Loss of major histocompatibility complex class II (MHCII) gene expression was associated with poor outcome in diffuse large B-cell lymphoma (DLBCL) in several studies.1–3 The mechanism for lost expression is unknown. MHCII gene expression is controlled by several transcription factors, including RFX (composed of RFXB, RFX5, and RFXAP), CREB, and NF-Y. These transcription factors interact with a master transactivator protein class II transactivator (CIITA) to form an enhanceosome complex.4 In the Bare Lymphocyte Syndrome (BLS), MHCII gene expression is absent due to small deletions or point mutations in the coding or splicing regions of either CIITA or an RFX subunit.5 This study investigated whether DLBCL with low MHCII expression had mutations similar to BLS.
DNA from 46 patient tumors, for which gene expression profiling was available, was obtained from the Lymphoma and Leukemia Molecular Profiling Project (LLMPP).1 These cases had been previously reviewed by a panel of expert hematopathologists and each contained a minimum of 70% tumor. Twenty-three were from de novo DLBCL in the lowest 10% of MHCII expression, 4 were in the 10–25% range, and 4 were transformed or relapsed DLBCL in the lowest 10%. Fifteen LLMPP cases were de novo MHCII+ samples in the upper 25–100% of expression. Fourteen additional samples were sequenced, including 1 MHCII+ DLBCL, 1 MHCII-DLBCL, and 2 MHCII-peripheral T-cell lymphomas (PTCL), unspecified. MHCII status for these samples was determined by immunohistochemistry. Seven reactive lymph nodes and 3 tonsils were considered positive controls. Ten cell lines (7 MHCII+, 3 MHCII−) were also sequenced.
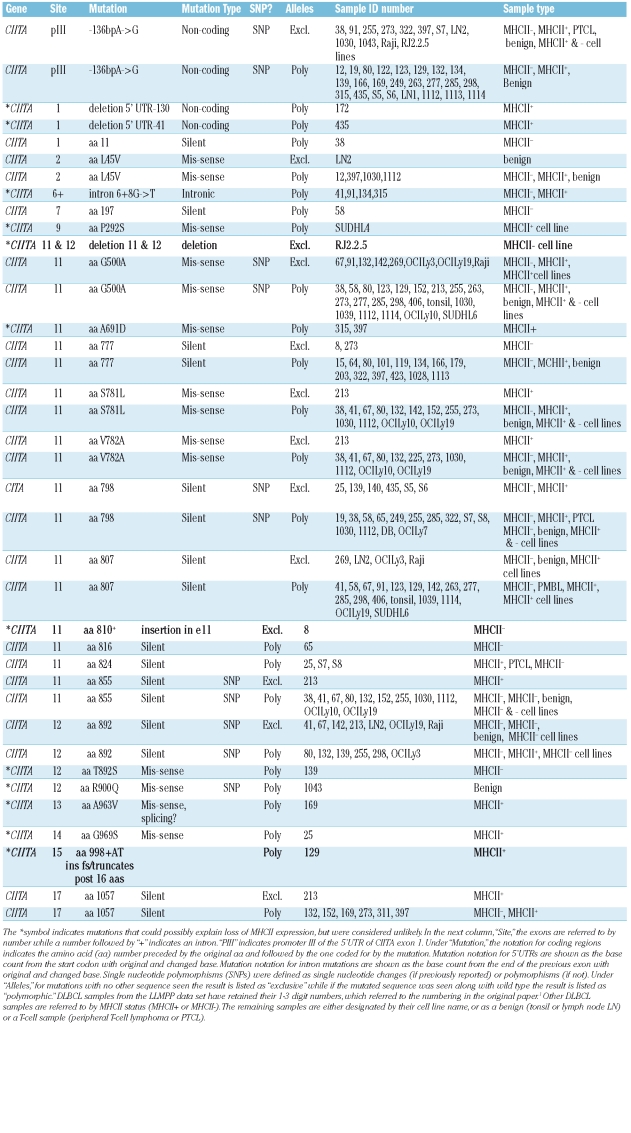
Six PCR reactions using the Multiplex PCR kit (Qiagen, Valencia, CA, USA) analyzed the coding regions of all 4 genes (primer sequences and conditions available on request) using the Multiplex PCR kit (Qiagen). Amplified DNA was purified with the Qiaquick PCR Purification Kit (Qiagen) and sequenced. Possible mutations were confirmed by sequencing the opposite strand. Gap software was used to compare sequences to NCBI genomic sequences. In 3 samples (129, 172, 435), 2 divergent overlapping sequences appeared. For these, the relevant amplicon from a separate, non-multiplex PCR was cloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced. Significant mutations were those resulting in a non-conserved change in amino acid sequence or change in a splice site and were not seen as the only allele in a known MHCII+ sample. Mutations were considered non-significant if they were silent mutations that caused no change in amino acid sequence, conserved mutations that caused no change in class of amino acid, or exclusive mutations in a sample known to express MHCII. Results summarized in Table 1. Only 3 mutations were considered unequivocal causes of loss of MHCII expression, by virtue of being present exclusively in MHCII-samples and by encoding mutations which would exclude required known functional regions of the coded proteins. In one case (in MHCII-DLBCL sample 8), an exclusive insertional mutation was identified in CIITA exon 11, 5’ of the codon for aa 810. This insertion would result in a truncated CIITA protein lacking C-terminal sequences necessary for function, including a nuclear localization sequence and the leucine rich repeat (LRR) regions.6,7 An exclusive Q252stop mutation was detected in exon 3 of RFXAP in one of the MHCII- PTCL samples (sample S8), which created a termination