not exhibit an ATM deletion (TP25). Nephroblastoma overexpressed precursor (NOV) is a critical regulator of human hematopoiesis33 and MYST histone acetyltransferase monocytic (MYST3) is a histone acetyltransferase commonly involved in translocations in acute myeloid leukemia.34 All these genes are, therefore, interesting targets for further analysis in the respective samples.
Deletion and mutation of the ATM gene is so far the only target gene identified in the commonly deleted region on chromosome 11 in T-PLL and other hematologic malignancies.6,7,18 The presence of a second tumor suppressor has been hypothesized in this region.31 Indeed, several other small circumscribed lesions similar to the focal ATM deletions were found on chromosome 11. The micro RNA hsa-mir-34b and hsa-mir 34c were involved in heterozygous deletions in a total of eight samples. Hsa-mir-34b and hsa-mir 34c are induced by p53 and are important regulators in p53-dependent pathways35 and may represent interesting targets, as reduced expression of miR-34s has been found in several tumors.36–38 Furthermore, the oncogenes ETS1 and FLI1 were encompassed in small confined deletions in two samples. Disruption of both of these genes has been determined to be an initiating event in malignant transformation of hematologic diseases and solid tumors.39,40
In the subsequent search for new common genomic lesions in T-PLL, the FOXP1 gene was detected in a small homozygous deletion in one sample and was heterozygously deleted in two other samples. FOXP1 is a member of the FOX family of transcription factors and is involved in the development of the heart, lungs and lymphocytes.41 Deletion and loss of expression in breast cancer confers a worse prognosis42 and this gene is targeted by recurrent chromosome translocations in mucosal-associated lymphoid tissue (MALT) lymphoma.43 We confirmed the deletion of this gene by quantitative real-time PCR. Although screening for mutations of this gene in samples containing heterozygous deletions showed no alterations in this respect, the FOXP1 gene could also be involved in a fusion gene, as recently detected in acute lymphoblastic leukemia.44 The underlying mechanism for accumulated gene fusions in T-PLL could be increased aberrant V(D)J recombination due to mutation of the ATM gene.45 The concept of perturbed V(D)J recombination has recently also been shown to be responsible for common deletions of the Ikaros gene in Philadelphia chromosome-positive acute lymphoblastic leukemia.46
Newly detected common lesions with recurrence in six or more samples were found on chromosomes 5p, 12p, 13q, 17 and 22. Loss of heterozygosity, either by heterozygous deletion or acquired UPD, in nine of 18 samples on both chromosome 17 and chromosome 22 suggests common lesions specific to T-PLL. Loss of chromosome 17p is a common phenomenon in chronic lymphocytic leukemia; it is known to confer a worse prognosis and bad response to chemotherapy, possibly through disruption of the p53 pathway.47 However, loss of heterozygosity of chromosome 17q at such high frequency has not been reported for leukemic diseases. This finding was further corroborated by the observation of a common region of acquired UPD in four of our samples on chromosome 17q. UPD can either arise through several mechanisms at the level of the gametes such as trisomy rescue, compensatory UPD or gametic complementation or can develop due to a somatic recombinational event.48 The UPD regions detected in our experiments are most probably acquired isodisomy that evolved due to somatic recombination events shown in our validation of acquired UPD, which demonstrated that loss of heterozygosity was only detectable in the tumor sample but not in the matched control. While acquired UPD has previously been described as a new genomic lesion in T-PLL,18 this is the first study to report a recurring (n=4) acquired UPD lesion on chromosome 17q.
In conclusion, the use of high density SNP arrays to genotype T-PLL has refined our knowledge of established genomic alterations and revealed numerous new candidate lesions by directly pinpointing affected genes for ongoing functional studies to elucidate the pathogenesis of T-PLL.
Supplementary Material
Footnotes
The online version of this article contains a supplementary appendix.
Authorship and Disclosures
DN analyzed the data, carried out validation experiments and wrote the paper. ELT and MHS designed the study, acquired samples and wrote the paper. NK designed this study, analyzed the data, acquired samples and wrote the paper. TA analyzed the data, MJD designed the study and acquired samples, WKH wrote the paper, SO designed the study, performed SNP array experiments, molecular allelokaryotyping and acquired samples, HPK designed the study, analyzed the data and wrote the paper.
The authors reported no potential conflicts of interest.
Funding: we thank the Parker Hughes Fund and National Institutes of Health for grants for supporting this study. DN is supported by a research grant from the Deutsche Forschungsgemeinschaft (DFG, NO 817/1-1), NK is supported by a fellowship from the Tower Cancer Research Foundation. HPK holds the Mark Goodson Chair in Oncology Research at Cedars Sinai Medical Center and is a member of the Jonsson Cancer Center and the Molecular Biology Institute of UCLA. This work was also supported by grant-in-aid from the Department of Health, Welfare and Labor and from MEXT of the Japanese government.
