Abstract
Analysis of time series of images can quantify plant growth and development, including the effects of genetic mutations (phenotypes) that give information about gene function. Here is demonstrated a software application named HYPOTrace that automatically extracts growth and shape information from electronic gray-scale images of Arabidopsis (Arabidopsis thaliana) seedlings. Key to the method is the iterative application of adaptive local principal components analysis to extract a set of ordered midline points (medial axis) from images of the seedling hypocotyl. Pixel intensity is weighted to avoid the medial axis being diverted by the cotyledons in areas where the two come in contact. An intensity feature useful for terminating the midline at the hypocotyl apex was isolated in each image by subtracting the baseline with a robust local regression algorithm. Applying the algorithm to time series of images of Arabidopsis seedlings responding to light resulted in automatic quantification of hypocotyl growth rate, apical hook opening, and phototropic bending with high spatiotemporal resolution. These functions are demonstrated here on wild-type, cryptochrome1, and phototropin1 seedlings for the purpose of showing that HYPOTrace generated expected results and to show how much richer the machine-vision description is compared to methods more typical in plant biology. HYPOTrace is expected to benefit seedling development research, particularly in the photomorphogenesis field, by replacing many tedious, error-prone manual measurements with a precise, largely automated computational tool.
The seedling hypocotyl has been the subject of much important plant biology research because its growth and development are profoundly modified by many environmental and endogenous (e.g. hormonal) factors. For example, breakthrough progress in photomorphogenesis came from the isolation of long hypocotyl mutants, which were later shown to have defective phytochrome and cryptochrome photoreceptor genes (Quail, 1998; Lin, 2000). Derivative screens isolated mutants with less severely affected hypocotyls requiring quantitative rather than qualitative separation from the wild type (Fankhauser and Staiger, 2002; Nagy and Schäfer, 2002). Study of these mutants helped define downstream signaling elements. Clearly, the experimental approach of measuring hypocotyl length with manual methods has produced much valuable information, but more detail-rich measurements can be expected to yield additional, qualitatively different information. Machine vision methods based on the acquisition of electronic images of hypocotyls and their analysis by computer algorithms offer the possibility of automatically capturing developmental dynamics with high spatial and temporal resolution. For example, Folta and Spalding (2001) showed by automatically measuring growth every 5 min that the phototropin 1 (phot1) receptor initiates hypocotyl growth suppression in response to blue light, and the cryptochrome 1 (cry1) receptor takes over the suppression after 30 min. But, due to the limited capabilities of the image analysis algorithm used in that study, information about hypocotyl elongation could only be obtained until hook opening began to change the form of the shoot apex, approximately 2 h after the onset of irradiation in those particular conditions.
Better would be a method that analyzed the midline or medial axis of the hypocotyl because this abstraction carries much information about stem length and shape. Methods based on midline extraction from images have been very useful in studies of root growth and bending (Ishikawa and Evans, 1997; Kimura et al., 1999; Lewis et al., 2007; Miller et al., 2007), but certain characteristics of the hypocotyl make midline extraction more difficult. Extreme curvature of the apical hook often results in the tips of the folded cotyledons contacting the hypocotyl, which complicates medial axis transformation or skeletonization methods of finding the midline. The midline-based method of Miller et al. (2007), while capable of high-resolution quantification of hypocotyl growth rate and apical hook opening in response to light, required manual image editing to separate the cotyledons from the hypocotyl in cases where they were appressed. Another major challenge posed by typical images of Arabidopsis (Arabidopsis thaliana) seedlings is locating the apical end of the hypocotyl to terminate the midline at the anatomically correct spot. The Miller et al. (2007) method terminates the midline at the tips of the cotyledons, rather than at the apex of the cotyledon. The method is effective, but the midline length measured includes the cotyledons and so changes over time are not attributable solely to hypocotyl growth.
An algorithm developed by Wang et al. (2008), employing a technique termed adaptive local principal components analysis (PCA), addressed the challenges of automating measurements of hypocotyl growth and shoot development. That basic algorithm has been refined and developed into a stand-alone piece of software equipped with a graphical user interface to form a tool capable of high-resolution measurements of seedling stem growth and shape from time series of electronic images. Here, the program called HYPOTrace is described and shown to measure light-induced hypocotyl inhibition, apical hook opening, phototropism, and nutation with a high degree of automation and resolution.