codon that would truncate RFXAP at aa252. This mutation would clearly disrupt RFXAP function, for the sequences of RFXAP C-terminal of aa 252 are required for productive MHCII transcription.8 A large deletion in exons 11 and 12 of CIITA was observed in the RJ2.2.5 line, a γ radiation induced MHCII-derivative of Raji that expresses a CIITA mRNA containing an 1811 base deletion at bp1122-2933. This deletion produced a truncated protein of 353 aa because of a frameshift at aa335 and a translational stop codon, creating a non-functional CIITA protein.9
Table 1.
Mutations found in CIITA and RFX, organized 5' to 3' down the lengths of the genes. Bolded mutations indicate those which were interpreted as likely to affect expression or function of the mutated protein.
In RFX5, 3 potentially significant mutations included a G to C mutation at +4 in intron 5 (sample 435), P409R in 2 samples (samples S8, D8), and an R470stop mutation identified in a PTCL MHCII-sample (sample S8) that encoded a truncation mutant. Other mutations were considered non-significant.
In summary, mutations in RFX and CIITA were infrequent in MHCII-DLBCL. These cases were clinical samples with heterogeneous cell populations and thus it is possible that mutations were under-identified. However, the most likely mechanism for loss of MHCII expression in DLBCL remains specific, co-ordinated downregulation of MHCII transcription, which could potentially be restored through therapeutic intervention.
Footnotes
Funding: grant from the American Cancer Society, #RSG0605501L1B. The authors have no relevant conflicts of interest to disclose.
References
- 1.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 2.Rimsza LM, LeBlanc ML, Unger JM, Miller TP, Grogan TM, Persky DO, et al. Gene expression predicts overall survival in paraffin-embedded tissues of diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2008;112:3425–33. doi: 10.1182/blood-2008-02-137372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang KC, Huang GC, Jones D, Lin YH. Distribution patterns of dendritic cells and T cells in diffuse large B-cell lymphomas correlate with prognoses. Clin Cancer Res. 2007;13:6666–72. doi: 10.1158/1078-0432.CCR-07-0504. [DOI] [PubMed] [Google Scholar]
- 4.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109 (Suppl):S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 5.Nekrep N, Fontes JD, Geyer M, Peterlin BM. When the lymphocyte loses its clothes. Immunity. 2003;18:453–7. doi: 10.1016/s1074-7613(03)00086-4. [DOI] [PubMed] [Google Scholar]
- 6.Harton JA, Ting JP. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol Cell Biol. 2000;20:6185–94. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown JA, Rogers EM, Boss JM. The MHC class II transactivator (CIITA) requires conserved leucine charged domains for interactions with the conserved W box promoter element. Nucleic Acids Res. 1998;26:4128–36. doi: 10.1093/nar/26.18.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long AB, Ferguson AM, Majumder P, Nagarajan UM, Boss JM. Conserved residues of the bare lymphocyte syndrome transcription factor RFXAP determine coordinate MHC class II expression. Mol Immunol. 2006;43:395–409. doi: 10.1016/j.molimm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Brown JA, He XF, Westerheide SD, Boss JM. Characterization of the expressed CIITA allele in the class II MHC transcriptional mutant RJ2.2.5. Immunogenetics. 1996;43:88–91. doi: 10.1007/BF00186611. [DOI] [PubMed] [Google Scholar]