References
- 1.Dearden CE. T-cell prolymphocytic leukemia. Med Oncol. 2006;23:17–22. doi: 10.1385/MO:23:1:17. [DOI] [PubMed] [Google Scholar]
- 2.Dearden CE, Matutes E, Cazin B, Tjonnfjord GE, Parreira A, Nomdedeu B, et al. High remission rate in T-cell prolymphocytic leukemia with CAMPATH-1H. Blood. 2001;98:1721–6. doi: 10.1182/blood.v98.6.1721. [DOI] [PubMed] [Google Scholar]
- 3.Keating MJ, Cazin B, Coutre S, Birhiray R, Kovacsovics T, Langer W, et al. Campath-1H treatment of T-cell prolymphocytic leukemia in patients for whom at least one prior chemotherapy regimen has failed. J Clin Oncol. 2002;20:205–13. doi: 10.1200/JCO.2002.20.1.205. [DOI] [PubMed] [Google Scholar]
- 4.Pekarsky Y, Hallas C, Croce CM. Molecular basis of mature T-cell leukemia. JAMA. 2001;286:2308–14. doi: 10.1001/jama.286.18.2308. [DOI] [PubMed] [Google Scholar]
- 5.Stern MH, Soulier J, Rosenzwajg M, Nakahara K, Canki-Klain N, Aurias A, et al. MTCP-1: a novel gene on the human chromosome Xq28 translocated to the T cell receptor alpha/delta locus in mature T cell proliferations. Oncogene. 1993;8:2475–83. [PubMed] [Google Scholar]
- 6.Stankovic T, Taylor AM, Yuille MR, Vorechovsky I. Recurrent ATM mutations in T-PLL on diverse haplotypes: no support for their germline origin. Blood. 2001;97:1517–8. doi: 10.1182/blood.v97.5.1517. [DOI] [PubMed] [Google Scholar]
- 7.Stilgenbauer S, Schaffner C, Litterst A, Liebisch P, Gilad S, Bar-Shira A, et al. Biallelic mutations in the ATM gene in T-prolymphocytic leukemia. Nat Med. 1997;3:1155–9. doi: 10.1038/nm1097-1155. [DOI] [PubMed] [Google Scholar]
- 8.Maljaei SH, Brito-Babapulle V, Hiorns LR, Catovsky D. Abnormalities of chromosomes 8, 11, 14, and X in T-prolymphocytic leukemia studied by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1998;103:110–6. doi: 10.1016/s0165-4608(97)00410-x. [DOI] [PubMed] [Google Scholar]
- 9.Sorour A, Brito-Babapulle V, Smedley D, Yuille M, Catovsky D. Unusual breakpoint distribution of 8p abnormalities in T-prolymphocytic leukemia: a study with YACS mapping to 8p11–p12. Cancer Genet Cytogenet. 2000;121:128–32. doi: 10.1016/s0165-4608(00)00239-9. [DOI] [PubMed] [Google Scholar]
- 10.Stoppa-Lyonnet D, Soulier J, Lauge A, Dastot H, Garand R, Sigaux F, et al. Inactivation of the ATM gene in T-cell prolymphocytic leukemias. Blood. 1998;91:3920–6. [PubMed] [Google Scholar]
- 11.Stoppa-Lyonnet D, Lauge A, Sigaux F, Stern MH. No germline ATM mutation in a series of 16 T-cell prolymphocytic leukemias. Blood. 2000;96:374–6. [PubMed] [Google Scholar]
- 12.Teitell MA. The TCL1 family of oncoproteins: co-activators of transformation. Nat Rev Cancer. 2005;5:640–8. doi: 10.1038/nrc1672. [DOI] [PubMed] [Google Scholar]
- 13.Despouy G, Joiner M, Le Toriellec E, Weil R, Stern MH. The TCL1 onco-protein inhibits activation-induced cell death by impairing PKCtheta and ERK pathways. Blood. 2007;110:4406–16. doi: 10.1182/blood-2006-11-059501. [DOI] [PubMed] [Google Scholar]
- 14.Herling M, Patel KA, Teitell MA, Konopleva M, Ravandi F, Kobayashi R, et al. High TCL1 expression and intact T-cell receptor signaling define a hyperproliferative subset of T-cell prolymphocytic leukemia. Blood. 2008;111:328–37. doi: 10.1182/blood-2007-07-101519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu Rev Genet. 1997;31:635–62. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- 16.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–58. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–8. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 18.Durig J, Bug S, Klein-Hitpass L, Boes T, Jons T, Martin-Subero JI, et al. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32) Leukemia. 2007;21:2153–63. doi: 10.1038/sj.leu.2404877. [DOI] [PubMed] [Google Scholar]
- 19.Le Toriellec E, Despouy G, Pierron G, Gaye N, Joiner M, Bellanger D, et al. Haploinsufficiency of CDKN1B contributes to leukemogenesis in T-cell prolymphocytic leukemia. Blood. 2008;111:2321–8. doi: 10.1182/blood-2007-06-095570. [DOI] [PubMed] [Google Scholar]