RESULTS AND DISCUSSION
Overview of HYPOTrace
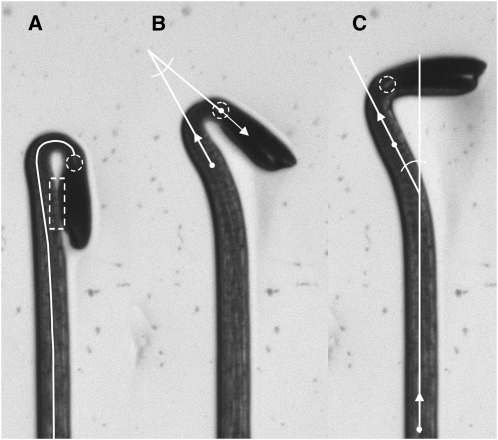
Figure 1 shows images selected from a time series of an Arabidopsis seedling undergoing de-etiolation in response to blue light. The seedling was growing along the surface of agar in a vertical petri plate. The plate was backlit with infrared radiation and the images were captured from the front by a charge-coupled device camera fitted with a close-focus zoom lens and a filter to prevent light other than the infrared from reaching the imaging sensor. The images display the high contrast between object and background and internal features required for automatic processing by the tool described here. The solid white midline or medial axis of the hypocotyl is generated by the tracing algorithm at the core of HYPOTrace. First, a thresholding operation binarizes the image to determine which pixels belong to the background and which are part of the seedling. Beginning at the base of the seedling, a circular region having a diameter slightly larger than the width of the hypocotyl is analyzed. The center of the circle (actually, the center of mass of the seedling pixels within the circle), defines the first midline point. PCA is performed on the vector representing seedling pixel positions and gray-scale values within the analysis circle. The resulting first eigenvector determines the direction of the medial axis at the position of the circle's center. The analysis window is moved one step in the direction of this vector and the process is repeated to determine the next midline point. The second eigenvalue produced by the PCA relates to the width of the seedling at circle center and is used to determine the radius of the next analysis window. Thus, the first eigenvector directs the circle's movement and the second eigenvalue adjusts its radius to achieve the appropriate, slight overfilling. In this manner, the analysis window is repositioned, resized, and its center found to produce an ordered set of midline points. This iterative process was termed adaptive local PCA (Wang et al., 2008).
Figure 1.
Images of a seedling responding to light with features related to the HYPOTrace method superimposed. A, Midline shown as a solid white line, hypocotyl terminus feature indicated with a dashed white circle, and region of cotyledons appressed to the hypocotyl indicated by dashed white rectangle. Changes in the midline length between images in a time series were used to quantify growth rate. B, Angle formed by the first principal component direction of the terminal point and the first principal component direction at a more basal reference point. This angle was used to quantify apical hook opening over time. C, Angle formed by the first principal component direction of a point at the hypocotyl base and the first principal component direction of the reference point shown in B. This hypocotyl direction angle was taken as a measure of phototropic bending.
The precision of the midline depends inversely on step size. Smaller steps give higher precision and a more faithful midline, especially in tightly curved regions, but at the cost of computation time. In straight regions, small steps would unnecessarily slow the computation. To achieve the appropriate balance between precision (midline point density) and computation speed, step size in HYPOTrace adapts to meet the local needs. Step size is set to 0.25 times the radius of the analysis window, which shortens the steps in the most tightly curved region of the hypocotyl because it is also the thinnest (Fig. 1A). Step size is further reduced by a factor that is related to local curvature as determined from changes in the directions of previous midline points. The result is an appropriate reduction in step size in regions where the midline is changing direction the most (i.e. where precision is most needed). The midline shown in Figure 1A would consist of approximately 80 points with the highest density being within the curved region of the apical hook. A spline is fit to the set of points to produce a smooth midline. Other details and mathematical description of midline tracing by adaptive local PCA can be found in Wang et al. (2008).
A major challenge to tracing the medial axis of the etiolated hypocotyl is preventing diversion of the midline toward the cotyledons in instances where they contact the hypocotyl (Fig. 1A, dashed rectangle). The method of Miller et al. (2007) employed a manual preprocessing step to create a strong boundary between hypocotyl and cotyledon in this common situation. A distinguishing feature of the tracing algorithm in HYPOTrace is a weighting function that takes advantage of the fact that cotyledon tissue is more opaque than hypocotyls and therefore appears darker. Down-weighting the darker regions reduces digression of the midline toward the cotyledons when they touch the hypocotyl.
An important and distinguishing feature of HYPOTrace is its automatic method of terminating the midline where the two cotyledon petioles join the hypocotyl. Above this junction (the cotyledonary node) lies the shoot apical meristem. Below the node is the hypocotyl. Ideally, the midline would be terminated at the cotyledonary node so that analysis of the midline would be anatomically specific to the hypocotyl. At the cotyledonary node, bifurcation of the two petioles produces a gap that appears in images as a local brightness, although not always detectable by eye. The dashed circle in Figure 1A encloses this region of interest. In many cases, HYPOTrace automatically locates this bifurcation point and accurately terminates the midline without user intervention by analyzing the intensity profile along the midline in the area where the hypocotyl narrows. The graphical user interface shown in Figure 2 provides the option of manually locating the appropriate terminus in cases where the intensity feature is not sufficiently robust for automatic detection.
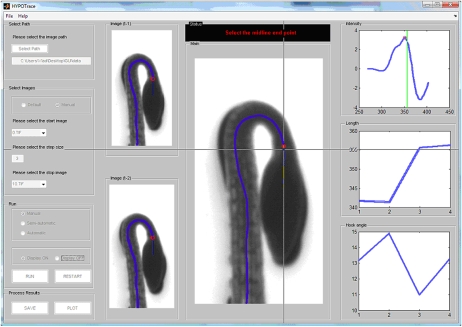
Figure 2.
A screenshot of the HYPOTrace graphical user interface.
Successful processing of a seedling in an image results in a set of midline point coordinates. From these, the length of the midline can be determined and therefore hypocotyl elongation rate can be determined from a time series of midlines. Each midline point also has a direction associated with it. A key direction is that of the terminal point, shown by the arrow extended from the point within the dashed circle in Figure 1B. The difference between the terminus direction and the direction of a hypocotyl reference point is taken as the apical hook angle (Fig. 1B). The reference point direction is located by traveling back 32 steps from the terminus and determining the average direction of three points before and after the reference point. The angle formed by the apical reference direction and a basal reference direction (Fig. 1C) gives information about the general direction of hypocotyl growth, suitable for characterizing tropisms. Thus, HYPOTrace measures hypocotyl length, apical hook angle, general stem angle, and rates of change of each parameter from time series of electronic gray-scale images.
The user interacts with the image data and the analysis functions through the HYPOTrace graphical user interface (Fig. 2). Upon running the program, the user is presented with the means to select the path to the data, which should be a series of sequentially numbered tagged image format (*.TIF) files. In the central window of the front panel, a binarized version of the final image in the series appears. The binary threshold slider is to be adjusted until the objects (seedlings) are fully white and the background fully black and the object contours as smooth as possible. After clicking run, the mouse is used to drag a box around the object to be analyzed (e.g. around one of the several seedlings that may be in the frame to initiate the tracing). The growing midline is superimposed on the binarized image of the object as it is calculated (unless the display-off option is selected to speed up the processing) and stops automatically once it reaches the cotyledon tip. If manual mode is selected, the program identifies one or two potential termination points (based on intensity features) on the midline and marks each with a red circle. The user selects the point that corresponds with the anatomical bifurcation point at the cotyledonary node. If none of the indicated points is acceptably close to the true bifurcation point, which may occur if the true bifurcation point is obscured for some reason, such as poor image quality, the user may manually select a more appropriate point along the midline. A plot of baseline-adjusted intensity along the midline, with labeled peaks corresponding to suggested termination points, is displayed as a green line in the top right intensity window of the front panel. If the user right-clicks on a potential termination point within the seedling image in the image window, the corresponding point in the intensity window is indicated, and the hypocotyl length and hook angle windows are updated. These functionalities help the user select a reliable termination point based on tracing history, geometry, and intensity features. After selecting a termination point, tracing of the same object in the next image of the series automatically begins and the process continues until the final image of the series is processed. Midline length and apical hook angle plots are updated in their respective windows after each image is processed. When the selected series of images is processed, time courses of hook angle, hypocotyl angle, and growth rate are saved in a spreadsheet. The data are saved after calibration into units calculated using conversion data (pixel/mm factors and time interval between images) entered into a separate notes text file located in the same directory as the image stack.