- 20.Dutt A, Beroukhim R. Single nucleotide polymorphism array analysis of cancer. Curr Opin Oncol. 2007;19:43–9. doi: 10.1097/CCO.0b013e328011a8c1. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of Affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–26. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, Hangaishi A, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–9. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 23.Weksberg R, Hughes S, Moldovan L, Bassett AS, Chow EW, Squire JA. A method for accurate detection of genomic microdeletions using real-time quantitative PCR. BMC Genomics. 2005;6:180. doi: 10.1186/1471-2164-6-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soulier J, Pierron G, Vecchione D, Garand R, Brizard F, Sigaux F, et al. A complex pattern of recurrent chromosomal losses and gains in T-cell prolymphocytic leukemia. Genes Chromosomes Cancer. 2001;31:248–54. doi: 10.1002/gcc.1141. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi H, Montgomery KT, Bohlander SK, Adra CN, Lim BL, Kucherlapati RS, et al. Fluorescence in situ hybridization mapping of translocations and deletions involving the short arm of human chromosome 12 in malignant hematologic diseases. Blood. 1994;84:3473–82. [PubMed] [Google Scholar]
- 26.Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 2005;15:162–74. doi: 10.1016/j.semcancer.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–84. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, et al. MiR-15a and miR-16–1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–71. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Constantinescu SN, Girardot M, Pecquet C. Mining for JAK-STAT mutations in cancer. Trends Biochem Sci. 2008;33:122–31. doi: 10.1016/j.tibs.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Lehmann S, Ogawa S, Raynaud SD, Sanada M, Nannya Y, Ticchioni M, et al. Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer. 2008;112:1296–305. doi: 10.1002/cncr.23270. [DOI] [PubMed] [Google Scholar]
- 32.Allam A, Marshall AJ. Role of the adaptor proteins Bam32, TAPP1 and TAPP2 in lymphocyte activation. Immunol Lett. 2005;97:7–17. doi: 10.1016/j.imlet.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 33.Gupta R, Hong D, Iborra F, Sarno S, Enver T. NOV (CCN3) functions as a regulator of human hematopoietic stem or progenitor cells. Science. 2007;316:590–3. doi: 10.1126/science.1136031. [DOI] [PubMed] [Google Scholar]
- 34.Troke PJ, Kindle KB, Collins HM, Heery DM. MOZ fusion proteins in acute myeloid leukaemia. Biochem Soc Symp. 2006:23–39. doi: 10.1042/bss0730023. [DOI] [PubMed] [Google Scholar]
- 35.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–8. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 36.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–22. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 37.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 39.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Truong AH, Ben-David Y. The role of Fli-1 in normal cell function and malignant transformation. Oncogene. 2000;19:6482–9. doi: 10.1038/sj.onc.1204042. [DOI] [PubMed] [Google Scholar]
- 41.Koon HB, Ippolito GC, Banham AH, Tucker PW. FOXP1: a potential therapeutic target in cancer. Expert Opin Ther Targets. 2007;11:955–65. doi: 10.1517/14728222.11.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fox SB, Brown P, Han C, Ashe S, Leek RD, Harris AL, et al. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor α and improved survival in primary human breast carcinomas. Clin Cancer Res. 2004;10:3521–7. doi: 10.1158/1078-0432.CCR-03-0461. [DOI] [PubMed] [Google Scholar]
- 43.Du MQ. MALT lymphoma: recent advances in aetiology and molecular genetics. J Clin Exp Hematop. 2007;47:31–42. doi: 10.3960/jslrt.47.31. [DOI] [PubMed] [Google Scholar]
- 44.Kawamata N, Ogawa S, Zimmermann M, Niebuhr B, Stocking C, Sanada M, et al. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc Natl Acad Sci USA. 2008;105:11921–6. doi: 10.1073/pnas.0711039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bredemeyer AL, Huang CY, Walker LM, Bassing CH, Sleckman BP. Aberrant V(D)J recombination in ataxia telangiectasia mutated-deficient lymphocytes is dependent on nonhomologous DNA end joining. J Immunol. 2008;181:2620–5. doi: 10.4049/jimmunol.181.4.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 47.Zenz T, Dohner H, Stilgenbauer S. Genetics and risk-stratified approach to therapy in chronic lymphocytic leukemia. Best Pract Res Clin Haematol. 2007;20:439–53. doi: 10.1016/j.beha.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Robinson WP. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays. 2000;22:452–9. doi: 10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.