HYPOTrace may also be operated in an automatic mode in which case the most robust intensity peak near the cotyledonary node is automatically selected in each image of the series, and the user has no opportunity to intervene. This mode provides hands-free processing of data, but requires high-quality images with a robust bifurcation point. An intermediate, semiautomatic mode is also available. In this mode, the user selects the termination point as previously described in the manual mode for the first four images. Growth rate information based on these initial manual selections is used to predict the correct termination point in future images. The program self-switches to automatic processing after a reliable bifurcation point is identified, but will pause for user selection of a termination point if the candidate detected on the basis of intensity does not agree with that predicted on the basis of growth rate.
Use Cases
A report on the algorithm design and mathematics underlying each of the HYPOTrace functions has appeared (Wang et al., 2008). Here, the stand-alone executable version equipped with a graphical user interface is demonstrated to be an effective tool for quantifying seedling photomorphogenesis with high resolution through the following use cases.
Growth Inhibition Induced by Blue Light
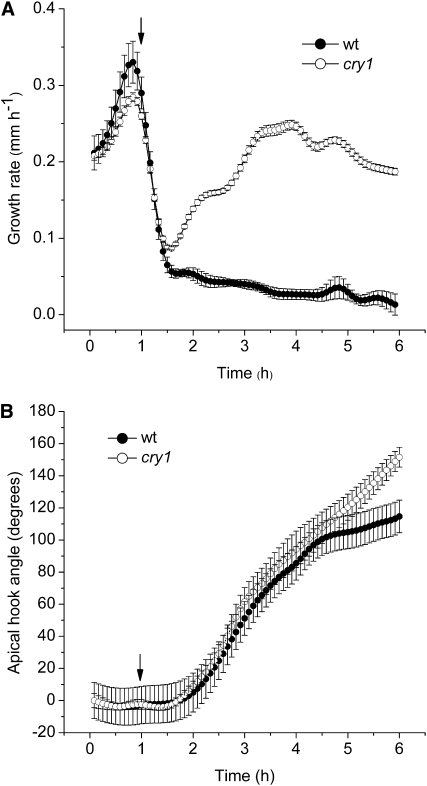
Light, particularly the blue waveband of the spectrum, rapidly suppresses elongation of etiolated seedling hypocotyls initially through activation of the phot1 photoreceptor and subsequently through coaction of the cry1, cry2, and phytochrome A (phyA) photoreceptors (Folta and Spalding, 2001). A set of imaging experiments was performed to test the ability of HYPOTrace to measure hypocotyl growth inhibition induced by blue light and whether the cry1 mutant phenotype recorded by HYPOTrace is in agreement with previous reports based on different, less automated, methodologies. Figure 3A shows that HYPOTrace recorded growth rate in the darkness of 0.2 to 0.3 mm h−1 in perfect agreement with two previous methods: one that could not monitor long-term growth (Folta and Spalding, 2001) and one that required manual editing of many of the images (Miller et al., 2007). Upon irradiation with blue light (50 μmol m−2 s−1), growth rate rapidly declined to approximately 20% of the dark rate within 30 min. This was also in perfect agreement with growth rate time courses measured by the other methods. The methods of Folta and Spalding (2001) and Miller et al. (2007) showed a slight recovery of growth rate for 1 to 2 h after the initial inhibition despite the continuous presence of blue light. This feature was less evident in the present experiment, perhaps due to subtle differences in the experimental conditions in the different studies or to the fact that HYPOTrace-determined midlines are more precisely hypocotyl-specific and less influenced by changes in cotyledon size or shape.
Figure 3.
Hypocotyl responses to blue light quantified by HYPOTrace. A, Inhibition of hypocotyl elongation rate by blue light in wild type (wt) and cry1. B, Opening of apical hook in wild type and cry1. Elongation (growth) rate and apical hook opening were measured simultaneously from the same series of images. In each panel, the arrow indicates the point of irradiation onset. The data points are mean values ± se; n = 15 for wild type; n = 16 for cry1.
When used to analyze cry1 seedlings, the same pattern in growth rate described by the other methods was observed. Growth rate declined initially as in wild type, due to the action of phot1 as found previously (Parks et al., 1998; Folta and Spalding, 2001). After approximately 30 min, growth rate escaped from inhibition and increased with some oscillations to a rate several-fold higher than wild type. This escape from recovery is responsible for the characteristic long-hypocotyl phenotype of the cry1 mutant after long-term growth in blue light. HYPOTrace operating automatically on approximately 75% of the seedlings was able to capture the details of the wild-type and mutant response development with high resolution over a long period of time. Thus, the gains in automation offered by HYPOTrace do not come at the expense of fidelity or resolution.
Apical Hook Opening Induced by Blue Light
By the method diagrammed in Figure 1, apical hook angle can be measured simultaneously with growth rate. Processing the images for growth rate (Fig. 3A) also generated a time course of hook angle opening in response to blue light (Fig. 3B). The data agree completely with those obtained by the less automatic method of Miller et al. (2007). Both methods showed a slowing of opening after the hook angle achieved 120 degrees and, in both studies, this required about 6 h of blue light. Both methods showed that hook opening lagged behind the initial peak of growth inhibition by approximately 30 min. Loss of cry1 did not affect the early stages of hook opening to any significant extent. Clearly, cry1 does not play a significant role in the blue-light-induced opening of the apical hook. Probable contributors to the initiation of hook opening are cry2 and phyA (Lin et al., 1998; Folta and Spalding, 2001).
Phototropism
The directional response of hypocotyl growth to unilateral light is mechanistically distinct from the growth inhibition and hook opening responses presented above. It is much more sensitive to light, being activated by orders of magnitude lower fluence rates than the high-irradiance suppression of elongation mediated by cry1 (Fig. 3A). Phot1 and phot2 are the photoreceptors responsible for mediating phototropism. This response is frequently measured in studies of seedling development, photoreceptor function, and auxin action. In fact, its study led to the discovery of auxin, the phototropin molecules, and other signal transduction pathway elements. It is an important environmental response model, but the typical manual techniques for measuring it are tedious and inaccurate due to the difficulty of judging the direction of the apical portion of a curved hypocotyl.
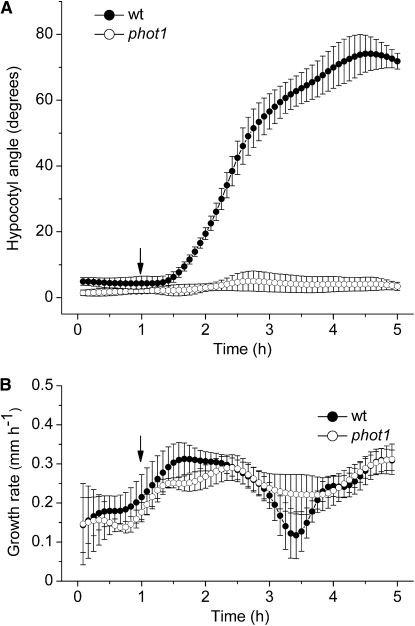
To test the ability of HYPOTrace to measure phototropic curvature development, images of seedlings responding to dim unilateral blue light (approximately 0.3 μmol m−2 s−1) were collected at 5-min intervals for 6 h. Figure 4A shows that approximately 30 min after the onset of unilateral blue light, hypocotyl angle increased steadily, although not linearly, for 3 h, reaching a value of approximately 75°. The complete lack of phototropism in the phot1 mutant is well captured by the method. Typically, phototropism is assayed by a single endpoint measurement or a few time points. Applying HYPOTrace to the analysis of a high-resolution time series of images generates information-rich datasets that increase the utility of phototropism as a plant stimulus-response model. Growth rate during phototropism was also quantified, enabling relationships between elongation rate and curvature to be examined. Figure 4B shows that growth rate was relatively constant while the hypocotyl was bending strongly in the case of wild type, or bending not at all in the case of phot1.
Figure 4.
Hypocotyl responses to dim unilateral blue light quantified by HYPOTrace. A, Phototropic curvature in wild type (wt) and phot1. B, Hypocotyl growth rate in wild type and phot1. In each panel, the arrow indicates the point of irradiation onset. The data points are mean values ± se; n = 7 for wild type; n = 4 for phot1.
An automated tool for making high-resolution measurements of Arabidopsis hypocotyls may find many uses in plant biology research and education. For this reason, HYPOTrace is freely available as an executable file downloadable from http://phytomorph.wisc.edu/HYPOTrace/download/index.htm, where a user guide that explains installation and operation is also available.
MATERIALS AND METHODS
Arabidopsis (Arabidopsis thaliana) seeds were planted on a 1% agar medium containing 1 mm KCl, 1 mm CaCl2, 5 mm MES, adjusted to pH 5.7 with BTP. After stratification at 4°C for 2 d, germination was induced by placing the seeds in 50 μmol m−2 s−1 white light for 30 to 60 min. The seeds were then grown on vertically oriented petri plates in darkness. Plates containing seedlings growing on the surface of the agar were mounted vertically and transverse to the optical axis of a charge-coupled device camera (Marlin F-146B; AVT Corp.) outfitted with a close-focus zoom lens (model R72; Tokina Co.). Seedlings adhered to the surface of the plate, remaining in the focal plane as they elongated. An infrared light source (model BL020201; Advanced Illumination Inc.), having a peak output at 948 nm, was placed behind the petri plate for back illumination. Detailed information about the image acquisition platform may be found at http://phytomorph.wisc.edu/parts_list.htm. Images were acquired at a resolution of 210 pixels mm−1 at 5-min intervals for 1 h in darkness before the indicated light treatment began. For the growth inhibition and apical hook measurements (Columbia wild type and cry1-304 mutants), blue light was supplied by at a fluence rate of 50 μmol m−2 s−1 blue light from a bank of light-emitting diodes (QB1310CS-470; Quantum Devices). For the phototropism experiments (Wassilewskija wild type and phot1-5 mutants), unilateral blue light at a fluence rate of 0.1 μmol m−2 s−1 was delivered from a 450-nm light-emitting diode (LED-LITE; World Precision Inc.) coupled to a fiber optic.
This work was supported by the National Science Foundation (grant no. DBI–0621702).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Edgar P. Spalding (spalding@wisc.edu).
Open access articles can be viewed online without a subscription.
References
- Fankhauser C, Staiger D (2002) Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 216 1–16 [DOI] [PubMed] [Google Scholar]
- Folta KM, Spalding EP (2001) Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J 26 471–478 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML (1997) Novel software for analysis of root gravitropism: comparative response patterns of Arabidopsis wild-type and axr1 seedlings. Plant Cell Environ 20 919–928 [DOI] [PubMed] [Google Scholar]
- Kimura K, Kikuchi S, Yamasaki S (1999) Accurate root length measurement by image analysis. Plant Soil 216 117–127 [Google Scholar]
- Lewis DR, Miller ND, Splitt BL, Wu G, Spalding EP (2007) Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis Multidrug Resistance-like ABC transporter genes. Plant Cell 19 1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C (2000) Plant blue-light receptors. Trends Plant Sci 5 337–342 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ND, Parks BM, Spalding EP (2007) Computer-vision analysis of seedling responses to light and gravity. Plant J 52 113–125 [DOI] [PubMed] [Google Scholar]
- Nagy F, Schäfer E (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol 53 329–355 [DOI] [PubMed] [Google Scholar]
- Parks BM, Cho MH, Spalding EP (1998) Two genetically separable phases of growth inhibition induced by blue light in Arabidopsis seedlings. Plant Physiol 118 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (1998) The phytochrome family: dissection of functional roles and signalling pathways among family members. Philos Trans R Soc Lond B Biol Sci 353 1399–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Assadi AH, Spalding EP (2008) Tracing branched curvilinear structures with a novel adaptive local PCA algorithm. In HR Arabnia, ed, Proceedings of the 2008 International Conference on Image Processing, Computer Vision, and Pattern Recognition, Ed 2. CSREA Press, Athens, GA, pp 557–563